Abstract
Salmonella is an important cause of bacterial food-borne gastroenteritis. Salmonella encounters multiple abiotic stresses during pathogen elimination methods used in food processing, and these stresses may influence its subsequent survivability within the host or in the environment. Upon ingestion, Salmonella is exposed to gastrointestinal acidity, a first line of the host innate defense system. This study tested the hypothesis that abiotic stresses encountered during food processing alter the metabolic mechanisms in Salmonella that enable survival and persistence during subsequent exposure to the host gastrointestinal acidic environment. Out of the four different abiotic stresses tested, viz., cold, peroxide, osmotic, and acid, preadaptation of the log-phase culture to cold stress (5°C for 5 h) significantly enhanced survival during subsequent acid stress (pH 4.0 for 90 min). The gene expression profile of Salmonella preadapted to cold stress revealed induction of multiple genes associated with amino acid metabolism, oxidative stress, and DNA repair, while only a few of the genes in the above-mentioned stress response and repair pathways were induced upon exposure to acid stress alone. Preadaptation to cold stress decreased the NAD+/NADH ratio and hydroxyl (OH·) radical formation compared with those achieved with the exposure to acid stress alone, indicating alteration of aerobic respiration and the oxidative state of the bacteria. The results from this study suggest that preadaptation to cold stress rescues Salmonella from the deleterious effect of subsequent acid stress exposure by induction of genes involved in stress response and repair pathways, by modification of aerobic respiration, and by redox modulation.
INTRODUCTION
Salmonella enterica serovar Typhimurium is a Gram-negative enteric pathogen that causes food-borne gastroenteritis in humans. According to the latest Morbidity and Mortality Weekly Report from CDC for surveillance of food-borne disease outbreaks, Salmonella leads to the most outbreak-related hospitalizations (1). S. Typhimurium has remained one of the most frequently isolated serotypes, according to the annual report on Salmonella from national enteric disease surveillance in 2011. S. Typhimurium continues to cause nationwide outbreaks which have been linked to contaminated food, including tomatoes (2006), peanut butter (2009), ground beef (2011), and live poultry and cantaloupe (2013) (2; http://www.cdc.gov/salmonella/outbreaks.html).
Salmonella encounters several abiotic stresses in the food processing environment or during storage. These stresses include (i) cold stress during storage of food; (ii) peroxide stress because hydrogen peroxide is commonly used in food processing due to its potent antimicrobial, oxidizing/reducing, and bleaching properties; (iii) osmotic stress because salt is used as one of the most common food preservatives; and (iv) acid stress because treatments with acidic solutions are used for preserving food quality and preventing microbial spoilage. In addition, when the host ingests contaminated food, Salmonella encounters gastric acidity as the host's first line of defense. The gut transit time is approximately 90 min (3, 4), and the pH of the stomach after consumption of food ranges from 3.0 to 5.0 in adults, while the pH of a child's stomach is between 4.0 and 5.0 (5, 6). Consequently, Salmonella must overcome the abiotic stresses encountered during food processing followed by acid stress (pH 4.0 for 90 min) during gastrointestinal (GI) infection. In virulent Salmonella strains, it has been shown that preadaptation to sublethal acidic conditions minimizes the lethal effects of subsequent extreme acidic conditions by inducing the genes in the acid tolerance response (ATR) system, and this induction is RpoS dependent (7). The objectives of this study were (i) to test if prior exposure of S. Typhimurium (which has a defective rpoS gene) to abiotic stresses, including mild acid stress, alters survival upon subsequent exposure to the host GI acidic environment and (ii) to understand the molecular mechanisms underlying altered survival in the host GI acidic environment.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
S. Typhimurium LT2 ATCC 700720 was used in this study. Prior to each experiment, a new vial of stock culture was thawed from −70°C storage, transferred twice in nutrient broth (NB; Difco, Detroit, MI), and grown at 37°C with shaking at 220 rpm for 12 to 16 h. Just prior to each experiment, log-phase bacteria were collected (∼5 h of growth) and used since they are more susceptible to stress conditions than stationary-phase bacteria.
Stress treatments.
A log-phase culture was centrifuged at 7,200 × g for 5 min and resuspended at an equal density (to achieve an optical density of 0.2 at 600 nm, or ∼108 CFU/ml) in NB prechilled to 5°C (cold stress), in NB with 1 mM H2O2 prewarmed to 37°C (oxidative stress), in NB with 5% (wt/vol) NaCl prewarmed at 37°C (osmotic stress), and in NB at pH 5.3 (adjusted with hydrochloric acid) prewarmed to 37°C (acid stress). These conditions fall within the range of the conditions used in food processing. These stress treatments for preadaptation were given for 0.5 h (shock) and 5 h (stress). Nonstressed Salmonella cultured in NB served as a negative control. Serial 10-fold dilutions of each pre- and poststress culture were plated for estimation of the number of CFU. An aliquot of each culture was collected for total RNA extraction and downstream gene expression analysis. After the respective shock and stress treatments, cultures were divided into two aliquots and centrifuged at 7,200 × g for 5 min. The first aliquot was subjected to subsequent acid stress by resuspending it in NB (pH 4.0), followed by incubation at 37°C for 90 min. The second aliquot served as a control and was resuspended in NB and incubated at 37°C for 90 min. An aliquot of each culture was again collected for the RNA extraction and downstream gene expression analysis and also plated for determination of the number of CFU, as described above.
We also tested Salmonella survival under long-term cold conditions. Briefly, a log-phase culture was centrifuged at 7,200 × g for 5 min and resuspended in NB prechilled to 5°C (to achieve an optical density of 0.2, or ∼108 CFU/ml), followed by incubation at 5°C for 336 h. A sample of the culture was also incubated at 37°C for 336 h and served as a control. Samples were collected at 0.5 h, 5 h, 48 h, and 240 h for enumeration of the CFU, as described above, and for global proteome profiling.
