Abstract
Species of the fungal genus Trichoderma (Hypocreales, Ascomycota) are well-known for their production of various secondary metabolites. Nonribosomal peptides and polyketides represent a major portion of these products. In a recent phylogenomic investigation of Trichoderma polyketide synthase (PKS)-encoding genes, the pks4 from T. reesei was shown to be an orthologue of pigment-forming PKSs involved in synthesis of aurofusarin and bikaverin in Fusarium spp. In this study, we show that deletion of this gene in T. reesei results in loss of green conidial pigmentation and in pigmentation alteration of teleomorph structures. It also has an impact on conidial cell wall stability and the antagonistic abilities of T. reesei against other fungi, including formation of inhibitory metabolites. In addition, deletion of pks4 significantly influences the expression of other PKS-encoding genes of T. reesei. To our knowledge, this is the first indication that a low-molecular-weight pigment-forming PKS is involved in defense, mechanical stability, and stress resistance in fungi.
INTRODUCTION
The economically important genus Trichoderma (Hypocreales, Ascomycota, Dikarya) is well-known for its mycotrophic lifestyle and for the broad range of biotrophic interactions with plants and animals. Moreover, it contains several cosmopolitan species characterized by their outstanding environmental opportunism. These properties have given rise to the use of several species in agriculture as biopesticides and biofertilizers, while T. reesei is utilized for production of bioenergy-related enzymes (1).
The molecular basis for the opportunistic success of Trichoderma is not yet well understood. While there is some evidence for a role of some secreted proteins (2, 3), less is known about a possible role(s) of secondary metabolites. In this respect, Trichoderma spp. are probably best known for production of peptaibols, which are nonribosomal peptides with antimicrobial and plant defense-stimulating activities (4). However, the role of polyketide synthases (PKSs) in Trichoderma ecophysiology is not well studied. Trichoderma spp. polyketides are produced by iterative PKSs, multifunctional enzymes consisting of several active sites capable of catalyzing the fusion of variable numbers of coenzyme A (CoA)-linked acyl monomers, such as acetyl-CoA and malonyl-CoA, into polymers known as polyketides. They can be further grouped into nonreducing (NR) and reducing (RD) PKSs according to their domain organization (5). Recently, Baker et al. (6) used a phylogenomic approach to study the PKS repertoire in T. reesei, T. atroviride, and T. virens, and their findings enabled the putative in silico prediction of some of the respective products. A total of 11 PKS-encoding genes were found in the T. reesei genome, among which 2 occur only in T. reesei and 9 have orthologues in T. virens or/and T. atroviride PKSs (6). pks4 (Trire2:82208, Triat2:79, and Trive2:77826 in T. reesei, T. atroviride, and T. virens, respectively), which encodes an enzyme of the nonreducing type (clade I), has been shown to have orthologues in other fungi, i.e., PKSs associated with synthesis of aurofusarin in Fusarium graminearum (7–9), bikaverin in F. fujikuroi (10, 11), and DHN melanin in Aspergillus spp. (12–19). It was therefore hypothesized that PKS4 would likewise be involved in the production of the characteristic green pigment of Trichoderma (6).
Pigment-forming PKSs are known to have functions beyond providing the color of conidia. For example, DHN-melanin is involved in virulence in Aspergillus spp. (15, 16, 20). In this study, we used a reverse genetic approach to examine the functions of pks4 in the ecophysiology of T. reesei. We found that PKS4 is indeed responsible for the pigmentation of conidia and the nonmelanized structures of fruiting bodies, but its loss of function also impacts the stability of the conidial wall and the antagonistic abilities of T. reesei against other fungi, including formation of inhibitory metabolites. In addition, we demonstrate that deletion of pks4 significantly changes the expression of other PKS-encoding genes of T. reesei. To our knowledge, this is the first indication that low-molecular-weight pigments can be involved in defense, mechanical stability, and stress resistance in fungi.
MATERIALS AND METHODS
Deletion of the pks4 gene in T. reesei.
