Abstract
The behavior of yeast cells during industrial processes such as the production of beer, wine, and bioethanol has been extensively studied. In contrast, our knowledge about yeast physiology during solid-state processes, such as bread dough, cheese, or cocoa fermentation, remains limited. We investigated changes in the transcriptomes of three genetically distinct Saccharomyces cerevisiae strains during bread dough fermentation. Our results show that regardless of the genetic background, all three strains exhibit similar changes in expression patterns. At the onset of fermentation, expression of glucose-regulated genes changes dramatically, and the osmotic stress response is activated. The middle fermentation phase is characterized by the induction of genes involved in amino acid metabolism. Finally, at the latest time point, cells suffer from nutrient depletion and activate pathways associated with starvation and stress responses. Further analysis shows that genes regulated by the high-osmolarity glycerol (HOG) pathway, the major pathway involved in the response to osmotic stress and glycerol homeostasis, are among the most differentially expressed genes at the onset of fermentation. More importantly, deletion of HOG1 and other genes of this pathway significantly reduces the fermentation capacity. Together, our results demonstrate that cells embedded in a solid matrix such as bread dough suffer severe osmotic stress and that a proper induction of the HOG pathway is critical for optimal fermentation.
INTRODUCTION
Solid-state fermentation (SSF) is defined as fermentation in the absence or near absence of free water, in which microbes grow on a moist solid substrate (1). SSF is commonly used to produce many different kinds of food, including cheese, soy sauce, and bread (2). Similarly, SSF has also been used successfully for the production of several enzymes (3–6) and secondary metabolites such as antibiotics, mycotoxins, and biosurfactants, etc. (7). Furthermore, there is a renewed interest in using SSF for bioethanol production from agricultural crops and their products (8–10).
The main difference between SSF and liquid fermentations such as those used in the production of beer and wine is the reduced level of free water in SSF. As a consequence, the movement of cells and diffusion of cellular metabolites and nutrients are limited, which may result in local gradients of temperature, moisture, pH, nutrients, and waste products (11). Moreover, the growth, gene expression profile, and metabolism of fungi during solid-state fermentation are different from those during liquid-state fermentation (7, 12–14). This might be due to a different cellular physiology of the fungi in the solid state compared to the liquid state.
Despite its industrial relevance, yeast physiology during solid-state fermentations is relatively understudied compared to liquid fermentations. This is especially the case for bread dough fermentation, where no data are available that describe yeast cell physiology during the fermentation process. This is at least partly due to the technical difficulties associated with isolating representative cell samples from the solid fermentation matrix. Some studies have attempted to gain insight into bread fermentation by mimicking the process in liquid (15–17). Whereas such studies definitely contributed to our knowledge, they change the single most important parameter of SSFs (the solid state) and are therefore not an optimal model for the industrial SSF process.
Global gene expression analysis approaches such as microarray analysis or transcriptome sequencing (RNA-seq) are powerful tools to obtain a snapshot of the cell's transcriptome and thereby gain insight into a cell's physiological state. Several studies have investigated the expression profile of yeast genes during liquid fermentation such as beer, wine, or even liquid model dough fermentation (15, 16, 18–23). Those studies showed that in general, the start of fermentation is associated with a loss of stress resistance due to the repression of stress-related genes by glucose or sucrose (24–26). Furthermore, in some cases, induction of pathways associated with osmotic stress has also been observed, presumably due to a high sugar concentration of the substrate (27).
Here, we optimized a method to isolate high-quality RNA samples from Saccharomyces cerevisiae cells embedded in fermenting bread dough and thus studied changes in the yeast transcriptome throughout the fermentation process. Using this method, we tracked changes in the transcriptomes of three genetically distinct S. cerevisiae strains during bread dough fermentation. Our results show that all strains show similar expression trends, with some subtle differences. In the first phase of fermentation, the cells need to adapt to the high osmolarity and nutrient concentration of the surrounding dough matrix. This initial adaptation phase is followed by an active fermentation phase, where cells first consume preferred sugars such as glucose and sucrose before switching to maltose. At the end of the short but vigorous fermentation, cells experience starvation and start building up stress resistance. By investigating the effect of the deletion of genes that are highly induced at the start of dough fermentation, we find that the high-osmolarity glycerol (HOG) pathway is instrumental for adaptation to solid-state conditions.
MATERIALS AND METHODS
Strains and microbial procedure.
Three isolated commercial strains from bread, wine, and the bioethanol industries were used for investigation of changes in the yeast transcriptome during dough fermentation. To select these 3 strains, we screened 24 genetically distinct strains for their dough fermentation capacity. The interdelta genetic fingerprinting assay was done as described previously by Legras and Karst (28). The method is based on the PCR amplification of interdelta regions using interdelta primers (delta 12 and delta 21) (28).
For follow-up on the results of the RNA-seq experiment, we made use of mutants with a single deletion of the genes of interest, available in the yeast deletion collection (S288c; Invitrogen). The deletion of each gene was confirmed by PCR.
Yeast cultures were grown under optimal conditions as described previously, and standard procedures for isolation and manipulation of DNA were used (29, 30). For dough preparation, the yeast cells were harvested at early stationary phase and washed with water before inoculation into dough.
Flour characterization, dough preparation, and fermentation.
Commercial flour obtained from Ceres (Vilvoorde, Belgium) was used throughout this study. Dough was prepared according to the straight-dough method described previously by Shogren and Finney (31), using the following formula: 100.0 g flour (on a 14% moisture basis), 6.0% (wt/wt) sucrose, 1.5% (wt/wt) sodium chloride, 52.0% (vol/wt) water, and 5.3% yeast. The ingredients were mixed in a 100-g pin bowl mixer (National Manufacturing, Lincoln, NE, USA) for 3 min 50 s. Next, the dough was divided into 10-g pieces, which were fermented in a fermentation cabinet at 30°C with a relative humidity (RH) of 90% for 30, 60, and 180 min.
Gas production measurement.
