Abstract
Members of the Glomeromycota form the arbuscular mycorrhiza (AM) symbiosis. They supply plants with inorganic nutrients, including nitrogen, from the soil. To gain insight into transporters potentially facilitating nitrogen transport processes, ammonium transporters (AMTs) of Geosiphon pyriformis, a glomeromycotan fungus forming a symbiosis with cyanobacteria, were studied. Three AMT genes were identified, and all three were expressed in the symbiotic stage. The localization and functional characterization of the proteins in a heterologous yeast system revealed distinct characteristics for each of them. AMT1 of G. pyriformis (GpAMT1) and GpAMT2 were both plasma membrane localized, but only GpAMT1 transported ammonium. Neither protein transported the ammonium analogue methylammonium. Unexpectedly, GpAMT3 was localized in the vacuolar membrane, and it has as-yet-unknown transport characteristics. An unusual cysteine residue in the AMT signature of GpAMT2 and GpAMT3 was identified, and the corresponding residue was demonstrated to play an important role in ammonium transport. Surprisingly, each of the three AMTs of G. pyriformis had very distinct features. The localization of an AMT in the yeast vacuolar membrane is novel, as is the described amino acid residue that clearly influences ammonium transport. The AMT characteristics might reflect adaptations to the lifestyle of glomeromycotan fungi.
INTRODUCTION
In nature, more than 90% of land plants undergo mutualistic root symbioses with various fungal partners, the so-called mycorrhizas (1). In ectomycorrhizal associations, which are predominantly formed by trees in temperate forests, the fungal partners stay outside the plant root cells, while in endomycorrhizal associations, the fungus penetrates the plant cells to form a very intimate association with its host (1, 2). The endomycorrhizal arbuscular mycorrhiza (AM) is a nearly ubiquitous plant-microorganism symbiosis in terrestrial ecosystems (3). Most likely, more than 80% of all terrestrial plants undergo AM with AM fungi (AMF) (4), which all belong to the distinct monophyletic fungal phylum Glomeromycota (5). Perfectly conserved AMF structures were found in 400-million-year-old silicified fossils (6, 7), and fossil AMF spores are dated to 460 million years ago (8). Therefore, the AM symbiosis is proposed to be a key acquisition of land plants that was crucial for their initial exploration of the terrestrial ecosystem, and the association must be considered an ancient and integral part of the life of land plants (9).
All AMF are obligate biotrophic organisms and depend on their photoautotrophic symbiosis partners for carbon supply (10). In the AM, the intraradical fungal hyphae penetrate cortical cells, where they form highly branched arbuscules (Latin arbuscula means “little tree”), which are the eponymous symbiotic structures of the symbiosis. They provide an extensive surface for nutrient exchange across the arbuscular plasma membrane of the fungus and the so-called periarbuscular membrane, which is continuous with the plasma membrane of the plant cell. The plant invests a significant portion of its photosynthetically fixed carbon (up to 20% [10]), delivered as monosaccharides to the fungus, which in exchange provides inorganic nutrients like phosphorus (P) and nitrogen (N), as well as inorganic micronutrients and water (11, 12).
While P nutrition has been one of the central topics in AM research in recent decades, N transport, which occurs in substantial amounts (13, 14), has been less investigated. It is known that AM fungi are able to take up N from the soil in various forms (15–18), but ammonium seems to be the favorite (13, 19, 20). The current model, based on 15N labeling studies (21–23), suggests that N is fixed over the glutamine synthetase/glutamate synthase (GS/GOGAT) pathway and transported as arginine along the fungal hyphae toward the host roots. In the arbuscule, arginine is cleaved by arginase, and N is believed to be released to the symbiotic interface and taken up by the host as NH4+ (12, 24). This model is supported by the characterization of a plant ammonium transporter that is preferentially expressed in arbusculated cells (25), as well as by the occurrence of fungal mRNA transcripts encoding enzymes that are involved in the suggested pathways in extra- and intraradical fungal structures (26, 27).
In most organisms, ammonium import is facilitated by proteins that belong to the conserved ammonium transporter/methylammonium permease/Rhesus (Amt/Mep/Rh) protein family (for a review, see reference 28). Its first characterized members were from Saccharomyces cerevisiae (29) and Arabidopsis thaliana (30). The transport mechanism of Amt/Mep/Rh family transporters, however, is still under debate and might even differ between closely related family members (31, 32). Today, more than 4,300 family members have been discovered (Pfam database), but only two of them are of glomeromycotan origin, namely, from the widely used model AMF Rhizophagus irregularis strain DAOM197198 (formerly Glomus intraradices DAOM197198 [33, 34]), ammonium transporter 1 (AMT1) of G. intraradices (GiAMT1) (35) and GiAMT2 (36). By complementation analysis of ammonium transport-deficient (mep1-3Δ) yeast mutant strains (37, 38), GiAMT1 was characterized as a typical high-affinity ammonium transporter (35), while GiAMT2 exhibited a lower ammonium transport capability (36). The lack of an established stable transformation system for any AMF (39), as well as their obligate biotrophy (10) and their coenocytic, multinucleate organization (40), hinder direct functional analysis in vivo.
Among the glomeromycotan fungi, Geosiphon pyriformis is unique by undergoing an endosymbiosis with a cyanobacterium, Nostoc punctiforme, in special, bladder-like aboveground vesicles (41). These symbiotic cyanobacteria form N2-fixing heterocysts and might feed the fungus with ammonium in N-free growth media, while in AM, the plant symbiont is dependent on nitrogen feeding by the fungal partner. However, a lack of increased heterocyst frequency in the Geosiphon symbiosis in comparison to the frequency of heterocysts in free-living N. punctiforme (41) points to the fact that the cyanobacteria are most likely not fixing excess N2 for feeding their host. The study of the Geosiphon symbiosis presents some advantages over “classical” AM. One such advantage is the fact that by using poly(A)-specific mRNA isolation methods, a cDNA expression library could be constructed that contains nearly exclusively symbiotically expressed fungal genes (42). This is not possible for the AM, where the availability of mRNA from symbiotic fungal structures is restricted by its dilution by plant root mRNA.
Here, we exploited the cDNA expression library to identify the symbiotic ammonium transporter complement of G. pyriformis. Using the well-defined heterologous S. cerevisiae system, we characterized three AMTs, all of which exhibited distinct and even unusual features with respect to subcellular localization and transport characteristics.
MATERIALS AND METHODS
E. coli strains, plasmids, and methods.
Standard procedures were used for the propagation and subcloning of plasmids in the Escherichia coli strain TOP10 (Invitrogen, Germany). The plasmids and primers used in this work are listed in Tables S1 and S2, respectively, in the supplemental material.
Yeast strains and culture conditions.
For the analyses in S. cerevisiae, the diploid ammonium transporter-deficient strain MLY131a/α (Mata/Matα mep1Δ::LEU2/mep1Δ::LEU2 mep2Δ::LEU2/mep2Δ::LEU2 mep3Δ::G418R/mep3Δ::G418R ura3-52/ura3-52) was used (38). Transformants were selected on Hartwell's complete medium (43) lacking uracil (HC-U). For uptake studies, expression studies, and microscopy, 5 ml of liquid HC-U medium was inoculated with single colonies, incubated at 30°C with shaking at 160 rpm overnight, and diluted into 50 ml of yeast nitrogen base (YNB) medium containing 2% (wt/vol) glucose as the carbon source and 0.1% (wt/vol) proline as the sole nitrogen source. Cells were incubated again at 30°C, 160 rpm overnight until an optical density at 600 nm (OD600) of 0.5 to 0.7 was reached. Cells were harvested and washed twice with water before analyses. All liquid yeast cultures were incubated in baffled flasks. Complementation studies were repeated with the isogenic haploid strain 31019b (37) and gave comparable results (data not shown).
Isolation of G. pyriformis ammonium transporter genes.
The cDNA expression library of Geosiphon pyriformis, with 6 × 105 primary-insert-containing clones (42), was screened for ammonium transporter genes by yeast complementation. The ammonium uptake-deficient yeast strain MLY131a/α (38) was transformed in several batches with 1 μg plasmid DNA each and a heat shock of 40 min at 42°C using the large-scale, high-efficiency yeast transformation protocol (44). In total, 1.5 × 106 transformants (yielding a theoretical coverage of 92%) were plated on YNB medium containing 0.5 or 0.1 mM (NH4)2SO4 as the sole nitrogen source. After 6 weeks of incubation at 30°C, 12 independent complementing clones were visible. Two of these were sequenced and found to contain identical inserts. The plasmid harboring AMT1 of G. pyriformis (GpAMT1) was designated pBL101.
