Abstract
Antimicrobial peptides produced by multicellular organisms as part of their innate system of defense against microorganisms are currently considered potential alternatives to conventional antibiotics in case of infection by multiresistant bacteria. However, while the mode of action of antimicrobial peptides is relatively well described, resistance mechanisms potentially induced or selected by these peptides are still poorly understood. In this work, we studied the mechanisms of action and resistance potentially induced by ApoEdpL-W, a new antimicrobial peptide derived from human apolipoprotein E. Investigation of the genetic response of Escherichia coli upon exposure to sublethal concentrations of ApoEdpL-W revealed that this antimicrobial peptide triggers activation of RcsCDB, CpxAR, and σE envelope stress pathways. This genetic response is not restricted to ApoEdpL-W, since several other antimicrobial peptides, including polymyxin B, melittin, LL-37, and modified S4 dermaseptin, also activate several E. coli envelope stress pathways. Finally, we demonstrate that induction of the CpxAR two-component system directly contributes to E. coli tolerance toward ApoEdpL-W, polymyxin B, and melittin. These results therefore show that E. coli senses and responds to different antimicrobial peptides by activation of the CpxAR pathway. While this study further extends the understanding of the array of peptide-induced stress signaling systems, it also provides insight into the contribution of Cpx envelope stress pathway to E. coli tolerance to antimicrobial peptides.
INTRODUCTION
Administration of antibiotics is the most efficient strategy for combatting pathogenic bacteria. However, decades of extensive use of antibiotics have led to the emergence of bacterial strains with higher or wider resistance spectra, causing increasing difficulty worldwide in management of bacterial infections (1). In parallel with research on new antibiotics, antimicrobial peptides (AMPs), mainly produced by epithelial surfaces of multicellular organisms as part of their innate defense system, have emerged as a plausible alternative to conventional antibiotics (2). Although AMPs vary in sequence, length, and structural conformation, they are mostly amphipathic compounds with spatially organized clusters of hydrophobic and cationic amino acids (3). The AMP net positive charge enables their binding to the negatively charged microbial surface, while the presence of hydrophobic residues promotes their insertion into membranes (3). Many AMPs form deleterious channels in bacterial membranes (4). Alternatively, AMPs can translocate across membranes into the cytoplasm, where they may inhibit essential processes such as nucleic acid, protein, enzyme, and cell wall syntheses (5–9).
In light of the wide distribution of AMPs in multicellular organisms and the long interplay between bacteria and their host during evolution, bacteria have acquired different mechanisms for minimizing the killing impact of AMPs (10). Mechanisms of resistance to AMPs can be classified into the following three major categories: (i) destruction/modification of AMPs by proteolytic cleavage (11–13); (ii) exclusion of AMPs from the cell via low-specificity efflux pumps (14–16); and (iii) reduction of bacterial susceptibility to AMPs by altering the membrane net charge, thereby impairing physicochemical interactions between cationic antimicrobial molecules and the negatively charged bacteria cell surface (17–20). Moreover, exposure to AMPs results in strong alterations in the bacterial gene transcription profile and in the induction of mostly nonspecific, poorly understood resistance mechanisms (21–25).
Recently, a new family of antimicrobial peptides, derived from human apolipoprotein E (ApoE) and acting via perturbation of the membrane lipid bilayer, was described (26, 27). The original ApoEdp peptide (sequence, LRKLRKRLLLRKLRKRLL) showed direct, broad anti-infective activity against bacteria and viruses (27). Replacement of all leucine residues with tryptophan amino acids in ApoEdpL-W (sequence, WRKWRKRWWWRKWRKRWW) led to production of a variant with increased potency and high antimicrobial activity against viruses, parasites, and Staphylococcus aureus, despite a slight decrease of antibacterial activity against Pseudomonas aeruginosa (28–30). To identify bacterial resistance potentially induced upon exposure to antimicrobial peptides, we studied the Escherichia coli genetic response to ApoEdpL-W and demonstrated the contribution of the CpxAR envelope stress signaling pathway to E. coli resistance to ApoEdpL-W and several other antimicrobial peptides.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are listed in Table 1.
Table 1.
Strains and plasmids used in this study
| E. coli strain or plasmid | Genotype or description | Antibiotic resistance(s) | Reference |
|---|---|---|---|
| Strains | |||
| MG1655 | K-12 wild-type strain | 61 | |
| TG1 ΔcpxR | ΔcpxR::Δfrt | 62 | |
| MG ΔcpxR::Kmfrt | P1 transduction from TG1 ΔcpxR into MG1655 | Kmr | This study |
| MG ΔrcsB::Kmfrt | P1 transduction from JW2205 (Keio collection) into MG1655 | Kmr | This study |
| MG ΔdegP::Kmfrt | P1 transduction from JW0157 (Keio collection) into MG1655 | Kmr | This study |
| MG1655 F′ | Biofilm-forming strain | Tetr | 37 |
| MG1655 F′tet ΔcpxR::Kmfrt | P1 transduction from MG ΔcpxR::Kmfrt into MG1655 F′ | Tetr Kmr | This study |
| MG1655F′tet ΔdegP::Kmfrt | P1 transduction from JW0157 (Keio collection) into MG1655 F′ | Tetr Kmr | This study |
| SK1938 (manC-lacZ) | MG1655_ΔΔlac_l(F(cpsB::lacZ)) | Kmr | 63 |
| SK1941 (manC-lacZ ΔrcsB) | MG1655_Δlac_l(F(cpsB::lacZ)) ΔrcsB::cat | Kmr Cmr | 64 |
| degP-lacZ | MC4100 ΔdegP::lacZ | 65 | |
| degP-lacZ ΔcpxR | P1 transduction from MG ΔcpxR::Kmfrt into degP-lacZ | Kmr | This study |
| BW25113 | Parental strain of the Keio collection mutants | 31 | |
| ΔcpxA | BW25113 ΔcpxA::Kmfrt | Kmr | 31 |
| ΔdegP | BW25113 ΔdegP::Kmfrt | Kmr | 31 |
| ΔmdtA | BW25113 ΔmdtA::Kmfrt | Kmr | 31 |
| Δspy | BW25113 Δspy::Kmfrt | Kmr | 31 |
| ΔompF | BW25113 ΔompF::Kmfrt | Kmr | 31 |
| ΔnanC | BW25113 ΔnanC::Kmfrt | Kmr | 31 |
| ΔompC | BW25113 ΔompC::Kmfrt | Kmr | 31 |
| ΔcpxP | BW25113 ΔcpxP::Kmfrt | Kmr | 31 |
| ΔacrD | BW25113 ΔacrD::Kmfrt | Kmr | 31 |
| ΔybaJ | BW25113 ΔybaJ::Kmfrt | Kmr | 31 |
| ΔydeH | BW25113 ΔydeH::Kmfrt | Kmr | 31 |
| Plasmids | |||
| pBAD18 | Cloning vector, arabinose-inducible promoter | Ampr | 66 |
| pBAD-nlpE | nlpE gene cloned into pBAD18 | Ampr | This study |
| pCA24N | High-copy-number plasmid with an IPTG-inducible promoter | Cmr | 32 |
| pCA24N-cpxR | E. coli cpxR cloned into pCA24N | Cmr | 32 |
| pCA24N-degP | E. coli degP cloned into pCA24N | Cmr | 32 |
| pCA24N-rseA | E. coli rseA cloned into pCA24N | Cmr | 32 |
The E. coli Keio collection was derived from wild-type BW25113 (31). pCA24N and derivatives were isolated from E. coli K-12 carrying different plasmids (ASKA Collection) (32). Deletion mutants were generated by P1 transduction from corresponding Keio mutants or mutants from our laboratory collection into E. coli MG1655 or E. coli MG1655 F′. pBAD18-nlpE was constructed by cloning the nlpE gene into the pBAD18 plasmid, containing an arabinose-inducible promoter. Primers used to clone nlpE or verify genetic constructions are listed in Table S1 in the supplemental material.