The volume of gas produced by different strains and mutants during dough fermentation was measured by using a Risograph instrument (National Manufacturing). Balls of dough were made as described above and were left to ferment for a maximum of 180 min at 30°C in the Risograph instrument. Gas production was measured continuously at 1-min intervals.
Sampling for RNA extraction.
Multiple pieces of dough were prepared and fermented in a fermentation cabinet (30°C and 90% relative humidity) for a maximum of 180 min. Samples were taken at 30, 60, and 180 min after the onset of fermentation. The samples were frozen by using liquid nitrogen and kept at −80°C before RNA extraction. Nonfermenting yeast cells right before mixing with dough were used as the “0-min” sample. The experiment was performed in parallel for two biological replicates of each strain.
RNA isolation.
We developed a method to isolate RNA from yeast cells embedded in bread dough based on standard methods for RNA isolation (29, 30) and a method described previously by Panadero et al. (32). Pieces of dough (0.5 g) were homogenized with 3.0 ml of ice-cold LETS buffer (0.2 M LiCl, 0.02 M EDTA, 0.02 mM Tris-HCl [pH 8.0], 0.4% SDS) by using an Ultra-Turrax T10 Basic disperser (IKA Werke GmbH, Staufen, Germany). Aliquots of 0.5 ml of the dough suspension were transferred into screw-cap microtubes containing 0.5 ml of phenol-chloroform-isoamyl alcohol (25:24:1) and 0.5 ml glass beads (acid-washed beads, 425 to 600 μm in diameter). The suspension was mixed vigorously three times for 20 s by using a Fast Prep homogenizer (MP Biomedicals). After centrifugation at 17,900 × g for 10 min at 4°C, the upper phases from 2 tubes were pooled in a microtube containing 0.5 ml chloroform-isoamyl alcohol (24:1). After thorough vortexing, phases were separated by centrifugation at 17,900 × g for 10 min (at 4°C). The upper phase was transferred into a clean microtube, and this step was repeated until the interface between the aqueous and organic layers was clear after centrifugation. Total nucleic acids were precipitated with 1 ml ice-cold 100% ethanol and 25 μl 40% potassium acetate and kept at −20°C overnight. Samples were then thawed on ice the next day and were centrifuged at 15,000 × g for 10 min. Each pellet was resuspended in 350 μl of RLT buffer (Qiagen GmbH, Hilden, Germany) plus 350 μl of 70% ethanol and was loaded onto an RNeasy minicolumn (Qiagen). Column eluents were pooled, the A260/A280 ratio was used to estimate RNA purity, and the quality of RNA was checked on a 1.2% agarose–Tris-acetate-EDTA (TAE)–formaldehyde gel. In the case of impurities, a second precipitation was done as described above, and the precipitate was washed by using 70% ethanol and resuspended in RNase-free water.
RNA-seq and data analysis.
Illumina next-generation sequencing with 100× coverage of the haploid S. cerevisiae genome was carried out for all samples, as described previously (33). After removal of the low-quality reads and adaptors, RNA-seq reads (50 bp, single end) were aligned to the Saccharomyces cerevisiae S288c reference genome (version EF4.66) by using TopHat 2.0.7 (34). Aligned reads were further visualized by using Tablet (35). Between 5 × 106 and 35 × 106 reads were aligned for each sample. RPKM (reads per kilobase per million mapped reads) expression values were calculated with Cufflinks 2.0.2 (36). Next, Cuffdiff was used to determine differential expression by comparing transcript abundances between pairs of duplicate experiments (see the supplemental material for details). Genes with significant differential expression (at least 2-fold changes and a false discovery rate [FDR] value of <0.0001) were selected for further analysis.
Hierarchical clustering for heat map.
After the removal of zero values, RPKM values were log2 transformed and quantile normalized. This was followed by hierarchical clustering with average linkage using Euclidian distance. Next, using the gPlots package in R, the log2-transformed values were plotted on a blue-yellow heat map, with saturation of colors at 5% extreme values.
Categorization of expression profiles and Gene Ontology.
Since we have a short time series, we used template-matching clustering of the expression data, using ORIClust (37). We performed the time-profile clustering on a specific filtered subset of genes, namely, those identified by edgeR to be differentially expressed in at least one time point compared to time point zero (38). We removed the rows where at least one of the samples had an RPKM of zero, log2 transformed the data, and performed a quantile normalization. This subset of genes was analyzed by using ORIClust (ORICC2 algorithm) to assign the genes to meaningful time-dependent categories. Next, we used the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov/) to search for enriched gene sets in different clusters (39). The Functional Annotation Tool in the online version of DAVID was run by using the default parameters, and the top 50 Gene Ontology (GO) terms in the Gene Ontology Biological Process category are reported (FDR < 0.01).
Physical interaction network.
A physical interaction network, G(V,E), models the interactome of an organism. In this network, the edges, E, represent the physical interactions between the different genes/gene products, V. Different layers of interactions are combined in this network, namely, protein-protein, protein-DNA, and phosphorylation interactions (40). Each interaction in the network is scored with a probability value. This value reflects the likelihood that the interaction truly exists in the organism's interactome (41, 42). To complement and annotate the interaction network, protein complex data were added to the network (43).
PheNetic.
PheNetic is an algorithm which extracts minimal connected subnetworks between a cause, a genetic perturbation, and its effects, differentially expressed genes (44). To this end, the algorithm selects from the network the edges which best connect the given cause to its effect through a regulatory path by maximizing the score S(D) in the equation below. A regulatory path is defined here as a path which can lead to differential expression from the given cause to an effect and thus a path in which the last edge is a protein-DNA edge.
In this equation, D is a subnetwork from G; I is the set of cause-effect pairs, where x corresponds to the cause and y corresponds to the effect; fr is a reward function based on the degree of differential expression; and P[regPath(x,y)|D] is the probability that a regulatory path exists in the selected subnetwork D from x to y. Finally, xC is a constant cost factor, which allows us to define the size of the selected network.
Here, PheNetic was used with a path length of 5, with the 20 best proofs for each of the 100 cause-effect pairs with the largest degree of differential expression (44). No adjustments of the probabilities in the network based on network centrality were performed.
