Abstract
We report here that stress experienced by bacteria due to aerosolization and air sampling can result in severe membrane impairment, leading to the release of DNA as free molecules. Escherichia coli and Bacillus atrophaeus bacteria were aerosolized and then either collected directly into liquid or collected using other collection media and then transferred into liquid. The amount of DNA released was quantified as the cell membrane damage index (ID), i.e., the number of 16S rRNA gene copies in the supernatant liquid relative to the total number in the bioaerosol sample. During aerosolization by a Collison nebulizer, the ID of E. coli and B. atrophaeus in the nebulizer suspension gradually increased during 60 min of continuous aerosolization. We found that the ID of bacteria during aerosolization was statistically significantly affected by the material of the Collison jar (glass > polycarbonate; P < 0.001) and by the bacterial species (E. coli > B. atrophaeus; P < 0.001). When E. coli was collected for 5 min by filtration, impaction, and impingement, its ID values were within the following ranges: 0.051 to 0.085, 0.16 to 0.37, and 0.068 to 0.23, respectively; when it was collected by electrostatic precipitation, the ID values (0.011 to 0.034) were significantly lower (P < 0.05) than those with other sampling methods. Air samples collected inside an equine facility for 2 h by filtration and impingement exhibited ID values in the range of 0.30 to 0.54. The data indicate that the amount of cell damage during bioaerosol sampling and the resulting release of DNA can be substantial and that this should be taken into account when analyzing bioaerosol samples.
INTRODUCTION
Investigation of the presence of airborne microorganisms (bioaerosols) in the ambient air is of interest due to their environmental and human health effects (1, 2). Numerous studies have shown that increased exposure to bioaerosols is positively correlated with the incidence of negative respiratory health effects, including lung irritation, asthma, rhinitis, allergy, and cough (1–4). It is generally accepted that health effects caused by exposure to bioaerosols depend not only on the organism and its concentration but also on its physiological status in the air, because viable and nonviable microorganisms have different potentials for causing adverse respiratory health effects (2, 5, 6).
Microorganisms in the airborne state may experience a variety of stressors, including unfavorable temperature and humidity, lack of nutrients, UV radiation, chemical pollutants, and other variables that affect their physiological status (7–11). Depending on that status, airborne cells can be classified as culturable, viable but not culturable, nonviable but maintaining membrane integrity, and cell fragments (12, 13). When bioaerosols are collected for environmental or health investigations or other purposes, it is desirable that the sampling method maintains their physiological status to minimize bias when quantifying and identifying microorganisms in the sample.
Numerous sampling devices have been developed and used to collect bioaerosols by filtration, impaction, impingement, electrostatic precipitation, and other methods. However, during each sampling process, the microorganisms are inevitably exposed to additional stress, which affects their viability and culturability. It has been observed that dehydration during sampling by filters (14, 15) and portable microbial impactors (16) may cause cell injury and loss of culturability, especially in sensitive species. While one of the liquid-based bioaerosol samplers, the BioSampler (SKC Inc., Eighty Four, PA), is considered to be a relatively low-stress sampling device for collecting bioaerosols (17), one study demonstrated that certain collection fluids, including glycerol and surfactant, greatly decreased the viability of Legionella pneumophila, presumably due to the elevated osmotic pressure (18). Stewart et al. observed that 49% of Pseudomonas fluorescens bacteria lost their culturability after impacting an agar surface at a speed of 40 m/s, most likely due to mechanical stress (19). Another study showed that the intactness of the genomic DNA was impaired due to the stress of impaction onto the collection surface (20). Recently, Zhao et al. (2011) found that sampling stress from a variety of bioaerosol samplers decreased the bacterial culturability (21, 22). In addition to the sampling process, microorganisms may also experience substantial stress during aerosolization. It was found in our earlier study that the viability of P. fluorescens bacteria aerosolized by a Collison nebulizer (BGI Inc., Waltham, MA) decreased by over 50% after 90 min of continuous aerosolization (23). Similarly, Thomas et al. indicated that 99.9% of an Escherichia coli population suffered sublethal injury after a 10-min aerosolization by a Collison nebulizer (24). They also concluded that the cell membrane was the major site of damage due to impaction and shear force stress that disturbed membrane homeostasis (24). In light of this study, we hypothesized that the cell membrane could also be a major site of damage during bioaerosol sampling when cells experience substantial mechanical stress, such as during impaction and impingement. In addition, the elevated osmotic stress resulting from nonmechanical sampling factors, such as desiccation, would make cell membranes more vulnerable to mechanical stress, possibly even leading to cell rupture.
In the past few years, quantitative PCR (qPCR) has gained popularity in bioaerosol research due to its capacity to rapidly quantify and identify microorganisms in air samples (25, 26). The collected microorganisms must first be lysed, but the method depends on how air samples are collected. Very often bioaerosols are collected directly into liquid by using impingers or first collected onto filters and then transferred into liquid. To concentrate such samples, liquid is centrifuged, and only the pelleted cells are used for DNA extraction, while the rest of the liquid is discarded (18, 27–30). However, if we consider that a large fraction of cells experience severe stress during aerosolization and air sampling, leading to the loss of their structural integrity, it becomes highly likely that genomic DNA from the ruptured cells is released into the liquid. If this DNA-rich liquid is discarded and not included as part of sample analysis, qPCR performed only on DNA extracted from the pelleted cells would lead to an underestimation of the collected bioaerosol quantity, resulting in an underestimation of the airborne concentration as well.
Thus, the goal of this study was to investigate the release of DNA as free molecules by membrane-damaged microorganisms due to the stress imposed on them during aerosolization and air sampling. To the best of our knowledge, this is the first study to investigate such an effect. The tests were performed with four bioaerosol sampling devices: the SKC Button aerosol sampler (SKC Inc., Eighty Four, PA), an Anderson-type impactor (BioStage; SKC Inc.), the BioSampler (SKC Inc.), and the newly developed electrostatic precipitator with a superhydrophobic surface (EPSS) (31–33). We also assessed the effects of various aerosolization and air sampling parameters on cell integrity and DNA release, including the material of the Collison nebulizer jar (either glass or polycarbonate), the sampling time for collection on a filter by the Button aerosol sampler, the jet-to-plate distance and jet velocity of an Anderson-type impactor, and the type of collection fluid used in the BioSampler. The tests were performed with both Gram-negative E. coli and Gram-positive Bacillus atrophaeus bacteria to investigate how different cell wall structures withstand aerosolization and air sampling stress. It is hoped that the results of this study will provide guidance for selecting appropriate sampling and aerosolization protocols so that the physiological status of bioaerosols is minimally affected, leading to a more accurate sample analysis in bioaerosol studies.
MATERIALS AND METHODS
Test microorganisms.
The sensitive Gram-negative bacterium E. coli (ATCC 15597) and the hardy Gram-positive bacterium B. atrophaeus (ATCC 49337) were selected as test microorganisms. These two organisms have been used widely in bioaerosol research to represent bacteria with different cell wall types and levels of hardiness (14, 18, 25, 34–36). Both organisms were cultivated on nutrient agar (Becton, Dickinson and Company, Sparks, MD) and stored at 4°C for less than 3 months prior to transfer. Prior to experiments, E. coli and B. atrophaeus were precultured in nutrient broth (Becton, Dickinson and Company, Sparks, MD) for 18 h at 37°C and 30°C, respectively. After growing for 18 h, both bacterial cultures were in stationary phase, and over 99% of B. atrophaeus organisms were present as vegetative cells, as verified by the Schaeffer-Fulton method for staining endospores (37). All freshly prepared test organisms were washed 3 times with sterile, deionized (DI) water (Millipore, Billerica, MA) by centrifugation at 7,000 × g for 5 min at 4°C (BR4; Jouan, Winchester, VA). The concentrated bacterial cells were then diluted with sterile DI water to prepare final bacterial suspensions with concentrations ranging from 1 × 108 to 3 × 108 cells/ml, as determined by microscopy.
Experimental setup.