GpAMT2 was isolated by degenerate PCR performed directly on the G. pyriformis cDNA library. For this, the S. cerevisiae Mep1 (ScMep1) sequence was screened by BLAST search (45) against the nonredundant protein collections of ascomycetes and of basidiomycetes. The first five hits of the two screens were aligned with ClustalW2 (46, 47), and degenerate primers were designed using the consensus-degenerate hybrid oligonucleotide primers (CODEHOP) strategy for distantly related protein sequences (48, 49). A seminested PCR approach using the primer pairs Mep1-50-FWD/Mep1-178-REV and Mep1-53-FWD/Mep1-178-REV yielded a 307-bp fragment of a novel gene, which was 59% identical to GiAMT1 on the nucleotide level. The 5′ and 3′ regions of GpAMT2 were also amplified by nested PCR approaches, using the primer pairs PMA5/GpAMT2-REV3 and PMA16/GpAMT2-REV5 and GpAMT2-FWD1/ADHclose and GpAMT2-FWD2/ADH109-REV, respectively. The two fragments were digested with SfiI and PstI and ligated in a three-fragment ligation into the SfiI sites of pDR196sfi to reconstitute the original cDNA plasmid (pBL102).
GpAMT3 was discovered as a 439-bp fragment after 454 pyrosequencing of the cDNA library (A. Schüßler, unpublished data). It was 73% identical to GiAMT1 at the nucleotide level. The 5′ and 3′ regions were amplified with primer pairs GpAMT3-clone-FWD/ADHclose and GpAMT3-clone-REV/PMA5, respectively. Both fragments were diluted 100-fold and then were pooled and used as the template for a PCR with the primer pair PMA5/ADHclose. The resulting amplicon was digested with SfiI and ligated into the SfiI sites of pDR196sfi (42) to reconstitute the original cDNA plasmid (pBL179).
The genomic versions of three AMT genes were sequenced from ϕ29-amplified G. pyriformis genomic DNA (42) after PCR with the primer pairs GpAMT1-sfi-FWD/GpAMT1-sfi-REV, GpAMT2-sfi-FWD/GpAMT2-sfi-REV, and GpAMT3-sfi-FWD/GpAMT3-sfi-REV and cloning into a TOPO vector (ZeroBlunt for sequencing; Invitrogen, Germany). For GpAMT3, three single nucleotide polymorphisms (SNPs) were detected between the genomic and the cDNA sequence, which possibly resulted from amplification errors during library preparation. Both versions (the genomic and the cDNA clone) were tested for ammonium removal in yeast on plasmids pBL180 and pBL183.
Amplification of the R. irregularis ammonium transporter gene GiAMT1.
Rhizophagus irregularis RNA was extracted from fungal mycelium using the NucleoSpin RNA plant kit (Macherey & Nagel, Germany), and cDNA was obtained with the SMART RACE cDNA amplification kit (Clontech, USA), following the manufacturer's instructions. GiAMT1 was amplified from this cDNA with the primer pair GiAMT1-sfi-FWD/GiAMT1-sfi-REV and cloned into the SfiI sites of pDR196sfi, yielding plasmid pBL151.
Expression constructs.
To create expression vectors not carrying untranslated regions (UTRs), the open reading frames (ORFs) of GpAMT1, GpAMT2, GpAMT3, ScMEP1, and ScMEP2 were amplified with the primer pairs GpAMT1-sfi-FWD/GpAMT1-sfi-REV, GpAMT2-sfi-FWD/GpAMT2-sfi-REV, GpAMT3-sfi-FWD/GpAMT3-sfi-REV, ScMEP1-sfi-FWD/ScMEP1-sfi-Rev, and ScMEP2-sfi-FWD/ScMEP2-sfi-REV, respectively. They were cloned into the SfiI sites of pDR196sfi, yielding plasmids pBL110, pBL111, pBL180, pBL103, and pBL104.
To obtain the version of GpAMT3 that carries the SNPs found in the genomic DNA (gDNA), the 3′ region was amplified from gDNA with the primer pair GpAMT3-gen-FWD/GpAMT3-sfi-REV and the 5′ region was amplified from the cDNA library (to exclude the intron) with the primer pair GpAMT3-sfi-FWD/GpAMT3-gen-REV. Both fragments were diluted 100-fold and pooled and were then amplified with the primer pair GpAMT3-sfi-FWD/GpAMT3-sfi-REV, digested with SfiI, and ligated into the SfiI sites of pDR196sfi, yielding plasmid pBL183.
For the enhanced green fluorescent protein (EGFP) fusion constructs, GpAMT1, GpAMT2, GpAMT3, and ScMEP1 were amplified with the primer pairs GpAMT1-sfi-FWD/GpAMT1-tag-sfi-REV, GpAMT2-sfi-FWD/GpAMT2-tag-sfi-REV, GpAMT3-sfi-FWD/GpAMT3-tag-sfi-REV, and ScMEP1-sfi-FWD/ScMEP1-tag-sfi-REV, respectively, yielding plasmids pBL107, pBL108, pBL186, and pBL109. EGFP was amplified from pRU4 (50) with the primers X-S-EGFP-FWD and EGFP-A-REV and cloned into the EagI and ApaI sites of pDR196sfi. As a control, the oligonucleotides ORF-FWD and ORF-REV were heat denatured, annealed, and cloned into the SfiI sites of PDR196sfi carrying EGFP, yielding plasmid pBL106.
For the serine-to-cysteine (StoC) or cysteine-to-serine (CtoS) variants of GpAMT1, GpAMT2, GpAMT3, and ScMep2, the 5′ and 3′ parts were amplified with the primer pairs GpAMT1-StoC-FWD/ADHclose and GpAMT1-StoC-REV/PMA5, GpAMT2-CtoS-FWD/ADHclose and GpAMT2-CtoS-REV/PMA5, GpAMT3-CtoS-FWD/ADHclose and GpAMT3-CtoS-REV/PMA5, and ScMEP2-StoC-FWD/ADHclose and ScMEP2-StoC-REV/PMA5, respectively. For each construct, the two fragments were diluted 100-fold, pooled, and used as the template for amplification with the primer pair PMA5/ADHclose. The PCR fragment was then digested with SfiI and ligated into the SfiI sites of pDR196sfi, yielding plasmid pBL130, pBL131, pBL181, or pBL134. By using pBL183 as the template with the primer pairs gGpAMT3-CtoS-FWD/ADHclose and gGpAMT3-CtoS-REV/PMA5, we obtained plasmid pBL185, containing the genomic version of GpAMT3 with the C-to-S mutation (gGpAMT3_C189S).
Sequence analyses.
Sequence analyses were performed using the CLC software package (CLCbio, Aarhus, Denmark) and BLAST (http://blast.ncbi.nlm.nih.gov) (45) and ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2) (47) algorithms. Transmembrane domains were predicted by using the TMHMM (http://www.cbs.dtu.dk/services/TMHMM) (51) and the SOSUI (http://bp.nuap.nagoya-u.ac.jp/sosui) (52) algorithms. For phylogenetic analysis, protein sequences were aligned with MAFFT (http://mafft.cbrc.jp/alignment/server) (53) using the L-INS-i setting favoring accuracy and the JTT substitution matrix (54) with a gap-opening penalty for group-to-group alignment of 1. The alignment was reduced to 361 unambiguously aligned positions, and maximum-likelihood phylogenetic trees were computed using RAxML (http://phylobench.vital-it.ch/raxml-bb/index.php) (55) using the GAMMA model, the JTT matrix, and empirical frequencies, with 1,000 bootstraps. All analyses were performed at the CIPRES scientific gateway (http://www.phylo.org/portal2).
Protein modeling.
A three-dimensional model of GpAMT2 was generated within the SWISS-MODEL workspace (http://swissmodel.expasy.org/workspace) (56) and processed with DeepView pdb-Viewer (57). For this, GpAMT2 was aligned to the PDB template 2b2h of Archaeoglobus fulgidus am-1.
Yeast growth assays.
For plate complementation tests, HC-U overnight cultures of yeast strains expressing transporter genes were diluted to an OD600 of 0.1. Amounts of around 2 μl each of a 5-fold dilution series were spotted on solid medium with a metal stamp. Plates were incubated at 30°C for the durations indicated in Fig. 3A, as well as in Fig. S3, S4C, and S6 in the supplemental material.
Fig 3.