Global proteome profiling.
Samples for proteome profiling were collected at 0.5 h, 5 h, 48 h, and 240 h for treatment (cold stress at 5°C) and as a control (no stress at 37°C) in triplicate, centrifuged at 7,200 × g for 5 min, washed twice with 0.9% sterile saline, and resuspended in 1 ml of distilled water. Bacteria were lysed in a Mini-Beadbeater (BioSpec Products, Bartlesville, OK) using 334 μg of microbeads of 106 μm in diameter and finer (Sigma, St. Louis, MO). The samples were centrifuged at 16,000 × g for 3 min. The supernatant (cell extract) was collected and stored in aliquots at −70°C. The total protein concentration was measured with a bicinchoninic acid protein assay kit (Pierce), and samples were submitted for proteome profiling using NanoACQUITY ultraperformance liquid chromatography-tandem mass spectrometry (MS; Waters, Manchester, United Kingdom) as described by Liang et al. (8) and Tang et al. (9). Briefly, each sample was digested with trypsin and introduced into a Symmetry C18 trapping column for analysis. The peptides were eluted from the trapping column before introduction into a mass spectrometer that was set for parallel fragmentation with scan times of 1.0 s and a low fragmentation energy of 5 V and a high fragmentation energy of 17 to 35 V. Fibrinopeptide B (Glu1) was used as the internal standard with the LockSpray dual electrospray ion source. Enolase was used as a spiked control. The Waters protein Lynx global server, version 2.2.5, was used to analyze the MS data set.
Gene expression profiling.
Samples for gene expression profiling for the stress treatments were collected upon preadaptation to each individual shock (0.5 h) and stress (5 h) treatment and also after exposure to the subsequent acid stress (pH 4.0 for 90 min) treatment. Sample preparation was performed as previously described (10, 11), with some modifications. Briefly, at various time points, the samples were collected as described for the stress treatments in two biological replicates and centrifuged at 7,200 × g for 5 min. The pellet was resuspended in TRIzol LS reagent (Invitrogen, Carlsbad, CA) and gently mixed, followed by centrifugation at 7,200 × g for 5 min. The TRIzol supernatant was stored in a clean tube for resuspension after proteinase K treatment. To the pellet, 1 ml of lysis enzyme cocktail containing 50 mg/ml of lysozyme (Sigma, St. Louis, MO) and 200 U/ml mutanolysin (Sigma) in TE buffer (10 mM Tris containing 1 mM EDTA, pH 8) was added. The solution was mixed gently and incubated at 37°C for 1 h, followed by centrifugation at 7,200 × g for 5 min. The supernatant was discarded and the pellet was resuspended in 250 μl of proteinase K buffer (100 mM Tris-HCl, 5 mM EDTA, 200 mM NaCl, 0.2% SDS, pH 8) containing 8 U/ml of proteinase K (Fermentas, Glen Burnie, MD). This was incubated at 55°C for 1 h with intermittent mixing, followed by resuspension in the TRIzol supernatant stored from the earlier step. RNA was obtained as described by the manufacturer's protocol (TRIzol LS; Invitrogen). RNA concentration and quality (A260/A280 and A260/A230, between 1.8 and 2.1) were measured using a NanoDrop spectrophotometer (Thermo Scientific, Waltham, MA). The RNA samples were analyzed for integrity using a 2100 BioAnalyzer (Agilent Technologies, Santa Clara, CA).
Total RNA (0.5 μg/μl) was reverse transcribed into cDNA with random hexamers and SuperScript II reverse transcriptase (Invitrogen), as described previously (10, 11), with some modifications. The reaction mixture was cleaned by using a QIAquick PCR purification kit (Qiagen, Valencia, CA) according to the manufacturer's instructions, except that the Tris-containing buffer was replaced with 75% ethanol for washing the column. The purified single-strand cDNA was eluted from the columns twice with a total of 100 μl of nuclease-free water (Ambion, Austin, TX).
cDNA fragmentation was done using DNase I (Promega, Madison, WI) according to the manufacturer's instructions. The fragmented cDNA was labeled using a GeneChip DNA labeling reagent (Affymetrix, Santa Clara, CA) and terminal transferase enzyme (New England BioLabs, Ipswich, MA). The samples were denatured prior to hybridization at 98°C for 10 min, followed by snap cooling at 4°C for 5 min.
Hybridization and normalization.
Labeled cDNA was hybridized onto a custom Affymetrix GeneChip designed to contain all the annotated coding sequences of S. Typhimurium LT2 ATCC 700720. Briefly, the array contained 9,852 probe sets, of which 4,735 probe sets were designed against S. Typhimurium. Each probe set contained 11 probes (25 bp). Labeled cDNA (500 ng) extracted from a pure culture of S. Typhimurium was hybridized to the chip as a positive control, which demonstrated that each probe set was specific for the intended open reading frame. The samples from the stress experiment were hybridized and scanned at the Center for Integrated BioSystems (Utah State University, Logan, UT) as per the manufacturer's protocols. Raw data (.cel files) were background corrected, quantile normalized, and summarized using the RMA-MS method (robust multiarray averaging, multispecies) (12) to take into account the genomes of the multiple species on the custom arrays. The resultant normalized log2-transformed intensity matrix was used for further statistical analysis.
NAD+/NADH quantification.