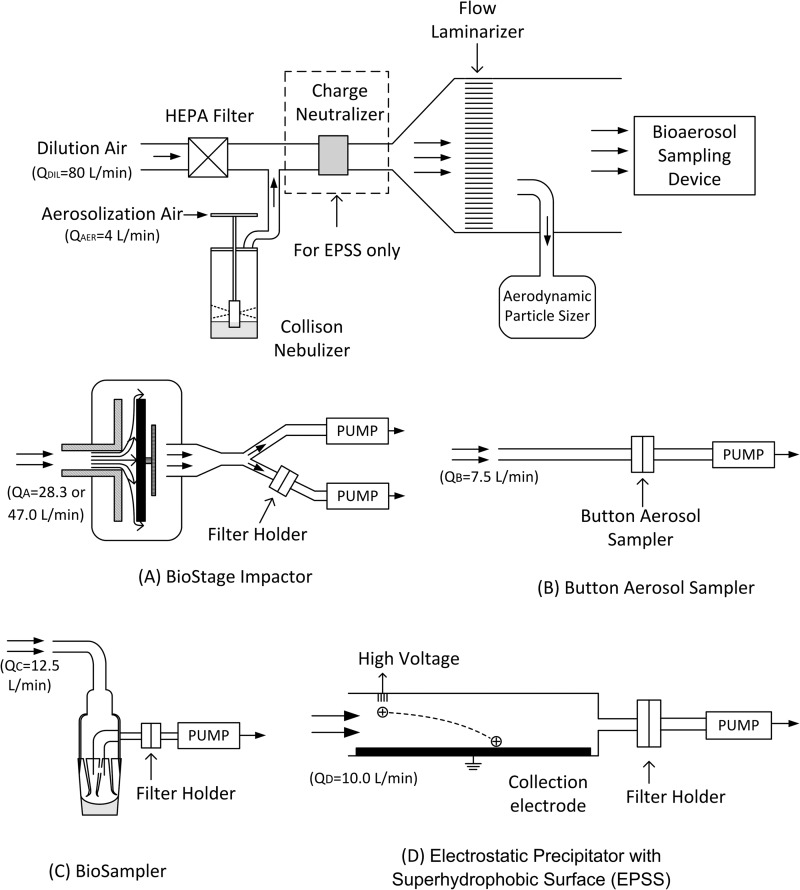
A schematic of the experimental setup is presented in Fig. 1. Bacterial suspensions were aerosolized using a three-jet Collison nebulizer (BGI Inc., Waltham, MA) with either a glass or polycarbonate jar by passing HEPA-filtered air at a flow rate (QAER) of 4 liters/min (pressure of 12 lb/in2). The relatively low aerosolization pressure and flow rate were chosen to minimize potential damage to the bacterial cells due to mechanical stress during aerosolization (24). The aerosolized test organisms were diluted with HEPA-filtered air at a flow rate (QDIL) of 80 liters/min and passed through a laminar flow-producing honeycomb inside the test chamber. For tests with the EPSS, a 2-mCi 210Po charge neutralizer was placed in the air stream before it entered the test chamber. During each test, the concentration of airborne microorganisms inside the chamber was monitored continuously by use of an aerodynamic particle sizer (APS 3321; TSI Inc., Shoreview, MN). The initial volume of freshly prepared culture within the Collison nebulizer was 20 ml during each experiment. The bioaerosol generated was collected by four air sampling devices, as described below. For each repeat with a particular sampling device, a fresh batch of the test bacteria was used. Samples were collected for 5 min immediately after starting the aerosolization to minimize cell damage. At least three repeats were conducted for each sampling device or sampling parameter. All experiments were performed inside a class II biosafety cabinet (Nuaire Inc., Plymouth, MN). Humidity and temperature inside the cabinet were monitored by a traceable hygrometer (Fisher Scientific, Pittsburgh, PA) during each test. The relative humidity (RH) ranged from 40% to 45%, depending on the day, while the temperature stayed in the range of 24 to 26°C.
Fig 1.
Experimental setup used to aerosolize and collect bioaerosols with the BioStage impactor (A), Button aerosol sampler (B), BioSampler (C), and electrostatic precipitator with superhydrophobic surface (EPSS) (D).
Bioaerosol collection system.
The aerosolized microorganisms were collected using a BioStage impactor (SKC Inc., Eighty Four, PA), a Button aerosol sampler (SKC Inc.), a BioSampler (SKC Inc.), or an EPSS as shown in Fig. 1.
A BioStage impactor was used to investigate the effect of impaction on cell integrity (Fig. 1A). We found the recovery of DNA from agar plates to be extremely low (a few percent [data not shown]). Thus, to facilitate effective recovery of collected bacteria and free DNA for analysis by qPCR, we used a sheet of aluminum foil positioned on an adjustable support pad inside a petri dish instead of agar as our collection surface. Since our previous study showed that jet velocity and jet-to-plate distance affect the collection efficiency of microbial impactors (38), we investigated whether these two factors affect the integrity of the E. coli cell structure as well. Thus, the impactor was operated for 5 min at different combinations of flow rate (QA; nominal flow rate of 28.3 liters/min with jet velocity of 23.7 m/s or increased flow rate of 47 liters/min with jet velocity of 39.3 m/s) and simulated agar volume (40 ml with jet-to-plate distance of 2.2 mm or 48 ml with jet-to-plate distance of 1.3 mm). To achieve the desired flow rates, two vacuum pumps were connected to the impactor by use of a Y splitter (Fig. 1A). A Millipore filter holder (Millipore, Billerica, MA) with a 0.45-μm-pore-size polycarbonate filter (Millipore) was connected at the inlet of one of the vacuum pumps to collect those particles that were smaller than the impactor's cutoff size (d50 [particle aerodynamic diameter for which the impactor's collection efficiency is 50%] = 0.6 μm per the manufacturer) or escaped due to bouncing from the aluminum foil surface. The total number of such particles was calculated based on the flow rate proportions of the sampling pumps. We were not able to connect the filter directly at the impactor's outlet due to a substantial pressure drop across the impactor and filter. After sampling, particles collected on the aluminum foil and filters were eluted by vortexing the collection media for 2 min in 10 ml and 5 ml of elution solution, respectively.
To investigate the effect of filter collection on bioaerosol cell integrity, a Button aerosol sampler was selected as a filter holder and used with a 0.6-μm-pore-size polycarbonate filter (Millipore, Billerica, MA). The sampler was operated at a flow rate (QB) of 7.5 liters/min, the maximum flow rate achieved in our setup (Fig. 1B). The Button aerosol sampler is designed to operate at 4 liters/min so that its inlet aspiration efficiency follows the inhalable sampling convention (39), but it has been used with flow rates as high as 10 liters/min (40), and the use of a 7.5-liter/min flow rate instead of the nominal 4 liters/min allowed us to collect more bacteria in a short time. The effect of sampling time on cell membrane integrity was investigated by using two different sampling protocols: (i) sampling of aerosolized E. coli for 5 min and (ii) sampling of aerosolized E. coli for 5 min followed by the passing of particle-free air though the filter for 2 h. Once the sampling was completed, the filter was removed from the sampler and placed into a 10-ml Tween mixture solution containing 0.1% peptone (Fisher, Fair Lawn, NJ), 0.01% Tween 80 (Fisher, Fair Lawn, NJ), and 0.005% Y-30 antifoam reagent (Sigma, St. Louis, MO) (18). Vortexing is generally accepted as an efficient way to elute bioaerosol particles from filters (41, 42), and our preliminary experiments showed that short-term vortexing (<2 min) did not result in significant release of DNA from freshly grown E. coli cells (data not shown). It was also shown that the use of ultrasonic agitation after vortexing improves the recovery of samples (14), but we were concerned that ultrasonic agitation might affect cell membrane integrity. To investigate this effect, some samples collected on filters were eluted by vortexing for 2 min, while the rest were first vortexed for 2 min and then further treated with ultrasonic agitation for 5 min, and the membrane integrity between the two methods was compared.
An SKC BioSampler with a 5-ml sampling cup was operated at a flow rate (QC) of 12.5 liters/min during each 5-min sampling period. A 0.45-μm-pore-size polycarbonate filter (Millipore) was connected to the BioSampler outlet to collect those particles that were either not collected or reaerosolized (Fig. 1C). Two types of collection fluid, namely, sterile DI water and Tween mixture solution (0.1% peptone, 0.01% Tween 80, and 0.005% antifoam reagent), which are commonly used in bioaerosol sampling (14, 18, 34, 43), were tested. During the BioSampler's operation, the bacteria were first impinged into the collection liquid and then subjected to centrifugal motion for the remainder of the sampling period. To test only the effect of centrifugal motion on the membrane integrity of bacteria, 5 ml of sampling fluid was spiked with a known number of E. coli cells and placed into a collection cup, and particle-free air was aspirated by the sampler for 5 min at 12.5 liters/min. In the control group, 5 ml of sampling fluid with the same number of E. coli bacteria was placed into a collection cup and kept static for 5 min.