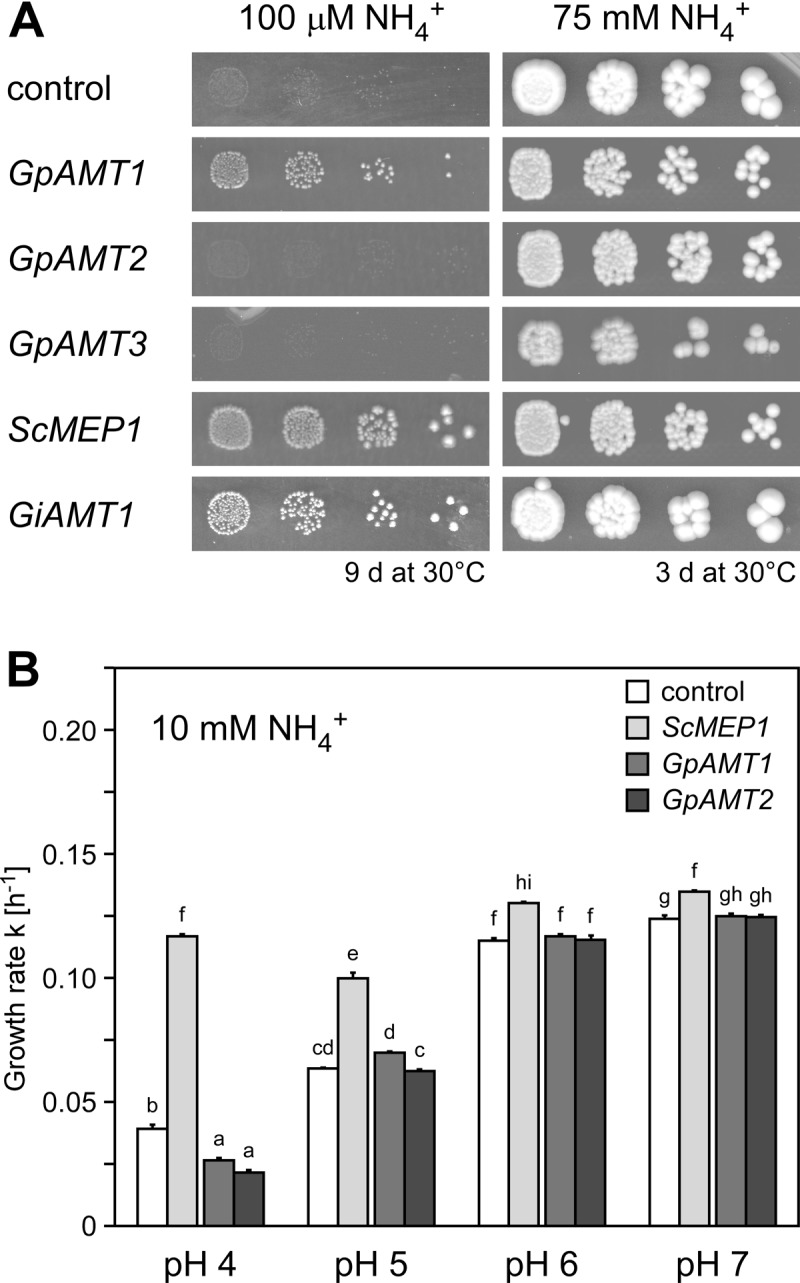
Functional complementation of yeast strain MLY131a/α (mep1-3Δ) with Geosiphon pyriformis AMT genes. The yeast was transformed with empty vector (negative control) or plasmids carrying GpAMT1, GpAMT2, GpAMT3, ScMEP1 (positive control), or GiAMT1 (positive control). (A) Growth on solid minimal medium containing 100 μM NH4+ as sole nitrogen source (left) and on HC-U medium containing 75 mM NH4+ (right). (B) Growth in buffered liquid minimal medium containing 10 mM NH4+. Only minor growth differences between cells expressing the G. pyriformis AMTs and the vector control could be observed during exponential growth. Error bars, ±standard deviation; n = 4 biological replicates. Different letters above bars indicate highly significant differences (P ≤ 0.001, ANOVA test) between pairwise comparisons.
Growth curves were recorded in 96-well microtiter plates in liquid medium. Overnight cultures of the respective strains were diluted to an OD600 of 0.01 in 200 μl of BA medium (58) buffered with 10 mM citrate at the pH indicated in Fig. 3B, as well as in Fig. S4A and B in the supplemental material, containing 3% (wt/vol) glucose as carbon source and either the (NH4)2SO4 concentration indicated in Fig. 3A and B, as well as in Fig. S3, S4, and S6 in the supplemental material, or 0.1% (wt/vol) proline as the sole nitrogen source. For pH 7, the medium was buffered with 10 mM MOPS (morpholinepropanesulfonic acid) instead of citrate. Various methylammonium concentrations were added. The plates were incubated at room temperature in a Sunrise microtiter plate reader (Tecan, Switzerland), adjusted to “wide” shaking, for 48 to 72 h. The OD620 was measured every 15 min. To calculate the growth rate k of logarithmically growing yeast cells, the first time point after the OD620 had passed 0.15 (t0) and the time point 4 h later (t1) were taken. The growth rate k was calculated by the following formula: k = {ln[OD620(t1)/OD620(t0)]}/4 h.
Uptake assays with [14C]methylammonium.
The initial rates of [14C]methylammonium uptake were measured as described previously (37). Briefly, yeast cells expressing GpAMT1, GpAMT2, ScMep1, or GiAMT1 or pDR196sfi-transformed cells were grown to an OD600 of 0.5 to 0.7 in YNB medium containing 0.1% (wt/vol) proline as the sole nitrogen source. Cells were harvested, washed twice, and resuspended in 20 mM phosphate buffer, pH 6 or 7, to a final OD600 of 8. One-hundred-microliter aliquots of cells were supplied with 0.1 M glucose 5 min prior to measurement and incubated at 30°C, 1,000 rpm in a Thermomixer comfort (Eppendorf, Germany). Reactions were started by the addition of 100 μl 40 mM phosphate buffer, pH 6 or 7, containing various concentrations (2 to 250 μM) of [14C]methylammonium (Hartmann Analytic, Germany). Cells were incubated at 30°C for 0.5 min, 1 min, 2 min, and 4 min, washed in 5 ml ice-cold 40 mM phosphate buffer, pH 6 or 7, containing 100 mM unlabeled methylammonium for 4 min in a multifiltration unit (Hölzel, Germany), and filtered through glass fiber filters. Filters were washed two times with 5 ml of 40 mM phosphate buffer, pH 6 or 7, and [14C]methylammonium content was measured by scintillation counting in RotiSzint (Carl Roth, Germany) or UltimaGold (PerkinElmer, USA) scintillation liquid in an LS500TD (Beckman Coulter, USA) or Tri-Carb 2100 (PerkinElmer, USA) scintillation counter.
Ammonium removal assays.
To directly determine the ammonium uptake rates of transformed yeast cells, ammonium removal assays were performed as described previously (59). Cells were grown overnight in 50 ml SD medium (43) with 0.1% (wt/vol) proline at 30°C, washed twice in water, and resuspended in 30 ml SD medium with 1 mM ammonium chloride to an OD600 of 2. The cultures were incubated at 30°C, 160 rpm, and 1-ml samples were taken at 0 min, 10 min, 30 min, 60 min, and then every 60 min for 6 h in total. The cells were pelleted, and 40 μl of the supernatant added to 760 μl OPA solution (0.2 M phosphate buffer, pH 7.3, containing 0.54% [wt/vol] o-phthal dialdehyde, 10% [vol/vol] ethanol, and 0.05% [vol/vol] β-mercaptoethanol) to quantify the remaining ammonium. After 25 min of incubation in the dark, extinction at 420 nm was measured. The system was calibrated with 0 to 2 mM NH4Cl solutions.
Statistical analyses.
Data sets were statistically analyzed either by t test or by analysis of variance (ANOVA), followed by a Tukey HSD (honestly significant difference) post hoc test using the R package (60) and the multcompView package therein (61).
CLSM.
For fluorescence microscopy, the cells were cultivated as described above. Confocal laser-scanning microscopy (CLSM) was performed using an upright TCS SP5 MP (Leica Microsystems, Germany). GFP was excited with the 488-nm line of the argon laser. Emitted light with a wavelength between 500 to 550 nm was detected.
Nucleotide sequence accession numbers.
Nucleotide sequences have been deposited in GenBank under the following accession numbers: GpAMT1, JX535577; GpAMT2, JX535578; and GpAMT3, JX535579.
RESULTS
Three ammonium transporter-like protein-coding transcripts are represented in the symbiotic transcriptome of Geosiphon pyriformis.
A cDNA library of symbiotic material (the so-called symbiotic bladders and young hyphae connected to these) of G. pyriformis (42) was screened for expressed genes with the potential to code for ammonium transporters (AMTs). By functional complementation of yeast mutants or degenerate PCR, two expressed genes sharing high similarity with the glomeromycotan AMTs GiAMT1 and GiAMT2 were identified (Fig. 1A; see also Fig. S1 in the supplemental material). A third gene was discovered after 454FLX-Titanium sequencing of the cDNA library (A. Schüßler, unpublished data).