Network visualization and analysis.
Visualization and analysis of the network were performed by using cytoscape (45), clueGO (46), and BiNGO (47). GO enrichments were performed by using a hypergeometric test and a Benjamini-Hochberg correction (48). GO annotations for S. cerevisiae were downloaded from the Gene Ontology (49) website (version 1.1600).
RNA-seq data.
Sequencing data were deposited in the GenBank database under bioproject identifier PRJNA212389.
RESULTS
Strain selection for dough fermentation and transcriptome analysis.
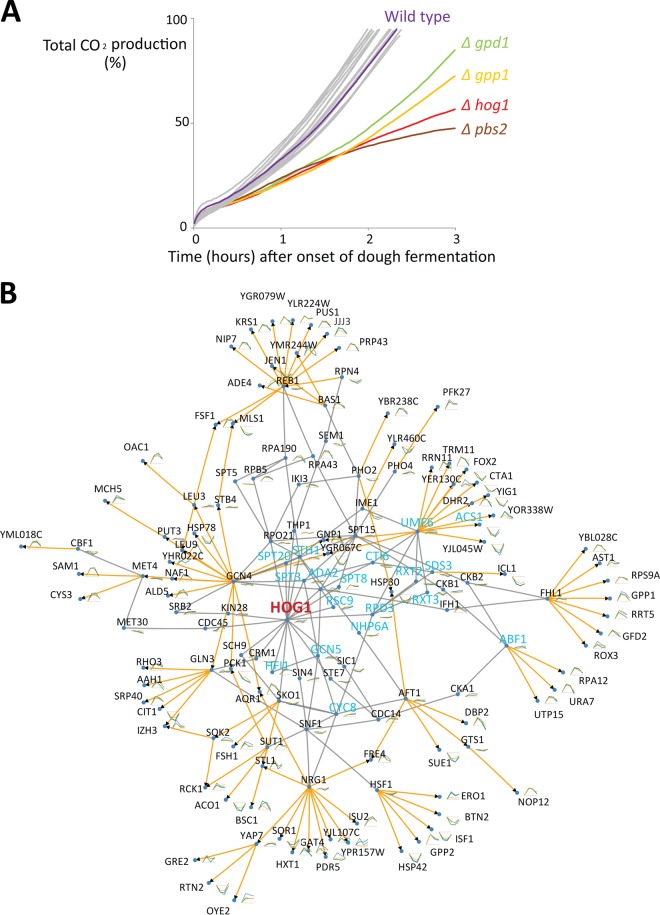
To obtain a general picture of yeast cell physiology during bread dough fermentation, we studied the transcriptome of yeast during this process. We selected three genetically distinct S. cerevisiae strains to investigate to what extent the responses are strain specific. To select these strains, we first screened 24 S. cerevisiae strains from different industrial applications (beer, wine, bioethanol, and bakery) with diverse genetic backgrounds (as estimated from an interdelta genetic fingerprinting assay [Fig. 1; see also Materials and Methods]) for CO2 production during dough fermentation as an indication of their fermentation capacity in dough. Based on the screening results, we selected three genetically distinct strains that showed a similar and sufficient capacity to ferment bread dough for further investigation. The three strains included a commonly used industrial baker's yeast strain as well as a bioethanol strain and a wine strain, which are genetically very different from the baker's yeast strain but yield similar levels of CO2 production during dough fermentation (Fig. 1A and B).
Fig 1.
Three genetically distinct strains from different fermentation industries (wine, bread, and bioethanol) with an acceptable dough fermentation capacity were selected. (A) The genetic relatedness of 24 strains from different fermentation industries was determined by an interdelta genetic fingerprinting assay. (B) CO2 production during dough fermentation indicates that all three strains can ferment dough at an acceptable rate.
Yeast cells show a transient response during dough fermentation.
Next, we investigated the transcriptional responses of the three selected strains throughout bread dough fermentation. We first optimized a method to extract high-quality RNA from yeast cells embedded in dough (see Materials and Methods for details). Next, we used RNA-seq to investigate changes in the transcriptomes of the strains at three different time points during the fermentation (30, 60, and 180 min after the start of fermentation) compared to the nonfermenting status (yeast cells harvested at the early stationary phase right before inoculation into the dough [0-min sample]). For each yeast strain, two independent biological replicates were analyzed.
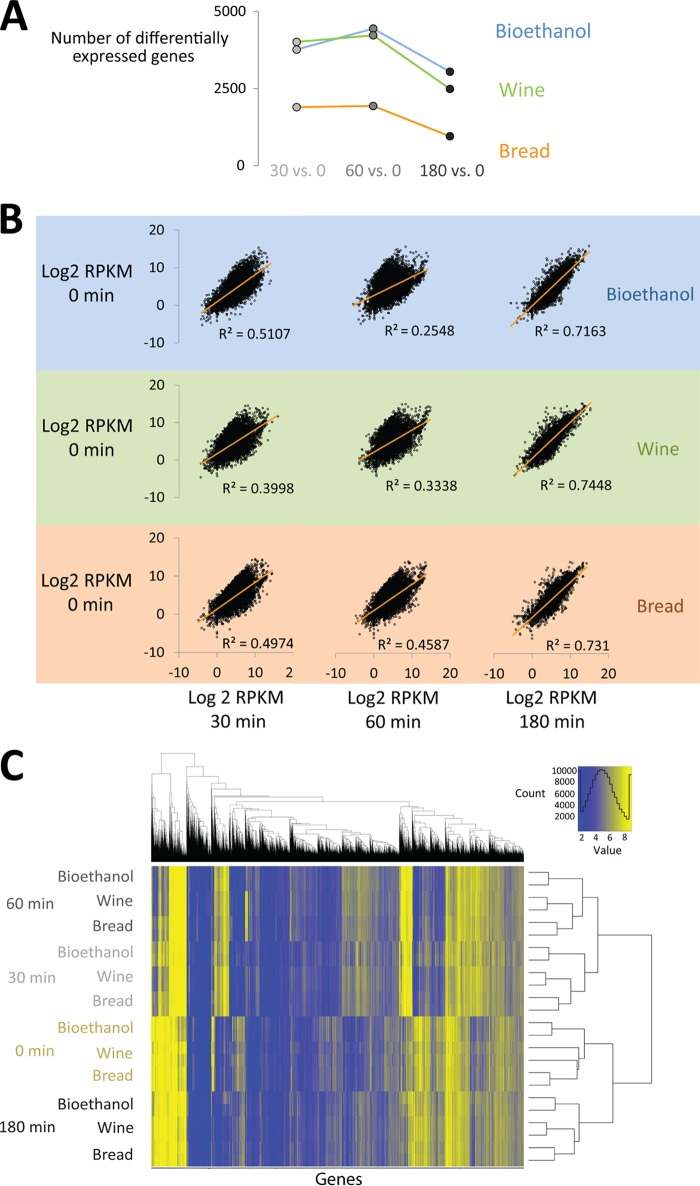
The transcriptome data obtained from this experiment indicated that yeast cells show a strong response to the changing conditions during the fermentation process, with around half of the total number of genes (protein or RNA coding) showing >2-fold changes in expression levels over the course of the fermentation process. Interestingly, the commercial baker's yeast strain on average showed smaller changes in its transcriptome, in terms of both the number of genes and the magnitude of changes, than the two other strains (Fig. 2A and B).
