Abstract
Changes in HIV tropism from R5 to non-R5 or development of drug resistance is often associated with virologic failure in patients treated with maraviroc, a CCR5 antagonist. We sought to examine changes in HIV envelope sequences and inferred tropism in patients who did not respond to maraviroc-based regimens. We selected 181 patients who experienced early virologic failure on maraviroc-containing therapy in the MOTIVATE trials. All patients had R5 HIV by the original Trofile assay before entry. We used population-based sequencing methods and the geno2pheno algorithm to examine changes in tropism and V3 sequences at the time of failure. Using deep sequencing, we assessed whether V3 sequences observed at failure emerged from preexisting subpopulations. From population genotyping data at failure, 90 patients had R5 results, and 91 had non-R5 results. Of the latter group, the geno2pheno false-positive rate (FPR) value fell from a median of 20 at screening to 1.1 at failure. By deep sequencing, the median percentage of non-R5 variants in these patients rose from 1.4% to 99.5% after a median of 4 weeks on maraviroc. In 70% of cases, deep sequencing could detect a pretreatment CXCR4-using subpopulation, which emerged at failure. Overall, there were two distinct patterns of failure of maraviroc. Patients failing with R5 generally had few V3 substitutions and low non-R5 prevalence by deep sequencing. Patients with non-R5 HIV who were failing developed very-high-prevalence non-R5 HIV (median, 99%) and had very low geno2pheno values.
INTRODUCTION
Successful antiretroviral treatment with the CCR5 antagonist maraviroc requires a tropism test to confirm that the patient's HIV uses the CCR5 coreceptor for cellular entry (R5 HIV) rather than CXCR4 (non-R5 HIV) (1–3). In phase III clinical trials of maraviroc, patients were screened for tropism status by using the original Trofile phenotypic coreceptor assay (OTA), which was subsequently replaced by the enhanced-sensitivity Trofile assay (ESTA) (4, 5). Recent rescreening of clinical trials of maraviroc has confirmed the utility of genotypic approaches for the determination of HIV tropism (6–10). Such approaches typically involve sequencing of the third variable (V3) region of the HIV envelope gene (11). Bioinformatic algorithms such as geno2pheno (12) are then used to infer the phenotypic tropism that is likely associated with a V3 genotype. geno2pheno converts an input V3 sequence into an output value in the form of a false-positive rate (FPR) ranging from 0 to 100. An FPR indicates how likely a sequence is to be incorrectly identified as a non-R5 sequence. Therefore, sequences yielding low false-positive rates have a high likelihood of being non-R5 sequences.
Historically, population-based sequencing has been the most commonly used genotypic approach for predicting coreceptor usage (11). However, more sensitive tropism determination methods can more accurately predict the response to maraviroc (5); thus, newer deep sequencing methods targeting the V3 loop are becoming increasingly common (7, 8, 13–16). These deep sequencing approaches can identify low-level non-R5 subpopulations in clinical samples, which may later emerge at much higher prevalences following treatment with maraviroc, thereby compromising treatment efficacy (16, 17).
There are several pathways by which patients may fail a maraviroc-containing therapy regimen. Most commonly, a minority non-R5 population in a patient's HIV population may expand under drug pressure, causing an overall change in observed tropism (3). Less commonly, the viral population may retain its CCR5 tropism while evolving the ability to use maraviroc-bound CCR5 protein for cellular entry, a form of maraviroc resistance (18). Third, the viral population may develop resistance to other agents in the background regimen in the absence of a change in susceptibility to maraviroc (19); this may be associated with either R5 or non-R5 tropism. Furthermore, as with other agents, adherence, absorption, and other patient-associated and pharmacokinetic factors can also lead to therapy failure.
Early detection of tropism shifts or maraviroc resistance can accelerate the decision to replace maraviroc with another antiretroviral agent and potentially prevent further accumulation of antiretroviral drug resistance to other agents in the regimen. Thus, we sampled patients relatively soon after they began maraviroc treatment to determine the utility of an early-monitoring approach.
In this study, we used both population-based and deep sequencing approaches to assess changes in tropism and V3 sequences among treatment-experienced, R5-infected patients who experienced virologic failure while receiving maraviroc in the MOTIVATE-1 and -2 studies (1, 3). Patients from the A4001029 study, which enrolled patients with non-R5 HIV (2), were not included in the current study; thus, all patients studied were determined to have exclusively R5 HIV by the OTA. Phylogenetic methods were also used to assess whether sequences present at failure were derived from preexisting minority subpopulations, and newer deep sequencing was used to assess changes in non-R5 prevalence after treatment with maraviroc. Previous studies (16, 17) noted the emergence of CXCR4-using virus from preexisting subpopulations, and CCR5 antagonists have been known to inhibit R5 only while selecting non-R5 subpopulations (3, 20), with such shifts appearing to occur very quickly (21). Furthermore, resistance to maraviroc has been associated with genotypic changes in the HIV envelope gene (18, 22). Thus, we hypothesized that there are distinct mechanisms of failure that can be identified by population-based and/or deep sequencing of the HIV V3 region.
(This work was presented in part at the International HIV and Hepatitis Virus Drug Resistance Workshop and Curative Strategies, Dubrovnik, Croatia, 8 to 12 June 2010.)
MATERIALS AND METHODS
Patients and sample composition.
A subset of patients who had suboptimal responses to maraviroc in the MOTIVATE trials was selected (n = 181). Patients were selected such that approximately the same proportion had non-R5 OTA results at failure as was reported for the MOTIVATE trials overall (57% in MOTIVATE and 58% in the current study) (3). All patients were treatment experienced, 100% of patients had R5 results by OTA at screening, and 69% had R5 results by ESTA (124/181). All patients received maraviroc (once or twice daily) plus an optimized background regimen of three to six other antiretroviral agents. All individuals gave written informed consent, including consent to allow other tropism testing to be performed on their samples. The University of British Columbia-Providence Health Care Research Ethics Board reviewed the research project and granted ethical approval.
Sequencing was performed on samples from two time points: one prior to receiving maraviroc (the screening sample) and one on treatment (the failure sample). This on-treatment failure sample was defined as the earliest available sample with a plasma viral load (pVL) of >500 HIV RNA copies/ml and an OTA result. The screening sample was drawn approximately 6 to 8 weeks prior to the beginning of maraviroc treatment; the failure sample was drawn a median of 4 weeks (interquartile range [IQR], 4 to 16 weeks) after the beginning of maraviroc treatment and a median of 2 weeks (IQR, 2 to 10 weeks) after the first viral load result of ≥500 copies/ml. While phenotypic tropism results were available for all samples, phenotypic maraviroc resistance assay results were not available for these samples. ESTA results were available at screening, but only OTA results were available at failure.
Genotypic tropism testing.