Nicotinamide nucleotide quantification was performed using an NAD+/NADH quantification kit (BioVision Research Products, Milpitas, CA) according to the manufacturer's instructions. Briefly, bacteria were centrifuged at 7,200 × g for 5 min, washed once with cold sterile phosphate-buffered saline (PBS; pH 7.2), and lysed using 334 μg of microbeads of 106 μm in diameter (Sigma, St. Louis, MO) in a Mini-Beadbeater. The samples were centrifuged (16,000 × g for 3 min) to collect the supernatant. The protein concentration of the cell extracts was measured, and the measurements of NAD+/NADH were normalized with respect to protein quantity.
Hydroxyl radical measurement.
Hydroxyl (OH·) radical measurement was performed using hydroxyphenyl fluorescein (HPF) (13) as described previously (14). Briefly, the bacterial cultures were centrifuged (7,200 × g, 5 min, 4°C) and washed once with an equal volume of sterile PBS. For the positive control, 20 mM H2O2 was used to treat the bacteria, whereas a nontreated bacterial suspension served as a negative control. The samples and assay controls were diluted to obtain 106 bacteria/ml, followed by addition of HPF at a final concentration of 5 μM per reaction mixture (15). The reaction mixtures were incubated for 60 min at 25°C in the dark. Fluorescence was measured using excitation at 488 nm and emission at 515 nm in a DTX 880 multimode detector (Beckman Coulter, Brea, CA).
Statistical analysis.
Stress survival, NAD+/NADH quantities, and OH· radical measurement data were analyzed using an unpaired Student t test. The difference was considered significant if P was ≤0.05. Data were collected at 0.5 h, 5 h, 48 h, and 240 h for the treatment (cold stress at 5°C) and control (no stress at 37°C) groups for proteome profiling. Because the amounts of some proteins were below the limit of detection in every sample, this led to several data points with values of zero, which impacted downstream statistical assessment. To account for this phenomenon while preserving interpretability, the expression level (Y) was transformed to a log(Y + 1+ε) scale, where ε is a very small random noise injected only when Y is equal to 0 (16). A two-class unpaired time course (repeated measures) with a corrected t statistic was used in significance analysis of microarrays (SAMs) (17) to calculate the q value. The difference was considered significant if q was ≤0.05. For gene expression data analysis, two-class unpaired with t statistic was used in the SAMs to calculate q values. The levels of expression of genes with q values of ≤0.05 were considered significantly different.
RESULTS
Stress survival of Salmonella.
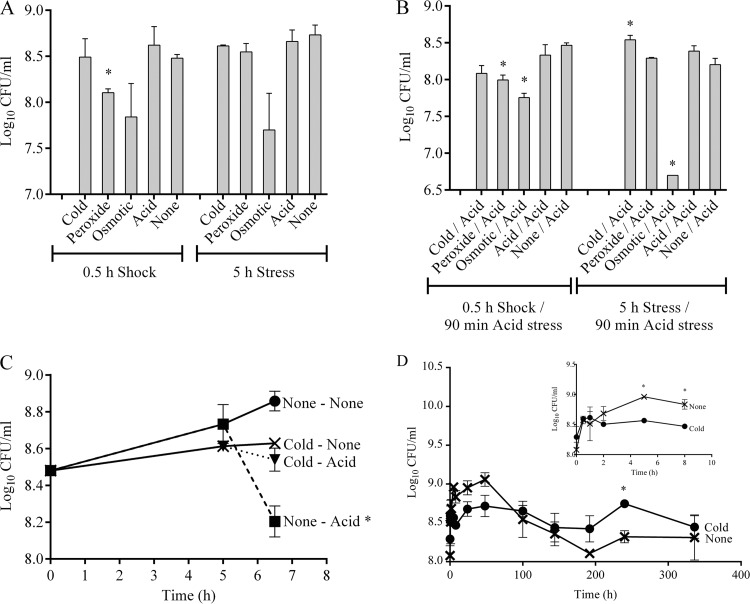
The response of S. Typhimurium to various conditions, such as cold, peroxide, osmotic, and acid, for 0.5 h (shock) and 5 h (stress) was tested. Only peroxide shock was found to significantly reduce the survival of S. Typhimurium (Fig. 1A). Consequently, the effect of preadaptation to the treatments described above on the survival of S. Typhimurium during subsequent acid stress exposure (pH 4.0 for 90 min) was examined. Preadaptation to peroxide shock, osmotic shock, and osmotic stress significantly reduced survival during subsequent acid stress exposure (Fig. 1B). Conversely, preadaptation to cold stress (5°C, 5 h) significantly increased survival during subsequent acid stress exposure (pH 4.0, 90 min) (Fig. 1B). This result was examined further, and it was found that preadaptation to cold stress rescued the bacteria from the effect of subsequent acid stress, leading to no reduction in cell density, as observed upon exposure to acid stress alone (Fig. 1C). In addition, the ability of S. Typhimurium to withstand exposure to low temperatures for long periods (336 h) was tested. During this time, survival at 37°C declined after 48 h, while at 5°C, significantly higher numbers of viable bacteria were observed between 48 h and 240 h. S. Typhimurium survived cold stress at a cell density of ∼108 CFU/ml, which was similar to the density of the original inoculum (Fig. 1D).
Fig 1.