Fig 2.
(A) A large number of genes are differentially expressed at different time points during bread dough fermentation compared to the 0-min sample. The commercial baker's yeast (“Bread”) strain induces fewer changes in the transcriptome in response to the new environment. Note that differentially expressed genes include both protein- and RNA-coding genes. (B) The transcriptome of cells at the latest fermentation time point shows the highest level of similarity to cells in stationary phase (0-min sample). Shown are pairwise scatter plots and correlation coefficients for all mRNA quantities (as estimated by RPKM values) of the nonfermenting sample (0 min) and the three fermentation time points (30, 60, and 180 min) for the three different yeast strains (bioethanol, wine, and bread yeast). Note that the correlation between biological replicates was very high (R2 of between 0.959 and 0.997) (data not shown). (C) Clustering of expression profiles shows that the different yeast strains display largely similar (but not identical) gene expression patterns during bread dough fermentation. The graph shows hierarchical clustering of all expression data (including biological replicates for each strain and each time point). Every horizontal line contains the expression data (RPKM values) for a given strain and sampling time (i.e., one sample). The horizontal lines are ordered based on their overall similarity, and the clustering tree on the right shows the similarity between the samples (see Materials and Methods).
Further analysis revealed that the three strains showed a remarkably similar transcriptional profile during fermentation. Specifically, when all transcriptome data were clustered based on similarity (see Materials and Methods), samples obtained from different strains at the same time point always clustered together (Fig. 2C). Moreover, all biological replicates always clustered together and yielded a very similar profile, suggesting that the analysis was robust and reproducible.
We calculated the average mRNA quantities, as estimated by the RPKM (reads per kilobase per million mapped reads) values, for each gene in replicate samples. Subsequently, the pairwise Euclidean distances for those values of samples grouped either by strain or by time point were calculated. A nonparametric Wilcoxon test showed that the distances between samples grouped by time point are smaller than the distances between samples grouped by strain (P < 10−5). This confirms that the three strains showed a similar response throughout the fermentation process, regardless of their genetic differences. This analysis also indicates that most genes show a transient response. Specifically, the samples from nonfermenting yeast (0 min) and the last fermentation time point (180 min) clustered together, as did the samples from intermediate time points (30 and 60 min) (Fig. 2C). We performed template-matching clustering of the expression data to divide differentially expressed genes into categories that show similar expression patterns throughout the fermentation process (Fig. 3). This analysis confirmed that the vast majority of genes with changed transcription levels during the fermentation process showed a transient response, either transient induction (clusters 2 and 3) (Fig. 3) or transient repression (clusters 5 and 6) (Fig. 3).
Fig 3.
The majority of differentially expressed genes display a transient response. Shown are the normalized expression levels for each gene across the fermentation process (black lines). The genes are grouped per yeast strain (commercial baker's yeast, wine yeast, or bioethanol yeast) based on similarities in expression patterns (i.e., the trend in gene expression from one time point to another). Note that number of genes in different expression categories for all 3 strains includes both protein-coding and RNA-coding genes.
The majority of differentially expressed genes are involved in metabolic shifts and the response to nutrient levels.
In order to obtain deeper insight into the transcriptional response of yeast cells during dough fermentation, we used the Gene Ontology classification to check for enrichment of specific functionally related gene groups within the different sets of coregulated genes (Fig. 3). The analysis showed that cluster 2, the group with transiently upregulated genes with maximum expression 30 min after the onset of fermentation, is enriched for genes involved in ribosome biogenesis, tRNA processing, and nuclear transport (P values for each GO category can be found in Table S1 in the supplemental material). Cluster 3, the group of transiently upregulated genes with maximum expression 60 min after the onset of fermentation, is enriched for genes involved in amino acid metabolism (especially aspartate family and sulfur amino acids), translation, and reproduction of transposable elements. On the other hand, the group of transiently repressed genes (clusters 5 and 6) (Fig. 3) contains genes involved in aerobic respiration, the tricarboxylic acid (TCA) cycle, protein catabolic processes, and autophagy. Together, these data indicate a shift from respiration to fermentation between 0 and 30 min and active fermentation and cell growth between 30 and 60 min, followed by increased stress and nutrient depletion toward the end of fermentation (180 min).
The HOG pathway is required to adapt to high osmolarity at the onset of dough fermentation.
Genes involved in glycerol homeostasis, such as GPD1, GPP1, and STL1, are among the most upregulated genes in the beginning of fermentation in all 3 strains. Homeostasis of glycerol, the major osmolyte that is produced to balance intracellular osmolarity in S. cerevisiae, is regulated by the high-osmolarity glycerol (HOG) pathway (the major pathway involved in the response to osmotic stress) (50, 51).