The V3 loop of the HIV envelope gene was amplified by nested reverse transcriptase PCR (RT-PCR) (3, 4). The screening samples were amplified and sequenced in triplicate; the failure samples had a single sequence generated per sample. Standard, population-based sequencing was performed on all screening and failure samples, as previously described (9). Deep V3 sequencing was also performed on all screening samples, plus a subset (n = 73) of failure samples, using methods described previously (7, 8). The 73 samples comprised the last batch of samples processed by population sequencing, with no targeted selection.
The tropism associated with the V3 loop sequences was inferred by using the geno2pheno algorithm (12), with FPR cutoffs of 5.75 for population-based sequencing and 3.5 for deep sequencing (23, 24), below which sequences were categorized as non-R5 sequences. These cutoffs were previously optimized for predicting the virologic response to maraviroc (23, 24). The percentage of non-R5 variants in the viral population was defined as the proportion of sequences scoring below or equal to an FPR of 3.5, as observed by deep sequencing, and previous studies have defined an R5 sample as having <2% non-R5 variants (7, 8).
The screening and failure sequences were assessed for amino acid changes that may have appeared following maraviroc-based therapy as well as for changes in the geno2pheno FPR value. Neighbor-joining phylogenetic trees were constructed with ClustalX by using deep sequencing data at screening and by using population-based or deep sequencing data at failure. Thus, it could be determined whether a sequence present at failure may have already been present prior to treatment with maraviroc. The change in the percentage of non-R5 variants between screening and failure was also examined by using the deep sequencing results. Sample phenotypes were obtained by OTA at all time points and by ESTA at screening.
Statistical analysis.
Statistical analyses performed included the Mann-Whitney U test and Kruskal-Wallis test for differences between medians (e.g., median plasma viral loads). The Fisher exact test was used for differences in proportions (e.g., the proportion of patients who had R5 HIV by ESTA at screening).
RESULTS
Patients and sample composition.
Patients in the current study were all treatment experienced and received maraviroc once daily (89 patients; 49%) or twice daily (92 patients; 51%), as per randomization at study entry. Most patients (91%) were enrolled in the North American MOTIVATE-1 trial (25), and 9% were enrolled in the MOTIVATE-2 trial, which had an identical study design. Of the 181 patients selected, 100 (55%) experienced virologic failure, 44 (24%) never achieved virologic suppression but completed 48 weeks of treatment, and 15 (8%) had a virologic rebound. Of the remaining 22 patients, 18 were lost to follow-up, 2 died, 1 experienced an adverse event, and 1 was withdrawn due to pregnancy. The mean age of subjects was 45 years (range, 19 to 70 years), and the proportion of males in the study was 91% (165/181). These data were similar to those for the maraviroc arms of the MOTIVATE trial overall (1). The proportion of patients of black race or ethnicity was 19% (34/181), which was slightly elevated relative to the larger trial overall (14%), and this was likely due to a higher number of maraviroc nonresponders who reported black race/ethnicity in the MOTIVATE trial (3) (Table 1).
Table 1.
Patient characteristicsa
| Parameter | Value for group |
P value for R5 vs non-R5 | |||
|---|---|---|---|---|---|
| MOTIVATE-1 and -2 MVC arms overall | Current study population |
||||
| Overall study population | Patients with non-R5 genotype by population-based sequencing at failure | Patients with R5 genotype by population-based sequencing at failure | |||
| No. of patients | 794 | 181 | 91 | 90 | |
| Mean age (yr) (range) | 46 (17–75)* | 45 (19–70) | 45 (19–70) | 45 (34–69) | NS |
| % male patients (no. of male patients/total no. of patients) | 89 (745/840)* | 91 (165/181) | 93 (85/91) | 89 (80/90) | NS |
| % female patients (no. of female patients/total no. of patients) | 11 (95/840)* | 9 (16/181) | 7 (6/91) | 11 (10/90) | |
| % white patients (no. of white patients/total no. of patients) | 83 (699/840)* | 80 (145/181) | 84 (76/91) | 77 (69/90) | NS |
| % black patients (no. of black patients/total no. of patients) | 14 (121/840)* | 19 (34/181) | 15 (14/91) | 22 (20/90) | |
| % R5 patients by ESTA at screening (no. of R5 patients/total no. of patients) | NA | 69 (124/181) | 48 (44/91) | 89 (80/90) | <0.001 |
| % R5 patients by original Trofile assay at failure (no. of R5 patients/total no. of patients) | 43 (57/133)* | 42 (76/181) | 2 (2/91) | 82 (74/90) | <0.001 |
| Median baseline pVL (log) (IQR) | 4.9 (4.4–5.3) | 5.1 (4.7–5.5) | 5.0 (4.6–5.4) | 5.2 (4.8–5.6) | 0.03 |
| Median failure pVL (log) (IQR) | NA | 4.1 (3.5–5.0) | 4.3 (3.6–5.1) | 4.0 (3.3–4.9) | 0.06 |
| Median baseline CD4 count (cells/mm3) (IQR) | 168 (74–289) | 72 (17–177) | 64 (15–174) | 79 (23–182) | NS |
| Median no. of active ARVs (wOBTss) (IQR) | 1.0 (0–2) | 0.5 (0–1.0) | 0.5 (0–1.0) | 1.0 (0–1.5) | 0.03 |
| Median geno2pheno FPR at screening (IQR) | 33.2 (14.7–56.5) | 31.1 (12.5–55.3) | 19.6 (6.9–41.4) | 41.9 (21.8–65.0) | <0.001 |
| Median geno2pheno FPR at failure (IQR) | NA | 5.3 (1.1–48.9) | 1.1 (0.4–1.8) | 48.9 (21.2–74.0) | <0.001 |
| Median % non-R5 variants by deep sequencing (screening, n = 181) (IQR) | 0 (0–0.3) | 0.1 (0–3.1) | 1.4 (0–15.2) | 0 (0–0.1) | <0.001 |
| Median % non-R5 variants by deep sequencing (failure, n = 73) (IQR) | NA | 0.8 (0–99.0) | 99.5 (94.8–99.9) | 0 (0–0.2) | <0.001 |
Shown are the baseline patient characteristics in the current study as well as those from the maraviroc (MVC) arms of the MOTIVATE-1 and -2 studies. Most values shown are median values, with the interquartile ranges (IQR) in parentheses, unless otherwise indicated. The data in the MOTIVATE column were derived from a previously reported data set (8) comprising a majority of maraviroc recipients in the MOTIVATE studies (94%; 788/840 patients). Due to the small numbers of patients, those with races/ethnicities other than white or black are not included in the table. pVL, plasma viral load; wOBTss, weighted optimized background therapy susceptibility score; FPR, false-positive rate; ARVs, antiretrovirals. Some values were derived from data reported previously (1, 3) and are marked with asterisks.
