Abstract
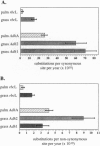
A number of studies have noted that nucleotide substitution rates at the chloroplast-encoded rbcL locus violate the molecular clock principle. Substitution rate variation at this plastid gene is particularly pronounced between palms and grasses; for example, a previous study estimated that substitution rates in rbcL sequences are approximately 5-fold faster in grasses than in palms. To determine whether a proportionate change in substitution rates also occurs in plant nuclear genes, we characterized nucleotide substitution rates in palm and grass sequences for the nuclear gene Adh. In this article, we report that palm sequences evolve at a rate of 2.61 x 10(-9) substitution per synonymous site per year, a rate which is slower than most plant nuclear genes. Grass Adh sequences evolve approximately 2.5-fold faster than palms at synonymous sites. Thus, synonymous rates in nuclear Adh genes show a marked decrease in palms relative to grasses, paralleling the pattern found at the plastid rbcL locus. This shared pattern indicates that synonymous rates are correlated between a nuclear and a plastid gene. Remarkably, nonsynonymous rates do not show this correlation. Nonsynonymous rates vary between two duplicated grass Adh loci, and nonsynonymous rates at the palm Adh locus are not markedly reduced relative to grasses.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akashi H. Synonymous codon usage in Drosophila melanogaster: natural selection and translational accuracy. Genetics. 1994 Mar;136(3):927–935. doi: 10.1093/genetics/136.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birky C. W., Jr, Maruyama T., Fuerst P. An approach to population and evolutionary genetic theory for genes in mitochondria and chloroplasts, and some results. Genetics. 1983 Mar;103(3):513–527. doi: 10.1093/genetics/103.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet J., Strauss S. H., Doerksen A. H., Price R. A. Extensive variation in evolutionary rate of rbcL gene sequences among seed plants. Proc Natl Acad Sci U S A. 1992 Aug 15;89(16):7844–7848. doi: 10.1073/pnas.89.16.7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten R. J. Rates of DNA sequence evolution differ between taxonomic groups. Science. 1986 Mar 21;231(4744):1393–1398. doi: 10.1126/science.3082006. [DOI] [PubMed] [Google Scholar]
- Dennis E. S., Sachs M. M., Gerlach W. L., Finnegan E. J., Peacock W. J. Molecular analysis of the alcohol dehydrogenase 2 (Adh2) gene of maize. Nucleic Acids Res. 1985 Feb 11;13(3):727–743. doi: 10.1093/nar/13.3.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut B. S., Clegg M. T. Molecular evolution of alcohol dehydrogenase 1 in members of the grass family. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2060–2064. doi: 10.1073/pnas.88.6.2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut B. S., Muse S. V., Clark W. D., Clegg M. T. Relative rates of nucleotide substitution at the rbcL locus of monocotyledonous plants. J Mol Evol. 1992 Oct;35(4):292–303. doi: 10.1007/BF00161167. [DOI] [PubMed] [Google Scholar]
- Goloubinoff P., Päbo S., Wilson A. C. Evolution of maize inferred from sequence diversity of an Adh2 gene segment from archaeological specimens. Proc Natl Acad Sci U S A. 1993 Mar 1;90(5):1997–2001. doi: 10.1073/pnas.90.5.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M., Kishino H. Heterogeneity of tempo and mode of mitochondrial DNA evolution among mammalian orders. Jpn J Genet. 1989 Aug;64(4):243–258. doi: 10.1266/jjg.64.243. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980 Dec;16(2):111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Li W. H., Ellsworth D. L., Krushkal J., Chang B. H., Hewett-Emmett D. Rates of nucleotide substitution in primates and rodents and the generation-time effect hypothesis. Mol Phylogenet Evol. 1996 Feb;5(1):182–187. doi: 10.1006/mpev.1996.0012. [DOI] [PubMed] [Google Scholar]
- Li W. H., Tanimura M., Sharp P. M. An evaluation of the molecular clock hypothesis using mammalian DNA sequences. J Mol Evol. 1987;25(4):330–342. doi: 10.1007/BF02603118. [DOI] [PubMed] [Google Scholar]
- Li W. H., Tanimura M. The molecular clock runs more slowly in man than in apes and monkeys. Nature. 1987 Mar 5;326(6108):93–96. doi: 10.1038/326093a0. [DOI] [PubMed] [Google Scholar]