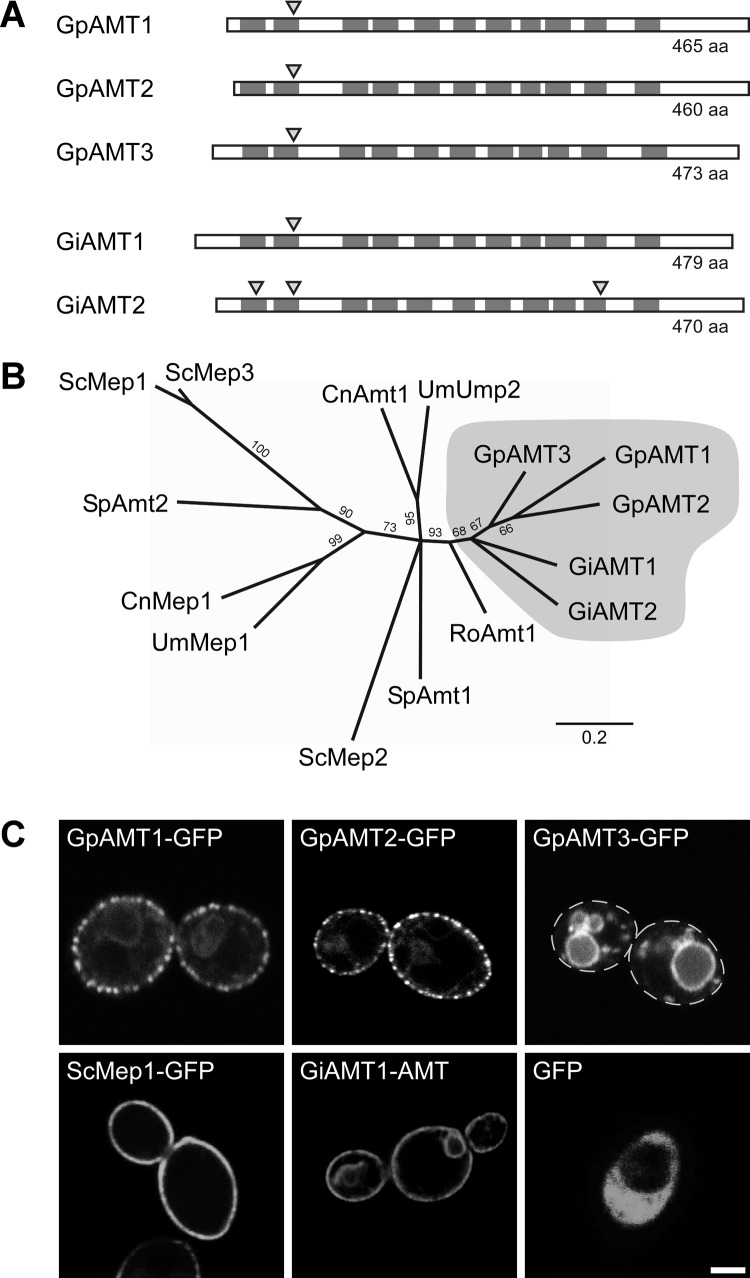
Fig 1.
Characteristics of Geosiphon pyriformis ammonium transporters (AMTs). (A) Transmembrane domain (TMD) topology and intron localization in comparison with those of the Rhizophagus irregularis ammonium transporters GiAMT1 (CAI54276) and GiAMT2 (CAX32490). Dark gray boxes indicate the positions of TMDs, and triangles mark intron positions. TMD positions and the position of one intron are highly conserved, while N and C termini differ in length and are less conserved. Schematic is to scale. aa, amino acids. (B) Unrooted phylogenetic tree computed by RAxML maximum-likelihood analysis (1,000 bootstraps) of selected representative fungal ammonium transporters. Branches with bootstrap support below 60% were collapsed to polytomies. Sequences with the following accession numbers were obtained from the GenBank database: Schizosaccharomyces pombe SpAmt1, NP_588424, and SpAmt2, XP_002175812; Saccharomyces cerevisiae ScMep1, P40260, ScMep2, P41948, and ScMep3, P53390; Ustilago maydis UmMep1, AAL08424, and UmUmp2, AAO42611; Cryptococcus neoformans CnMep1, XP_566614, and CnAmt1, XP_567361; and Rhizopus oryzae RoAmt1, EIE87911. Bootstrap support is indicated at the branches, scale bar indicates substitutions per site, and gray shading marks glomeromycotan AMTs. (C) Heterologous expression of soluble GFP or GFP-tagged ammonium transporter proteins in the ammonium transporter-deficient S. cerevisiae mutant strain MLY131a/α. Confocal laser-scanning microscopy (CLSM) results are shown; dashed lines in GpAMT3-GFP image indicate cell borders. Bar, 2 μm.
All three putative gene products share the highly conserved characteristics of AMTs. They consist of 464 (GpAMT1), 459 (GpAMT2), and 472 (GpAMT3) amino acids, respectively, show the AMT-typical localization of the 11 transmembrane domains (Fig. 1A; see also Fig. S2 in the supplemental material) predicted by TMHMM (51, 62) and SOSUI (63), and possess all 14 conserved and functionally important amino acid residues supposed to form the pore of AMTs (see Fig. S2) (28, 64–68). The coding sequences (CDS) of all three genes have a low GC content of approximately 40%, which is comparable to the mean of 38% among previously identified coding regions from R. irregularis.
Phylogenetic analysis of more than 340 AMT sequences (69) revealed a distinct clade formed by the five glomeromycotan AMTs, while ascomycotan and basidiomycotan AMTs divide into subgroups. The analysis of a subset of sequences is shown in Fig. 1B. The branching order of the R. irregularis and G. pyriformis AMTs cannot yet be convincingly resolved. The three Geosiphon transporters appear monophyletic but with low support. A monophyletic relation of GiAMT1 and GiAMT2 is even less supported (bootstrap support, <60%). The common phylogenetic origin of all five glomeromycotan AMT genes is reflected by a conserved intron (Fig. 1A) in the respective genomic loci, positioned between triplets coding for two conserved amino acids (a glutamine and a tryptophan; see Fig. S2 in the supplemental material).
GpAMT1, GpAMT2, and GpAMT3 subcellular localization in Saccharomyces cerevisiae.
For functional characterization, an ammonium transport-deficient yeast mutant was transformed using plasmids for the expression of either soluble GFP or GFP-tagged versions of GpAMT1, GpAMT2, or GpAMT3. GFP-tagged versions of the functional ammonium transporters ScMep1 and GiAMT1 served as positive controls. Confocal laser-scanning microscopy (CLSM) revealed that soluble GFP localized to the yeast cytoplasm, while the GFP-tagged transporters GpAMT1 and GpAMT2 localized to the yeast plasma membrane (Fig. 1C). GpAMT3 unexpectedly localized to the vacuolar membrane (Fig. 1C). The plasma membrane localization of GpAMT1-GFP and GpAMT2-GFP was in distinct patches, which has been reported before for many membrane proteins expressed in yeast (70, 71). GFP fusion proteins of ammonium-transporting and plasma membrane-located AMTs were functional, as their expression complemented the yeast mutant (see Fig. S3 in the supplemental material).
GpAMT1 and GpAMT2 do not efficiently transport methylammonium.
To quantify the substrate affinity and transport capacity of the plasma membrane-localized ammonium transporters GpAMT1 and GpAMT2, we performed yeast methylammonium (MA) uptake assays with radiolabeled [14C]MA. As negative controls, we included empty-vector-transformed yeast cells, and as positive controls, ScMep1- and GiAMT1-expressing cells. Both positive controls showed MA uptake in the expected ranges but with slightly higher Km values than in published data (Fig. 2A). Surprisingly, both GpAMT1- and GpAMT2-expressing cells showed no MA uptake (Fig. 2A). Such a lack of MA transport had been reported in fungi only for one AMT that is, interestingly, from a member of the Glomeromycota (36), and for the structurally different AMT-like protein MepC of Aspergillus nidulans (72).
Fig 2.
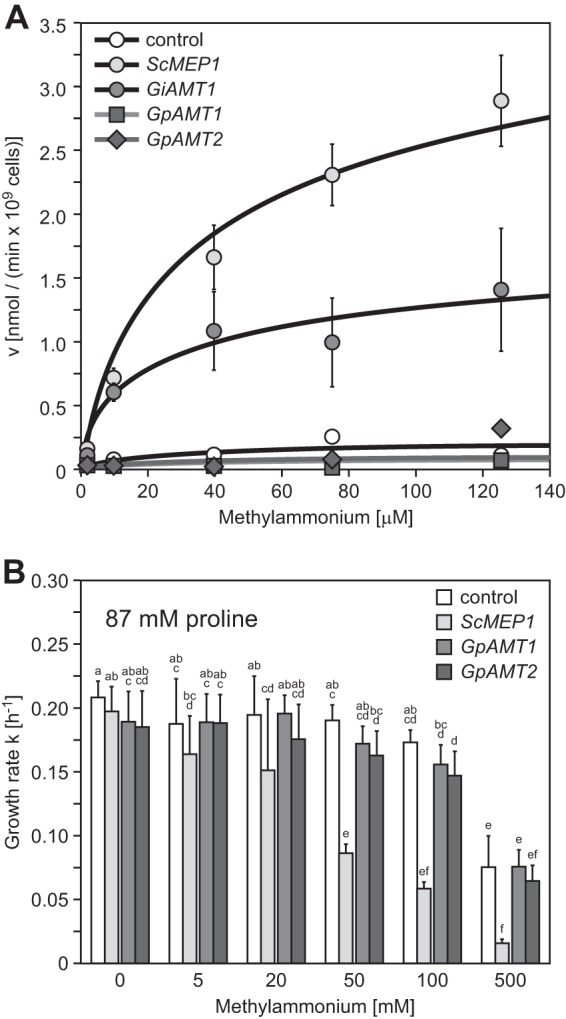
Methylammonium transport of yeast strain MLY131a/α (mep1-3Δ) transformed with Geosiphon pyriformis AMTs. (A) Concentration-dependent [14C]methylammonium uptake of yeast cells transformed with empty vector (negative control), ScMEP1 (positive control), GiAMT1, GpAMT1, or GpAMT2. ScMep1, Km = 40.0 μM, Vmax = 3.5 nmol min−1 for 109 cells; GiAMT1, Km = 11.1 μM, Vmax = 1.3 nmol min−1 for 109 cells. Error bars, ±standard deviation; n = 3 to 6 biological replicates. (B) Growth of the same transformants in buffered liquid medium containing 0.1% proline as sole nitrogen source and different concentrations of cytotoxic methylammonium. Error bars, ±standard deviation; n = 6 biological replicates. Different letters above bars indicate highly significant differences (P ≤ 0.001, ANOVA test) between pairwise comparisons.