As expected for a study of patients who experience failure of therapy, the patients in the current study had higher plasma viral loads, lower CD4 cell counts, and fewer active drugs in their background regimens than patients in the MOTIVATE studies (1, 8) overall (Table 1).
The failure sample was taken as the earliest available on-treatment sample with both a viral load of >500 copies/ml and an OTA result from the same time point. Samples with viral loads of <500 copies were not tested by OTA and were therefore excluded from the study. The median viral load at failure was 4.1 log copies/ml (IQR, 3.5 to 5.0 log copies/ml), ranging from a minimum of 670 copies/ml to a maximum of 10 million copies/ml.
Most patients (55%) in the study experienced protocol-defined virologic failure (PDVF) over the 48 weeks of the MOTIVATE trials. However, the samples tested were generally from earlier time points than the week where PDVF was met. The median times to PDVF were approximately 17 weeks and 25 weeks for the groups failing with non-R5 and R5 OTA phenotypes, respectively (3). In comparison, the samples in the current study were from medians of 4 weeks and 4 weeks for patients with non-R5 and those with R5 phenotypes, respectively, since we intentionally selected the earliest available failure samples.
Performance of population genotyping for determining HIV tropism.
At screening, all patients had R5 HIV as determined by OTA, but 12% and 31% had non-R5 results by population-based sequencing and ESTA, respectively. At the failure time point, 91 patients had genotypic non-R5 results (50%), and 90 patients had genotypic R5 results (50%) by population sequencing. In comparison, the proportions determined by the phenotypic OTA were 105 non-R5 (58%) and 76 R5 (42%) patients. Approximately half of the patients had non-R5 HIV by both genotypic and phenotypic methods at failure (89 patients; 49%). Of the remaining patients, 41% had R5 HIV as determined by both methods (n = 74), and 10% (n = 18) had discordant results (with 16/18 having R5 HIV as determined by genotyping but non-R5 HIV as determined by OTA).
Changes in V3 sequences and geno2pheno values after maraviroc treatment.
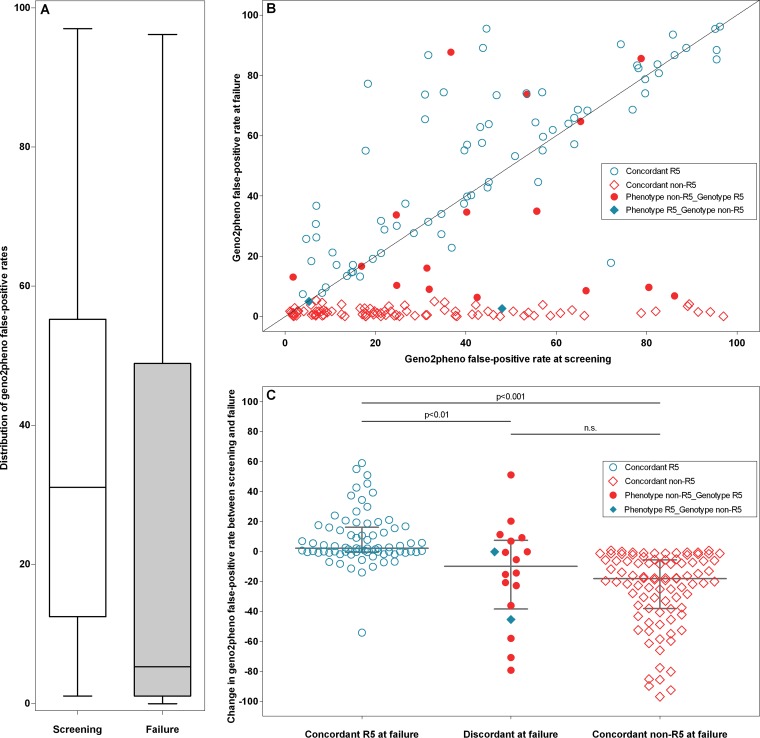
The median FPR for all patients regardless of tropism status fell from 31.0 at screening to 5.3 at failure (Fig. 1A), owing to the large number of patients with non-R5 HIV who were failing. Importantly, these patients fell into two distinct categories: those who maintained essentially the same geno2pheno FPR and those for whom a large decrease in the FPR value between screening and failure was observed (Fig. 1B). The overall drop in the geno2pheno FPR was driven by an increase in the number of patients with non-R5 genotypes, with this number increasing over 4-fold, from 21 patients at screening to 91 patients at failure (12% to 50%). Between screening and failure, the geno2pheno FPR fell by a median of 18.2 (IQR, −38.0 to −5.7) for those patients with concordant non-R5 results (Fig. 1C). These patients had extremely low geno2pheno false-positive rates at failure, with a median FPR of 1.1 (IQR, 0.4 to 1.7). In comparison, the median FPR of these same patients at screening was 20 (IQR, 6.9 to 38). In contrast, there were negligible changes in the geno2pheno FPR in patients failing treatment who had concordant R5 results. For these patients, the median FPR change was 2.2 (IQR, −0.5 to 16) (Fig. 1C and Table 1).
Fig 1.
(A) Overall decrease in the geno2pheno false-positive rate (FPR) values between screening and failure. The distribution of geno2pheno FPR values is shown for the screening (left) and failure (right) sequences. Boxes indicate the interquartile ranges of the values, with the median values indicated by a solid horizontal line. Whiskers correspond to 1.5 times the interquartile range. (B) geno2pheno false-positive rates at screening and failure. Shown is a scatter plot of the geno2pheno FPRs for all patients with coordinates at two time points, with screening values on the horizontal axis and failure values on the vertical axis. Points are marked by whether tropism results at failure were concordant between phenotype and genotype determinations (see key). The geno2pheno FPR decreased by a large amount between screening and failure for patients in the non-R5 group but changed very little for those in the R5 group. (C) Change in geno2pheno false-positive rates between screening and failure. Shown is a scatter plot of the false-positive rate change between screening and failure. Horizontal lines denote the median values, with error bars indicating the interquartile ranges. Patients with concordant R5 tropism at failure had a median FPR change of 2 (IQR, −1 to 16), versus a median decline of 18 FPR units (IQR, −38 to −6) in the concordant non-R5 group (P < 0.001). Patients with discordant results at failure had an overall intermediate FPR decline (median FPR change, 10; IQR, −39 to 7). Points are marked by whether tropism results at failure were concordant between phenotype and genotype determinations (see key).
Not surprisingly, the most common emergent amino acid substitutions among patients with non-R5 genotypes were substitutions to basic amino acids: 11R (36 patients; 40%), 13R (23 patients; 25%), and 25K (20 patients; 22%). Consequently, use of the 11/25 rule (26) identified a substantial proportion of sequences as non-R5 sequences (57/91; 63%) at the time of failure.
Limited evidence of maraviroc resistance.