Survival after shock and stress treatments. (A) Survival of S. Typhimurium after cold (5°C), peroxide (1 mM H2O2), osmotic (5% NaCl), and acid (pH 5.3) shock (0.5 h) and stress (5 h) treatments. *, significant difference (P < 0.05) compared to no shock and no stress. (B) Survival of S. Typhimurium after preadaptation to cold (5°C), peroxide (1 mM H2O2), osmotic (5% NaCl), and acid (pH 5.3) shock (0.5 h) and stress (5 h) treatments, followed by exposure to acid stress (pH 4.0, 90 min). *, significant difference (P < 0.05) compared to no shock/acid (pH 4.0, 90 min) and no stress/acid (pH 4.0, 90 min). (C) Survival after cold stress (5°C, 5 h) alone and after preadaptation to cold stress (5°C, 5 h) followed by exposure to acid stress (pH 4.0, 90 min). *, significant difference (P < 0.05) compared to no stress. (D) Survival of S. Typhimurium during cold stress (5°C) and no stress (37°C) for 336 h. (Inset) Growth between 0 h and 8 h. *, significant difference (P < 0.05) compared to no stress.
Global proteome profiling of cold-stressed Salmonella.
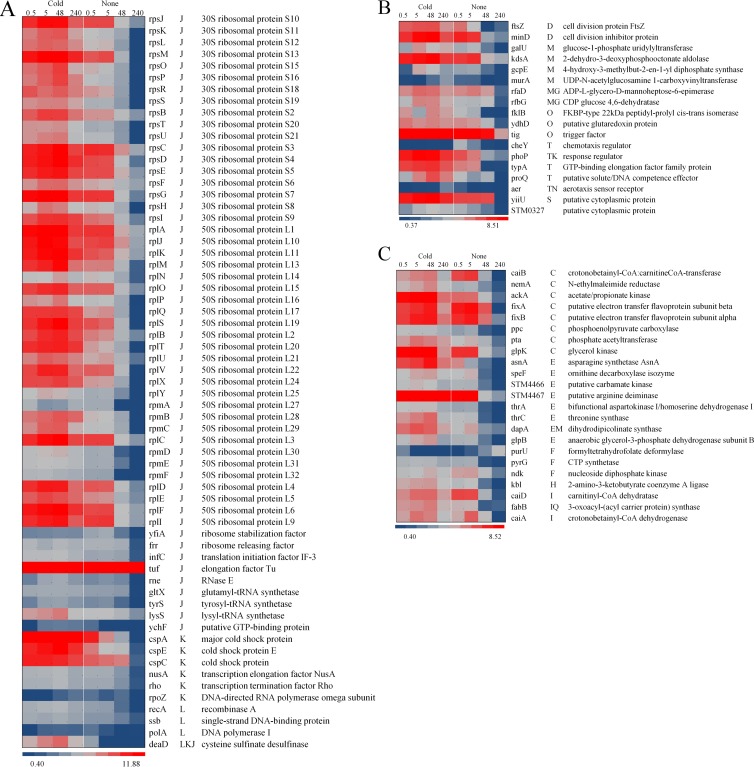
The proteome profile was determined primarily to confirm the differential expression of proteins in response to short-term (5 h) and long-term (up to 336 h) cold stress. This analysis showed that exposure to cold stress led to differential regulation of 104 proteins. The differentially regulated proteins were grouped into three categories and 16 clusters of orthologous groups (COGs), which included (i) information storage and processing (COGs J, K, and L), (ii) cellular processes (COGs D, O, M, N, P, and T), and (iii) metabolism (COGs C, G, E, F, H, I, and Q), where the definitions of the abbreviations for the COG categories are as follows: J, translation; K, transcription; L, DNA replication, recombination, and repair; D, cell division and chromosome partitioning; O, posttranslational modification, protein turnover, chaperones; M, cell envelope biogenesis, outer membrane; N, cell motility and secretion; P, inorganic ion transport and metabolism; T, signal transduction mechanisms; C, energy production and conversion; G, carbohydrate transport and metabolism; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; H, coenzyme metabolism; I, lipid metabolism; and Q, secondary metabolite biosynthesis, transport, and catabolism. Protein expression significantly declined after 48 h in the control group, whereas it remained consistently induced in cold-stressed bacteria during the entire treatment period. This observation correlated with the trend for the viable bacteria (in numbers of CFU/ml) in the control group versus the cold-stressed bacteria (Fig. 1D). Of the ribosomal proteins detected, 77% (44/57) remained induced during the entire cold stress exposure. Cold shock proteins CspA, CspE, and CspC were also induced, and their levels remained constant during the cold stress treatment but declined after 48 h during incubation at 37°C. Two DNA recombination and repair proteins (RecA and Ssb) were induced with cold stress (Fig. 2A). Importantly, the cell division proteins FtsZ and MinD remained induced at 5°C but declined after 48 h in the control group (Fig. 2B). The proteins associated with metabolism also followed the trend of the other proteins which were induced by cold stress and remained induced for 336 h under conditions of cold stress (Fig. 2C). This expression trend was pervasive among the proteins found between the cold stress and the control groups, except for PurU (formyltetrahydrofolate deformylase), which was repressed during cold stress (Fig. 2C).
Fig 2.
Heat map representations of the 104 differentially regulated proteins (q < 0.05) due to cold stress (5°C) over time (0.5 h, 5 h, 48 h, and 240 h, as indicated above the map). The proteins are grouped on the basis of the COG functional annotation. (A) Proteins related to information storage and processing (COGs J, K, and L); (B) proteins related to cellular processes (COGs D, O, M, N, P, and T). (C) Proteins related to metabolism (COGs C, E, F, H, G, I, and Q). The blue-gray-red color scale corresponds to low expression intensity to high expression intensity. CoA, coenzyme A.
The differential regulation of the proteins in response to cold stress was observed from 5 h through 240 h. However, the method used for proteome profiling here is more sensitive to detection of highly abundant proteins and therefore could potentially miss some of the low-abundance proteins. Consequently, we used global gene expression technology to define the specific regulatory and metabolism changes in response to preadaptation of S. Typhimurium to cold stress, which rescued the bacteria from the deleterious effects of subsequent exposure to acid stress, as was observed earlier (Fig. 1C).