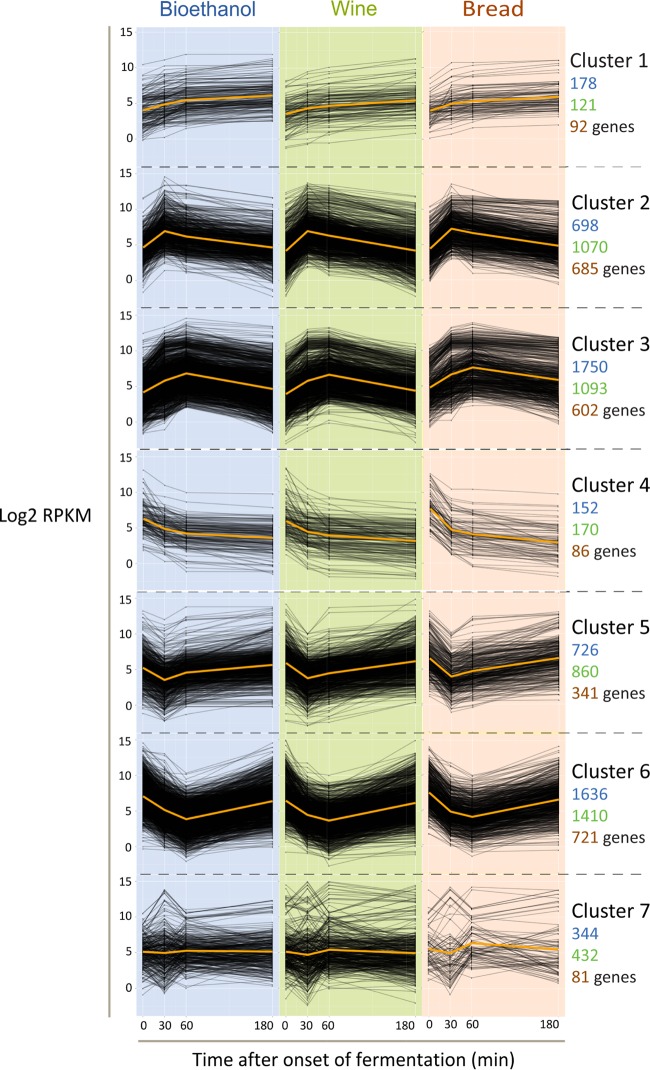
To investigate if the upregulation of these genes is important for efficient dough fermentation, we measured the dough fermentation performance of a set of 20 mutants (in the S288c background) carrying deletions of these genes as well as other key genes involved in the response to osmotic stress and several other genes that were highly upregulated at the onset of dough fermentation (see Table S2 in the supplemental material). The results indicate that deletion of key genes involved in the response to osmotic stress, namely, HOG1, PBS2, GPD1, and GPP1, results in impaired dough fermentation and low activity of the mutants in dough fermentation. However, for the other tested mutants, no significant effect was observed (Fig. 4A).
Fig 4.
(A) Deletion of key genes involved in the osmotic response results in impaired dough fermentation. Shown is CO2 production during dough fermentation, which serves as a proxy for the fermentation efficiency. Note that none of these mutants showed differences in activity compared to the wild type during fermentation with standard rich medium (yeast extract-peptone-dextrose) (see Fig. S1 in the supplemental material). (B) The HOG pathway triggers changes in the transcriptome through chromatin modification. Shown is a subnetwork selected by PheNetic for the 100 most differentially expressed genes for each strain at the first time point. Orange edges indicate protein-DNA interactions, and gray edges indicate protein-protein/phosphorylation interactions. The differential expression per node is visualized in a line chart in which the blue lines indicate expression for the bioethanol strain, the green lines indicate expression for the wine strain, and the orange lines indicate expression for the bread strain. Genes involved in chromatin modification are shown in blue.
To gain further mechanistic insight into how the HOG pathway could affect cellular responses during dough fermentation, a minimal common subnetwork selection algorithm, PheNetic (44), was applied to the expression data. Briefly, a molecular interaction network representing the known interactome of yeast was compiled from publicly available data. This network was used to identify the minimal common subnetwork that connects a perturbation to the differentially expressed genes through regulatory paths by using PheNetic (see Materials and Methods). The inferred subnetwork identifies the intermediary genes involved in signaling mechanisms, which do not necessarily show altered expression levels but mediate the cellular response in the organism.
As the major changes in osmotic pressure occur when the cells are inoculated in dough, we used the expression data obtained after 30 min of fermentation for this analysis. The minimal common subnetwork between HOG1, the downstream effector of the HOG pathway, and the 100 most differentially expressed genes for each analyzed strain at the 30-min time point was generated by using PheNetic.
The resulting subnetwork consists of 144 genes connected by 195 interactions. Of 100 differentially expressed genes, 89 were selected into the resulting network, which implies that PheNetic selected an additional 55 intermediary genes to connect HOG1 to the corresponding differentially expressed genes.
Clear enrichments in genes involved in chromatin modification (GO:16586) with corrected P values of <1e−7 can be found (Fig. 4B). Additionally, the selected network displays enrichment of genes related to stress response regulation (GO:80134) (P value of 4e−3), oxoacid metabolism (GO:6114) (P value of 1e−2), and glycerol biosynthesis (GO:6114) (P value of 2e−4). Together, this analysis further confirmed the HOG pathway as a central regulator of the early response to SSF conditions and identified a massive transcriptional reprogramming mechanism that may involve chromatin modification.
DISCUSSION
This study is the first to investigate the changes in the yeast transcriptome during dough fermentation under the actual conditions encountered during bread production (solid state). Our results demonstrate that the yeast transcriptome changes dramatically during dough fermentation. A large number of protein- and RNA-coding genes are differentially expressed at the early, middle, and late time points during dough fermentation. Moreover, although genetically different strains show slight differences, for example, in the expression levels of genes involved in biosynthesis of vitamins or amino acids, the three strains induced similar transient changes in their transcriptomes, which suggests that the overall response is general regardless of the genetic background. The fact that the commercial baker's yeast strain induced less-pronounced changes in its transcriptome, in terms of both the number of genes and the magnitude of changes, than the two other strains might indicate a higher degree of adaptation of this strain for dough fermentation, although this remains speculative.