In contrast to patients with non-R5 failure genotypes, patients who maintained genotypic R5 HIV through the study period (n = 90) exhibited no clear accumulation of mutations at the failure time point. Among these patients, the median geno2pheno FPRs at screening and failure showed very little change, 41.9 and 48.9, respectively. For 21 patients (23%), no V3 amino acid changes were observed following maraviroc treatment, while substitutions in the remaining 70 patients (77%) were restricted to partial amino acid changes (mixtures). Among these patients without a tropism change, the sites with the highest rates of substitutions were codons 10, 13, 14, 18, and 25. The most common substitutions at these positions were 10R, 13H/P, 14I/M, 18R, and 25D. Importantly, however, the prevalence of these substitutions, including ones previously documented to be associated with maraviroc resistance, was very low in this population, ranging between 8 and 14 samples (9 to 16%), depending on the substitution. These may indeed be simply natural polymorphisms unrelated to maraviroc resistance. Furthermore, since most of these samples did not have phenotypic maraviroc resistance assay results, the ability to interpret the implications of these substitutions is limited, and many patients with R5 viruses may simply have been nonadherent or had viruses that were resistant to other components of their regimens.
Change in the non-R5 viral population as determined by deep sequencing.
The viral population present prior to treatment with maraviroc was assessed by deep sequencing of the screening plasma samples. The deep sequencing data were then investigated for both the change in the percentage of non-R5 variants as well as the phylogenetic relationship between the screening and failure V3 sequences.
All screening samples underwent deep V3 sequencing, as did a subset of 73 failure samples. At screening, the median percentage of non-R5 variants per patient was 0.1% (IQR, 0 to 3.1%), reflective of the R5 phenotypes of all patients. However, a majority of patients had at least some level of non-R5 sequences present at screening (96/181; 53%), including one patient for whom 99.9% of recovered sequences were interpreted as being non-R5 sequences at screening, who had R5 HIV as determined by OTA and ESTA at screening but experienced virologic failure with dual-mixed results by OTA at week 4. A total of 50 patients (28%) had ≥2% non-R5 HIV variants according to their deep sequencing screening results, over twice as many as were detected at screening by population-based sequencing, despite all patients having R5 OTA phenotypes at screening.
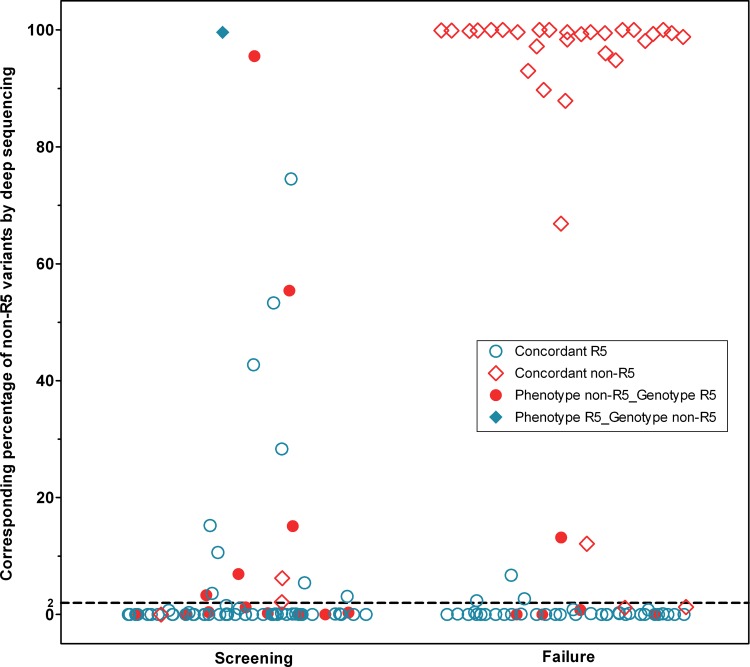
Of the 50 patients with non-R5 variants present at a ≥2% prevalence as determined by deep sequencing, 42 (84%) were confirmed to have non-R5 variants at failure by population-based genotyping. Where deep sequencing results were available at both time points (n = 73), the overall median percentage of non-R5 variants rose slightly from 0% (IQR, 0 to 1.2%) at screening to 0.8% (0 to 98.8%) at failure. When these patients were restricted to those with non-R5 HIV at failure as determined by population-based sequencing, the median percentage of non-R5 variants rose to 99.4% (IQR, 95.4 to 99.9%) at failure (Fig. 2).
Fig 2.
Percentages of non-R5 variants by deep sequencing results at screening and failure. Shown is a scatter plot of the percent non-R5 variants for all patients with deep sequencing results at screening and failure. Points are marked by whether the tropism results for the same time point were concordant between phenotype and population-based genotype determinations (see key). The failure column illustrates how the majority of failure samples had very high or very low non-R5 prevalence. Determination of phenotypes was performed by ESTA at screening and by the original Trofile assay (OTA) at failure. A dashed line at a 2% non-R5 prevalence represents a cutoff for deep sequencing, above which a sample was classified as having non-R5 tropism.
Strikingly, the distribution of non-R5 variants in patients treated with maraviroc was nearly completely dichotomous. The vast majority of patients (65/73; 89%) had treatment failure with either <5% non-R5 variants or >95% non-R5 variants according to deep sequencing results, with very few patients falling in between (Fig. 2). As mentioned above, the population-based sequencing results were also quite unambiguous in their interpretation. Of those patients with non-R5 population sequencing results at failure, over three-quarters had extremely low geno2pheno FPRs, 2 or lower (70/91; 77%), indicative of “highly” non-R5 virus (23).
Phylogenetic relationship between screening and failure sequences.
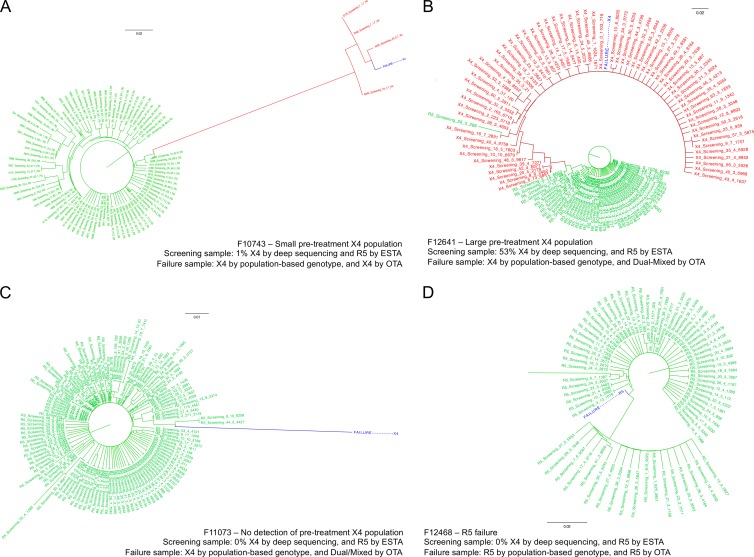
Phylogenetic trees were generated by using the screening deep sequencing data and the failure population-based sequence data. A set of representative example trees is given in Fig. 3 and in the supplemental material. For many patients, a distinct minority subpopulation of non-R5 variants was detected by deep sequencing at screening. This minority subpopulation often emerged following treatment and was detected with standard population-based sequencing methods. The trees underwent manual inspection to assess the degree of the phylogenetic relationship between the failure V3 sequence and sequences detected by deep sequencing prior to maraviroc treatment. Overall, 70% of patients (64/91) with non-R5 HIV at failure had a closely related non-R5 subpopulation present prior to treatment with maraviroc, confirming previous reports of the selection of pretreatment non-R5 reservoirs by maraviroc (16, 17). These CXCR4-using subpopulations were present despite patients being prescreened as having R5 HIV by OTA, and a number were missed by population-based sequencing as well.