Peptides and growth conditions.
ApoEdpL-W and Fluo-ApoEdpL-W used in this study were manufactured by Alta Bioscience (Birmingham, United Kingdom) and resuspended in water plus 5% dimethyl sulfoxide (DMSO). Melittin, LL-37, and polymyxin B were purchased from Sigma and diluted in water. K4K20-S4 dermaseptin was also diluted in water (33).
All experiments were carried out in Mueller-Hinton (MH) medium at 37°C, except for those using LL-37, which were conducted in 0.4% glucose-M63B1 minimal medium (M63B1-G). When required, antibiotics were added to the medium at the following final concentrations: kanamycin (Km), 50 mg/liter; chloramphenicol (Cm), 25 mg/liter; ampicillin (Amp), 100 mg/liter; and tetracycline (Tet), 7.5 mg/liter.
MIC determination.
Strains cultured for 6 h in MH or M63B1-G medium were inoculated into fresh medium at an optical density at 600 nm (OD600) of 0.0001. Peptides were diluted in water, and 10-μl samples of peptide solution (a concentration 10 times higher than the final concentration) were placed in wells of 96-well polystyrene microtiter plates. Ninety-microliter samples of the diluted bacterial culture (see above) were added to wells. Microtiter plates were incubated overnight at 37°C. The MIC of the antimicrobial peptide was defined as the lowest concentration that inhibited bacterial growth.
Bacterial killing assay.
Overnight cultures were diluted in fresh medium containing isopropyl-β-d-thiogalactopyranoside (IPTG) and chloramphenicol, when necessary, at an OD600 of 0.005 and incubated at 37°C with aeration until reaching an OD600 of 0.1. Bacteria were then exposed to different concentrations of peptide and incubated at 37°C. At different times, samples were taken, centrifuged, and resuspended in equivalent volume of 1× phosphate-buffered saline (PBS). They were then serially diluted and plated on appropriate LB agar plates to enumerate viable colonies (CFU).
RNA isolation.
Overnight cultures were diluted in fresh MH medium at an OD600 of 0.005 and incubated at 37°C until reaching an OD600 of 0.1. Bacteria were then exposed or not to 3 μM ApoEdpL-W for 30 min at 37°C with agitation. The OD600 of the cultures was adjusted to 3 with corresponding used medium (3-ml final volume), and 2 volumes of RNA Protect reagent (Qiagen, Valencia, CA) was added (6 ml) to maintain the stability and integrity of the RNA. Bacterial cells were centrifuged at 8,000 rpm for 10 min at 4°C, and total RNA was then extracted using a modification of the RNeasy kit (Qiagen) protocol described below. Both conditions were compared by using three biological replicates. Briefly, bacterial pellets were resuspended in 200 μl of TE buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, diethyl pyrocarbonate [DEPC] [0.1% {vol/vol}]) containing 1 mg/ml of lysozyme and incubated for 5 min at room temperature. The cells were then homogenized with 700 μl RLT lysis buffer (Qiagen), and the homogenate was vortexed for approximately 1 min. Five hundred microliters of ethanol (100%) was then added, and the solution was applied to an RNeasy column. From this point on, instructions from the manufacturer were followed, including on-column DNase treatment. Purified total RNA concentrations were measured using a Nanodrop spectrophotometer at 260 nm.
Microarray and data analysis.
Three biological replicates of RNA extracts for each condition were treated as described in the Affymetrix GeneChip expression analysis technical manual (P/N 702232 rev. 2). Quality of the sample hybridizations was checked on Agilent RNA Nano LabChips (Agilent Technologies) before performing data analysis. Data analysis was performed using R software based on the Bioconductor package. For each experimental condition, probe intensities from three independent biological replicates were analyzed. Preprocessing of the gene expression array was carried out using the model-based robust multichip average (RMA) algorithm (34), enabling global background correction, quantile normalization, and summarization of the 11 probe values into a single probe set. The local pooled error (LPE) test was used for pairwise expression comparison (35), followed by the Benjamini and Hochberg P value adjustment method (threshold P value of <0.05). Only fold changes (FC) superior or inferior to 2 were considered to identify induced or repressed genes, respectively.
β-Galactosidase assays.
β-Galactosidase activities were measured as described below. Overnight cultures were diluted in fresh MH or M63B1-G medium containing antibiotics and IPTG, when necessary (as indicated in the figure legends), to an OD600 of 0.005 and incubated at 37°C with agitation until reaching an OD600 of 0.1. One-milliliter samples were transferred to a 12-well plate containing or not containing an antimicrobial peptide. Microtiter plates were then incubated for 30 min with agitation at 37°C. β-Galactosidase activity was assayed in duplicate for each strain, as previously described (36).
Fluorescence microscopy.
Exponentially growing bacterial cells (OD600 = 0.1) were exposed to Fluo-ApoEdpL-W (3 μM) in MH medium for 30 min. The cells (500 μl) were centrifuged, washed 2 times with 1× PBS, and then resuspended in 100 μl of 1× PBS. Thirty microliters of bacterial cells was loaded on an 8-well black epoxy slide previously treated with 0.1% poly-l-lysine for 2 min. Bacteria were fixed with a 3% paraformaldehyde solution and stained with 25 μl of DAPI (4′,6-diamidino-2-phenylindole) at 20 mg/liter for 45 min at room temperature. Fluorescence microscopy was performed using a Nikon Eclipse E4000 microscope, and images were taken using a 100× lens.
Evaluation of ApoEdpL-W antimicrobial activity on biofilm bacteria.
E. coli biofilms were formed in MH medium in 96-well polyvinyl chloride (PVC) microtiter plates at 37°C. Twenty-four-hour biofilms were washed once with 1× PBS, using a multichannel pipette to remove unattached cells. One-hundred-microliter samples of ApoEdpL-W diluted in fresh MH medium at different concentrations were added to the biofilms, and microtiter plates were reincubated for 24 h. The biofilms were then washed with 1× PBS once before being resuspended in 100 μl of 1× PBS. Effects of the peptide were determined by CFU enumeration before adding the peptide (T0) and after 24 h of treatment (T24).
Statistical analysis.
Two-tailed unpaired Student's t test analyses were performed using Prism 5.0 for Mac OS X (GraphPad Software). Each experiment was performed at least three times (NS, not significant; *, P ≤ 0.05; **, P ≤ 0.01; and ***, P ≤ 0.001).
RESULTS
ApoEdpL-W AMP activity in E. coli planktonic and biofilm bacteria.