This result was verified by an MA toxicity experiment; MA taken up by yeast cells is toxic and leads to growth reduction. Yeast cells expressing either GpAMT1 or GpAMT2, as well as the vector control, did not show a significant growth reduction in liquid medium up to a certain threshold concentration of MA, while ScMep1-expressing cells showed a clear growth reduction already at much lower concentrations (Fig. 2B). The growth difference in the presence of toxic MA was independent of pH at pHs of <7, while at pH 7, passive methylamine influx (73) limited the growth of all strains (see Fig. S4A in the supplemental material). The medium pH itself had no effect on growth in the investigated range of pH 3 to pH 7 (see Fig. S4B in the supplemental material), which excludes the possibility that the AMTs had different pH optima for MA transport.
A growth assay on MA-containing plates gave similar results: ScMep1-expressing (positive-control) cells showed a severe growth defect in comparison to the growth of the vector control (see Fig. S4C in the supplemental material). In this assay, the GpAMT1- and GpAMT2-expressing cells exhibited only slightly reduced growth rates, suggesting that minimal amounts of MA might be transported after the 5-day prolonged incubation (see Fig. S4C in the supplemental material). Thus, neither GpAMT1 nor GpAMT2 is able to transport MA across the yeast plasma membrane in substantial amounts.
GpAMT1 but not GpAMT2 or GpAMT3 is a functional plasma membrane AMT in Saccharomyces cerevisiae.
GpAMT1 was discovered as a functional ammonium transporter in an S. cerevisiae complementation screen with the G. pyriformis cDNA library. The transport efficiency of this protein across the yeast plasma membrane is significantly lower than that of ScMep1, an endogenous AMT of S. cerevisiae, as is evident by lower growth rates (Fig. 3A and B). Since GpAMT2 was isolated in a degenerate PCR approach and GpAMT3 after partial 454 pyrosequencing of the cDNA library, both were also tested for functionality in S. cerevisiae by transformation into the same ammonium uptake-deficient yeast mutant that was used for functional complementation.
The expression of GpAMT2 or GpAMT3 did not provide growth of the ammonium transport-deficient yeast mutant above the background level (Fig. 3A). While no ammonium transport across the plasma membrane was expected for GpAMT3, since it is localized to the vacuolar membrane in S. cerevisiae, the lack of ammonium transport capability of plasma membrane-localized GpAMT2 was surprising. As AMTs may display different pH optima (31), we determined the growth rate k (see Materials and Methods) of yeast cells expressing GpAMT1, GpAMT2, or the endogenous AMT ScMep1 by growing the cells in buffered liquid culture medium at pH values from 4 to 7 during exponential growth (Fig. 3B). Under these assay conditions, neither GpAMT1 nor GpAMT2 expression led to growth above the background rate. The pH-dependent growth enhancement due to the passive influx of larger amounts of uncharged NH3 with higher pH, which diminishes the growth benefit by ScMep1 expression (Fig. 3B), was evident.
For confirmation of the ammonium transport capacities of the G. pyriformis AMTs, ammonium removal assays (59) were performed. Dense yeast cultures (OD600 of 2) were incubated in relatively high ammonium concentrations (1 mM) for several hours, and the ammonium content in the medium was recorded over time. This assay allows estimation of the overall ammonium transport, and the results confirmed those of the previous yeast complementation assays. GpAMT1 is a functional ammonium transporter but does not transport methylammonium. Neither GpAMT2 nor GpAMT3 was able to remove ammonium from the medium (Fig. 4).
Fig 4.
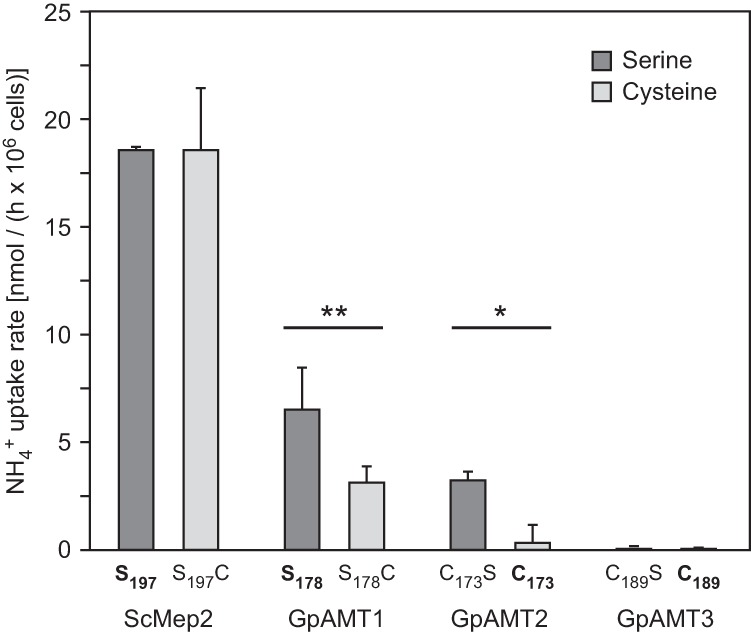
Influence of mutation of a conserved serine residue on ammonium uptake rate of yeast strain MLY131a/α (mep1-3Δ) expressing yeast (control) or Geosiphon pyriformis AMTs. The respective wild-type constructs are indicated in boldface. Serine-to-cysteine mutation of GpAMT1 reduced its transport capacity, while cysteine-to-serine mutation of GpAMT2 increased its transport capacity; as expected, no effect of cysteine-to-serine mutation was detected for GpAMT3, which is located at the vacuole membrane. Uptake rates were determined by an ammonium removal assay in the presence of 1 mM NH4+. Error bars, ±standard deviation; n = 3 biological replicates. Statistical differences were calculated using Student's t test (*, P ≤ 0.05; **, P ≤ 0.01). No significant differences were found between ScMEP2 S197 and S197C or GpAMT3 C189S and C189 strains.
Ammonium transport capability of GpAMT2 is dependent on a single amino acid exchange.
Comparison of the GpAMT2 and GpAMT3 amino acid sequences with the sequences of other fungal AMTs revealed the presence of all amino acids reported to be directly involved in the ammonium transport process (see Fig. S2 in the supplemental material). However, in the AMT signature (ATS, Prosite entry PS01219) in transmembrane domain V (74), an amino acid difference was detected. A position usually occupied by a serine, alanine, or glycine amino acid is replaced by a cysteine in GpAMT2 and GpAMT3 (see Fig. S2 in the supplemental material). This residue lies in an α-helical region in proximity to one of the histidines supposed to play a crucial role in ammonium conduction through the pore (68). We hypothesized that the cysteine might influence the proton-accepting capability of the respective histidine (see Fig. S5 in the supplemental material) and tested this by mutation of C173 to serine (C173S) in GpAMT2; reciprocally, S178 in GpAMT1 was changed to cysteine (S178C). Ammonium removal assays revealed that the C173S mutation made GpAMT2 competent for ammonium transport, while the ammonium transport capability of the mutated GpAMT1 S178C was reduced (Fig. 4). As controls, a C189S mutant of GpAMT3 and an S197C version of ScMep2 were used. The GpAMT3 C189S mutant did not show ammonium transport, which was expected because of its vacuolar localization in S. cerevisiae. The S197C version of ScMep2, an S. cerevisiae ammonium permease also known for its function in signaling during nutrient limitation (38, 75, 76), did not affect ammonium transport capability in ammonium removal assays. When tested for invasive pseudohyphal growth, the morphological reaction in response to nitrogen starvation, both the wild-type and the mutated version of ScMep2 were equally able to mediate this signal (see Fig. S6 in the supplemental material).
DISCUSSION
Geosiphon pyriformis ammonium transporters show distinct characteristics.
We present three AMT (-like) genes present in the symbiotic transcriptome of G. pyriformis. Neither by functional yeast complementation nor by degenerate PCR were additional AMT genes found. In the 454 sequencing data from the cDNA library, which were low coverage only (approximately 90,000 individual reads from a one-quarter 454FLX-Titanium run; A. Schüßler, unpublished data), three hits for GpAMT2 and one hit for GpAMT3 were found. GpAMT1 was not found in this data set. In a recent higher-coverage sequencing of the cDNA library (approximately 17 million individual sequences from an Illumina MiSeq run; A. Brachmann, unpublished data) 59, 106, and 77 hits were found for GpAMT1, GpAMT2, and GpAMT3, respectively. All three genes are thus expressed at comparable levels in the functional symbiosis with cyanobacteria.