Fig 3.
Representative phylogenetic trees from four patients who experienced virologic failure All four panels show phylogenetic trees generated from the deep-sequencing data at screening and the population-based genotyping at failure. Screening R5 sequences are shown in green, X4 sequences are shown in red, and the failure sequence is shown in blue. (A) Failure due to a small pretreatment X4 population. (B) Failure due to a large pretreatment X4 population. (C) Screening sample where pretreatment X4 sequences were not detected but failure was with a non-R5 genotype and phenotype. (D) Example of a patient who experienced failure with an R5 genotype and phenotype. One outlier branch has been truncated for display purposes. All four of these samples were R5 samples by both the original and enhanced sensitivity Trofile assays at screening. Higher-resolution versions of these panels can be found in the supplemental material.
Comparison of tropism methods.
The performance of population-based sequencing was assessed at both screening and failure by comparing the results to deep sequencing as the “gold standard.” ESTA results were available for comparison at screening, and OTA results were available for comparison at failure.
When the two methods for determining genotypic tropism were compared at screening, population-based sequencing had 30% sensitivity (15/50 non-R5 samples) and 95% specificity (125/131 R5 samples) relative to deep sequencing. However, the performance of population-based sequencing was dramatically better at failure. This method achieved 88% sensitivity (29/33 called non-R5) and 95% specificity (38/40 called R5) relative to deep sequencing, likely due to the higher proportions of non-R5 variants after maraviroc treatment.
The genotypes were also compared to the phenotypes. At screening, population-based sequencing had 19% sensitivity (11/57) and 92% specificity (114/124) relative to ESTA. Deep sequencing had 53% sensitivity (30/57) and 84% specificity (104/124) relative to ESTA at screening. At failure, when population-based genotypes were compared to the OTA phenotypes at the same time point, the assays were 90% concordant (163/181 samples). The overall sensitivity of genotyping compared to phenotyping was 85% (89/105 non-R5), with 97% specificity (74/76 R5) for these failure samples. This performance is comparable to the performance of deep sequencing relative to OTA at failure: 83% sensitivity (30/36) and 92% specificity (34/37).
Virologic response to maraviroc.
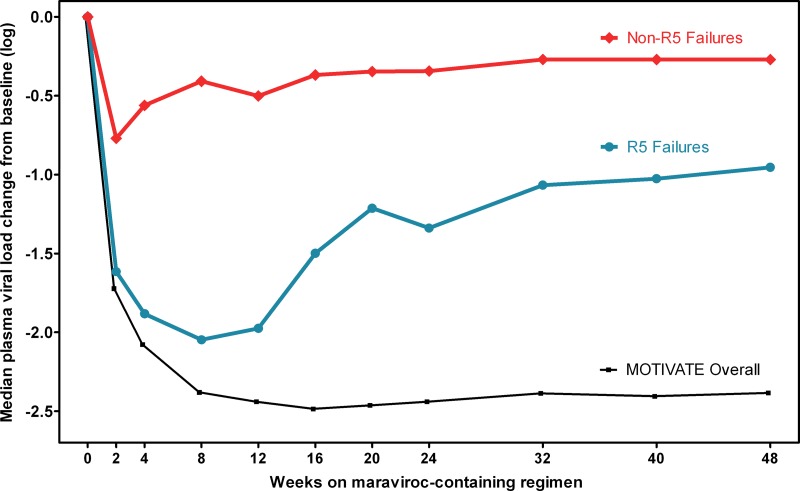
While all patients had R5 HIV as determined by OTA at screening, they could be stratified by their genotypic tropism results at their failure visit. Patients with a non-R5 genotype determined by population-based sequencing at failure had overall poorer virologic responses to maraviroc (Fig. 4). At week 8, the median decline in plasma viral load (pVL) from baseline was 2.0 logs for those with R5 HIV but 0.4 logs for those with non-R5 HIV at failure (P < 0.001). In contrast, the median change in pVL at week 8 for the maraviroc arms in the MOTIVATE trials overall was approximately 2.4 logs, which is larger than the viral load decreases for either group in the current study (P < 0.01) (Fig. 4).
Fig 4.
Virologic responses were reduced among patients with non-R5 genotype results at failure. The median change in plasma viral load from baseline among maraviroc recipients is shown. Patients are stratified according to whether their first available failure sample had an R5 or non-R5 population genotype. For comparison, the median viral load change of the maraviroc arms in the MOTIVATE trials overall is also shown.
A total of 88% of patients with non-R5 population genotypes at failure (80/91) failed to achieve an undetectable viral load during the study, versus 77% (69/90) of those with R5 genotypes at failure. Protocol-defined virologic failure was documented for 69% of patients with non-R5 genotypes at failure (63/91), compared to 41% of patients with R5 genotypes at failure (37/90). Patients with R5 genotypes at failure had higher rates of virologic rebound than did those with non-R5 genotypes: 14% (13/90) versus 2% (2/91). They were also twice as likely to have never achieved virologic suppression throughout the study but to remain enrolled: 32% (29/90) versus 16% (15/91).
Comparison to the enhanced-sensitivity Trofile assay.
As stated above, sensitivities of population-based sequencing and deep sequencing were 19% and 53% relative to ESTA at screening, respectively, with concordance values of 69% and 74% (Fig. 2). Of those patients where rescreening by ESTA indicated pretreatment non-R5 phenotypes, 50 of 57 (88%) were confirmed to have non-R5 phenotypes determined by OTA at failure, similar to the results from genotyping (47 of 57 patients; 82%). Patients with pretreatment R5 phenotypes determined by both OTA and ESTA were more likely to fail therapy with R5 phenotypes or genotypes (56% or 65%) than those with non-R5 phenotypes (44% or 35%). All follow-up results were tested by OTA, but it is important to note that this study indicates that very-low-minority non-R5 variants are not commonly associated with maraviroc failure. Deep sequencing analysis demonstrated that when phenotypic tropism changes occurred, they were generally accompanied by a very high non-R5 prevalence (Fig. 2). Accordingly, our results are likely to be unaffected by the fact that the failure phenotypes were determined by using OTA rather than ESTA. Furthermore, we found largely similar results when we restricted our analyses to patients with R5 phenotypes determined by ESTA at screening only (see Fig. S1 in the supplemental material).