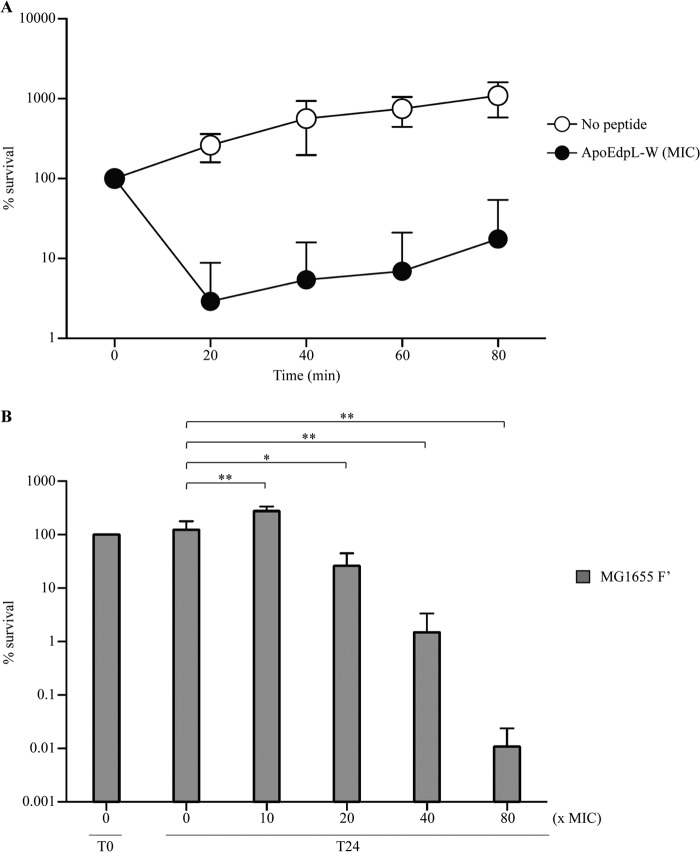
The ApoEdpL-W antimicrobial peptide was previously shown to inhibit growth of both P. aeruginosa and S. aureus when added at micromolar concentrations on planktonic cultures (26). We tested the activity of ApoEdpL-W on planktonic and biofilm E. coli K-12 bacteria by using either strain MG1655 or its biofilm-forming isogenic derivative, MG1655 F′ (37). Using increasing concentrations of ApoEdpL-W, we determined that the ApoEdpL-W MIC on exponential-phase planktonic E. coli was 5 μM. Accordingly, exponential-phase cultures displayed a strong drop in viability (1 to 2 log CFU) after 20 min of treatment with 5 μM ApoEdpL-W (Fig. 1A). In order to test ApoEdpL-W activity on biofilm bacteria, which are characterized by high levels of tolerance toward antibiotics, peptide concentrations ranging from 10- to 80-fold higher than the MIC were applied to 24-h E. coli K-12 MG1655 F′ biofilms formed in microtiter wells. Under these conditions, a slight growth stimulation was observed using ApoEdpL-W at 10-fold higher than the MIC (50 μM), which could be due to the release of nutrients upon low levels of bacterial lysis. However, use of higher ApoEdpL-W concentrations significantly reduced biofilm bacterial viability, with a 4-log CFU reduction after 24 h at 80-fold higher than the MIC (400 μM) (Fig. 1B).
Fig 1.
Impact of ApoEdpL-W on planktonic and biofilm E. coli bacteria. (A) Growing E. coli cells were exposed to 0 or 5 μM ApoEdpL-W for 80 min, during which samples were taken every 20 min, serially diluted, and plated on LB plates. Percent survival was calculated by CFU counting and compared to numbers obtained at 0 min. (B) A 24-h MG1655 F′ biofilm was treated with increasing concentrations of ApoEdpL-W for 24 h. Viable cells of the treated biofilm population were quantified by CFU enumeration and were compared to numbers obtained prior to ApoEdpL-W treatment. Percent survival values represent at least 3 replicates. *, P < 0.05; **, P < 0.01 by two-tailed unpaired Student's t test.
ApoEdpL-W AMP is localized in the cell envelope.
To further explore ApoEdpL-W activity in E. coli, we used a fluorescently tagged derivative of ApoEdpL-W (Fluo-ApoEdpL-W) and first determined that Fluo-ApoEdpL-W and ApoEdpL-W have the same MIC on E. coli K-12 MG1655 (5 μM) (data not shown). To monitor Fluo-ApoEdpL-W localization in bacteria exposed to the peptide, the cells were exposed to sublethal concentrations of ApoEdpL-W in order to keep cell lysis to a minimum. E. coli MG1655 bacteria in early exponential phase were incubated for 30 min in the presence of the sublethal concentration of 3 μM Fluo-ApoEdpL-W, previously shown to have a mild effect on bacterial growth and viability (see Fig. S1 in the supplemental material). Epifluorescence microscopy analysis indicated that the Fluo-ApoEdpL-W peptide was localized at the periphery of the treated bacteria but was not detected in their cytoplasm, consistent with a potential cell envelope site of action for ApoEdpL-W (Fig. 2) (26).
Fig 2.
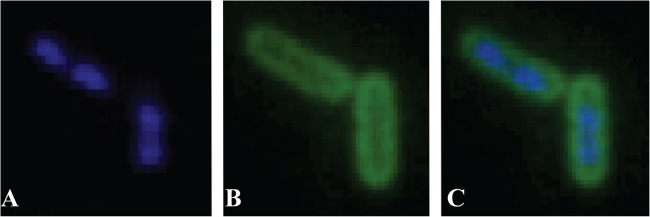
Localization of fluorescent ApoEdpL-W in the envelope of E. coli cells. Growing E. coli cells were exposed to 3 μM Fluo-ApoEdpL-W for 30 min and observed by epifluorescence microscopy (Eclipse E400; Nikon). (A) DAPI staining. (B) Fluo-ApoEdpL-W. (C) Merged images.
E. coli genetic responses upon exposure to ApoEdpL-W antimicrobial peptide.
To investigate the mode of action of ApoEdpL-W and potential mechanisms of bacterial resistance to this antimicrobial peptide, we studied the genetic response of E. coli upon exposure to subinhibitory concentrations of ApoEdpL-W by using an E. coli DNA microarray. Exponentially growing E. coli bacteria were exposed for only 30 min to a 3 μM sublethal concentration of ApoEdpL-W AMP in order to avoid massive cell lysis (see Fig. S1 in the supplemental material). RNAs corresponding to these experimental conditions were extracted, and responses induced by ApoEdpL-W were analyzed using Affymetrix DNA chips. A total of 175 genes were found to be differentially expressed in response to ApoEdpL-W (see Tables S2 and S3), including 69 downregulated genes involved in general cell metabolism and energetics. Among the 106 upregulated genes, many were related to iron acquisition (fepA, fes, entABCEF, fhuA, fhuF, sufA, and exbD) and surface polysaccharide synthesis (manA, galU, yjbEGH, rcsA, and otsB). In particular, 17 of the 19 genes coding for colanic acid synthesis (wza, wzc, wcaABCDEF, gmd, wcaGHI, manC, wcaJ, wzxC, and wcaKL) were identified (see Tables S2 and S3). Several genes involved in bacterial stress responses were also upregulated by ApoEdpL-W. They included genes induced by osmotic shock (osmB, osmC, and osmY) or oxidative stress (katE) or encoding chaperones from the general stress response (dnaK, htpG, lolA, clpB, ipbA, and ipbB). Strikingly, 51 of 106 genes identified as being induced upon exposure to ApoEdpL-W were part of regulons involved in E. coli envelope stress, such as the RcsCDB (39 genes) and CpxAR (13 genes) two-component systems and the σE pathway (9 genes) (38, 39) (see Tables S2 and S3). Considering the membrane localization and potential mode of action of ApoEdpL-W, we focused the rest of our analyses on the contributions of envelope stress factors to resistance to ApoEdpL-W.
The ApoEdpL-W peptide induces E. coli envelope stress responses.