The Amt/Mep/Rh protein family consists of homologous membrane proteins with 450 to 500 amino acids and 10 to 12 (mostly 11) transmembrane domains. A typical ammonium transporter signature (74) and 14 amino acid residues that are supposed to be crucial in the formation of the hydrophilic pore and in the facilitation of ammonium transport in the E. coli AMT AmtB (EcAmtB) have been reported (28, 32, 77). All three G. pyriformis AMT protein sequences possess these characteristics of typical AMTs (see Fig. S2 in the supplemental material).
AMTs are usually functionally characterized by measuring the kinetics and capacity for uptake of radiolabeled methylammonium (MA), in most cases in the heterologous yeast system that was also used in this study. The first cloned and characterized fungal AMT, ScMep1 of S. cerevisiae (29), was named methylammonium permease (MEP) based on its ability to transport MA, and a few years later, ScMep2 and ScMep3 were discovered in mutagenesis screens for MA-resistant yeast mutants (37). Outside the fungal kingdom, two plant AMTs, from Arabidopsis thaliana (78, 79) and Lotus japonicus (80), and the mammalian Rh proteins, which are low-affinity AMTs (28), were reported to not transport MA, demonstrating that MA transport is not a universal feature of AMTs. Until recently, MepC of A. nidulans, which is only distantly related (31% to 35% identity) to the other AMTs of A. nidulans (72), was the only fungal AMT lacking MA transport capability. The second one, GiAMT2, was recently described from an AMF (36). Thus, interestingly, of the three glomeromycotan AMTs functionally active in yeast (GiAMT1, GiAMT2, and GpAMT1), only one (GiAMT1) transports MA. This casts the classical approach of using MA transport for the biochemical characterization of AMTs into doubt, as it becomes clear that ammonium transport activity may not be directly related to MA transport capability.
All members of the Amt/Mep/Rh family studied until now, including the previously characterized glomeromycotan AMTs GiAMT1 and GiAMT2, are plasma membrane-localized functional AMTs. Although the five glomeromycotan AMTs are highly similar on both the amino acid and DNA level (see Fig. S1 and S2 in the supplemental material), very distinct ammonium and MA transport characteristics were uncovered for all three G. pyriformis AMTs in this study. Only GpAMT1 was able to complement the ammonium uptake-deficient yeast mutant; GpAMT2 and GpAMT3 were not. For GpAMT3, this could easily be explained by its unexpected vacuolar localization. However, GpAMT2 localizes to the plasma membrane but appears not to transport ammonium into the cell. It is known that under certain conditions, some AMTs may transport ammonium bidirectionally (81), although these transporters are nowadays thought to be gas channels for NH3 (28). For GpAMT2, we can exclude this possibility, because when grown under slightly acidic to neutral pH conditions, which lead to higher growth rates of the yeast mutant due to enhanced passive influx of NH3, complementation with GpAMT2 did not enhance growth (Fig. 3B).
Although GpAMT2 did not functionally complement the ammonium uptake-deficient yeast mutant, the possibility that it is a functional AMT in the homologous system cannot be excluded. Its transport activity could be dependent on oligomerization or interaction with membrane lipids or proteins. Alternatively, GpAMT2 could be an ammonium sensor, which occur in the Amt/Mep/Rh family. In yeast, ScMep2 is involved in the nutrient limitation-triggered morphological switch to pseudohyphal growth (75). However, sensor function is usually mediated by functional (ammonium-transporting) AMTs and GpAMT2 was not able to complement for pseudohyphal growth when expressed in ammonium transporter-deficient yeast cells.
However, a single exchange of an amino acid that was identified as a nonconserved cysteine located in close proximity to the two histidine residues (see Fig. S5 in the supplemental material) that are involved in ammonium conductance through the pore (68) was sufficient to turn GpAMT2 into a functional AMT in yeast. The corresponding inverse mutation in GpAMT1 decreased its transport capacity. This adds this amino acid position to the list of residues already known to directly influence ammonium transport. In addition, it points to the interesting fact that GpAMT2 has retained all features of a functional AMT. The hydrophilic pore seems to be present, but ammonium transport in yeast is inhibited by C173. On the other hand, the corresponding inverse mutation in the endogenous yeast transporter ScMep2 had no negative effect, indicating varying impact of the serine residue on transport capacity due to different transport mechanisms or due to differences in the structure of the hydrophilic pore. This makes it conceivable to speculate that, in the homologous system, GpAMT2 could have a slightly changed structural topology and be a functional AMT.
Potential biological roles of the newly characterized AMTs.
Geosiphon pyriformis forms a symbiosis with Nostoc punctiforme cyanobacteria that form heterocysts (also in the symbiosis) and are capable of atmospheric N2 fixation (59). The three AMTs characterized here should play a role in ammonium uptake from the environment or from the symbiosome space, where ammonium might be released by the cyanobacterial symbiont. GpAMT3 could be responsible for the transport of ammonium into or from the vacuole (or both) in the homologous system. Due to its vacuolar localization in yeast, GpAMT3 was not further characterized by the approaches used in this study. However, its localization is highly interesting. In the AM symbiosis, large amounts of arginine are cleaved in the arbuscules within the plant root cells to provide the plant with ammonium (26). The expression of the respective arginine anabolism genes was also identified in the symbiotic-bladder cDNA library (A. Schüßler, unpublished data), indicating that similar mechanisms also exist in Geosiphon. GpAMT3-like transporter types could therefore be responsible for ammonium release from vacuoles or for pumping ammonium into vacuoles, potentially regulating ammonium release and ammonium levels in the cytoplasm.
The vacuolar localization of GpAMT3 in yeast is also interesting with regard to the fact that no fungal vacuolar AMTs are known. To our knowledge, only for Dictyostelium discoideum were vacuolar AMTs characterized. AmtB is specifically located at the contractile vacuole and exports excess ammonium (82). AmtA localizes to stalk cell vacuoles and may play a role in the control of stalk cell differentiation (83). However, it is unknown whether these transporters would localize to the vacuolar membrane in the yeast system. One should also note that the contractile vacuole is not a typical vacuolar membrane, and it is partly continuous with the plasma membrane. Similar characteristics are present for the perisymbiotic membrane in the Geosiphon symbiosis, which is derived from an invaginated plasma membrane and homologous to the arbuscular plasma membrane. Likely it has a very specific protein complement that is different from that of the “normal” hyphal plasma membrane. The role of GpAMT3 might be the export of ammonium from the fungal cytoplasm, perhaps to deliver ammonium to the photoautotrophic symbiosis partners in glomeromycotan symbioses or for vacuolar ammonium storage and release, perhaps also related to detoxification of high ammonium concentrations. It will be very interesting to study such questions in the future.
Outlook.
In the yeast system, GpAMT1 was shown to represent a plasma membrane AMT not transporting MA, GpAMT2 to be a plasma membrane transporter transporting neither ammonium nor MA, and GpAMT3 to be a vacuolar membrane protein with yet-unknown transport characteristics. Thus, all three AMTs possess very distinct properties, and future studies should uncover potential orthologous genes and proteins in other AM-forming glomeromycotan fungi. Moreover, a characterization in the homologous Geosiphon symbiosis could be performed by analysis of protein expression after injection of synthetic nucleic acids into the symbiotic bladders, although this interesting symbiotic model system is currently not cultivated in the laboratory. However, a revival of studies on Geosiphon using modern methods would surely allow fundamental new insights into this interesting system and also into AM.
Besides P, N is a very important and often limiting nutrient that is transported by AMF from the soil to the plant host. Recent studies on a Medicago truncatula mutant lacking the mycorrhiza-specific phosphate transporter M. truncatula PT4 (MtPT4) revealed that N transport alone might be sufficient for the establishment of functional AM (84). This result provides further evidence for the strict regulation of nutrient exchange between AM symbiotic partners in order to preserve an evolutionarily stable mutualism. To get deeper insights into the underlying processes, more-detailed studies appear necessary. The identification and characterization of additional glomeromycotan AMTs and their localization in the AM symbiosis will be steps to reveal the glomeromycotan AMT complement and specific functions and, eventually, will help to understand the globally significant symbiotic N fluxes in the AM.
Supplementary Material
ACKNOWLEDGMENTS
We thank Giuliana Lott for the generation of transporter mutant constructs. We acknowledge the continuous support of Martin Parniske, head of Genetics at the LMU Biocenters.
This work was supported by a fellowship to M.E. of the Universität Bayern e.V.
We are not aware of any conflicts of interest.
Footnotes
Published ahead of print 20 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00139-13.