DISCUSSION
In this study, genotypic analysis indicated that failure on maraviroc followed two distinct pathways. Those patients who experienced an HIV tropism shift to non-R5 phenotypes had a large decline in the geno2pheno false-positive rate, accumulated V3 substitutions at multiple codons, and had a large increase in the prevalence of non-R5 variants to a median of 99% according to deep sequencing. Patients with R5 results at failure tended to have geno2pheno values very similar to their screening values and accumulated few amino acid substitutions in V3 compared to the pretreatment sequences. In 71% of patients, deep sequencing was able to detect a pretreatment non-R5 subpopulation which emerged at failure.
We also showed that standard population-based sequencing is capable of identifying on-treatment tropism changes accompanying maraviroc failure with high sensitivity (85%) relative to phenotyping. The sensitivity reported here is even much higher than previous sensitivities reported by our group for the same population at screening (9).
The sensitivity of population-based sequencing more than tripled after patients began treatment with maraviroc (88% sensitivity on treatment versus 24% at screening compared to deep sequencing). Other studies have typically reported much lower sensitivities for population-based sequencing (9, 27–30). The high sensitivity reported for the failure samples in the current study is likely attributable to the selective effect of maraviroc treatment on patient HIV. For those who failed maraviroc-based therapy with non-R5 HIV, the average percentage of non-R5 variants rose to 99% according to deep sequencing, increasing the ability of population-based sequencing to give a non-R5 result. Under these circumstances, population-based methods performed better than usual since non-R5 prevalence is usually masked by a predominantly R5 viral population, thus limiting sensitivity. This rapid emergence of high-prevalence non-R5 variants has also been reported in cases of treatment failure with other CCR5 antagonists (21, 31).
Maraviroc recipients with non-R5 HIV infection at failure had poorer virologic responses to the medication than those whose virus did not change tropism, even among this population of patients who failed maraviroc-based therapy. This is likely due to the additional loss of maraviroc activity in patients who fail therapy with non-R5 HIV, whereas failure with R5 HIV may have been due to a number of reasons, including maraviroc resistance, resistance to the other background antiretroviral agents, and/or poor adherence. However, the response to antiretroviral therapy in general may also be impacted by the presence of non-R5 HIV infection (32, 33).
Some limitations of this study should be acknowledged. It is difficult to extend these findings generally to all maraviroc-treated populations, as this study was conducted in a selected population prescreened for R5 HIV by OTA. However, the proportion of non-R5 OTA results at failure in our study (57%) was quite reflective of the proportion seen in the MOTIVATE trials overall (58%) (3), so our results are likely generalizable to the larger MOTIVATE study population. While ESTA results were available at screening, determination of the follow-up phenotypes was performed by using OTA. However, the additional sensitivity of ESTA over OTA (reported detection limit of 0.3%, versus 10% for non-R5 HIV [4]) likely had very little effect on our results, since genotypic analyses indicated that phenotypic tropism changes of patients on maraviroc treatment were associated with extremely high non-R5 prevalence well above the 10% detection limit of the original Trofile assay. Despite its high sensitivity, deep sequencing could not identify preexisting non-R5 populations in 30% of patients with non-R5 HIV at failure. Thus, this approach may still lack sufficient sensitivity, or the sampling volume may have been insufficient for detection of minority variants. Alternatively, non-R5 variants may evolve from R5 populations more rapidly during maraviroc treatment or may have emerged between screening and enrollment, as previously reported for 8% of MOTIVATE participants (3). Non-R5 variants may also emerge from compartments other than blood plasma. Among the studied population, 25 patients had switched to non-R5 phenotypes at maraviroc initiation (14%). This study was also limited in its ability to better characterize maraviroc resistance. To date, reduced maximal percent inhibition (MPI) in a phenotypic assay assessing susceptibility of the patient virus to maraviroc is the only consistent characteristic of maraviroc resistance (34); no signature mutations have been observed for maraviroc (35, 36) or other CCR5 antagonists (37). Similar to those previous findings, no consistent patterns of mutations that were associated with virologic failure while maintaining an R5 population were noted. This may be due to a number of factors, such as insufficient time on the medication to induce resistance-associated mutations, the possibility that mutations may emerge outside the V3 loop, and/or the possibility that maraviroc resistance mutations are patient specific and difficult to generalize. Furthermore, only a small number of patients who experienced failure on maraviroc with R5 viruses have actually been shown to have maraviroc-resistant HIV in phenotypic assays (35). For such patients, other factors, such as adherence or resistance to the other agents in their regimens, may be involved.
Our analyses indicate that maraviroc treatment dichotomized V3 sequences and their inferred coreceptor usage. Genotypic tropism analyses demonstrated large decreases in geno2pheno values and large increases in the percentage of non-R5 variants. Patients with non-R5 HIV at failure experienced suboptimal virologic responses to maraviroc in this study, likely driven by their non-R5 status. However, genotypic analysis in patients with failure R5 results was not informative, with little change in geno2pheno values and few amino acid substitutions that might be attributed to maraviroc resistance. In contrast, the results for the non-R5 population were unambiguous and striking, suggesting that both deep and population-based sequencing approaches are useful tools for monitoring patients receiving maraviroc.
Supplementary Material
Footnotes
Published ahead of print 30 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01534-13.