A novel electrostatic precipitator with a superhydrophobic surface (EPSS) was designed in our laboratory and was used as the fourth sampling device (Fig. 1D). The device is described in detail elsewhere (31, 32), but briefly, it has the shape of a closed half-cylinder positioned at an angle to the horizontal, with the round top part containing an ionizer and the flat bottom plate holding a narrow collection electrode covered by a superhydrophobic substance positioned slightly below the surface. Particles that enter the sampler are electrically charged and then deposited onto the collection electrode by electrostatic forces. Once the sampling is completed, a 40-μl water droplet is introduced at the top of the collection electrode. Due to gravitational force, the droplet rolls down and gathers the deposited particles. The droplet containing the particles is collected in a vial and then diluted by adding 960 μl of sterile deionized water for subsequent analysis. The EPSS was operated at a flow rate (QD) of 10.0 liters/min for 5 min. A Millipore filter holder (Millipore) with a 0.45-μm-pore-size polycarbonate filter (Millipore) was placed downstream of the EPSS to collect those particles that were not captured.
The four sampling devices were also used to collect the Gram-positive bacterium B. atrophaeus to test the effects of the sampling methods on its membrane integrity. The experimental conditions were chosen only to assess the stress due to one of the four tested collection methods, without considering other variables such as sampling time, filter elution with ultrasonic agitation, and the use of a Tween mixture. Thus, the aerosolized B. atrophaeus cells were collected under the following experimental conditions: (i) the Button aerosol sampler with filter was operated for 5 min, and the collected bacteria were eluted from the filter by vortexing for 2 min; (ii) the BioStage impactor was operated for 5 min with a jet velocity of 39.3 m/s and jet-to-plate distance of 1.3 mm; (iii) the BioSampler was operated with 5 ml of DI water for 5 min at a sampling flow rate of 12.5 liters/min; and (iv) the EPSS was operated for 5 min at a sampling flow rate of 10.0 liters/min.
Cell membrane damage index.
To assess cell membrane damage during aerosolization and air sampling, 1 ml of solution taken from a liquid sample or sample eluted from a filter was centrifuged at 16,100 × g for 5 min at 4°C. Next, 950 μl of supernatant liquid was carefully transferred to a new 1.5-ml centrifuge tube by gentle pipetting, while the remaining 50 μl of liquid containing the pellet was mixed with 950 μl of sterile DI water. We assumed that in each sample, the DNA in the supernatant liquid originated from the bacterial cells that lost membrane integrity, while the DNA in the pellet represented cells that maintained membrane structure.
In order to validate our assumption, experiments were carried out in triplicate by spiking a known quantity of either freshly grown E. coli cells (7 × 107) or E. coli genomic DNA (6.5 × 106 copies of the E. coli genome) or their mixture into 1 ml of sterile DI water. After vortexing for 30 s, the samples were processed using the same procedures as those described above. The E. coli cells in the pellet and DNA in the supernatant liquid were quantified by microscopic counting and qPCR, respectively. When the E. coli cells and E. coli genomic DNA were spiked separately, their recovery was 101.0% ± 6.7% and 97.1% ± 10.9%, respectively. When they were spiked together into the same 1-ml sterile DI water sample, the recoveries for E. coli cells and E. coli genomic DNA were 106.7% ± 11.2% and 117.8% ± 18.9%, respectively. This indicates that free DNA in the supernatant liquid could be separated efficiently from the DNA in the pellet cells by our method.
The extent of membrane damage for different sampling conditions was calculated as the cell membrane damage index (ID), i.e., the ratio of 16S rRNA gene copies in the supernatant liquid to the entire number of 16S rRNA gene copies in the sample:
| (1) |
where NS (number of copies/ml) is the concentration of target 16S rRNA gene copies in the supernatant phase of the liquid sample after centrifugation, as determined by qPCR; and NP (number of copies/ml) is the concentration of target 16S rRNA gene copies in the pellet sample, determined using cell counts from epifluorescence microscopy and the number of 16S rRNA genes for a specific bacterial genome. The determination of NS and NP is described below. Depending on the stress that the bacteria experienced, ID values could range from 0 to 1, with higher values indicating more damage.
When sampling bacteria with the BioStage impactor, particles smaller than the impactor's cutoff size (d50 = 0.6 μm per the manufacturer) as well as particles that bounced off the collection surface (aluminum foil) escaped the impactor and were collected on the filter mounted at the inlet of one of the vacuum pumps (Fig. 1A). Therefore, in determining NS and NP, we considered that the bacteria and their fragments collected not only on the aluminum foil in the impactor but also on the pump filter:
| (2) |
| (3) |
where CS_filter (number of copies/ml) and CS_foil (number of copies/ml) are the concentrations of target 16S rRNA gene copies in the supernatant phase of liquid samples eluted from the filter and aluminum foil, respectively. CP_filter and CP_foil are the concentrations of intact cells in the resuspended pellet samples from the filter and aluminum foil, respectively. Vfilter and Vfoil are the solution volumes into which the bacteria were eluted from the filter and aluminum foil (5 ml and 10 ml, respectively). n is the number of target gene copies per cell, and η is the airflow fraction passing through the filter:
| (4) |
where Qfilter (liters/min) is the flow rate though the filter, and QA (liters/min) is the BioStage impactor sampling flow rate as shown in Fig. 1A. Both Qfilter and QA were measured by a mass flow meter (TSI Inc., Shoreview, MN).
Counting by microscopy.
The concentration of bacteria in liquid or resuspended cell pellets was determined by epifluorescence microscopy using an Axioskop 20 microscope (Carl Zeiss Inc., Thornwood, NY) according to a previously published method (25). Depending on the initial concentration of bacteria in each sample, a dilution factor was chosen to yield 10 to 40 stained bacteria per microscope view field. At least 40 random fields were counted for each sample, and the concentration of bacteria, CBacteria (number of cells/ml), was calculated as follows:
| (5) |
where N is the average number of bacteria per microscope view field, X is the number of fields for the entire filter, F is the dilution factor, and V is the volume of liquid sample used to prepare the microscope slide (ml). NP (number of cells/ml) was then determined as follows:
| (6) |
where n is the number of target gene copies per cell (n = 7 for both E. coli and B. atrophaeus [GenBank accession numbers NC_010473.1 and NC_014639, respectively]).
DNA extraction and quantitative PCR.
Quantitative PCR was performed on an iCycler iQ5 RT-PCR detection system (Bio-Rad Laboratories, Hercules, CA). As in previous studies, the universal primer pairs (forward, 5′-TCCTACGGGAGGCAGCAGT-3′; and reverse, 5′-GGACTACCAGGGTATCTAATCCTGTT-3′) for the bacterial 16S rRNA gene were selected with target amplicon sizes of 466 bp and 467 bp for E. coli and B. atrophaeus, respectively (25, 44). Reaction mixtures were prepared by combining 10 μl of 2× SYBR green supermix (Bio-Rad Laboratories, Hercules, CA), 2 μl of each 2.5 μM primer, 5 μl of template DNA, and 1 μl PCR-grade water, for a total volume of 20 μl for each reaction. The amplification reaction was performed with an iCycler iQ thermal cycler (Bio-Rad Laboratories, Hercules, CA) using the following temperature program: 10 min of denaturation at 95°C and 40 cycles of 15 s of denaturation at 95°C and 1 min of annealing/extension at 60°C. Data analysis was performed using iCycler iQ real-time detection system software. After completion of PCR amplification in each reaction mixture, a melting curve test was performed to check the purity of the generated amplicons.
To prepare standard curves for qPCRs, a batch of freshly harvested cells was first quantified by epifluorescence microscopy, and genomic DNA was extracted from a known number of cells by using the DNeasy Blood & Tissue kit protocol (Qiagen, Valencia, CA) for qPCR quantification. Standard curves were prepared by plotting each cycle threshold (CT) value against the log target gene copy number (equation 6). The number of 16S rRNA gene copies present in the supernatant liquid (NS) was determined by purifying the DNA fragments present in the supernatant according to the DNeasy Blood & Tissue kit protocol (Qiagen, Valencia, CA), performing qPCR, and applying the standard curves.