REFERENCES
- 1. Smith SE, Read DJ. 2008. Mycorrhizal symbiosis, 3rd ed. Elsevier, New York, NY [Google Scholar]
- 2. Genre A, Chabaud M, Faccio A, Barker D, Bonfante P. 2008. Prepenetration apparatus assembly precedes and predicts the colonization patterns of arbuscular mycorrhizal fungi within the root cortex of both Medicago truncatula and Daucus carota. Plant Cell 20:1407–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brachmann A, Parniske M. 2006. The most widespread symbiosis on Earth. PLoS Biol. 4:e239. 10.1371/journal.pbio.0040239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bougoure J, Ludwig M, Brundrett M, Grierson P. 2009. Identity and specificity of the fungi forming mycorrhizas with the rare mycoheterotrophic orchid Rhizanthella gardneri. Mycol. Res. 113:1097–1106 [DOI] [PubMed] [Google Scholar]
- 5. Schüßler A, Schwarzott D, Walker C. 2002. A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycol. Res. 105:1413–1421 [Google Scholar]
- 6. Dotzler N, Walker C, Krings M, Hass H, Kerp H, Taylor TN, Agerer R. 2009. Acaulosporoid glomeromycotan spores with a germination shield from the 400-million-year-old Rhynie chert. Mycol. Prog. 8:9–18 [Google Scholar]
- 7. Remy W, Taylor T, Hass H, Kerp H. 1994. Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc. Natl. Acad. Sci. U. S. A. 91:11841–11843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Redecker D, Kodner R, Graham LE. 2000. Glomalean fungi from the Ordovician. Science 289:1920–1921 [DOI] [PubMed] [Google Scholar]
- 9. Schüßler A, Walker C. 2011. Evolution of the ‘plant-symbiotic' fungal phylum, Glomeromycota, p 163–185 In Pöggeler S, Wöstemeyer J. (ed), The Mycota XIV—evolution of fungi and fungal-like organisms. Springer, Heidelberg, Germany [Google Scholar]
- 10. Bago B, Pfeffer P, Abubaker J, Jun J, Allen J, Brouillette J, Douds D, Lammers P, Shachar-Hill Y. 2003. Carbon export from arbuscular mycorrhizal roots involves the translocation of carbohydrate as well as lipid. Plant Physiol. 131:1496–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Finlay R. 2008. Ecological aspects of mycorrhizal symbiosis: with special emphasis on the functional diversity of interactions involving the extraradical mycelium. J. Exp. Bot. 59:1115–1126 [DOI] [PubMed] [Google Scholar]
- 12. Govindarajulu M, Pfeffer P, Jin H, Abubaker J, Douds D, Allen J, Bücking H, Lammers P, Shachar-Hill Y. 2005. Nitrogen transfer in the arbuscular mycorrhizal symbiosis. Nature 435:819–823 [DOI] [PubMed] [Google Scholar]
- 13. Toussaint J, St-Arnaud M, Charest C. 2004. Nitrogen transfer and assimilation between the arbuscular mycorrhizal fungus Glomus intraradices Schenck & Smith and Ri T-DNA roots of Daucus carota L. in an in vitro compartmented system. Can. J. Microbiol. 50:251–260 [DOI] [PubMed] [Google Scholar]
- 14. Tanaka Y, Yano K. 2005. Nitrogen delivery to maize via mycorrhizal hyphae depends on the form of N supplied. Plant Cell Environ. 28:1247–1254 [Google Scholar]
- 15. Tobar RM, Azcón R, Barea JM. 1994. The improvement of plant N acquisition from an ammonium-treated, drought-stressed soil by the fungal symbiont in arbuscular mycorrhizae. Mycorrhiza 4:105–108 [Google Scholar]
- 16. Johansen A, Jakobsen I, Jensen ES. 1992. Hyphal transport of 15N-labelled nitrogen by a vesicular-arbuscular mycorrhizal fungus and its effect on depletion of inorganic soil N. New Phytol. 122:281–288 [DOI] [PubMed] [Google Scholar]
- 17. Johansen A, Jakobsen I, Jensen ES. 1993. External hyphae of vesicular-arbuscular mycorrhizal fungi associated with Trifolium subterraneum L. New Phytol. 124:61–68 [Google Scholar]
- 18. Fellbaum CR, Gachomo EW, Beesetty Y, Choudhari S, Strahan GD, Pfeffer PE, Kiers ET, Bucking H. 2012. Carbon availability triggers fungal nitrogen uptake and transport in arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. U. S. A. 109:2666–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hawkins H-J, Johansen A, George E. 2000. Uptake and transport of organic and inorganic nitrogen by arbuscular mycorrhizal fungi. Plant Soil 226:275–285 [Google Scholar]
- 20. Villegas J, Williams R, Nantais L, Archambault J, Fortin J. 1996. Effects of N source on pH and nutrient exchange of extramatrical mycelium in a mycorrhizal Ri T-DNA transformed root system. Mycorrhiza 6:247–251 [Google Scholar]
- 21. Jin H, Pfeffer P, Douds D, Piotrowski E, Lammers P, Shachar-Hill Y. 2005. The uptake, metabolism, transport and transfer of nitrogen in an arbuscular mycorrhizal symbiosis. New Phytol. 168:687–696 [DOI] [PubMed] [Google Scholar]
- 22. Cruz C, Egsgaard H, Trujillo C, Ambus P, Requena N, Martins-Loução M, Jakobsen I. 2007. Enzymatic evidence for the key role of arginine in nitrogen translocation by arbuscular mycorrhizal fungi. Plant Physiol. 144:782–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Johansen A, Finlay R, Olsson P. 1996. Nitrogen metabolism of external hyphae of the arbuscular mycorrhizal fungus Glomus intraradices. New Phytol. 133:705–712 [Google Scholar]
- 24. Bago B, Pfeffer P, Shachar-Hill Y. 2001. Could the urea cycle be translocating nitrogen in the arbuscular mycorrhizal symbiosis? New Phytol. 149:4–8 [DOI] [PubMed] [Google Scholar]
- 25. Guether M, Neuhauser B, Balestrini R, Dynowski M, Ludewig U, Bonfante P. 2009. A mycorrhizal-specific ammonium transporter from Lotus japonicus acquires nitrogen released by arbuscular mycorrhizal fungi. Plant Physiol. 150:73–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tian C, Kasiborski B, Koul R, Lammers PJ, Bucking H, Shachar-Hill Y. 2010. Regulation of the nitrogen transfer pathway in the arbuscular mycorrhizal symbiosis: gene characterization and the coordination of expression with nitrogen flux. Plant Physiol. 153:1175–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gomez SK, Javot H, Deewatthanawong P, Torres-Jerez I, Tang Y, Blancaflor EB, Udvardi MK, Harrison MJ. 2009. Medicago truncatula and Glomus intraradices gene expression in cortical cells harboring arbuscules in the arbuscular mycorrhizal symbiosis. BMC Plant Biol. 9:10. 10.1186/1471-2229-9-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Andrade S, Einsle O. 2007. The Amt/Mep/Rh family of ammonium transport proteins. Mol. Membrane Biol. 24:357–365 [DOI] [PubMed] [Google Scholar]
- 29. Marini A, Vissers S, Urrestarazu A, André B. 1994. Cloning and expression of the MEP1 gene encoding an ammonium transporter in Saccharomyces cerevisiae. EMBO J. 13:3456–3463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ninnemann O, Jauniaux JC, Frommer WB. 1994. Identification of a high affinity NH4+ transporter from plants. EMBO J. 13:3464–3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Boeckstaens M, André B, Marini A. 2008. Distinct transport mechanisms in yeast ammonium transport/sensor proteins of the Mep/Amt/Rh family and impact on filamentation. J. Biol. Chem. 283:21362–21370 [DOI] [PubMed] [Google Scholar]
- 32. Lamoureux G, Javelle A, Baday S, Wang S, Berneche S. 2010. Transport mechanisms in the ammonium transporter family. Transfus. Clin. Biol. 17:168–175 [DOI] [PubMed] [Google Scholar]
- 33. Schüßler A, Walker C. 2010. The Glomeromycota: a species list with new families and genera. Schüßler A, Walker C, Gloucester, United Kingdom: Available at www.amf-phylogeny.com [Google Scholar]
- 34. Stockinger H, Walker C, Schussler A. 2009. ‘Glomus intraradices DAOM197198′, a model fungus in arbuscular mycorrhiza research, is not Glomus intraradices. New Phytol. 183:1176–1187 [DOI] [PubMed] [Google Scholar]
- 35. López-Pedrosa A, González-Guerrero M, Valderas A, Azcón-Aguilar C, Ferrol N. 2006. GiAMT1 encodes a functional high-affinity ammonium transporter that is expressed in the extraradical mycelium of Glomus intraradices. Fungal Gen. Biol. 43:102–110 [DOI] [PubMed] [Google Scholar]
- 36. Perez-Tienda J, Testillano PS, Balestrini R, Fiorilli V, Azcon-Aguilar C, Ferrol N. 2011. GiAMT2, a new member of the ammonium transporter family in the arbuscular mycorrhizal fungus Glomus intraradices. Fungal Gen. Biol. 48:1044–1055 [DOI] [PubMed] [Google Scholar]
- 37. Marini A, Soussi-Boudekou S, Vissers S, Andre B. 1997. A family of ammonium transporters in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:4282–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lorenz MC, Heitman J. 1998. The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 17:1236–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Helber N, Requena N. 2008. Expression of the fluorescence markers DsRed and GFP fused to a nuclear localization signal in the arbuscular mycorrhizal fungus Glomus intraradices. New Phytol. 177:537–548 [DOI] [PubMed] [Google Scholar]