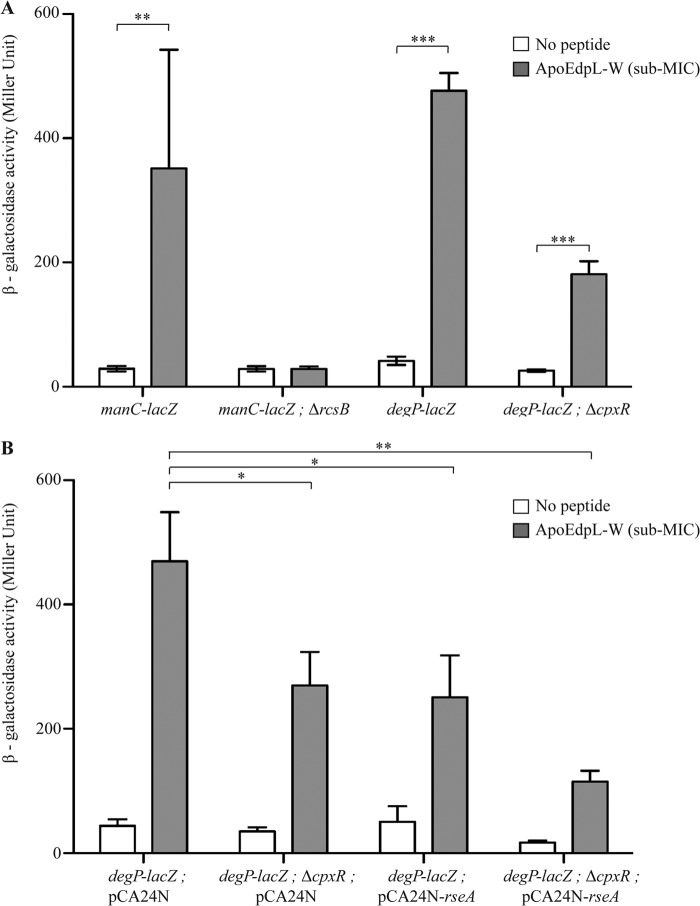
Reporter gene fusions between lacZ and the manC promoter (to monitor induction of the RcsCDB regulon) and between lacZ and the degP promoter (to monitor induction of the CpxAR and σE regulons) were used to confirm the DNA array analysis. Comparison of β-galactosidase activities in bacteria exposed or not to a 3 μM sublethal concentration of ApoEdpL-W showed that both RcsCDB and CpxAR were induced by ApoEdpL-W (Fig. 3A). Introduction of a cpxR mutation did not completely prevent degP-lacZ induction, therefore suggesting that ApoEdpL-W could also induce the σE system (Fig. 3A). Since the σE gene (rpoE) is an essential gene in E. coli, the possible induction of the σE pathway by ApoEdpL-W was evaluated by overexpressing the anti-σE factor RseA, which is known to sequester σE (40). The extent of degP-lacZ induction by ApoEdpL-W was reduced by the introduction of pCA24N-rseA into E. coli degP-lacZ and E. coli ΔcpxR degP-lacZ reporter strains, therefore confirming the induction of the σE pathway by ApoEdpL-W (Fig. 3B). Taken together, these results demonstrate that E. coli induces RcsCDB, CpxAR, and σE envelope stress pathways upon exposure to ApoEdpL-W.
Fig 3.
Induction of RcsCDB, CpxAR, and σE pathways in response to ApoEdpL-W. (A) β-Galactosidase activity measurements of lacZ transcriptional fusions in genes belonging to each regulon, with and without the regulator deletion. (B) β-Galactosidase activity measurements of degP-lacZ fusion with and without cpxR deletion and/or a decrease of σE activity by overexpressing the repressor RseA. In the latter case, experiments were carried out in MH medium plus chloramphenicol and IPTG at 0.01 mM. Growing E. coli cells carrying the different reporter fusions were exposed to 0 or 3 μM ApoEdpL-W for 30 min, and β-galactosidase activities were measured as described in Materials and Methods. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by two-tailed unpaired Student's t test.
The CpxAR pathway, but not the RcsCDB or σE pathway, is involved in resistance to ApoEdpL-W.
Induction of the Rcs, Cpx, and σE envelope stress pathways upon E. coli exposure to subinhibitory concentrations of ApoEdpL-W could result from membrane perturbation induced by ApoEdpL-W insertion into E. coli membranes. To test whether these genetic responses could contribute to resistance to the ApoEdpL-W peptide, we inactivated rcsB and observed that absence of a functional Rcs system did not alter E. coli's sensitivity to ApoEdpL-W (data not shown). Similarly, introduction of the pCA24N-rseA plasmid and the associated reduction in σE activity increased sensitivity to ApoEdpL-W, although not to a statistically significant level (see Fig. S2A in the supplemental material). In contrast, inactivation of cpxR lowered E. coli's ApoEdpL-W tolerance, whereas introduction of pCA24N-cpxR restored the wild-type phenotype (Fig. 4). This result suggested that induction of the Cpx signaling system could reduce E. coli susceptibility to the ApoEdpL-W antimicrobial peptide. To further identify Cpx-regulated genes involved in resistance to ApoEdpL-W, we took advantage of the Keio collection of ordered single-gene deletion mutants performed in E. coli K-12 strain BW25113 (31). Because the ApoEdpL-W MICs were similar for strains MG1655 and BW25113, the MICs of 10 BW25113 mutants in genes belonging to the Cpx regulon, including 3 genes upregulated in the presence of ApoEdpL-W (degP, spy, and cpxP), were determined (Table 2; see Table S2). Among these 10 mutants, only the degP mutant displayed a 2-fold increased susceptibility to ApoEdpL-W (Table 2). Consistently, MG1655ΔdegP also displayed a similar increased susceptibility to ApoEdpL-W, which could be partially complemented by pCA24N-degP (see Fig. S3). In contrast, no restoration of the wild-type phenotype was observed in a cpxR mutant complemented by the pCA24N-degP plasmid (data not shown); moreover, introduction of a pCA24N-rseA plasmid leading to decreased σE activity in the cpxR mutant did not significantly increase its susceptibility to ApoEdpL-W (see Fig. S2B). To confirm that the induction of the Cpx pathway contributes to E. coli tolerance to ApoEdpL-W, the nlpE gene, encoding the lipoprotein NlpE, which is known to induce the Cpx system (41), was overexpressed. The impact of nlpE overexpression in wild-type and cpxR mutant strains on E. coli survival after exposure to ApoEdpL-W was evaluated, and this assay revealed that NlpE-dependent induction of the Cpx pathway led to a 7-fold increase of E. coli's ApoEdpL-W tolerance (see Fig. S4).
Fig 4.
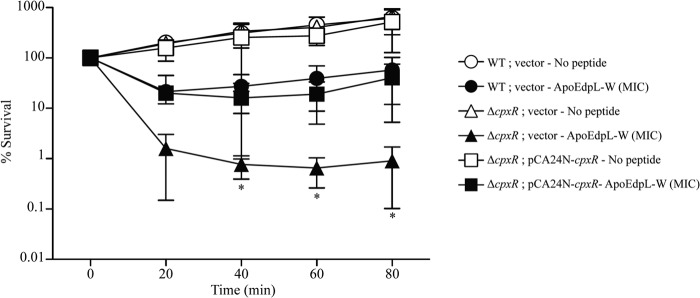
Role of the CpxAR system in the tolerance of planktonic bacteria to ApoEdpL-W. The wild-type (WT) strain, its corresponding cpxR mutant, and the complemented strain were grown in MH medium plus chloramphenicol and IPTG (0.01 mM) until reaching an OD600 of 0.1. The strains were exposed to 0 or 5 μM ApoEdpL-W for 80 min, during which samples were taken every 20 min. Survival of each strain was estimated by CFU counting and compared to numbers obtained prior to ApoEdpL-W treatment. Asterisks indicate values significantly different from those of the wild-type strain by two-tailed unpaired Student's t test. *, P < 0.05.
Table 2.
Impact of ApoEdpL-W on growth of E. coli mutants impaired in the Cpx pathway response
| Strain/mutation | Inactivated function | MIC (μM) |
|---|---|---|
| BW25113 | Wild type (Keio) | 5 |
| ΔacrD | Component of the AcrAD-TolC multidrug efflux transport system | 5 |
| ΔcpxP | Negative regulator of Cpx response | 5 |
| ΔdegP | Periplasmic serine endoprotease | 2.5 |
| ΔmdtA | Component of the MdtABC multidrug efflux transport system | 5 |
| ΔnanC | N-Acetylneuraminic acid outer membrane channel | 5 |
| ΔompC | Outer membrane porin C | 5 |
| ΔompF | Outer membrane porin F | 5 |
| Δspy | Periplasmic protein related to spheroblast formation | 5 |
| ΔtomB (ΔybaJ) | Hha toxin overexpression modulator | 5 |
| ΔydeH | Diguanylate cyclase | 5 |
Finally, comparison of sensitivities to ApoEdpL-W of 24-h biofilms formed by the E. coli MG1655 F′ wild type and its cpxR and degP mutants showed that both mutants exhibited increased susceptibility to ApoEdpL-W (Fig. 5).