Analysis of the function of the various differentially expressed genes yields insight into the physiological state of yeast cells as the dough fermentation process progresses. Overall, the cells showed a strong response to changing nutrient levels. At the onset of fermentation, the upregulation of ribosome biogenesis (RiBi) genes and the downregulation of genes associated with resistance to stress, such as genes involved in trehalose biosynthesis, indicate a response to nutrient level and glucose regulation (24–26). These changes at the beginning of the fermentation process, along with the downregulation of genes involved in the TCA cycle, aerobic respiration, and oxidative phosphorylation, reflect a shift in the nutrient-depleted stationary-phase cells to active fermentation.
Generally, when faced with low water activity or high osmolarity, microbes accumulate different compatible solutes, such as ions, amino acids, or polyols, to prevent water loss (52–54). In S. cerevisiae, the response to hyperosmotic stress is regulated through a mitogen-activated protein (MAP) kinase pathway, the HOG pathway, which results in accumulation of glycerol (50, 51). Consistent with this, our analysis shows that genes involved in biosynthesis of glycerol are among the most strongly upregulated genes at the onset of dough fermentation, suggesting that the cells experience a rather significant osmotic shock.
AQR1 was the most upregulated gene in all three strains at the earliest time point. This gene codes for a cell membrane transporter that mediates excretion of amino acids under conditions in which, despite an abundant availability of carbon, cell growth is limited by a second factor such as the lack of an essential compound (55). In bread dough fermentation, osmotic stress is likely to be the growth-restrictive factor that leads to a striking upregulation of AQR1. The observed induction of (retroviral) Ty elements at the beginning of fermentation is more puzzling. Activation of transposable elements in response to stress conditions was previously reported for different organisms (56–58). This activation has been proposed to stimulate genetic variability that could help the cell to adapt to environmental changes (59).
The middle fermentation phase is associated with increased expression levels of genes involved in amino acid and protein biosynthesis. These changes indicate induction of cell growth and may also imply further adaptation to osmotic stress (60, 61). Finally, at the last stage of the fermentation process, cells upregulate genes involved in maltose metabolism, trehalose and glycogen biosynthesis, autophagy, and protein catabolic processes. Together, these changes are indicative of glucose depletion and decreased nutrient levels during dough fermentation. This observation, together with the upregulation of genes involved in the response to heat at the latest time point, indicates a gradient of nutrients and heat of the fermentation process in the microenvironment surrounding cells as a result of poor diffusion of solutes and low thermal conductivity of the solid substrate.
Apart from genes with known function, it is important to mention that a large group of differentially expressed genes encoded putative proteins of unknown functions. This has also been observed by other studies of stress conditions (62, 63).
Finally, we show that deletion of key genes or targets of the HOG pathway results in impaired dough fermentation. This is consistent with previously reported work that showed that mutations in central genes of this pathway, namely, PBS2 or HOG1, cause osmosensitivity and accumulation of reduced levels of glycerol (64). Moreover, network analysis showed that the HOG pathway plays a central role in transcriptome changes at the onset of dough fermentation. The enrichment of the genes involved in chromatin modification in the network is consistent with a known role of chromatin-modifying enzymes and histone modifications in the regulation of transcriptional changes in response to environmental changes and stress (65, 66).
Together, our results indicate that cells embedded in a solid matrix such as bread dough suffer severe osmotic stress and that a proper induction of the HOG pathway is critical for optimal bread dough fermentation. These results indicate that this pathway and its targets may be good candidates for genetic modification or screening in order to obtain improved strains for SSF in general and dough fermentation in particular.
Supplementary Material
ACKNOWLEDGMENTS
We thank Verstrepen and Courtin laboratory members for their help and suggestions.
Research in the laboratory of K.J.V. is supported by ERC starting grant 241426, the VIB, the EMBO YIP program, HFSP research grant RGP0050/2013, the Fonds voor Wetenschappelijk Onderzoek-Vlaanderen (FWO) (Brussels, Belgium), and the IWT. We specifically acknowledge financial support from KU Leuven IDO project IDO/12/011 and from the FWO for the postdoctoral fellowship of K.V. and E.D. and project funding (G.0544.10N).
Footnotes
Published ahead of print 20 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02649-13.