Collection of environmental air samples.
An air sample was collected inside an equine facility at the Rutgers Equine Science Center, NJ. Three samplers, one Button aerosol sampler and two BioSamplers, were operated concurrently for 2 h inside a stall with no horse present. The samplers were placed 0.6 m above the stall bedding. The Button aerosol sampler was used with a 0.6-μm-pore-size polycarbonate filter (25-mm diameter; Millipore, Billerica, MA) to collect an air sample at a flow rate of 6.5 liters/min. Two SKC BioSamplers with 5-ml sampling cups were operated at a flow rate of 12.5 liters/min for 2 h. One BioSampler used sterile DI water, and another used a Tween solution as collection liquid. Due to liquid evaporation during sampling, the collection liquid was refilled to 5 ml every 15 min for both BioSamplers. After sampling, filters and liquid samples were immediately placed in a cooler, transported within minutes to the laboratory, and immediately processed as follows.
Particles collected on a filter were eluted by vortexing for 2 min in 5 ml sterile DI water. Liquid suspensions from each BioSampler were transferred to 50-ml sterile tubes. Five milliliters of sterile DI water or Tween mixture was added to each BioSampler, and the sampler was vigorously shaken for 15 s to remove any particles that remained on the inner walls. Liquid suspensions from the second wash were then combined with the initial samples. One milliliter of liquid was then taken from the pooled sample from each sampler for subsequent analysis. Specifically, the samples were centrifuged at 16,100 × g for 5 min at 4°C, and 950 μl supernatant liquid was transferred to a new 1.5-ml centrifuge tube by gentle pipetting. The DNAs in the pellet sample and supernatant liquid were extracted and purified by using a DNeasy Blood & Tissue kit (Qiagen, Valencia, CA). The 16S rRNA gene copy number in each sample was then determined by qPCR.
Statistical analysis.
Statistical analysis was performed using Statistica software, version 10.0 (StatSoft Inc., Tulsa, OK). Factorial analysis of variance (ANOVA) was used to analyze the ID as a function of the Collison nebulizer jar material, bacterial species, and aerosolization time. For samples collected by the Button aerosol sampler and BioStage impactor, factorial ANOVA was performed to analyze ID as a function of the sample collection/filter elution method and jet-to-plate distance/jet velocity, respectively. For samples collected by the BioSampler, single-factor ANOVA was conducted to analyze the ID as a function of the collection fluid type. For each sample collection device, Student's t test was applied to compare the results between E. coli and B. atrophaeus. For each bacterial species, comparisons between the EPSS and the other three collection devices were made with Student's t test. For all tests, a statistically significant difference was defined as one having a P value of <0.05.
RESULTS
Aerosolization by Collison nebulizer.
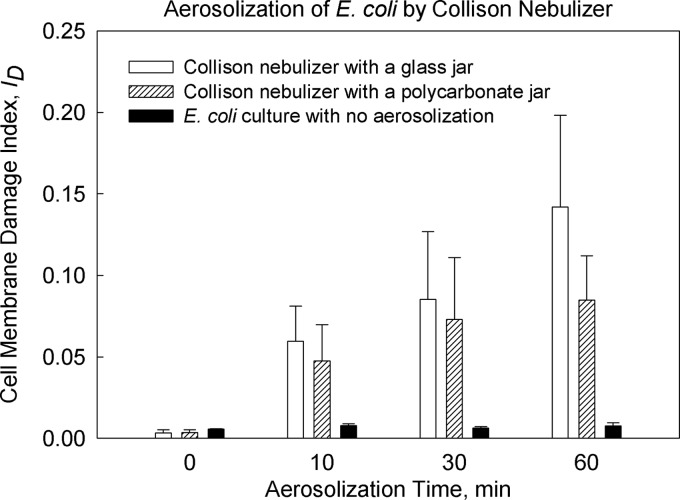
Figure 2 presents the cell membrane damage index (ID) for E. coli bacteria as a function of aerosolization time. When a pure E. coli culture was suspended in water for 0 to 60 min without aerosolization, the ID stayed below 0.01, showing no significant effect of time (P = 0.16). When the Collison nebulizer was in operation, the ID of the E. coli culture in the nebulizer's reservoir exhibited a clear increase over time, and the increase depended on the Collison jar material (glass versus polycarbonate). For the Collison nebulizer with a glass jar, the ID values were 0.003 ± 0.002, 0.060 ± 0.022, 0.085 ± 0.042, and 0.142 ± 0.056 for 0, 10, 30, and 60 min of nebulization time, respectively. When a polycarbonate jar was used, the ID values were 0.003 ± 0.002, 0.048 ± 0.022, 0.073 ± 0.038, and 0.085 ± 0.027 for 0, 10, 30, and 60 min of nebulization time, respectively. For each aerosolization time of >0 min, the ID values with a polycarbonate jar were significantly lower than those with a glass jar (P < 0.001). In order to minimize the mechanical stress imposed on E. coli bacteria during aerosolization in subsequent experiments, we chose to use a Collison nebulizer with a polycarbonate jar and an aerosolization time of 5 min.
Fig 2.
Effect of aerosolization time on E. coli cell integrity using the Collison nebulizer. Each bar shows the average for triplicate samples, and error bars show 1 standard deviation.
Sampling by filtration.
The ID values of samples collected by filtration using a Button aerosol sampler are shown in Fig. 3. In method A, the bacteria were collected on a filter for 5 min and then eluted from it by either vortexing for 2 min or vortexing for 2 min followed by 5 min of ultrasonic agitation. The ID value was 0.051 ± 0.014 when only vortexing was used, but it increased to 0.063 ± 0.019 when ultrasonic agitation was applied after vortexing. In method B, the bacteria were sampled for 5 min, and then particle-free air was pulled through the filter for 2 h to test the effect of extended sampling time on cell membrane integrity. In this case, the ID value for samples treated by vortexing was 0.058 ± 0.018, while the combination of vortexing and ultrasonic agitation increased the ID to 0.085 ± 0.022. For both filter elution methods, ID values were greater after the exposure of collected bacteria to particle-free air for 2 h than without such exposure. Factorial ANOVA showed that both sampling and filter elution methods had significant effects on the ID of E. coli bacteria: P = 0.007 for the sampling method, and P = 0.001 for the filter elution method. No significant interaction between these two factors was found (P = 0.189).
Fig 3.
Effects of sampling and filter elution methods on cell integrity of E. coli cells collected on filters. Method A, sampling for 5 min; method B, sampling for 5 min followed by the passing of particle-free air for 2 h. Each bar shows the average for triplicate samples, and error bars show 1 standard deviation.
Sampling by impaction.
Two factors, jet velocity and jet-to-plate distance, were investigated for their effects on cell membrane integrity of E. coli collected by a BioStage impactor. The standard air sampling flow rate of 28.3 liters/min resulted in a jet velocity of 23.7 m/s, while the higher sampling flow rate of 47 liters/min yielded a jet velocity of 39.3 m/s. Two simulated agar volumes, 48 ml and 40 ml, yielded jet-to-plate distances of 1.3 and 2.2 mm, respectively. As shown in Fig. 4, a jet velocity of 23.7 m/s with a jet-to-plate distance of 2.2 mm resulted in an ID value of 0.159 ± 0.008. However, the ID increased to 0.175 ± 0.017 when the jet velocity was increased to 39.3 m/s for the same jet-to-plate distance. When the jet velocity was maintained at 23.7 m/s but the jet-to-plate distance was decreased from 2.2 mm to 1.3 mm, the ID increased from 0.159 ± 0.008 to 0.205 ± 0.010. The highest ID was observed when the jet velocity was increased to 39.3 m/s with the lower jet-to-plate distance of 1.3 mm, with the value reaching as high as 0.368 ± 0.009. According to factorial ANOVA, both jet-to-plate distance and jet velocity had significant effects on cell membrane damage (P < 0.001), and there was also a significant interaction between these two factors (P < 0.001).
Fig 4.