- Martin A. P., Palumbi S. R. Body size, metabolic rate, generation time, and the molecular clock. Proc Natl Acad Sci U S A. 1993 May 1;90(9):4087–4091. doi: 10.1073/pnas.90.9.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T., Birky C. W., Jr Effects of periodic selection on gene diversity in organelle genomes and other systems without recombination. Genetics. 1991 Feb;127(2):449–451. doi: 10.1093/genetics/127.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse S. V., Gaut B. S. A likelihood approach for comparing synonymous and nonsynonymous nucleotide substitution rates, with application to the chloroplast genome. Mol Biol Evol. 1994 Sep;11(5):715–724. doi: 10.1093/oxfordjournals.molbev.a040152. [DOI] [PubMed] [Google Scholar]
- Muse S. V., Weir B. S. Testing for equality of evolutionary rates. Genetics. 1992 Sep;132(1):269–276. doi: 10.1093/genetics/132.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986 Sep;3(5):418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Nei M., Stephens J. C., Saitou N. Methods for computing the standard errors of branching points in an evolutionary tree and their application to molecular data from humans and apes. Mol Biol Evol. 1985 Jan;2(1):66–85. doi: 10.1093/oxfordjournals.molbev.a040333. [DOI] [PubMed] [Google Scholar]
- Nickrent D. L., Starr E. M. High rates of nucleotide substitution in nuclear small-subunit (18S) rDNA from holoparasitic flowering plants. J Mol Evol. 1994 Jul;39(1):62–70. doi: 10.1007/BF00178250. [DOI] [PubMed] [Google Scholar]
- Ohta T. Pattern of nucleotide substitutions in growth hormone-prolactin gene family: a paradigm for evolution by gene duplication. Genetics. 1993 Aug;134(4):1271–1276. doi: 10.1093/genetics/134.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota T., Kimura M. On the constancy of the evolutionary rate of cistrons. J Mol Evol. 1971;1(1):18–25. doi: 10.1007/BF01659391. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sala F., Galli M. G., Levi M., Burroni D., Parisi B., Pedrali-Noy G., Spadari S. Functional roles of the plant alpha-like and gamma-like DNA polymerases. FEBS Lett. 1981 Feb 9;124(1):112–118. doi: 10.1016/0014-5793(81)80064-6. [DOI] [PubMed] [Google Scholar]
- Sharp P. M. Determinants of DNA sequence divergence between Escherichia coli and Salmonella typhimurium: codon usage, map position, and concerted evolution. J Mol Evol. 1991 Jul;33(1):23–33. doi: 10.1007/BF02100192. [DOI] [PubMed] [Google Scholar]
- Vawter L., Brown W. M. Nuclear and mitochondrial DNA comparisons reveal extreme rate variation in the molecular clock. Science. 1986 Oct 10;234(4773):194–196. doi: 10.1126/science.3018931. [DOI] [PubMed] [Google Scholar]
- Waters E. R. The molecular evolution of the small heat-shock proteins in plants. Genetics. 1995 Oct;141(2):785–795. doi: 10.1093/genetics/141.2.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M. A., Gaut B., Clegg M. T. Chloroplast DNA evolves slowly in the palm family (Arecaceae). Mol Biol Evol. 1990 Jul;7(4):303–314. doi: 10.1093/oxfordjournals.molbev.a040605. [DOI] [PubMed] [Google Scholar]
- Wolfe K. H., Gouy M., Yang Y. W., Sharp P. M., Li W. H. Date of the monocot-dicot divergence estimated from chloroplast DNA sequence data. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6201–6205. doi: 10.1073/pnas.86.16.6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe K. H., Li W. H., Sharp P. M. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci U S A. 1987 Dec;84(24):9054–9058. doi: 10.1073/pnas.84.24.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe K. H., Sharp P. M. Mammalian gene evolution: nucleotide sequence divergence between mouse and rat. J Mol Evol. 1993 Oct;37(4):441–456. doi: 10.1007/BF00178874. [DOI] [PubMed] [Google Scholar]
- Wu C. I., Li W. H. Evidence for higher rates of nucleotide substitution in rodents than in man. Proc Natl Acad Sci U S A. 1985 Mar;82(6):1741–1745. doi: 10.1073/pnas.82.6.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S., Harry D. E. Molecular phylogeny and evolutionary rates of alcohol dehydrogenases in vertebrates and plants. Mol Biol Evol. 1993 Nov;10(6):1215–1226. doi: 10.1093/oxfordjournals.molbev.a040073. [DOI] [PubMed] [Google Scholar]