REFERENCES
- 1. Gulick RM, Lalezari J, Goodrich J, Clumeck N, DeJesus E, Horban A, Nadler J, Clotet B, Karlsson A, Wohlfeiler M, Montana JB, McHale M, Sullivan J, Ridgway C, Felstead S, Dunne MW, van der Ryst E, Mayer H. 2008. Maraviroc for previously treated patients with R5 HIV-1 infection. N. Engl. J. Med. 359:1429–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Saag M, Goodrich J, Fätkenheuer G, Clotet B, Clumeck N, Sullivan J, Westby M, van der Ryst E, Mayer H. 2009. A double-blind, placebo-controlled trial of maraviroc in treatment-experienced patients infected with non-R5 HIV-1. J. Infect. Dis. 199:1638–1647 [DOI] [PubMed] [Google Scholar]
- 3. Fätkenheuer G, Nelson M, Lazzarin A, Konourina I, Hoepelman AIM, Lampiris H, Hirschel B, Tebas P, Raffi F, Trottier B, Bellos N, Saag M, Cooper DA, Westby M, Tawadrous M, Sullivan JF, Ridgway C, Dunne MW, Felstead S, Mayer H, van der Ryst E. 2008. Subgroup analyses of maraviroc in previously treated R5 HIV-1 infection. N. Engl. J. Med. 359:1442–1455 [DOI] [PubMed] [Google Scholar]
- 4. Wilkin TJ, Goetz MB, Leduc R, Skowron G, Su Z, Chan ES, Heera J, Chapman D, Spritzler J, Reeves JD, Gulick RM, Coakley E. 2011. Reanalysis of coreceptor tropism in HIV-1-infected adults using a phenotypic assay with enhanced sensitivity. Clin. Infect. Dis. 52:925–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cooper D, Heera J, Goodrich J, Tawadrous M, Saag M, Dejesus E, Clumeck N, Walmsley S, Ting N, Coakley E, Reeves JD, Reyes-Teran G, Westby M, Van Der Ryst E, Ive P, Mohapi L, Mingrone H, Horban A, Hackman F, Sullivan J, Mayer H. 2010. Maraviroc versus efavirenz, both in combination with zidovudine-lamivudine, for the treatment of antiretroviral-naive subjects with CCR5-tropic HIV-1 infection. J. Infect. Dis. 201:803–813 [DOI] [PubMed] [Google Scholar]
- 6. McGovern RA, Thielen A, Portsmouth S, Mo T, Dong W, Woods CK, Zhong X, Brumme CJ, Chapman D, Lewis M, James I, Heera J, Valdez H, Harrigan PR. 2012. Population-based sequencing of the V3-loop can predict the virological response to maraviroc in treatment-naive patients of the MERIT trial. J. Acquir. Immune Defic. Syndr. 61:279–286 [DOI] [PubMed] [Google Scholar]
- 7. Swenson LC, Mo T, Dong WWY, Zhong X, Woods CK, Thielen A, Jensen M, Knapp DJHF, Chapman D, Portsmouth S, Lewis M, James I, Heera J, Valdez H, Harrigan PR. 2011. Deep V3 sequencing for HIV type 1 tropism in treatment-naive patients: a reanalysis of the MERIT trial of maraviroc. Clin. Infect. Dis. 53:732–742 [DOI] [PubMed] [Google Scholar]
- 8. Swenson LC, Mo T, Dong WWY, Zhong X, Woods CK, Jensen MA, Thielen A, Chapman D, Lewis M, James I, Heera J, Valdez H, Harrigan PR. 2011. Deep sequencing to infer HIV-1 co-receptor usage: application to three clinical trials of maraviroc in treatment-experienced patients. J. Infect. Dis. 203:237–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McGovern RA, Thielen A, Mo T, Dong W, Woods CK, Chapman D, Lewis M, James I, Heera J, Valdez H, Harrigan PR. 2010. Population-based V3 genotypic tropism assay: a retrospective analysis using screening samples from the A4001029 and MOTIVATE studies. AIDS 24:2517–2525 [DOI] [PubMed] [Google Scholar]
- 10. Kagan RM, Johnson EP, Siaw M, Biswas P, Chapman DS, Su Z, Platt JL, Pesano RL. 2012. A genotypic test for HIV-1 tropism combining Sanger sequencing with ultradeep sequencing predicts virologic response in treatment-experienced patients. PLoS One 7:e46334. 10.1371/journal.pone.0046334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Swenson LC, Boehme R, Thielen A, McGovern RA, Harrigan PR. 2010. Genotypic determination of HIV-1 tropism in the clinical setting. HIV Ther. 4:293–303 [Google Scholar]
- 12. Sing T, Low AJ, Beerenwinkel N, Sander O, Cheung PK, Domingues FS, Büch J, Däumer M, Kaiser R, Lengauer T, Harrigan PR. 2007. Predicting HIV coreceptor usage on the basis of genetic and clinical covariates. Antivir. Ther. 12:1097–1106 [PubMed] [Google Scholar]
- 13. Swenson LC, Däumer M, Paredes R. 2012. Next-generation sequencing to assess HIV tropism. Curr. Opin. HIV AIDS 7:478–485 [DOI] [PubMed] [Google Scholar]
- 14. Raymond S, Saliou A, Nicot F, Delobel P, Dubois M, Cazabat M, Sandres-Sauné K, Marchou B, Massip P, Izopet J. 2011. Frequency of CXCR4-using viruses in primary HIV-1 infections using ultra-deep pyrosequencing. AIDS 25:1668–1673 [DOI] [PubMed] [Google Scholar]
- 15. Archer J, Weber J, Henry K, Winner D, Gibson R, Lee L, Paxinos E, Arts EJ, Robertson DL, Mimms L, Quiñones-Mateu ME. 2012. Use of four next-generation sequencing platforms to determine HIV-1 coreceptor tropism. PLoS One 7:e49602. 10.1371/journal.pone.0049602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Archer J, Braverman MS, Taillon BE, Desany B, James I, Harrigan PR, Lewis M, Robertson DL. 2009. Detection of low-frequency pretherapy chemokine (CXC motif) receptor 4 (CXCR4)-using HIV-1 with ultra-deep pyrosequencing. AIDS 23:1209–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Westby M, Lewis M, Whitcomb J, Youle M, Pozniak AL, James IT, Jenkins TM, Perros M, Van Der Ryst E. 2006. Emergence of CXCR4-using human immunodeficiency virus type 1 (HIV-1) variants in a minority of HIV-1-infected patients following treatment with the CCR5 antagonist maraviroc is from a pretreatment CXCR4-using virus reservoir. J. Virol. 80:4909–4920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tilton JC, Wilen CB, Didigu CA, Sinha R, Harrison JE, Agrawal-Gamse C, Henning EA, Bushman FD, Martin JN, Deeks SG, Doms RW. 2010. A maraviroc-resistant HIV-1 with narrow cross-resistance to other CCR5 antagonists depends on both N-terminal and extracellular loop domains of drug-bound CCR5. J. Virol. 84:10863–10876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schapiro JM, Boucher CA, Kuritzkes DR, van de Vijver DA, Llibre JM, Lewis M, Simpson P, Delogne C, McFadyen L, Chapman D, Perros M, Valdez H, van der Ryst E, Westby M. 2011. Baseline CD4+ T-cell counts and weighted background susceptibility scores strongly predict response to maraviroc regimens in treatment-experienced patients. Antivir. Ther. 16:395–404 [DOI] [PubMed] [Google Scholar]