REFERENCES
- 1.Pandey A. 1992. Recent process developments in solid-state fermentation. Process Biochem. 27:109–117 [Google Scholar]
- 2.Pandey A, Soccol CR, Rodriguez-Leon J, Nigam P. 2001. Solid-state fermentation in biotechnology: fundamentals and applications, 1st ed. Asiatech Publishers, New Delhi, India [Google Scholar]
- 3.Pandey A, Selvakumar P, Soccol CR, Nigam P. 1999. Solid state fermentation for the production of industrial enzymes. Curr. Sci. 77:149–162 [Google Scholar]
- 4.Selvakumar P, Ashakumary L, Pandey A. 1998. Biosynthesis of glucoamylase from Aspergillus niger by solid-state fermentation using tea waste as the basis of a solid substrate. Bioresour. Technol. 65:83–85 [Google Scholar]
- 5.Cen P, Xia L. 1999. Production of cellulase by solid-state fermentation, p 69–92 In Scheper T, Tsao GT. (ed), Recent progress in bioconversion of lignocellulosics. Springer, Berlin, Germany [Google Scholar]
- 6.Singh-Nee Nigam P, Singh D. 1996. Processing of agricultural wastes in solid state fermentation for microbial protein production. J. Sci. Ind. Res. 55:373–380 [Google Scholar]
- 7.Robinson T, Singh D, Nigam P. 2001. Solid-state fermentation: a promising microbial technology for secondary metabolite production. Appl. Microbiol. Biotechnol. 55:284–289 [DOI] [PubMed] [Google Scholar]
- 8.Mohanty SK, Behera S, Swain MR, Ray RC. 2009. Bioethanol production from mahula (Madhuca latifolia L.) flowers by solid-state fermentation. Appl. Energy 86:640–644 [Google Scholar]
- 9.Rodríguez L, Toro M, Vazquez F, Correa-Daneri M, Gouiric S, Vallejo M. 2010. Bioethanol production from grape and sugar beet pomaces by solid-state fermentation. Int. J. Hydrogen Energy 35:5914–5917 [Google Scholar]
- 10.Mazaheri D, Shojaosadati SA, Mousavi SM, Hejazi P, Saharkhiz S. 2012. Bioethanol production from carob pods by solid-state fermentation with Zymomonas mobilis. Appl. Energy 99:372–378 [Google Scholar]
- 11.Hölker U, Höfer M, Lenz J. 2004. Biotechnological advantages of laboratory-scale solid-state fermentation with fungi. Appl. Microbiol. Biotechnol. 64:175–186 [DOI] [PubMed] [Google Scholar]
- 12.Pandey A, Soccol CR, Mitchell D. 2000. New developments in solid state fermentation. I. Bioprocesses and products. Process Biochem. 35:1153–1169 [Google Scholar]
- 13.Pandey A, Ashakumary L, Selvakumar P, Vijayalakshmi K. 1994. Influence of water activity on growth and activity of Aspergillus niger for glycoamylase production in solid-state fermentation. World J. Microbiol. Biotechnol. 10:485–486 [DOI] [PubMed] [Google Scholar]
- 14.López-Calleja A, Cuadra T, Barrios-González J, Fierro F, Fernández F. 2012. Solid-state and submerged fermentations show different gene expression profiles in cephalosporin C production by Acremonium chrysogenum. J. Mol. Microbiol. Biotechnol. 22:126–134 [DOI] [PubMed] [Google Scholar]
- 15.Backhus LE, DeRisi J, Brown PO, Bisson LF. 2001. Functional genomic analysis of a commercial wine strain of Saccharomyces cerevisiae under differing nitrogen conditions. FEMS Yeast Res. 1:111–125 [DOI] [PubMed] [Google Scholar]
- 16.Pérez-Torrado R, Panadero J, Hernández-López MJ, Prieto JA, Randez-Gil F. 2010. Global expression studies in baker's yeast reveal target genes for the improvement of industrially-relevant traits: the cases of CAF16 and ORC2. Microb. Cell Fact. 9:56. 10.1186/1475-2859-9-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cholet O, Hénaut A, Casaregola S, Bonnarme P. 2007. Gene expression and biochemical analysis of cheese-ripening yeasts: focus on catabolism of l-methionine, lactate, and lactose. Appl. Environ. Microbiol. 73:2561–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.James T, Campbell S, Donnelly D, Bond U. 2003. Transcription profile of brewery yeast under fermentation conditions. J. Appl. Microbiol. 94:432–448 [DOI] [PubMed] [Google Scholar]
- 19.Olesen K, Felding T, Gjermansen C, Hansen J. 2002. The dynamics of the Saccharomyces carlsbergensis brewing yeast transcriptome during a production-scale lager beer fermentation. FEMS Yeast Res. 2:563–573 [DOI] [PubMed] [Google Scholar]
- 20.Marks VD, Merwe GK, Vuuren HJ. 2003. Transcriptional profiling of wine yeast in fermenting grape juice: regulatory effect of diammonium phosphate. FEMS Yeast Res. 3:269–287 [DOI] [PubMed] [Google Scholar]
- 21.Rossignol T, Dulau L, Julien A, Blondin B. 2003. Genome-wide monitoring of wine yeast gene expression during alcoholic fermentation. Yeast 20:1369–1385 [DOI] [PubMed] [Google Scholar]
- 22.Zuzuarregui A, Olmo M. 2004. Expression of stress response genes in wine strains with different fermentative behavior. FEMS Yeast Res. 4:699–710 [DOI] [PubMed] [Google Scholar]
- 23.Tanaka F, Ando A, Nakamura T, Takagi H, Shima J. 2006. Functional genomic analysis of commercial baker's yeast during initial stages of model dough-fermentation. Food Microbiol. 23:717–728 [DOI] [PubMed] [Google Scholar]
- 24.Van Dijck P, Colavizza D, Smet P, Thevelein JM. 1995. Differential importance of trehalose in stress resistance in fermenting and nonfermenting Saccharomyces cerevisiae cells. Appl. Environ. Microbiol. 61:109–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Versele M, Thevelein JM, Van Dijck P. 2004. The high general stress resistance of the Saccharomyces cerevisiae fil1 adenylate cyclase mutant (Cyr1Lys1682) is only partially dependent on trehalose, Hsp104 and overexpression of Msn2/4-regulated genes. Yeast 21:75–86 [DOI] [PubMed] [Google Scholar]
- 26.Verstrepen KJ, Iserentant D, Malcorps P, Derdelinckx G, Van Dijck P, Winderickx J, Pretorius IS, Thevelein JM, Delvaux FR. 2004. Glucose and sucrose: hazardous fast-food for industrial yeast? Trends Biotechnol. 22:531–537 [DOI] [PubMed] [Google Scholar]
- 27.Remize F, Cambon B, Barnavon L, Dequin S. 2003. Glycerol formation during wine fermentation is mainly linked to Gpd1p and is only partially controlled by the HOG pathway. Yeast 20:1243–1253 [DOI] [PubMed] [Google Scholar]
- 28.Legras JL, Karst F. 2003. Optimisation of interdelta analysis for Saccharomyces cerevisiae strain characterisation. FEMS Microbiol. Lett. 221:249–255 [DOI] [PubMed] [Google Scholar]
- 29.Abelson JN, Simon MI, Guthrie C, Fink GR. (ed). 2004. Guide to yeast genetics and molecular biology. Elsevier, London, United Kingdom [Google Scholar]