Effects of different jet-to-plate distances and jet velocities on E. coli cell integrity for sampling with the BioStage impactor. Each bar shows the average for triplicate samples, and error bars show 1 standard deviation.
Sampling by impingement.
In experiments with the BioSampler, a filter was placed at the sampler's outlet to capture particles or their fragments that were not collected or were reaerosolized and escaped the sampler. Neither free DNA nor intact whole cells were detected on those filters, indicating that the particle escape rate was low and thus could be neglected in our study. This finding was consistent with those of other studies (34, 45). An effect of collection fluid type (sterile DI water or Tween mixture) on ID values after a 5-min collection time is shown in Fig. 5. When sterile DI water was used, the ID was 0.068 ± 0.029. However, when a Tween mixture was used, the value increased to 0.234 ± 0.088, and the increase was statistically significant (P = 0.009).
Fig 5.
Effect on E. coli cell integrity of using the BioSampler under three conditions: static E. coli culture in the BioSampler, particle-free air aspirated into the BioSampler with the E. coli culture, and aspiration of aerosolized E. coli into the collection fluid (sterile DI water or Tween mixture). Each bar shows the average for triplicate samples, and error bars show 1 standard deviation.
In order to further assess the effect of collection fluid on the cell integrity of E. coli bacteria, a known number of E. coli cells from a fresh culture was added to BioSampler cups filled with either 5 ml of DI water or 5 ml of Tween mixture. The solutions were kept static for 5 min, and then aliquots of liquid samples were taken out for analysis. As shown in Fig. 5, the ID value was 0.008 ± 0.002 for E. coli suspended in DI water but increased to 0.012 ± 0.002 for cells suspended in the Tween mixture (P = 0.01). In the next step, a known number of E. coli cells from a fresh culture was added to two 5-ml collection cups filled with DI water or Tween mixture, and then two BioSamplers aspirated particle-free air at 12.5 liters/min for 5 min. As a result, the ID was found to be 0.014 ± 0.005 for the sample that was suspended in DI water but increased significantly, to 0.066 ± 0.004, for the sample that was suspended in the Tween mixture (P < 0.001). Also, when particle-free air was aspirated into the BioSampler, both ID values were significantly higher than those when the BioSampler sampling cups were kept static (P = 0.034 for DI water and P < 0.001 for Tween mixture) but significantly lower than the ID values obtained when the aerosolized E. coli cells were actively collected by the BioSampler (P = 0.008 for DI water and P = 0.004 for Tween mixture).
Sampling by electrostatic precipitation.
The ID value for E. coli bacteria collected by the EPSS was 0.016 ± 0.016. Similar to the results with the BioSampler, neither the free DNA nor intact E. coli cells were detected on the filter downstream of the sampler. This result indicates that few particles escaped from the EPSS, which was consistent with a previously demonstrated high collection efficiency for this newly designed sampler (32, 33).
Aerosolization of the Gram-positive bacterium B. atrophaeus.
Figure 6 shows the effect of aerosolization time on the extent of cell membrane damage of a B. atrophaeus culture suspension in a Collison nebulizer. When we used a glass jar, the ID values were 0.004 ± 0.002, 0.043 ± 0.001, 0.040 ± 0.007, and 0.031 ± 0.007 for 0, 10, 30, and 60 min of aerosolization, respectively. However, when we used a polycarbonate jar, the ID values were significantly lower for the same aerosolization times: 0.001 ± 0.001, 0.005 ± 0.001, 0.008 ± 0.005, and 0.013 ± 0.009 for 0, 10, 30, and 60 min of aerosolization, respectively (P < 0.001). These results were similar to the findings for E. coli bacteria, supporting our conclusion that the polycarbonate jar induced less damage to cell membranes than the glass jar. For comparison, the ID values for B. atrophaeus bacteria kept in a liquid reservoir without aerosolization were 0.000 ± 0.000, 0.000 ± 0.000, 0.001 ± 0.000, and 0.002 ± 0.000 for 0, 10, 30, and 60 min, respectively. When the two bacterial species were compared, the ID values for B. atrophaeus bacteria were significantly lower than those for E. coli bacteria for either a glass jar (P < 0.001) or a polycarbonate jar (P < 0.001).
Fig 6.
Effect of aerosolization time on B. atrophaeus cell integrity for sampling using the Collision nebulizer. Each bar shows the average for triplicate samples, and error bars show 1 standard deviation.
Collection of B. atrophaeus by four different collection devices.
The cell membrane damage indexes of B. atrophaeus bacteria were compared with those of E. coli for collection with the four tested devices. The sampling conditions and results are presented in Fig. 7. The ID values for B. atrophaeus ranged from 0.002 ± 0.003 for sampling with the EPSS to 0.052 ± 0.008 for sampling with the BioSampler. The ID values for E. coli ranged from 0.016 ± 0.016 for sampling with the EPSS to 0.368 ± 0.009 for sampling with the BioStage impactor. According to the t test, there was no significant difference between ID values for E. coli and B. atrophaeus collected using the Button aerosol sampler (P = 0.961), the BioSampler (P = 0.234), or the EPSS (P = 0.213). However, the ID value for B. atrophaeus was significantly lower than that for E. coli when collected using the BioStage impactor (P < 0.001).
Fig 7.
Comparison of cell membrane damage indexes (ID) for B. atrophaeus and E. coli bacteria collected using four different samplers for 5 min. For the Button aerosol sampler, bacteria were collected on a filter and eluted by vortexing for only 2 min; for the BioStage impactor, the jet velocity was 39.3 m/s, and the jet-to-plate distance was 1.28 mm; for the BioSampler, 5 ml of sterile DI water was used as collection fluid; and for the EPSS, the sampling flow rate was 10.0 liters/min. Each bar shows the average for triplicate samples, and error bars show 1 standard deviation.
Among the four sampling devices, the EPSS showed the lowest average ID value for both microorganisms. For E. coli, this result was statistically significant for all samplers: the BioSampler (P = 0.041), the Button aerosol sampler (P = 0.026), and the BioStage impactor (P = 0.014). For B. atrophaeus, the ID value for sampling with the EPSS was significantly lower than that for sampling using the BioSampler (P = 0.010) and the Button aerosol sampler (P = 0.001) but was not significantly different from that for sampling with the BioStage impactor (P = 0.109).
Detection of free DNA in environmental aerosol samples.
Quantities of DNA in environmental samples were determined by qPCR, using the E. coli 16S rRNA gene to create a standard curve. The PCR efficiencies of the E. coli 16S rRNA gene and the environmental samples were between 90% and 105%, and no inhibitor effect was observed. The qPCR results were converted to numbers of bacteria per m3 by assuming four 16S rRNA gene copies per bacterial genome (46). When only the pellet sample was considered, the airborne bacterial concentration inside the equine facility was found to be 2.8 × 106, 5.8 × 106, and 1.4 × 107 bacteria/m3 for samples collected by the BioSampler with the Tween mixture, the BioSampler with water, and the Button aerosol sampler, respectively. Comparable amounts of DNA were detected in all three supernatant liquid samples, whose bacterial concentrations were determined to be 1.2 × 106, 6.7 × 106, and 1.5 × 107 bacteria/m3 for samples collected by the BioSampler with the Tween mixture, the BioSampler with water, and the Button aerosol sampler, respectively. Accordingly, the ID values were calculated to be 0.30, 0.54, and 0.52, respectively, for samples collected by the three devices.
DISCUSSION
Numerous studies have reported that bacteria experience stress during aerosolization and collection, due to mechanical forces (19, 24) and, possibly, exposure to dry air (14, 16). Bacteria that sustain sublethal injury could easily become viable but nonculturable or even lose their viability (24, 47, 48). Here we demonstrate for the first time that under certain conditions, the stress of aerosolization and the air sampling process is strong enough to break cell membranes and release the genomic DNA as free molecules. Furthermore, we introduce the concept of the cell membrane damage index (ID) to reflect the magnitude of membrane damage that is experienced by bacteria. The ID value can range from “0,” indicating no damage, to “1,” indicating that all bacteria in a sample have lost their cell membrane integrity. This index could be used as an indicator of the physiological status of the collected bacteria, and it could also provide a useful way to evaluate sampling protocols and adjust the design parameters of bioaerosol samplers, with the goal of minimizing damage to bioaerosol samples.