- 40. Bécard GP, Pfeffer PE. 1993. Status of nuclear division in arbuscular mycorrhizal fungi during in vitro development. Protoplasma 174:62–68 [Google Scholar]
- 41. Schüßler A. 2012. The Geosiphon-Nostoc endosymbiosis and its role as a model for arbuscular mycorrhiza research, p 77–91 In Hook B. (ed), The Mycota IX—fungal associations. Springer-Verlag, Heidelberg, Germany [Google Scholar]
- 42. Schüßler A, Martin H, Cohen D, Fitz M, Wipf D. 2006. Characterization of a carbohydrate transporter from symbiotic glomeromycotan fungi. Nature 444:933–936 [DOI] [PubMed] [Google Scholar]
- 43. Amberg DC, Burke DJ, Strathern JN. 2005. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 44. Gietz RD, Schiestl RH. 2007. Large-scale high-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2:38–41 [DOI] [PubMed] [Google Scholar]
- 45. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 46. Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 48. Rose T, Henikoff J, Henikoff S. 2003. CODEHOP (COnsensus-DEgenerate Hybrid Oligonucleotide Primer) PCR primer design. Nucleic Acids Res. 31:3763–3766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rose T, Schultz E, Henikoff J, Pietrokovski S, McCallum C, Henikoff S. 1998. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 26:1628–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Brachmann A, Weinzierl G, Kamper J, Kahmann R. 2001. Identification of genes in the bW/bE regulatory cascade in Ustilago maydis. Mol. Microbiol. 42:1047–1063 [DOI] [PubMed] [Google Scholar]
- 51. Sonnhammer E, von Heijne G, Krogh A. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6:175–182 [PubMed] [Google Scholar]
- 52. Hirokawa T, Boon-Chieng S, Mitaku S. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378–379 [DOI] [PubMed] [Google Scholar]
- 53. Katoh K, Kuma K, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33:511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jones D, Taylor W, Thornton J. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8:275–282 [DOI] [PubMed] [Google Scholar]
- 55. Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML Web-Servers. Syst. Biol. 75:758–771 [DOI] [PubMed] [Google Scholar]
- 56. Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201 [DOI] [PubMed] [Google Scholar]
- 57. Guex N, Peitsch M. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723 [DOI] [PubMed] [Google Scholar]
- 58. Jacobs P, Jauniaux J, Grenson M. 1980. A cis-dominant regulatory mutation linked to the argB-argC gene cluster in Saccharomyces cerevisiae. J. Mol. Biol. 139:691–704 [DOI] [PubMed] [Google Scholar]
- 59. Neuhäuser B, Dunkel N, Satheesh SV, Morschhauser J. 2011. Role of the Npr1 kinase in ammonium transport and signaling by the ammonium permease Mep2 in Candida albicans. Eukaryot. Cell 10:332–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.R Core Team 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing. http://www.R-project.org/
- 61. Graves S, Piepho H-P, Selzer L, Dorai-Raj S. 2012. multcompView: visualizations of paired comparisons. R package version 0.1-5. http://CRAN.R-project.org/package=multcompView
- 62. Krogh A, Larsson B, von Heijne G, Sonnhammer E. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580 [DOI] [PubMed] [Google Scholar]
- 63. Mitaku S, Hirokawa T, Tsuji T. 2002. Amphiphilicity index of polar amino acids as an aid in the characterization of amino acid preference at membrane-water interfaces. Bioinformatics 18:608–616 [DOI] [PubMed] [Google Scholar]
- 64. Zheng L, Kostrewa D, Berneche S, Winkler FK, Li XD. 2004. The mechanism of ammonia transport based on the crystal structure of AmtB of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 101:17090–17095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Khademi S, O'Connell J, III, Remis J, Robles-Colmenares Y, Miercke LJ, Stroud RM. 2004. Mechanism of ammonia transport by Amt/MEP/Rh: structure of AmtB at 1.35 A. Science 305:1587–1594 [DOI] [PubMed] [Google Scholar]
- 66. Andrade S, Dickmanns A, Ficner R, Einsle O. 2005. Crystal structure of the archaeal ammonium transporter Amt-1 from Archaeoglobus fulgidus. Proc. Natl. Acad. Sci. U. S. A. 102:14994–14999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Andrade SL, Dickmanns A, Ficner R, Einsle O. 2005. Expression, purification and crystallization of the ammonium transporter Amt-1 from Archaeoglobus fulgidus. Acta Crystallogr Sect F Struct Biol Cryst Commun. 61:861–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wang S, Orabi EA, Baday S, Berneche S, Lamoureux G. 2012. Ammonium transporters achieve charge transfer by fragmenting their substrate. J. Am. Chem. Soc. 134:10419–10427 [DOI] [PubMed] [Google Scholar]
- 69. McDonald TR, Dietrich FS, Lutzoni F. 2012. Multiple horizontal gene transfers of ammonium transporters/ammonia permeases from prokaryotes to eukaryotes: toward a new functional and evolutionary classification. Mol. Biol. Evol. 29:51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Grossmann G, Opekarová M, Malinsky J, Weig-Meckl I, Tanner W. 2007. Membrane potential governs lateral segregation of plasma membrane proteins and lipids in yeast. EMBO J. 26:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Grossmann G, Opekarova M, Novakova L, Stolz J, Tanner W. 2006. Lipid raft-based membrane compartmentation of a plant transport protein expressed in Saccharomyces cerevisiae. Eukaryot. Cell 5:945–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Monahan BJ, Askin MC, Hynes MJ, Davis MA. 2006. Differential expression of Aspergillus nidulans ammonium permease genes is regulated by GATA transcription factor AreA. Eukaryot. Cell 5:226–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Soupene E, Ramirez RM, Kustu S. 2001. Evidence that fungal MEP proteins mediate diffusion of the uncharged species NH(3) across the cytoplasmic membrane. Mol. Cell. Biol. 21:5733–5741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. von Wirén N, Merrick M. 2004. Regulation and function of ammonium carriers in bacteria, fungi, and plants, p 95–120 In Boles E, Kramer R. (ed), Topics in current genetics, vol 9 Molecular mechanisms controlling transmembrane transport. Springer-Verlag, Berlin, Germany [Google Scholar]
- 75. Lorenz MC, Heitman J. 1998. Regulators of pseudohyphal differentiation in Saccharomyces cerevisiae identified through multicopy suppressor analysis in ammonium permease mutant strains. Genetics 150:1443–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Van Nuland A, Vandormael P, Donaton M, Alenquer M, Lourenço A, Quintino E, Versele M, Thevelein JM. 2006. Ammonium permease-based sensing mechanism for rapid ammonium activation of the protein kinase A pathway in yeast. Mol. Microbiol. 59:1485–1505 [DOI] [PubMed] [Google Scholar]
- 77. Pantoja O. 2012. High affinity ammonium transporters: molecular mechanism of action. Front. Plant Sci. 3:34. 10.3389/fpls.2012.00034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sohlenkamp C, Wood C, Roeb G, Udvardi M. 2002. Characterization of Arabidopsis AtAMT2, a high-affinity ammonium transporter of the plasma membrane. Plant Physiol. 130:1788–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sohlenkamp C, Shelden M, Howitt S, Udvardi M. 2000. Characterization of Arabidopsis AtAMT2, a novel ammonium transporter in plants. FEBS Lett. 467:273–278 [DOI] [PubMed] [Google Scholar]
- 80. Simon-Rosin U, Wood C, Udvardi MK. 2003. Molecular and cellular characterisation of LjAMT2;1, an ammonium transporter from the model legume Lotus japonicus. Plant Mol. Biol. 51:99–108 [DOI] [PubMed] [Google Scholar]
- 81. Soupene E, Lee H, Kustu S. 2002. Ammonium/methylammonium transport (Amt) proteins facilitate diffusion of NH3 bidirectionally. Proc. Natl. Acad. Sci. U. S. A. 99:3926–3931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kirsten JH, Xiong Y, Davis CT, Singleton CK. 2008. Subcellular localization of ammonium transporters in Dictyostelium discoideum. BMC Cell Biol. 9:71. 10.1186/1471-2121-9-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Uchikawa T, Yamamoto A, Inouye K. 2011. Origin and function of the stalk-cell vacuole in Dictyostelium. Dev. Biol. 352:48–57 [DOI] [PubMed] [Google Scholar]
- 84. Javot H, Penmetsa RV, Breuillin F, Bhattarai KK, Noar RD, Gomez SK, Zhang Q, Cook DR, Harrison MJ. 2011. Medicago truncatula mtpt4 mutants reveal a role for nitrogen in the regulation of arbuscule degeneration in arbuscular mycorrhizal symbiosis. Plant J. 68:954–965 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.