Gene expression during stress.
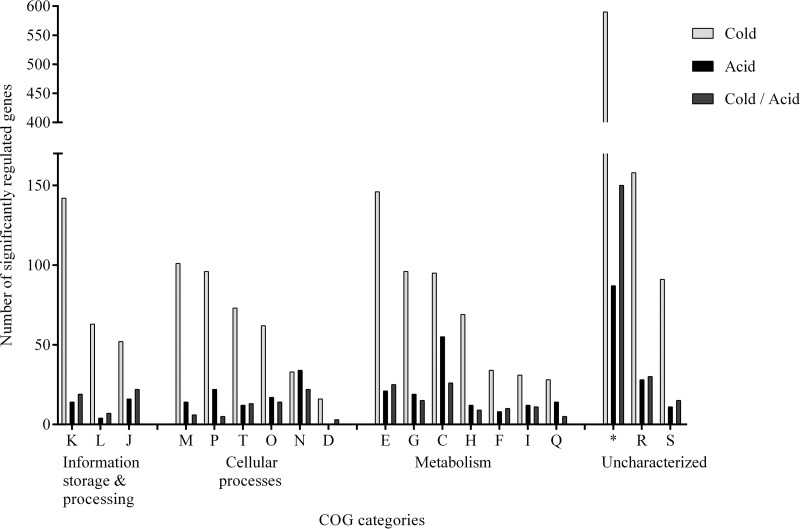
Gene expression analysis resulted in the identification of 1,809 significantly differentially expressed genes due to preadaptation to cold stress (5°C, 5 h) and 357 significantly differentially expressed genes due to acid stress (pH 4.0, 90 min) alone. The top three COG categories regulated due to preadaptation to cold stress were transcription (COG K), membrane biogenesis (COG M), and amino acid metabolism (COG E), while acid stress regulated the COG categories for energy metabolism (COG C), cell motility (COG N), and ion transport (COG P) (Fig. 3).
Fig 3.
Distribution in COG categories of 1,809 genes significantly (q < 0.05) regulated due to cold stress (5°C, 5 h) and 357 genes significantly (q < 0.05) regulated due to subsequent acid stress (pH 4.0, 90 min) alone.
(i) Differentially regulated genes in response to stress.
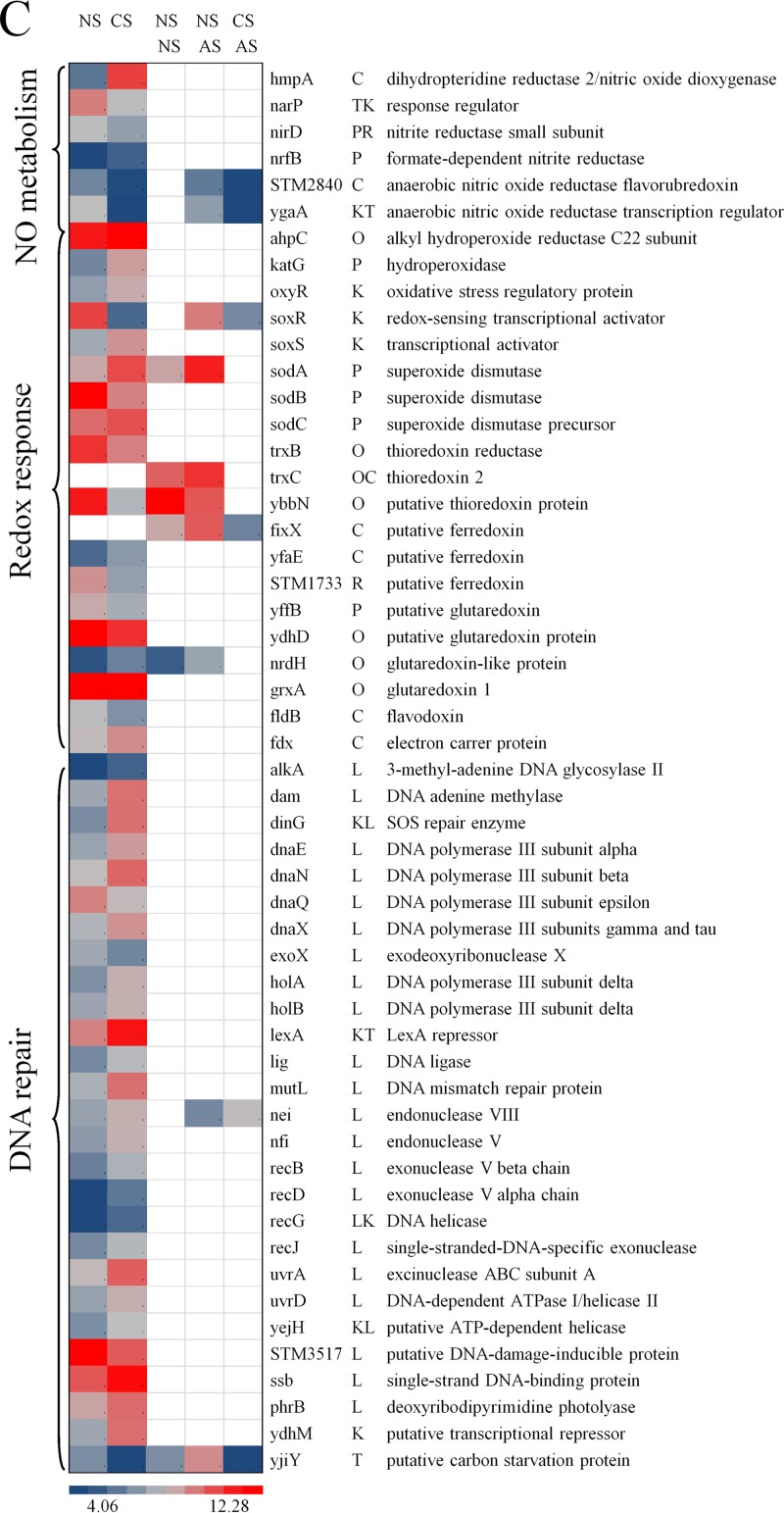
Cold stress genes and proteins regulate membrane fluidity and resume protein synthesis (18), and this effect was reflected by induction of cold stress genes (cspB and cspD) and proteins (CspA, CspE, and CspC) in response to preadaptation to cold stress. COG analysis also revealed that a high number of genes associated with membrane biogenesis (COG M) and transcription (COG K) were differentially regulated (Fig. 3). Among the genes associated with amino acid metabolism (COG E), multiple genes related to glutamate, glutamine, arginine, and lysine transport and metabolism, in response to preadaptation to cold stress, were significantly upregulated. The downregulated genes included STM2186, guaA, STM4463, STM4464, STM4467, and yggA (Fig. 4B). Glutamate-, arginine-, and lysine-dependent systems provide resistance to acidic conditions by regulating and maintaining the internal pH homeostasis via production of ammonia and carbon dioxide (19–22). Moreover, lysine decarboxylases (cadA and ldcC) and lysine/cadaverine transport (cadB) were significantly induced with cold stress (Fig. 4B). In addition, the oxidative stress genes ahpC, katG, oxyR, soxS, sodA, sodC, and grxA were significantly induced in response to preadaptation to cold stress, which may provide increased protection against hydrogen peroxide and hydroxyl and superoxide radicals (23) (Fig. 4C). Multiple genes involved in DNA repair were induced due to preadaptation to cold stress, except for the genes dnaQ, exoX, STM3517, and yjiY, which were repressed (Fig. 4C). On the contrary, the genes involved in proteolysis were significantly repressed, except for the genes clpA and clpS, which were induced (Fig. 4B).
Fig 4.
Heat map representations of genes differentially regulated (q < 0.05) due to cold stress (5°C, 5 h) and acid stress (pH 4.0, 90 min) and due to preadaptation to cold stress (5°C, 5 h) followed by subsequent exposure to acid stress (pH 4.0, 90 min). Abbreviations: NS, no stress; CS, cold stress; AS, acid stress. In each heat map, the first two columns represent significant (q < 0.05) differences in gene expression due to cold stress (5°C, 5 h) alone; the third and fourth columns represent significant (q < 0.05) differences in gene expression due to acid stress (pH 4.0, 90 min) alone; the fourth and fifth columns represent significant (q < 0.05) differences in gene expression due to preadaptation to cold stress and subsequent exposure to acid stress (pH 4.0, 90 min). The blue-gray-red color scale corresponds to low expression intensity to high expression intensity. An empty white block or no color means that the particular gene for that respective comparison was not significantly (q < 0.05) differently regulated.