Fig 5.
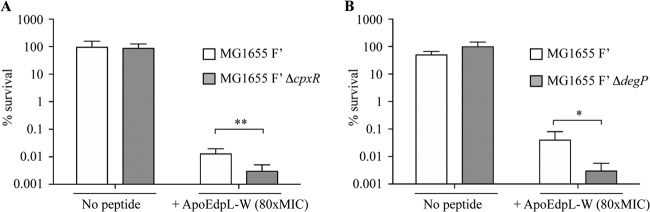
Impact of the CpxAR system on E. coli biofilm ApoEdpL-W tolerance. Twenty-four-hour biofilms formed by the wild-type strain and corresponding cpxR and degP mutants were treated with 0 or 400 μM ApoEdpL-W (80× MIC). Bacterial survival was estimated by viable cell counts after 24 h of treatment. Percent survival represents viable cells after 24 h of treatment compared to those in untreated biofilm prior to addition of ApoEdpL-W. *, P < 0.05; **, P < 0.01 by two-tailed unpaired Student's t test.
Taken together, these results show that ApoEdpL-W-dependent induction of the CpxAR pathway and degP expression contributed to E. coli tolerance to this antimicrobial peptide.
The CpxAR system is induced by different AMPs and contributes to E. coli's polymyxin B and melittin tolerance.
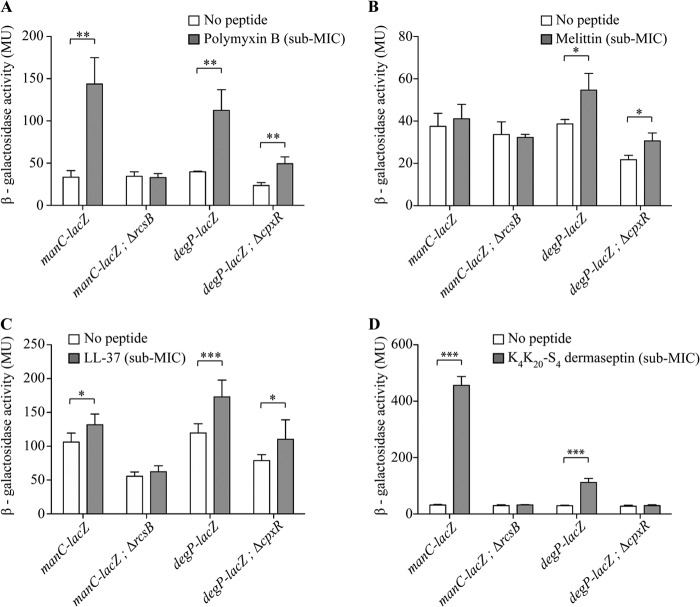
To investigate whether AMPs unrelated to ApoEdpL-W and targeting cell membranes could also induce envelope stress response pathways, four different AMPs known or predicted to disrupt or permeabilize bacterial membranes were selected for analysis: (i) polymyxin B, a cationic cyclic peptide derived from Bacillus polymyxa that binds to lipid A of the lipopolysaccharide (LPS), destabilizing and disrupting outer and inner membranes (42, 43); (ii) melittin, extracted from bee venom (44, 45); (iii) the human cathelicidin-derived antimicrobial peptide LL-37 (46, 47); and (iv) K4K20-S4 dermaseptin, derived from dermaseptin S4, isolated from frog skin and proposed to act on bacterial membranes (48, 49). Melittin and LL-37 are both α-helical peptides and are proposed to form similar types of transmembrane pores in a lipid bilayer by a toroidal pore mechanism where peptides and lipids together form well-defined pores (45, 47). After MIC determination for these different AMPs, E. coli bacteria were exposed to sublethal and nonlytic concentrations of each compound (ranging from 0.5× MIC to 0.8× MIC) to monitor genetic responses induced by the chosen AMP. As done previously, manC-lacZ and degP-lacZ reporter gene fusions were used to monitor expression of the Rcs, Cpx, and σE pathways in the presence of polymyxin B (0.1 mg/liter), melittin (5 mg/liter), LL-37 (4 mg/liter), and K4K20-S4 dermaseptin (0.1 mg/liter). While both the Rcs and Cpx systems were induced upon exposure to polymyxin B, K4K20-S4 dermaseptin, and LL-37, melittin only slightly induced the Cpx system (Fig. 6). Moreover, induction of degP expression upon exposure to polymyxin B, LL-37, and melittin was likely mediated by both the Cpx and σE pathways, whereas K4K20-S4 dermaseptin activated only the Cpx system, since a cpxR mutation completely abolished degP expression upon exposure to this peptide. We then tested the impact of the Rcs and Cpx pathways on E. coli's polymyxin B tolerance, which induces both pathways, and melittin, which activates only the Cpx pathway. While the susceptibilities of the rcsB mutant to polymyxin B and melittin were unchanged, a cpxR mutant displayed a significantly reduced tolerance regarding both peptides compared to the wild-type strain, suggesting a CpxAR system involvement in AMP tolerance (Fig. 7).
Fig 6.
Induction of RcsCDB and/or CpxAR pathway in response to different cationic antimicrobial peptides. β-Galactosidase activity measurements are shown for lacZ transcriptional fusions with genes belonging to each regulon, with and without the regulator deletion, after exposure for 30 min to 0.1 mg/liter polymyxin B (0.8× MIC) (A), 5 mg/liter melittin (0.8× MIC) (B), 4 mg/liter LL-37 (0.65× MIC) (C), and 0.1 mg/liter K4K20-S4 dermaseptin (0.5× MIC) (D). MU, Miller units. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by two-tailed unpaired Student's t test.
Fig 7.
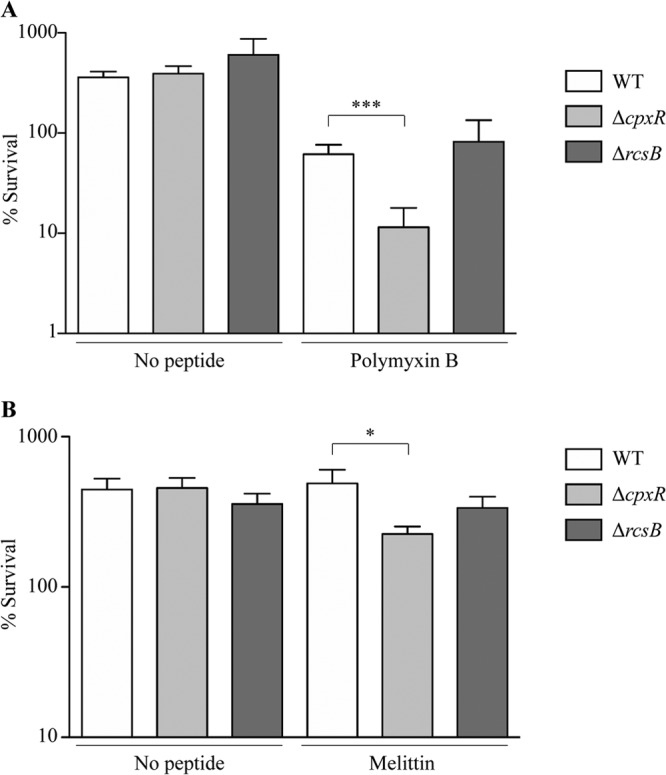
Impact of RcsCDB and/or CpxAR system on the tolerance of E. coli planktonic bacteria to different cationic antimicrobial peptides. The wild-type strain and corresponding cpxR and rcsB mutants were grown in MH medium until reaching an OD600 of 0.1. They were then exposed for 60 min to 0× or 4× MIC of polymyxin B (A) or 0× or 2× MIC of melittin (B). Survival of each strain was estimated by CFU counting and compared to values obtained prior to AMP treatment. *, P < 0.05; ***, P < 0.001 by two-tailed unpaired Student's t test.