- 20. McGovern RA, Symons J, Poon AFY, Harrigan PR, van Lelyveld SFL, Hoepelman AIM, van Ham PM, Dong W, Wensing AMJ, Nijhuis M. 2013. Maraviroc treatment in non-R5-HIV-1-infected patients results in the selection of extreme CXCR4-using variants with limited effect on the total viral setpoint. J. Antimicrob. Chemother. 68:2007–2014 [DOI] [PubMed] [Google Scholar]
- 21. Tsibris AMN, Korber B, Arnaout R, Russ C, Lo C-C, Leitner T, Gaschen B, Theiler J, Paredes R, Su Z, Hughes MD, Gulick RM, Greaves W, Coakley E, Flexner C, Nusbaum C, Kuritzkes DR. 2009. Quantitative deep sequencing reveals dynamic HIV-1 escape and large population shifts during CCR5 antagonist therapy in vivo. PLoS One 4:e5683. 10.1371/journal.pone.0005683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Roche M, Salimi H, Duncan R, Wilkinson BL, Chikere K, Moore MS, Webb NE, Zappi H, Sterjovski J, Flynn JK, Ellett A, Gray LR, Lee B, Jubb B, Westby M, Ramsland PA, Lewin SR, Payne RJ, Churchill MJ, Gorry PR. 2013. A common mechanism of clinical HIV-1 resistance to the CCR5 antagonist maraviroc despite divergent resistance levels and lack of common gp120 resistance mutations. Retrovirology 10:43. 10.1186/1742-4690-10-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McGovern RA, Dong W, Mo T, Woods C, Zhong X, Thielen A, Jensen M, Heera J, Ellery S, Lewis M, James I, Biswas P, Chapman D, Valdez H, Harrigan R. 2009. Optimization of clinically relevant cutpoints for the determination of HIV co-receptor usage to predict maraviroc responses in treatment experienced (TE) patients using population V3 genotyping, poster PE3.4/8. In Abstr. 12th Eur. AIDS Conf. European AIDS Clinical Society, Cologne, Germany [Google Scholar]
- 24. Swenson LC, Dong W, Mo T, Woods C, Zhong X, Thielen A, Jensen M, Biswas P, Ellery S, Lewis M, James I, Chapman D, Valdez H, Harrigan R. 2009. “Deep” sequencing to identify treatment-experienced patients who respond to maraviroc (MVC), poster PE3.3/2. In Abstr. 12th Eur. AIDS Conf. European AIDS Clinical Society, Cologne, Germany [Google Scholar]
- 25. Lalezari J, Goodrich J, DeJesus E, Lampiris H, Gulick R, Saag M, Ridgway C, McHale M, van der Ryst E, Mayer H. 2007. Efficacy and safety of maraviroc plus optimized background therapy in viremic, ART-experienced patients infected with CCR5-tropic HIV-1: 24-week results of phase 2b/3 studies, abstr 104bLB. In Abstr. 14th Conf. Retroviruses Opportun. Infect., Los Angeles, CA [Google Scholar]
- 26. Resch W, Hoffman N, Swanstrom R. 2001. Improved success of phenotype prediction of the human immunodeficiency virus type 1 from envelope variable loop 3 sequence using neural networks. Virology 288:51–62 [DOI] [PubMed] [Google Scholar]
- 27. Obermeier M, Symons J, Wensing AMJ. 2012. HIV population genotypic tropism testing and its clinical significance. Curr. Opin. HIV AIDS 7:470–477 [DOI] [PubMed] [Google Scholar]
- 28. Low AJ, Dong W, Chan D, Sing T, Swanstrom R, Jensen M, Pillai S, Good B, Harrigan PR. 2007. Current V3 genotyping algorithms are inadequate for predicting X4 co-receptor usage in clinical isolates. AIDS 21:F17–F24. 10.1097/QAD.0b013e3282ef81ea [DOI] [PubMed] [Google Scholar]
- 29. De Mendoza C, Van Baelen K, Poveda E, Rondelez E, Zahonero N, Stuyver L, Garrido C, Villacian J, Soriano V. 2008. Performance of a population-based HIV-1 tropism phenotypic assay and correlation with V3 genotypic prediction tools in recent HIV-1 seroconverters. J. Acquir. Immune Defic. Syndr. 48:241–244 [DOI] [PubMed] [Google Scholar]
- 30. Skrabal K, Low AJ, Dong W, Sing T, Cheung PK, Mammano F, Harrigan PR. 2007. Determining human immunodeficiency virus coreceptor use in a clinical setting: degree of correlation between two phenotypic assays and a bioinformatic model. J. Clin. Microbiol. 45:279–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Demarest JF, Amrine-Madsen H, Irlbeck DM, Kitrinos KM. 2009. Virologic failure in first-line human immunodeficiency virus therapy with a CCR5 entry inhibitor, aplaviroc, plus a fixed-dose combination of lamivudine-zidovudine: nucleoside reverse transcriptase inhibitor resistance regardless of envelope tropism. Antimicrob. Agents Chemother. 53:1116–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brumme ZL, Dong WWY, Yip B, Wynhoven B, Hoffman NG, Swanstrom R, Jensen MA, Mullins JI, Hogg RS, Montaner JSG, Harrigan PR. 2004. Clinical and immunological impact of HIV envelope V3 sequence variation after starting initial triple antiretroviral therapy. AIDS 18:F1. 10.1097/00002030-200403050-00001 [DOI] [PubMed] [Google Scholar]
- 33. Brumme ZL, Goodrich J, Mayer HB, Brumme CJ, Henrick BM, Wynhoven B, Asselin JJ, Cheung PK, Hogg RS, Montaner JSG, Harrigan PR. 2005. Molecular and clinical epidemiology of CXCR4-using HIV-1 in a large population of antiretroviral-naive individuals. J. Infect. Dis. 192:466–474 [DOI] [PubMed] [Google Scholar]
- 34. Westby M, Smith-Burchnell C, Mori J, Lewis M, Mosley M, Stockdale M, Dorr P, Ciaramella G, Perros M. 2007. Reduced maximal inhibition in phenotypic susceptibility assays indicates that viral strains resistant to the CCR5 antagonist maraviroc utilize inhibitor-bound receptor for entry. J. Virol. 81:2359–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moore JP, Kuritzkes DR. 2009. A pièce de resistance: how HIV-1 escapes small molecule CCR5 inhibitors. Curr. Opin. HIV AIDS 4:118–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Delobel P, Cazabat M, Saliou A, Loiseau C, Coassin L, Raymond S, Requena M, Marchou B, Massip P, Izopet J 21 June 2013. Primary resistance of CCR5-tropic HIV-1 to maraviroc cannot be predicted by the V3 sequence. J. Antimicrob. Chemother. [Epub ahead of print.]. 10.1093/jac/dkt249 [DOI] [PubMed] [Google Scholar]
- 37. McNicholas P, Vilchez RA, Greaves W, Kumar S, Onyebuchi C, Black T, Strizki JM. 2012. Detection of HIV-1 CXCR4 tropism and resistance in treatment experienced subjects receiving CCR5 antagonist-vicriviroc. J. Clin. Virol. 55:134–139 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.