Interestingly, acid stress alone induced the genes glnA, glnQ, gltI, and fliB, which are related to glutamate and lysine transport and metabolism (Fig. 4B); induced the genes sodA and trxC, which are related to oxidative stress; repressed the genes hslV, dnaJ, clpB, and clpP, which are involved in proteolysis; and did not differentially regulate any of the genes related to DNA repair (Fig. 4C).
Only a few of the above-mentioned genes, the majority of which are responsible for proteolysis, were regulated in S. Typhimurium that was preadapted to cold stress and subsequently exposed to acid stress. All the genes, including the genes involved in proteolysis, were repressed and followed a trend similar to that seen in response to preadaptation to cold stress alone (Fig. 4B and C).
(ii) Differentially regulated genes involved in the TCA cycle.
The citric acid (tricarboxylic acid [TCA]) cycle feeds substrates, such as NADH and succinate, to the electron transfer chain (ETC) to produce energy. Acid stress regulated a large number of genes involved in energy production and conversion (COG C) (Fig. 3). The genes involved in the TCA cycle, such as icdA, sucAD, acnB, and mdh, were induced due to acid stress, whereas preadaptation to cold stress induced the genes acnA, icdA, and sucAD (Fig. 4A), suggesting a difference in NADH production and energy requirement in response to preadaptation to cold stress and exposure to acid stress alone. None of the above-mentioned genes involved in the TCA cycle were differentially regulated in S. Typhimurium preadapted to cold stress and subsequently exposed to acid stress.
(iii) Differentially regulated genes involved in ETC.
The NADH and succinate generated in the TCA cycle are oxidized in the ETC, and NAD+ and fumarate are regenerated, thus providing energy to power ATP synthase. Genes encoding the four complexes of ETC were regulated very differently in response to preadaptation to cold stress in comparison with their regulation in response to exposure to acid stress. Multiple genes involved in complex I (NADH dehydrogenase) of the ETC were induced due to preadaptation to cold stress, whereas only two of them, nuoG and nuoH, were induced due to acid stress (Fig. 4A). Multiple genes involved in complex II (succinate dehydrogenase) of the ETC were induced due to acid stress, whereas preadaptation to cold stress did not significantly regulate this complex. Genes encoding fumarate reductase subunits (frdA to frdD) and cytochrome o ubiquinol oxidase subunits (cyoA and cyoB) were repressed due to preadaptation to cold stress, except for two genes, cyoD and cydB. On the other hand, acid stress induced cytochrome o ubiquinol oxidase subunits (cyoA to cyoD) and ATP synthase subunits (atpA, atpD, and atpF to atpH). None of the above-mentioned genes, except cyoA, cyoB, and atpA, involved in complexes of the ETC were differentially regulated in S. Typhimurium preadapted to cold stress and subsequently exposed to acid stress. These observations indicate differences in NADH consumption and conversion to NAD+ in the ETC in response to preadaptation to cold stress and exposure to acid stress alone.