The Collison nebulizer has been used widely to generate bioaerosols in laboratory experiments, even though studies have suggested that the recirculation of culture suspension exerts a strong stress on the bacteria due to shear forces and impaction onto the inside wall of the container (24, 49). As a result, loss of culturability and fragmentation of cells were frequently observed for the aerosolized bacteria (23, 49, 50). In a recent study, the cell membrane was suggested as a major site of damage during aerosolization by the Collison nebulizer (24). Bacterial cells under aerosolization stress were discovered to have a loss of respiratory enzymatic activities, membrane depolarization, or even a loss of membrane integrity (24). Here we confirm that the Collison nebulizer can cause severe damage to the bacterial cell membrane and report that the release of genomic DNA was observed, presumably due to the mechanical stress of shear force and wall impaction. Interestingly, our findings show that a Collison nebulizer container made of polycarbonate material induces less stress on bacterial cultures than one made of glass. We speculate that a greater amount of the kinetic energy of the impacting bacteria is transferred to the polycarbonate material than with glass, presumably due to the greater deformation of polycarbonate material (51). Accordingly, when cells are impacted onto the polycarbonate surface, less of the remaining energy acts back onto the biological particles, thus resulting in less damage.
Jet-to-plate distance and jet velocity are two important factors that determine the collection efficiency of impaction-based aerosol samplers. Our experiments with the BioStage impactor demonstrated that an increase in jet velocity and a decrease in jet-to-plate distance result in an increase of the cell membrane damage index. The experiments also showed that E. coli bacteria experience more stress than B. atrophaeus. Considering that the viability of bacteria is highly correlated with the integrity of the cell membrane, our results confirm an earlier study that suggested that jet-to-plate distance and jet velocity affect the culturability of microorganisms collected by impaction (38). According to that study, an increase in jet-to-plate distance leads to the dissipation of air jets, which means that there is a lower jet velocity and impaction of bacteria onto the collection surface with a lower kinetic energy. Since the recovery of DNA from agar plates was found to be extremely low, aluminum foil was used as a collection surface to facilitate effective recovery of collected bacteria and free DNA. As a result, a large fraction of impact energy was transferred back to the bacteria, causing damage to their membranes. When bacteria are collected onto a semisolid surface such as agar instead of onto a hard surface, damage to the cell membrane is likely to be lower, since the agar will absorb some of the impact energy. On the other hand, even for collection on agar by use of impaction, the damage to bacteria is still considerable, as demonstrated by earlier studies (19).
The stress on bacteria due to collection by the BioSampler comes from a variety of sources, including, but not limited to, the impingement of bacteria into the collection fluid, particle bounce and reaerosolization due to high-speed centrifugal motion, and the possible detrimental effects of substances present in the collection fluid. First, we hypothesized that the Tween mixture might be toxic to E. coli and thus decrease cell membrane integrity. This was demonstrated by a higher ID for E. coli bacteria that were added into the Tween mixture and kept static than for bacteria that were added into sterile deionized water and also kept static. An antifoam agent present in the Tween mixture has been found to reduce the growth of Helicobacter pylori (52) and Hyphomicrobium zavrzinii ZV 580 (53). Moreover, our finding was similar to that of a previous study showing that DI water preserved the viability of L. pneumophila better than the Tween mixture when sampling by use of a BioSampler (18). Second, centrifugal motion during the sampling process could add to the damage to the cell structure. It has been shown that liquid loss during BioSampler operation increases the chance of particle bounce and reaerosolization, which add extra stress to the bacterial cells and impair their membrane structure (18, 34). In our study, after 5 min of sampling, the Tween mixture lost 1.3 ml, which was greater than the loss of DI water (0.9 ml), based on the initial volume of 5 ml for both fluids. This greater volume loss of the Tween mixture than of DI water and the resulting increase in particle bounce could partially explain the much higher ID value observed for the Tween mixture than for DI water. Nonetheless, the ID value when the particle-free air was aspirated into the BioSampler cup containing E. coli culture was still much lower than the value observed when airborne E. coli bacteria were actively collected using the BioSampler, indicating additional stress from either the aerosolization process, impingement, or, most likely, a combination of both.
In addition to DNA release due to stress from mechanical processes, we also found that a nonmechanical stress, such as desiccation, also facilitated the release of DNA by the impaired bacterial cells. Studies applying filtration for bioaerosol collection have used sampling times ranging from a few minutes to several hours (27, 54), or even as long as 24 h (55, 56). It has been shown that prolonged sampling periods by impaction-based samplers increase the risk of microorganism viability loss (16, 57, 58). The desiccation of the already collected bioaerosols, together with the desiccation of agar media, contributed to decreased microorganism recovery (16). We found no reports on the impact of dehydration on cell membrane integrity; however, the protein coating of airborne Gumboro virus was reported to be damaged at a lower humidity level (59). The data presented in our study clearly show that cell membrane rupture is more substantial with prolonged sampling periods, which was demonstrated by higher ID values for samples exposed to dry air for an extended sampling period than for those without such exposure. Thus, the sampling time should be as short as reasonably possible in order to reduce the desiccation effects on the collected samples when operating both filtration- and impaction-based samplers.
We also found that the filter elution method can contribute to cell membrane damage. An increased recovery of bacteria from filter samples has been reported by adding the extra step of ultrasonic agitation (14), but our findings show that this treatment can add more stress to the collected bacteria and result in an increased release of DNA. On the other hand, vortexing is generally accepted as an efficient way to elute bioaerosol particles from filters (41, 42), and we found that short-term vortexing (<2 min) did not result in significant release of DNA from freshly grown E. coli cells (data not shown). Thus, to minimize the stress on the bacteria collected on the filter, including for molecular analysis techniques, sampling protocols should also consider filter elution methods.
Our findings also showed that for the same aerosolization and air sampling conditions, the Gram-positive bacterium B. atrophaeus was less susceptible to mechanical stress, such as that from impaction and shear forces, than the Gram-negative bacterium E. coli. The higher resistance of B. atrophaeus to stress is likely due to its thicker and more rigid peptidoglycan layer, which is responsible for its cell wall strength, than those of more sensitive Gram-negative bacteria (19). This result also suggests that in sampling ambient microorganisms using inertia-based methods, e.g., impaction, we could selectively enrich Gram-positive bacteria over Gram-negative bacteria if we apply an enumeration method based on intact cells or do not take into account the released DNA. Consequently, this would bias our information regarding the relative abundances of various bacterial species within the complex airborne microbial community.
In comparing the four bioaerosol sampling devices used in our study, we found that samples collected by our newly developed EPSS had the lowest ID, followed by the BioSampler with DI water and the Button aerosol sampler when only vortexing was used to extract bacteria. In contrast, E. coli bacteria collected by the BioStage impactor or the BioSampler with the Tween mixture seemed to be exposed to greater stress, which was high enough to affect the cell membrane integrity and release as much as one-third of the total amount of DNA material. Electrostatic collection results in a low velocity of bacterial deposition onto the collection surface, which is conducive to cell membrane preservation. This particular sampler prototype collects bioaerosols on a superhydrophobic surface and then concentrates them into small volumes of liquid (40 μl or less), thus allowing one to achieve very high concentration rates (31, 33). Our earlier study showed that exposure to strong electrostatic fields while airborne does not induce appreciable cell damage (60). These features of the new electrostatic precipitator—a high sample concentration rate and an ability to maintain cell integrity—should be valuable for bioaerosol detection, especially when high sensitivity and a low detection limit are desired.
It should be noted that the B. atrophaeus organisms used in our study were mostly vegetative cells, while in the natural environment some Gram-positive bacteria, e.g., Bacillus and Clostridium spp., often exist in spore form (61). Those spores are known to have resistant structures which protect bacteria from unfavorable environmental conditions, e.g., desiccation (62) and mechanical stress (63). In addition, the aerosolized bacteria were mostly single cells, as verified by the measured bioaerosol size distribution. However, airborne microorganisms in the natural environment often form aggregates of multiple cells or attach to particulate matter (64). Thus, it was of great interest to test how these factors affect the susceptibility of natural bioaerosols to sampling stress. Our samples collected by a filter sampler and two BioSamplers inside an equine facility for 2 h accumulated a sufficient amount of bacteria for subsequent analysis by qPCR. The data showed that free DNA was detected in air samples collected by all three devices. It was rather surprising to find that the amount of free DNA was comparable to or even higher than that in intact cells, depending on the sampling method. While our sampling protocol did not separately determine what fraction of free DNA was captured directly from air and what fraction was released due to sampling stress, our findings do indicate that a substantial amount (∼50%) of the DNA in a processed sample could be free DNA. This demonstrates that commonly used protocols for bioaerosol sampling and sample processing could underestimate the presence of airborne microbial content in the natural environment by a large fraction (up to 50%).