- 30.Amberg DC, Burke DJ, Strathern JN. 2005. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual, 2005 ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 31.Shogren M, Finney K. 1984. Bread-making test for 10 grams of flour. Cereal Chem. 61:418–423 [Google Scholar]
- 32.Panadero J, Randez-Gil F, Prieto JA. 2005. Validation of a flour-free model dough system for throughput studies of baker's yeast. Appl. Environ. Microbiol. 71:1142–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou X, Ren L, Meng Q, Li Y, Yu Y, Yu J. 2010. The next-generation sequencing technology and application. Protein Cell 1:520–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Milne I, Stephen G, Bayer M, Cock PJ, Pritchard L, Cardle L, Shaw PD, Marshall D. 2013. Using Tablet for visual exploration of second-generation sequencing data. Brief. Bioinformatics 14:193–202 [DOI] [PubMed] [Google Scholar]
- 36.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7:562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu T, Lin N, Shi N, Zhang B. 2009. Information criterion-based clustering with order-restricted candidate profiles in short time-course microarray experiments. BMC Bioinformatics 10:146. 10.1186/1471-2105-10-146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. 2003. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 4:P3. 10.1186/gb-2003-4-5-p3 [DOI] [PubMed] [Google Scholar]
- 40.Voordeckers K, De Maeyer D, Zande E, Vinces MD, Meert W, Cloots L, Ryan O, Marchal K, Verstrepen KJ. 2012. Identification of a complex genetic network underlying Saccharomyces cerevisiae colony morphology. Mol. Microbiol. 86:225–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yeang C-H, Ideker T, Jaakkola T. 2004. Physical network models. J. Comput. Biol. 11:243–262 [DOI] [PubMed] [Google Scholar]
- 42.Yeger-Lotem E, Riva L, Su LJ, Gitler AD, Cashikar AG, King OD, Auluck PK, Geddie ML, Valastyan JS, Karger DR. 2009. Bridging high-throughput genetic and transcriptional data reveals cellular responses to alpha-synuclein toxicity. Nat. Genet. 41:316–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pu S, Wong J, Turner B, Cho E, Wodak SJ. 2009. Up-to-date catalogues of yeast protein complexes. Nucleic Acids Res. 37:825–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Maeyer D, Renkens J, Cloots L, De Raedt L, Marchal K. 2013. PheNetic: network-based interpretation of unstructured gene lists in E. coli. Mol. Biosyst. 9:1594–1603 [DOI] [PubMed] [Google Scholar]
- 45.Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T. 2011. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27:431–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman W-H, Pagès F, Trajanoski Z, Galon J. 2009. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25:1091–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maere S, Heymans K, Kuiper M. 2005. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21:3448–3449 [DOI] [PubMed] [Google Scholar]
- 48.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B Stat. Methodol. 57:289–300 [Google Scholar]
- 49.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT. 2000. Gene ontology: tool for the unification of biology. Nat. Genet. 25:25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gustin MC, Albertyn J, Alexander M, Davenport K. 1998. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62:1264–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hohmann S. 2002. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66:300–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Welsh DT. 2000. Ecological significance of compatible solute accumulation by micro-organisms: from single cells to global climate. FEMS Microbiol. Rev. 24:263–290 [DOI] [PubMed] [Google Scholar]
- 53.Ruijter GJ, Visser J, Rinzema A. 2004. Polyol accumulation by Aspergillus oryzae at low water activity in solid-state fermentation. Microbiology 150:1095–1101 [DOI] [PubMed] [Google Scholar]
- 54.Kets E, Teunissen P, De Bont J. 1996. Effect of compatible solutes on survival of lactic acid bacteria subjected to drying. Appl. Environ. Microbiol. 62:259–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Velasco I, Tenreiro S, Calderon IL, André B. 2004. Saccharomyces cerevisiae Aqr1 is an internal-membrane transporter involved in excretion of amino acids. Eukaryot. Cell 3:1492–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grandbastien M-A, Audeon C, Bonnivard E, Casacuberta J, Chalhoub B, Costa A-P, Le Q, Melayah D, Petit M, Poncet C. 2005. Stress activation and genomic impact of Tnt1 retrotransposons in Solanaceae. Cytogenet. Genome Res. 110:229–241 [DOI] [PubMed] [Google Scholar]
- 57.Lesage P, Todeschini A. 2005. Happy together: the life and times of Ty retrotransposons and their hosts. Cytogenet. Genome Res. 110:70–90 [DOI] [PubMed] [Google Scholar]
- 58.Oliver KR, Greene WK. 2009. Transposable elements: powerful facilitators of evolution. Bioessays 31:703–714 [DOI] [PubMed] [Google Scholar]
- 59.McClintock B. 1984. The significance of responses of the genome to challenge. Science 226:792–801 [DOI] [PubMed] [Google Scholar]
- 60.Uesono Y, Toh-e A. 2002. Transient inhibition of translation initiation by osmotic stress. J. Biol. Chem. 277:13848–13855 [DOI] [PubMed] [Google Scholar]
- 61.Rep M, Krantz M, Thevelein JM, Hohmann S. 2000. The transcriptional response of Saccharomyces cerevisiae to osmotic shock: Hot1p and Msn2p/Msn4p are required for the induction of subsets of high osmolarity glycerol pathway-dependent genes. J. Biol. Chem. 275:8290–8300 [DOI] [PubMed] [Google Scholar]
- 62.Alexandre H, Ansanay-Galeote V, Dequin S, Blondin B. 2001. Global gene expression during short-term ethanol stress in Saccharomyces cerevisiae. FEBS Lett. 498:98–103 [DOI] [PubMed] [Google Scholar]
- 63.Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brewster JL, de Valoir T, Dwyer ND, Winter E, Gustin MC. 1993. An osmosensing signal transduction pathway in yeast. Science 259:1760–1763 [DOI] [PubMed] [Google Scholar]
- 65.Alejandro-Osorio AL, Huebert DJ, Porcaro DT, Sonntag ME, Nillasithanukroh S, Will JL, Gasch AP. 2009. The histone deacetylase Rpd3p is required for transient changes in genomic expression in response to stress. Genome Biol. 10:R57. 10.1186/gb-2009-10-5-r57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shivaswamy S, Iyer VR. 2008. Stress-dependent dynamics of global chromatin remodeling in yeast: dual role for SWI/SNF in the heat shock stress response. Mol. Cell. Biol. 28:2221–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.