Oxidative state.
The preadaptation to cold stress alone significantly decreased the ratio of NAD+/NADH and exposure to acid stress alone significantly increased the ratio compared with that for their respective controls. When S. Typhimurium was preadapted to cold stress and subsequently exposed to acid stress, the result was a significant reduction of the NAD+/NADH ratio compared with that after exposure to acid stress alone and no stress (Fig. 5A). Conversion of NADH to NAD+ in the ETC leads to superoxide generation; thus, a sudden spike in the NAD+/NADH ratio leads to increased superoxide generation and, hence, oxidative damage (15). When the capacity to dismutate superoxide is not sufficient, free superoxide radicals damage the Fe-S clusters, releasing Fe2+ ions. The free Fe2+ ions react with hydrogen peroxide to produce OH· radicals, as described in the Fenton reaction (24, 25), leading to DNA damage and cell death (15, 25, 26). The OH· radical formation significantly decreased with preadaptation to cold stress. When S. Typhimurium was preadapted to cold stress and subsequently exposed to acid stress, OH· radical formation further declined (Fig. 5B). The exposure to acid stress alone did not significantly change the amount of OH· radicals formed. These oxidative state results combined with the gene expression profiles suggest that the preadaptation to cold stress induced multiple metabolic cascades that relieved the redox damage when the bacteria were subsequently exposed to acid stress.
Fig 5.
(A, B) NAD+/NADH ratio measurements (A) and OH· radical measurements performed using HPF (B) upon exposure to cold stress (5°C, 5 h) and acid stress (pH 4.0, 90 min) and upon preadaptation to cold stress (5°C, 5 h) before exposure to subsequent acid stress (pH 4.0, 90 min). *, significant difference (P < 0.05) compared to the respective no-stress control; •, significant difference compared to no stress followed by acid stress treatment (pH 4.0, 90 min); RFU, relative fluorescence units.
DISCUSSION
The viable cell count data showed that the preadaptation of S. Typhimurium to cold stress (5°C, 5 h) significantly increased its survival during subsequent acid stress exposure (pH 4.0, 90 min), despite the fact that S. Typhimurium LT2 has a rare UUG start codon for rpoS (27), which leads to the inability of S. Typhimurium to elicit a sustained acid tolerance response (7). The COG analysis of the gene expression data provided an advantage over single-gene analysis, in revealing the major functions that were being regulated in response to preadaptation to cold stress and that rescued the bacteria from the effects of subsequent exposure to acid stress. Preadaptation to cold stress induced the genes associated with glutamate-, arginine-, and lysine-dependent systems that function in maintaining pH homeostasis (7, 20); genes associated with oxidative stress that function in protecting against secondary oxidative damage from acid exposure (7, 20); and genes associated with DNA repair, all of which are usually expected to be induced upon exposure to acid stress in an rpoS-dependent manner in other Salmonella strains (18–23). These observations suggested that exposure to cold stress bypassed the rpoS requirement in eliciting the response required for protection against mild acidic conditions and its secondary effects. In contrast to the acid stress response (19, 20, 22, 28), the response to cold stress does not require sigma factor. While discovery of the specific transcription initiation factor for the induced genes or the mechanism of gene regulation in response to cold stress in S. Typhimurium was beyond the scope of this study, it is evident that preadaptation to cold stress was able to activate that factor or regulate the expression of the genes mentioned above, which in turn rescued the bacteria from the effects of subsequent acid stress exposure and its secondary effects.
Further investigations into the expression profiles of the genes in the COG category regulated by acid stress (COG C) revealed a metabolic link to the TCA cycle, which feeds substrates such as NADH and succinate into the ETC for oxidation and energy production. The genes involved in the TCA cycle and the ETC were regulated very differently in response to preadaptation to cold stress and in response to acid stress alone, indicating differences in NAD+/NADH and redox modulation. This study demonstrated that exposure to acid stress increased the NAD+/NADH ratio. However, preadaptation to cold stress before exposure to subsequent acid stress decreased the NAD+/NADH ratio and, in turn, decreased OH· radical formation, rescuing the bacteria from secondary oxidative damage due to acid exposure.
The results from this study suggest that preadaptation to cold (abiotic) stress regulates mechanisms in S. Typhimurium that enable survival and persistence during subsequent acid exposure (biotic stress). The mechanism revealed in this study can hold true for any other Salmonella strain, since it can be speculated to occur either simultaneously with the rpoS activation cascade or independently of rpoS activation. Further studies with multiple Salmonella strains will be needed to discern the additional detailed mechanisms that will help in developing alternative strategies for pathogen elimination and to increase food safety.