In conclusion, our results strongly suggest that bioaerosol quantification using molecular methods, such as qPCR, should include not only DNA in intact cells but also DNA released by cells damaged during aerosolization and air sampling, i.e., the free DNA from the supernatant should not be discarded but should be included in the sample analysis. Otherwise, bioaerosol concentrations might be substantially underestimated. A negative bias of >20% was observed when a BioSampler containing a Tween mixture sampling solution was used to sample bacteria for only 5 min. A negative bias as high as 50% was observed in environmental bioaerosol samples collected and processed by commonly used bioaerosol protocols. It is hoped that this study will provide guidance for selecting bioaerosol aerosolization and sampling methods and their analysis protocols that minimize bioaerosol quantification bias using molecular tools.
ACKNOWLEDGMENTS
This publication was supported by grant R01-OH009783 (“Advanced Sampler for Measuring Exposure to Biological Aerosols”) from the CDC/NIOSH and by project 07160, funded by the New Jersey Agricultural Experiment Station (NJAES) at Rutgers, The State University of New Jersey.
The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official view of the CDC/NIOSH or the NJAES.
Footnotes
Published ahead of print 4 October 2013
REFERENCES
- 1.Burge H. 1990. Bioaerosols: prevalence and health effects in the indoor environment. J. Allergy Clin. Immunol. 86:687–701 [DOI] [PubMed] [Google Scholar]
- 2.Douwes J, Thorne P, Pearce N, Heederik D. 2003. Bioaerosol health effects and exposure assessment: progress and prospects. Ann. Occup. Hyg. 47:187–200 [DOI] [PubMed] [Google Scholar]
- 3.Fung F, Hughson W. 2003. Health effects of indoor fungal bioaerosol exposure. Appl. Occup. Environ. Hyg. 18:535–545 [DOI] [PubMed] [Google Scholar]
- 4.Herr CE, Zur Nieden A, Jankofsky M, Stilianakis NI, Boedeker RH, Eikmann TF. 2003. Effects of bioaerosol polluted outdoor air on airways of residents: a cross sectional study. Occup. Environ. Med. 60:336–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirvonen MR, Ruotsalainen M, Savolainen K, Nevalainen A. 1997. Effect of viability of actinomycete spores on their ability to stimulate production of nitric oxide and reactive oxygen species in RAW264.7 macrophages. Toxicology 124:105–114 [DOI] [PubMed] [Google Scholar]
- 6.Lee T, Grinshpun SA, Martuzevicius D, Adhikari A, Crawford CM, Reponen T. 2006. Culturability and concentration of indoor and outdoor airborne fungi in six single-family homes. Atmos. Environ. 40:2902–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Handley BA, Webster AJF. 1995. Some factors affecting the airborne survival of bacteria outdoors. J. Appl. Bacteriol. 79:368–378 [DOI] [PubMed] [Google Scholar]
- 8.Hatch MT, Dimmick RL. 1966. Physiological responses of airborne bacteria to shifts in relative humidity. Bacteriol. Rev. 30:597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang JW. 2009. The effect of environmental parameters on the survival of airborne infectious agents. J. R. Soc. Interface 6:S697–S702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tong T, Lighthart B. 1997. Solar radiation has a lethal effect on natural populations of culturable outdoor atmospheric bacteria. Atmos. Environ. 31:897–900 [Google Scholar]
- 11.Zentner RJ. 1966. Physical and chemical stresses of aerosolisation. Bacteriol. Rev. 30:551–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caron GN, Stephens P, Badley RA. 1998. Assessment of bacterial viability status by flow cytometry and single cell sorting. J. Appl. Microbiol. 84:988–998 [DOI] [PubMed] [Google Scholar]
- 13.Oliver JD. 2005. The viable but nonculturable state in bacteria. J. Microbiol. 43:93–100 [PubMed] [Google Scholar]
- 14.Wang Z, Reponen T, Grinshpun SA, Gorny RL, Willeke K. 2001. Effect of sampling time and air humidity on the bioefficiency of filter samplers for bioaerosol collection. J. Aerosol Sci. 32:661–674 [Google Scholar]
- 15.Willeke K, Macher JM. 1999. Air sampling, p 11-1–11-25 In Macher J. (ed), Bioaerosols: assessment and control. American Conference of Governmental Industrial Hygienists, Cincinnati, OH [Google Scholar]
- 16.Mainelis G, Tabayoyong M. 2010. The effect of sampling time and the overall performance of portable microbial impactors. Aerosol Sci. Technol. 44:75–82 [Google Scholar]
- 17.Nevalainen A, Willeke K, Liebhaber F, Pastuszka J, Burge H, Henningson E. 1993. Bioaerosol sampling: aerosol measurement principles, techniques, and applications. Van Nostrand Reinhold, New York, NY [Google Scholar]
- 18.Chang C-W, Chou F-C. 2011. Assessment of bioaerosol sampling techniques for viable Legionella pneumophila by ethidium monoazide quantitative PCR. Aerosol Sci. Technol. 45:343–351 [Google Scholar]
- 19.Stewart SL, Grinshpun SA, Willeke K, Terzieva S, Ulevicius V, Donnelly J. 1995. Effect of impact stress on microbial recovery on an agar surface. Appl. Environ. Microbiol. 61:1232–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King MD, McFarland AR. 2011. Bioaerosol sampling with a wetted wall cyclone: cell culturability and DNA integrity of Escherichia coli bacteria. Aerosol Sci. Technol. 46:82–93 [Google Scholar]
- 21.Zhao Y, Aarnink AJA, Doornenbal P, Huynh TTT, Groot Koerkamp PWG, de Jong MCM, Landman WJM. 2011. Investigation of the efficiencies of bioaerosol samplers for collecting aerosolized bacteria using a fluorescent tracer. I. Effects of non-sampling processes on bacterial culturability. Aerosol Sci. Technol. 45:423–431 [Google Scholar]
- 22.Zhao Y, Aarnink AJA, Doornenbal P, Huynh TTT, Groot Koerkamp PWG, Landman WJM, de Jong MCM. 2011. Investigation of the efficiencies of bioaerosol samplers for collecting aerosolized bacteria using a fluorescent tracer. II. Sampling efficiency and half-life time. Aerosol Sci. Technol. 45:432–442 [Google Scholar]
- 23.Mainelis G, Berry D, An HR, Yao MS, DeVoe K, Fennell DE, Jaeger R. 2005. Design and performance of a single-pass bubbling bioaerosol generator. Atmos. Environ. 39:3521–3533 [Google Scholar]
- 24.Thomas RJ, Webber D, Hopkins R, Frost A, Laws T, Jayasekera PN, Atkins T. 2011. The cell membrane as a major site of damage during aerosolization of Escherichia coli. Appl. Environ. Microbiol. 77:920–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.An HR, Mainelis G, White L. 2006. Development and calibration of real-time PCR for quantification of airborne microorganisms in air samples. Atmos. Environ. 40:7924–7939 [Google Scholar]
- 26.Peccia J, Hernandez M. 2006. Incorporating polymerase chain reaction-based identification, population characterization, and quantification of microorganisms into aerosol science: a review. Atmos. Environ. 40:3941–3961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen PS, Li CS. 2005. Quantification of airborne Mycobacterium tuberculosis in health care setting using real-time qPCR coupled to an air-sampling filter method. Aerosol Sci. Technol. 39:371–376 [Google Scholar]
- 28.Pascual L, Perez-Luz S, Moreno C, Apraiz D, Catalan V. 2001. Detection of Legionella pneumophila in bioaerosols by polymerase chain reaction. Can. J. Microbiol. 47:341–347 [PubMed] [Google Scholar]
- 29.Schafer MP, Martinez KF, Mathews ES. 2003. Rapid detection and determination of the aerodynamic size range of airborne mycobacteria associated with whirlpools. Appl. Occup. Environ. Hyg. 18:41–50 [DOI] [PubMed] [Google Scholar]