DISCUSSION
In the context of the emergence of antibiotic resistance, antimicrobial peptides stand as a plausible alternative against bacterial infections in some clinical situations (2). In this study, we investigated resistance mechanisms potentially induced by a new antimicrobial peptide derived from human apolipoprotein E, ApoEdpL-W. ApoEdpL-W is an aromatic-substituted peptide previously reported to be active against P. aeruginosa and S. aureus pathogens and to possibly interact with membranes (26). We showed that E. coli exposure to ApoEdpL-W induces Rcs, Cpx, and σE pathways, three regulatory pathways known to sense envelope stress. Interestingly, transcriptome analyses revealed strong similarities between responses to ApoEdpL-W and those to peptidoglycan-disrupting β-lactams (cefsulodin and amdinocillin). Indeed, expression of Rcs-regulated genes was increased in response to cefsulodin and amdinocillin, while Cpx and σE were induced upon exposure to amdinocillin and amdinocillin plus cefsulodin (50). While antimicrobial peptides do not target peptidoglycan, the similarities observed between E. coli responses to β-lactams and to ApoEdpL-W, both inducing regulatory systems involved in sensing envelope perturbations, suggest that ApoEdpL-W targets the cell envelope, which is consistent with the demonstrated localization of this peptide in the bacterial cell envelope.
Bacterial two-component systems sense and respond to different stimuli, including membrane stresses caused by antimicrobial peptides. In some cases, they were shown to play a role in tolerance to the recognized AMP. For instance, in Salmonella enterica serovar Typhimurium, AMP binding to the PhoQ sensor directly activates the PhoP/PhoQ two-component system, which then contributes to tolerance toward different AMPs, notably by regulating genes involved in LPS modifications (18, 24, 51). Recently, the RcsCDB pathway was shown to perceive the action of polymyxin B in S. enterica and to contribute to the intrinsic tolerance of bacteria to this antibiotic (25, 52, 53). In this work, we found that whereas exposure to ApoEdpL-W induces all Rcs, Cpx, and σE E. coli envelope stress responses, only the Cpx two-component system contributes to the tolerance of E. coli to this peptide. Indeed, while the Rcs system is induced upon exposure to ApoEdpL-W or polymyxin B, it is not required for the tolerance of E. coli to these peptides. This indicates that in E. coli, while a general cell envelope perturbation is sensed upon the presence of ApoEdpL-W and leads to induction of multiple cell envelope stress pathways, not all upregulated genes are involved in E. coli's ApoEdpL-W tolerance. The Cpx system senses envelope perturbations such as protein misfolding and accumulation and bacterial contact with surfaces (39, 54). It responds to different physicochemical stimuli by activating expression of genes encoding periplasmic proteases and chaperone proteins (39). In contrast, its role in antimicrobial resistance is less well understood. The CpxAR system was shown to be involved in Salmonella resistance to protamine and several α-helical AMPs (55). More recently, the CpxAR system was shown to be induced upon exposure to an antimicrobial cationic polyethylenimine in E. coli (56) and to confer resistance to several β-lactams and chloramphenicol in Klebsiella pneumoniae (57). Here we showed that inactivation of cpxR increased the sensitivity of E. coli to ApoEdpL-W. In addition, induction of the Cpx pathway upon NlpE overexpression increased E. coli's ApoEdpL-W tolerance in a CpxR-dependent manner. Interestingly, the overexpression of NlpE in a cpxR mutant also led to an increased susceptibility to ApoEdpL-W; under these conditions, the cpxR mutant could be unable to control the important envelope stress induced by perturbations generated by both ApoEdpL-W and the overexpression of NlpE, explaining this increased susceptibility to ApoEdpL-W treatment. Furthermore, we determined that the cpx-regulated gene degP is involved in E. coli tolerance to ApoEdpL-W. degP encodes a periplasmic endopeptidase of the ATP-independent serine protease family and presents both chaperone and proteolytic activities (temperature-dependent switch from chaperone to protease activity) (58). Extracytoplasmic proteases were previously associated with antimicrobial tolerance. For instance, the outer membrane OmpT protease is involved in E. coli tolerance to protamine by degrading this membrane-permeabilizing peptide (12), while PgtE, a Salmonella OmpT protease homolog (46% identity and 65% similarity), contributes to resistance to several α-helical antimicrobial peptides (11). DegP has also been shown to be involved in E. coli's lactoferricin B tolerance, probably by proteolytic degradation of this antimicrobial peptide (59). DegP preferentially cleaves after valine and isoleucine residues, even when additional determinants (sequence and structure) are involved in the cleavage (60). As ApoEdpL-W does not contain either of these two amino acids, this suggests that the peptide might not be a substrate for DegP. Consistently, no effect of DegP on the fluorescence signal was observed in bacteria exposed to the Fluo-ApoEdpL-W peptide (data not shown). Hence, while the precise role of DegP in E. coli's ApoEdpL-W tolerance remains to be investigated further, we speculate that induction of DegP expression could reduce damages induced by ApoEdpL-W in the periplasm.
We also showed that decreased σE activity upon overexpression of RseA led to reduced levels of degP expression upon exposure to ApoEdpL-W, as in the cpxR mutant. However, only the cpxR mutation, not depletion of σE activity, increased E. coli sensitivity to ApoEdpL-W. These results indicate that although degP is required for E. coli's ApoEdpL-W tolerance, increased susceptibility of the cpxR mutant is DegP independent and involves others cpx-regulated factors, which is consistent with the absence of complementation of the cpxR mutant by overexpressing degP.
Mechanisms of resistance to AMPs are mostly nonspecific, and antimicrobial peptide tolerance often relies on ill-understood bacterial adaptation to each peptide rather than on specific resistance mechanisms (10). Here we showed that different tested peptides induced specific sets of envelope stress pathways, depending on their global charge, structure, and/or mechanism of action: polymyxin B, ApoEdpL-W, and, to a lesser extent, LL-37, all predicted to target membranes, activated Rcs, Cpx, and σE envelope stress pathways, whereas K4K20-S4 dermaseptin activated both Rcs and Cpx systems but not the σE pathway. Finally, melittin led to only weak activation of Cpx and σE pathways. These results are consistent with a recent study showing that in S. enterica serovar Typhimurium, the noncanonical Rcs pathway is activated by polymyxin B and several other AMPs but not by envelope-permeabilizing agents (SDS, EDTA, and Triton X-100) or polyamines, suggesting that Rcs activation requires detection of specific outer membrane alterations induced by peptides rather than mere global membrane permeabilization (53).
Antimicrobial peptides represent a promising source of novel anti-infection molecules (2). Using several AMPs, we demonstrated that the Cpx pathway is involved in resistance to ApoEdpL-W, polymyxin B, or melittin, and potentially other predicted membrane-acting antimicrobial peptides. These results therefore show that the study of peptide-specific genetic responses induced by different antimicrobial peptides improves our fundamental understanding of peptides' modes of action, while providing key insights into potential bacterial resistance mechanisms to antimicrobial peptides.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sylvie Létoffé for helpful discussions. We also thank Mike Birch, Ai2 Ltd., as well as members of the NPARI consortium for support during the course of this project.
This work was supported by a grant from the European Commission (grant LSHE-CT-2006-037692) and the French Government's Investissement d'Avenir program Laboratoire d'Excellence “Integrative Biology of Emerging Infectious Diseases” (grant ANR-10-LABX-62-IBEID).