Footnotes
Published ahead of print 20 September 2013
REFERENCES
- 1.Centers for Disease Control and Prevention 2013. Surveillance for foodborne disease outbreaks—United States, 2009-2010. MMWR Morb. Mortal. Wkly. Rep. 62:41–47 [PMC free article] [PubMed] [Google Scholar]
- 2.Maki DG. 2009. Coming to grips with foodborne infection—peanut butter, peppers, and nationwide Salmonella outbreaks. N. Engl. J. Med. 360:949–953 [DOI] [PubMed] [Google Scholar]
- 3.Camilleri M, Colemont LJ, Phillips SF, Brown ML, Thomforde GM, Chapman N, Zinsmeister AR. 1989. Human gastric emptying and colonic filling of solids characterized by a new method. Am. J. Physiol. 257:G284–G290 [DOI] [PubMed] [Google Scholar]
- 4.Cann PA, Read NW, Cammack J, Childs H, Holden S, Kashman R, Longmore J, Nix S, Simms N, Swallow K, Weller J. 1983. Psychological stress and the passage of a standard meal through the stomach and small intestine in man. Gut 24:236–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agunod M, Yamaguchi N, Lopez R, Luhby A, Glass G. 1969. Correlative study of hydrochloric acid, pepsin, and intrinsic factor secretion in newborns and infants. Dig. Dis. Sci. 14:400–414 [DOI] [PubMed] [Google Scholar]
- 6.Lonnerdal B. 2002. Expression of human milk proteins in plants. J. Am. Coll. Nutr. 21:218S–221S [DOI] [PubMed] [Google Scholar]
- 7.Lee IS, Lin J, Hall HK, Bearson B, Foster JW. 1995. The stationary-phase sigma factor sigma S (RpoS) is required for a sustained acid tolerance response in virulent Salmonella typhimurium. Mol. Microbiol. 17:155–167 [DOI] [PubMed] [Google Scholar]
- 8.Liang Y, Gardner DR, Miller CD, Chen D, Anderson AJ, Weimer BC, Sims RC. 2006. Study of biochemical pathways and enzymes involved in pyrene degradation by Mycobacterium sp. strain KMS. Appl. Environ. Microbiol. 72:7821–7828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang CT, Guo Y, Lu M, Wang YN, Peng JY, Wu WB, Li WH, Weimer BC, Chen D. 2009. Development of larval Schistosoma japonicum blocked in Oncomelania hupensis by pre-infection with larval Exorchis sp. J. Parasitol. 95:1321–1325 [DOI] [PubMed] [Google Scholar]
- 10.Ganesan B, Dobrowolski P, Weimer BC. 2006. Identification of the leucine-to-2-methylbutyric acid catabolic pathway of Lactococcus lactis. Appl. Environ. Microbiol. 72:4264–4273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie Y, Chou LS, Cutler A, Weimer B. 2004. DNA Macroarray profiling of Lactococcus lactis subsp. lactis IL1403 gene expression during environmental stresses. Appl. Environ. Microbiol. 70:6738–6747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stevens JR, Ganesan B, Desai P, Rajan S, Weimer BC. 2008. Statistical issues in the normalization of multi-species microarray data, p 47–62 In Proceedings of the Conference on Applied Statistics in Agriculture. Kansas State University, Manhattan, KS [Google Scholar]
- 13.Setsukinai K, Urano Y, Kakinuma K, Majima HJ, Nagano T. 2003. Development of novel fluorescence probes that can reliably detect reactive oxygen species and distinguish specific species. J. Biol. Chem. 278:3170–3175 [DOI] [PubMed] [Google Scholar]
- 14.Mols M, van Kranenburg R, Tempelaars MH, van Schaik W, Moezelaar R, Abee T. 2010. Comparative analysis of transcriptional and physiological responses of Bacillus cereus to organic and inorganic acid shocks. Int. J. Food Microbiol. 137:13–21 [DOI] [PubMed] [Google Scholar]
- 15.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810 [DOI] [PubMed] [Google Scholar]
- 16.Stevens JR. 2013. Statistical methods in metabolomics. Woodhead Publishing Series in Food Science. Woodhead Publishing Ltd., Cambridge, United Kingdom [Google Scholar]
- 17.Tusher VG, Tibshirani R, Chu G. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 98:5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamanaka K. 1999. Cold shock response in Escherichia coli. J. Mol. Microbiol. Biotechnol. 1:193–202 [PubMed] [Google Scholar]
- 19.Kieboom J, Abee T. 2006. Arginine-dependent acid resistance in Salmonella enterica serovar Typhimurium. J. Bacteriol. 188:5650–5653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin J, Smith M, Chapin K, Baik H, Bennett G, Foster J. 1996. Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl. Environ. Microbiol. 62:3094–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moreau PL. 2007. The lysine decarboxylase CadA protects Escherichia coli starved of phosphate against fermentation acids. J. Bacteriol. 189:2249–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richard H, Foster JW. 2004. Escherichia coli glutamate- and arginine-dependent acid resistance systems increase internal pH and reverse transmembrane potential. J. Bacteriol. 186:6032–6041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farr SB, Kogoma T. 1991. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol. Mol. Biol. Rev. 55:561–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imlay JA. 2006. Iron-sulphur clusters and the problem with oxygen. Mol. Microbiol. 59:1073–1082 [DOI] [PubMed] [Google Scholar]
- 25.Imlay JA, Linn S. 1988. DNA damage and oxygen radical toxicity. Science 240:1302–1309 [DOI] [PubMed] [Google Scholar]
- 26.Mols M, van Kranenburg R, van Melis CC, Moezelaar R, Abee T. 2010. Analysis of acid-stressed Bacillus cereus reveals a major oxidative response and inactivation-associated radical formation. Environ. Microbiol. 12:873–885 [DOI] [PubMed] [Google Scholar]
- 27.Wilmes-Riesenberg MR, Foster JW, Curtiss R., III 1997. An altered rpoS allele contributes to the avirulence of Salmonella typhimurium LT2. Infect. Immun. 65:203–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park YK, Bearson B, Bang SH, Bang IS, Foster JW. 1996. Internal pH crisis, lysine decarboxylase and the acid tolerance response of Salmonella typhimurium. Mol. Microbiol. 20:605–611 [DOI] [PubMed] [Google Scholar]