- 30.Wilson KH. 2002. High-density microarray of small subunit ribosomal DNA probes. Appl. Environ. Microbiol. 68:2535–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han T, An HR, Mainelis G. 2010. Performance of an electrostatic precipitator with superhydrophobic surface when collecting airborne bacteria. Aerosol Sci. Technol. 44:339–348 [Google Scholar]
- 32.Han T, Mainelis G. 2008. Design and development of an electrostatic sampler for bioaerosols with high concentration rate. J. Aerosol Sci. 39:1066–1078 [Google Scholar]
- 33.Han T, Nazarenko Y, Lioy PJ, Mainelis G. 2011. Collection efficiencies of an electrostatic sampler with superhydrophobic surface for fungal bioaerosols. Indoor Air 21:110–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han T, Mainelis G. 2012. Investigation of inherent and latent internal losses in liquid-based bioaerosol samplers. J. Aerosol Sci. 45:58–68 [Google Scholar]
- 35.Hospodsky D, Yamamoto N, Peccia J. 2010. Accuracy, precision, and method detection limits of quantitative PCR for airborne bacteria and fungi. Appl. Environ. Microbiol. 76:7004–7012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee BU, Kim SS. 2003. Sampling E. coli and B. subtilis bacteria bioaerosols by a new type of impactor with a cooled impaction plate. J. Aerosol Sci. 34:1097–1100 [Google Scholar]
- 37.Gerhardt P, Murray RGE, Wood WA, Krieg NR. 1994. Methods for general and molecular bacteriology. ASM Press, Washington, DC [Google Scholar]
- 38.Yao M, Mainelis G. 2006. Effect of physical and biological parameters on enumeration of bioaerosols by portable microbial impactors. J. Aerosol Sci. 37:1467–1483 [Google Scholar]
- 39.Aizenberg V, Grinshpun SA, Willeke K, Smith JP, Baron PA. 2000. Performance characteristics of the button personal inhalable aerosol sampler. Am. Ind. Hyg. Assoc. J. 61:398–404 [DOI] [PubMed] [Google Scholar]
- 40.Hauck BC, Grinshpun SA, Reponen A, Reponen T, Willeke K, Bornschein RL. 1997. Field testing of new aerosol sampling method with a porous curved surface as inlet. Am. Ind. Hyg. Assoc. J. 58:713–719 [DOI] [PubMed] [Google Scholar]
- 41.Burton NC, Adhikari A, Grinshpun S, Hornung R, Reponen T. 2005. The effect of filter material on bioaerosol collection of Bacillus subtilis spores used as a Bacillus anthracis simulant. J. Environ. Monit. 7:475–480 [DOI] [PubMed] [Google Scholar]
- 42.Wang Z, Reponen T, Willeke K, Grinshpun SA. 1999. Survival of bacteria on respirator filters. Aerosol Sci. Technol. 30:300–308 [Google Scholar]
- 43.Willeke K, Lin X, Grinshpun SA. 1998. Improved aerosol collection by combined impaction and centrifugal motion. Aerosol Sci. Technol. 28:439–456 [Google Scholar]
- 44.Nadkarni MA, Martin FE, Jacques NA, Hunter N. 2002. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 148:257–266 [DOI] [PubMed] [Google Scholar]
- 45.Lin X, Reponen TA, Willeke K, Grinshpun SA, Foarde KK, Ensor DS. 1999. Long-term sampling of airborne bacteria and fungi into a non-evaporating liquid. Atmos. Environ. 33:4291–4298 [Google Scholar]
- 46.Lee ZM, Bussema C, 3rd, Schmidt TM. 2009. rrnDB: documenting the number of rRNA and tRNA genes in bacteria and archaea. Nucleic Acids Res. 37:D489–D493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Colwell RR, Brayton PR, Grimes DJ, Roszak DB, Huq SA, Palmer LM. 1985. Viable but non-culturable Vibrio cholerae and related pathogens in the environment: implications for release of genetically engineered microorganisms. Biotechnology 3:817–820 [Google Scholar]
- 48.Heidelberg JF, Shahamat M, Levin M, Rahman I, Stelma G, Grim C, Colwell RR. 1997. Effect of aerosolization on culturability and viability of gram-negative bacteria. Appl. Environ. Microbiol. 63:3585–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reponen T, Willeke K, Ulevicius V, Grinshpun SA, Donnelly J. 1997. Techniques for dispersion of microorganisms into air. Aerosol Sci. Technol. 27:405–421 [Google Scholar]
- 50.Mainelis G, Willeke K, Baron P, Reponen T, Grinshpun SA, Gorny RL, Trakumas S. 2001. Electrical charges on airborne microorganisms. J. Aerosol Sci. 32:1087–1110 [Google Scholar]
- 51.Wall S, John W, Wang HC, Goren SL. 1990. Measurements of kinetic energy loss for particles impacting surfaces. Aerosol Sci. Technol. 12:926–946 [Google Scholar]
- 52.Kane AV, Plaut AG. 1996. Unique susceptibility of Helicobacter pylori to simethicone emulsifiers in alimentary therapeutic agents. Antimicrob. Agents Chemother. 40:500–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jerome V, Hermann M, Hilbrig F, Freitag R. 2007. Development of a fed-batch process for the production of a dye-linked formaldehyde dehydrogenase in Hyphomicrobium zavarzinii ZV 580. Appl. Microbiol. Biotechnol. 77:779–788 [DOI] [PubMed] [Google Scholar]
- 54.Wu Z, Blomquist G, Westermark S, Wang XR. 2002. Application of PCR and probe hybridization techniques in detection of airborne fungal spores in environmental samples. J. Environ. Monit. 4:673–678 [DOI] [PubMed] [Google Scholar]
- 55.Brodie EL, DeSantis TZ, Parker JPM, Zubietta IX, Piceno YM, Andersen GL. 2007. Urban aerosols harbor diverse and dynamic bacterial populations. Proc. Natl. Acad. Sci. U. S. A. 104:299–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maher N, Dillon HK, Vermund SH, Unnasch TR. 2001. Magnetic bead capture eliminates PCR inhibitors in samples collected from the airborne environment, permitting detection of Pneumocystis carinii DNA. Appl. Environ. Microbiol. 67:449–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hensel A, Petzoldt K. 1995. Biological and biochemical analysis of bacteria and viruses, p 335–360 In Cox CS, Wathes CM. (ed), Bioaerosols handbook. Lewis Publishers, New York, NY [Google Scholar]
- 58.Juozaitis A, Willeke K, Grinshpun SA, Donnelly J. 1994. Impaction onto a glass slide or agar versus impingement into a liquid for the collection and recovery of airborne microorganisms. Appl. Environ. Microbiol. 60:861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao Y, Aarnink AJA, Dijkman R, Fabri T, de Jong MCM, Groot Koerkamp PWG. 2012. Effects of temperature, relative humidity, absolute humidity, and evaporation potential on survival of airborne Gumboro vaccine virus. Appl. Environ. Microbiol. 78:1048–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yao MS, Mainelis G, An HR. 2005. Inactivation of microorganisms using electrostatic fields. Environ. Sci. Technol. 39:3338–3344 [DOI] [PubMed] [Google Scholar]
- 61.Leggett MJ, McDonnell G, Denyer SP, Setlow P, Maillard JY. 2012. Bacterial spore structures and their protective role in biocide resistance. J. Appl. Microbiol. 113:485–498 [DOI] [PubMed] [Google Scholar]
- 62.Cano RJ, Borucki MK. 1995. Revival and identification of bacterial spores in 25- to 40-million-year-old Dominican amber. Science 268:1060–1064 [DOI] [PubMed] [Google Scholar]
- 63.Jones CA, Padula NL, Setlow P. 2005. Effect of mechanical abrasion on the viability, disruption and germination of spores of Bacillus subtilis. J. Appl. Microbiol. 99:1484–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Adhikari A, Reponen A, Grinshpun SA, Martuzevicius D, LeMasters G. 2006. Correlation of ambient inhalable bioaerosols with particulate matter and ozone: a two-year study. Environ. Pollut. 140:16–28 [DOI] [PubMed] [Google Scholar]