Footnotes
Published ahead of print 4 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02593-13.
REFERENCES
- 1.Martinez JL, Fajardo A, Garmendia L, Hernandez A, Linares JF, Martínez-Solano L, Sánchez MB. 2009. A global view of antibiotic resistance. FEMS Microbiol. Rev. 33:44–65 [DOI] [PubMed] [Google Scholar]
- 2.Jenssen H, Hamill P, Hancock RE. 2006. Peptide antimicrobial agents. Clin. Microbiol. Rev. 19:491–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zasloff M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389–395 [DOI] [PubMed] [Google Scholar]
- 4.Brogden KA. 2005. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3:238–250 [DOI] [PubMed] [Google Scholar]
- 5.Brotz H, Bierbaum G, Leopold K, Reynolds PE, Sahl HG. 1998. The lantibiotic mersacidin inhibits peptidoglycan synthesis by targeting lipid II. Antimicrob. Agents Chemother. 42:154–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kragol G, Lovas S, Varadi G, Condie BA, Hoffmann R, Otvos L., Jr 2001. The antibacterial peptide pyrrhocoricin inhibits the ATPase actions of DnaK and prevents chaperone-assisted protein folding. Biochemistry 40:3016–3026 [DOI] [PubMed] [Google Scholar]
- 7.Park CB, Kim HS, Kim SC. 1998. Mechanism of action of the antimicrobial peptide buforin II: buforin II kills microorganisms by penetrating the cell membrane and inhibiting cellular functions. Biochem. Biophys. Res. Commun. 244:253–257 [DOI] [PubMed] [Google Scholar]
- 8.Patrzykat A, Friedrich CL, Zhang L, Mendoza V, Hancock RE. 2002. Sublethal concentrations of pleurocidin-derived antimicrobial peptides inhibit macromolecular synthesis in Escherichia coli. Antimicrob. Agents Chemother. 46:605–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Srinivas N, Jetter P, Ueberbacher BJ, Werneburg M, Zerbe K, Steinmann J, Van der Meijden B, Bernardini F, Lederer A, Dias RL, Misson PE, Henze H, Zumbrunn J, Gombert FO, Obrecht D, Hunziker P, Schauer S, Ziegler U, Kach A, Eberl L, Riedel K, DeMarco SJ, Robinson JA. 2010. Peptidomimetic antibiotics target outer-membrane biogenesis in Pseudomonas aeruginosa. Science 327:1010–1013 [DOI] [PubMed] [Google Scholar]
- 10.Anaya-Lopez JL, Lopez-Meza JE, Ochoa-Zarzosa A. 2013. Bacterial resistance to cationic antimicrobial peptides. Crit. Rev. Microbiol. 39:180–195 [DOI] [PubMed] [Google Scholar]
- 11.Guina T, Yi EC, Wang H, Hackett M, Miller SI. 2000. A PhoP-regulated outer membrane protease of Salmonella enterica serovar Typhimurium promotes resistance to alpha-helical antimicrobial peptides. J. Bacteriol. 182:4077–4086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stumpe S, Schmid R, Stephens DL, Georgiou G, Bakker EP. 1998. Identification of OmpT as the protease that hydrolyzes the antimicrobial peptide protamine before it enters growing cells of Escherichia coli. J. Bacteriol. 180:4002–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sieprawska-Lupa M, Mydel P, Krawczyk K, Wojcik K, Puklo M, Lupa B, Suder P, Silberring J, Reed M, Pohl J, Shafer W, McAleese F, Foster T, Travis J, Potempa J. 2004. Degradation of human antimicrobial peptide LL-37 by Staphylococcus aureus-derived proteinases. Antimicrob. Agents Chemother. 48:4673–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shafer WM, Qu X, Waring AJ, Lehrer RI. 1998. Modulation of Neisseria gonorrhoeae susceptibility to vertebrate antibacterial peptides due to a member of the resistance/nodulation/division efflux pump family. Proc. Natl. Acad. Sci. U. S. A. 95:1829–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Otto M, Peschel A, Gotz F. 1998. Producer self-protection against the lantibiotic epidermin by the ABC transporter EpiFEG of Staphylococcus epidermidis Tu3298. FEMS Microbiol. Lett. 166:203–211 [DOI] [PubMed] [Google Scholar]
- 16.Bengoechea JA, Skurnik M. 2000. Temperature-regulated efflux pump/potassium antiporter system mediates resistance to cationic antimicrobial peptides in Yersinia. Mol. Microbiol. 37:67–80 [DOI] [PubMed] [Google Scholar]
- 17.Peschel A, Otto M, Jack RW, Kalbacher H, Jung G, Gotz F. 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274:8405–8410 [DOI] [PubMed] [Google Scholar]
- 18.Guo L, Lim KB, Poduje CM, Daniel M, Gunn JS, Hackett M, Miller SI. 1998. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell 95:189–198 [DOI] [PubMed] [Google Scholar]
- 19.Gunn JS, Lim KB, Krueger J, Kim K, Guo L, Hackett M, Miller SI. 1998. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol. 27:1171–1182 [DOI] [PubMed] [Google Scholar]
- 20.Otto M. 2009. Bacterial sensing of antimicrobial peptides. Contrib. Microbiol. 16:136–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oh JT, Cajal Y, Dhurjati PS, Van Dyk TK, Jain MK. 1998. Cecropins induce the hyperosmotic stress response in Escherichia coli. Biochim. Biophys. Acta 1415:235–245 [DOI] [PubMed] [Google Scholar]
- 22.Hong RW, Shchepetov M, Weiser JN, Axelsen PH. 2003. Transcriptional profile of the Escherichia coli response to the antimicrobial insect peptide cecropin A. Antimicrob. Agents Chemother. 47:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomasinsig L, Scocchi M, Mettulio R, Zanetti M. 2004. Genome-wide transcriptional profiling of the Escherichia coli response to a proline-rich antimicrobial peptide. Antimicrob. Agents Chemother. 48:3260–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, Klevit RE, Le Moual H, Miller SI. 2005. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122:461–472 [DOI] [PubMed] [Google Scholar]
- 25.Erickson KD, Detweiler CS. 2006. The Rcs phosphorelay system is specific to enteric pathogens/commensals and activates ydeI, a gene important for persistent Salmonella infection of mice. Mol. Microbiol. 62:883–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelly BA, Neil SJ, McKnight A, Santos JM, Sinnis P, Jack ER, Middleton DA, Dobson CB. 2007. Apolipoprotein E-derived antimicrobial peptide analogues with altered membrane affinity and increased potency and breadth of activity. FEBS J. 274:4511–4525 [DOI] [PubMed] [Google Scholar]
- 27.Dobson CB, Sales SD, Hoggard P, Wozniak MA, Crutcher KA. 2006. The receptor-binding region of human apolipoprotein E has direct anti-infective activity. J. Infect. Dis. 193:442–450 [DOI] [PubMed] [Google Scholar]
- 28.Forbes S, McBain AJ, Felton-Smith S, Jowitt TA, Birchenough HL, Dobson CB. 2013. Comparative surface antimicrobial properties of synthetic biocides and novel human apolipoprotein E derived antimicrobial peptides. Biomaterials 34:5453–5464 [DOI] [PubMed] [Google Scholar]
- 29.Rossignol T, Kelly B, Dobson C, d'Enfert C. 2011. Endocytosis-mediated vacuolar accumulation of the human ApoE apolipoprotein-derived ApoEdpL-W antimicrobial peptide contributes to its antifungal activity in Candida albicans. Antimicrob. Agents Chemother. 55:4670–4681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelly BA, Harrison I, McKnight A, Dobson CB. 2010. Anti-infective activity of apolipoprotein domain derived peptides in vitro: identification of novel antimicrobial peptides related to apolipoprotein B with anti-HIV activity. BMC Immunol. 11:13. 10.1186/1471-2172-11-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. 2005. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 12:291–299 [DOI] [PubMed] [Google Scholar]
- 33.Bergaoui I, Zairi A, Tangy F, Aouni M, Selmi B, Hani K. 2013. In vitro antiviral activity of dermaseptin S(4) and derivatives from amphibian skin against herpes simplex virus type 2. J. Med. Virol. 85:272–281 [DOI] [PubMed] [Google Scholar]
- 34.Bolstad BM, Irizarry RA, Astrand M, Speed TP. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185–193 [DOI] [PubMed] [Google Scholar]
- 35.Jain N, Thatte J, Braciale T, Ley K, O'Connell M, Lee JK. 2003. Local-pooled-error test for identifying differentially expressed genes with a small number of replicated microarrays. Bioinformatics 19:1945–1951 [DOI] [PubMed] [Google Scholar]
- 36.Miller JH. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 37.Ghigo JM. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442–445 [DOI] [PubMed] [Google Scholar]
- 38.Raivio TL, Silhavy TJ. 2001. Periplasmic stress and ECF sigma factors. Annu. Rev. Microbiol. 55:591–624 [DOI] [PubMed] [Google Scholar]
- 39.Majdalani N, Gottesman S. 2005. The Rcs phosphorelay: a complex signal transduction system. Annu. Rev. Microbiol. 59:379–405 [DOI] [PubMed] [Google Scholar]
- 40.Missiakas D, Mayer MP, Lemaire M, Georgopoulos C, Raina S. 1997. Modulation of the Escherichia coli σE (RpoE) heat-shock transcription-factor activity by the RseA, RseB and RseC proteins. Mol. Microbiol. 24:355–371 [DOI] [PubMed] [Google Scholar]
- 41.Snyder WB, Davis LJ, Danese PN, Cosma CL, Silhavy TJ. 1995. Overproduction of NlpE, a new outer membrane lipoprotein, suppresses the toxicity of periplasmic LacZ by activation of the Cpx signal transduction pathway. J. Bacteriol. 177:4216–4223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morrison DC, Jacobs DM. 1976. Binding of polymyxin B to the lipid A portion of bacterial lipopolysaccharides. Immunochemistry 13:813–818 [DOI] [PubMed] [Google Scholar]
- 43.Storm DR, Rosenthal KS, Swanson PE. 1977. Polymyxin and related peptide antibiotics. Annu. Rev. Biochem. 46:723–763 [DOI] [PubMed] [Google Scholar]
- 44.Habermann E. 1972. Bee and wasp venoms. Science 177:314–322 [DOI] [PubMed] [Google Scholar]
- 45.Yang L, Harroun TA, Weiss TM, Ding L, Huang HW. 2001. Barrel-stave model or toroidal model? A case study on melittin pores. Biophys. J. 81:1475–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sorensen O, Arnljots K, Cowland JB, Bainton DF, Borregaard N. 1997. The human antibacterial cathelicidin, hCAP-18, is synthesized in myelocytes and metamyelocytes and localized to specific granules in neutrophils. Blood 90:2796–2803 [PubMed] [Google Scholar]
- 47.Henzler-Wildman KA, Martinez GV, Brown MF, Ramamoorthy A. 2004. Perturbation of the hydrophobic core of lipid bilayers by the human antimicrobial peptide LL-37. Biochemistry 43:8459–8469 [DOI] [PubMed] [Google Scholar]
- 48.Feder R, Dagan A, Mor A. 2000. Structure-activity relationship study of antimicrobial dermaseptin S4 showing the consequences of peptide oligomerization on selective cytotoxicity. J. Biol. Chem. 275:4230–4238 [DOI] [PubMed] [Google Scholar]
- 49.Navon-Venezia S, Feder R, Gaidukov L, Carmeli Y, Mor A. 2002. Antibacterial properties of dermaseptin S4 derivatives with in vivo activity. Antimicrob. Agents Chemother. 46:689–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laubacher ME, Ades SE. 2008. The Rcs phosphorelay is a cell envelope stress response activated by peptidoglycan stress and contributes to intrinsic antibiotic resistance. J. Bacteriol. 190:2065–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gunn JS, Miller SI. 1996. PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J. Bacteriol. 178:6857–6864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pilonieta MC, Erickson KD, Ernst RK, Detweiler CS. 2009. A protein important for antimicrobial peptide resistance, YdeI/OmdA, is in the periplasm and interacts with OmpD/NmpC. J. Bacteriol. 191:7243–7252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Farris C, Sanowar S, Bader MW, Pfuetzner R, Miller SI. 2010. Antimicrobial peptides activate the Rcs regulon through the outer membrane lipoprotein RcsF. J. Bacteriol. 192:4894–4903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang YH, Ferrieres L, Clarke DJ. 2006. The role of the Rcs phosphorelay in Enterobacteriaceae. Res. Microbiol. 157:206–212 [DOI] [PubMed] [Google Scholar]
- 55.Weatherspoon-Griffin N, Zhao G, Kong W, Kong Y, Morigen, Andrews-Polymenis H, McClelland M, Shi Y. 2011. The CpxR/CpxA two-component system up-regulates two Tat-dependent peptidoglycan amidases to confer bacterial resistance to antimicrobial peptide. J. Biol. Chem. 286:5529–5539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lander BA, Checchi KD, Koplin SA, Smith VF, Domanski TL, Isaac DD, Lin S. 2012. Extracytoplasmic stress responses induced by antimicrobial cationic polyethylenimines. Curr. Microbiol. 65:488–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Srinivasan VB, Vaidyanathan V, Mondal A, Rajamohan G. 2012. Role of the two component signal transduction system CpxAR in conferring cefepime and chloramphenicol resistance in Klebsiella pneumoniae NTUH-K2044. PLoS One 7:e33777. 10.1371/journal.pone.0033777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spiess C, Beil A, Ehrmann M. 1999. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 97:339–347 [DOI] [PubMed] [Google Scholar]
- 59.Ulvatne H, Haukland HH, Samuelsen O, Kramer M, Vorland LH. 2002. Proteases in Escherichia coli and Staphylococcus aureus confer reduced susceptibility to lactoferricin B. J. Antimicrob. Chemother. 50:461–467 [DOI] [PubMed] [Google Scholar]
- 60.Kolmar H, Waller PR, Sauer RT. 1996. The DegP and DegQ periplasmic endoproteases of Escherichia coli: specificity for cleavage sites and substrate conformation. J. Bacteriol. 178:5925–5929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guyer MS, Reed RR, Steitz JA, Low KB. 1981. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harbor Symp. Quant. Biol. 45:135–140 [DOI] [PubMed] [Google Scholar]
- 62.Beloin C, Valle J, Latour-Lambert P, Faure P, Kzreminski M, Balestrino D, Haagensen JA, Molin S, Prensier G, Arbeille B, Ghigo JM. 2004. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol. Microbiol. 51:659–674 [DOI] [PubMed] [Google Scholar]
- 63.Davalos-Garcia M, Conter A, Toesca I, Gutierrez C, Cam K. 2001. Regulation of osmC gene expression by the two-component system rcsB-rcsC in Escherichia coli. J. Bacteriol. 183:5870–5876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Castanie-Cornet MP, Treffandier H, Francez-Charlot A, Gutierrez C, Cam K. 2007. The glutamate-dependent acid resistance system in Escherichia coli: essential and dual role of the His-Asp phosphorelay RcsCDB/AF. Microbiology 153:238–246 [DOI] [PubMed] [Google Scholar]
- 65.Raina S, Missiakas D, Georgopoulos C. 1995. The rpoE gene encoding the sigma E (sigma 24) heat shock sigma factor of Escherichia coli. EMBO J. 14:1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.