The pks4 gene was deleted by utilizing a double-joint PCR method as described by Yu et al. (21). Briefly, DNA fragments of 5′- and 3′-flanking regions of pks4 were fused with a hygromycin B (hyg) selection marker. Amplification of the 5′-flanking sequence was done using primers F1 and R3 (CAATGGCCGAATGTTCTAGC and GGAACAAGTTGAGCCAGAGC, respectively), the 3′-flanking region was amplified with primers F4 and R6 (GCAATACACGGTGAGAACGA and TGCGGAGGATCGAGACTATT, respectively), and the hygromycin B sequence was amplified with the primers hygF and hygR (GCTGGAGCTAGTGGAGGTCA and CGGTCGGCATCTACTCTATT, respectively). In a second PCR, the fragments were assembled into a single linear construct (21). The third PCR amplification of the final construct was performed using nested primers F2 and R5 (AGGTACGCATGGAGACAACA and TACACACGCACTCACGCATA, respectively), leaving F1 and R6 available for downstream knockout verification. Similar to previously published methods (22), a protoplast polyethylene glycol-mediated transformation and selection scheme was utilized for introduction of the linear transforming DNA construct and subsequent selection on hygromycin. Albino transformants were selected and examined for double-crossover-mediated pks4 replacement with the hygromycin cassette by PCR amplification using primer pairs F1/R6 and F2/R5.
Verification of the pks4 absence in the genome of T. reesei Δpks4 was tested by a specific quantitative PCR (qPCR) with primers for pks4 (Table 1) and the following amplification protocol: initial denaturation step for 3 min at 95°C, followed by 40 cycles of denaturation for 15 s at 95°C, annealing for 20 s at 54°C, and extension at 72°C for 15 s.
Table 1.
Trichoderma reesei repertoire of PKS-encoding genes, their most closely related orthologs for which products or functions have been identified, and the primers and annealing temperatures used for the expression analysis
| Protein IDa | Nameb | R or NR and clade(s) | No. of amino acids | PKS | Identity (%) | E-value | Accession no.(s) | NCBI Blastp identification | Reference(s) or source | Forward/reverse primers (5′–3′) | Tm (°C) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Tr82208 | PKS4 | NR clade I | 2,146 | Yellow-green conidial pigment polyketide synthase | 47 | 0.0 | ACJ13039 | alb1 from Aspergillus fumigatus | 28 | CCAGCAGACAGATACAAC/AACAGTCCAGGCTCATTA | 54 |
| Tr81964 | PKS8 | NR clade I, II | 1,863 | Uncharacterized protein | 47 | 0.0 | JN257714 | adaA from A. niger or vrtA from Penicillium aethiopicum involved in anthracenone and naphthacenedione production, respectively | 29, 30 | ATAACACTGGCGTCACAT/CAATCAGCACAGCAATCTC | 54.7 |
| Tr105804 | PKS3 | NR clade III | 2,116 | PKS involved in lipid metabolism | 54 | 0.0 | XP_003719468 | PKS16 from Botryotinia fuckeliana | Botrytis cinerea T4 Genome Consortium | CCGTATCTCTGCTGTATC/GTGAACCATCTTGAAGGA | NAc |
| Tr73621 | Singlet, PKS1S | NR clade III | 2,633 | Uncharacterized protein | 37 | 0.0 | EGE04288 | Phenolpthiocerol synthesis (ppsB) from Trichophyton equinum | Trichophyton equinum CBS 127.97 Genome Consortium | AGCATAAGCGGAATACATC/AGCCTGAGAAGAGTTGAT | 50.2 |
| Tr65172 | PKS1 | RD clade I, lovastatin/citrinin diketide | 2,598 | Uncharacterized lovastatin/citrinin-like diketide synthase | 35/40 | 0.0/2E-93 | BAC20566/AAP32477 | MlcB from P. citrinum synthesizes diketide portion of lovastatin/PKS for ochratoxin A from A. ochraceus | 31, 32 | AACATCAATCTCAACATC/ACACATCGGTATAAGTATA | 57 |
| Tr65891 | PKS2 | 2,374 | 37 | 0.0 | ELQ36243 | 6-Methylsalicylic acid synthase from Magnaporthe oryzae | 33 | GGACATATTCAACAGGATTCTC/GGTGGCAACATCTTCAAG | 54 | ||
| Tr60118 | PKS6 | 2,415 | 46 | 0.0 | AAR90259 | Uncharacterized PKS from Cochliobolus heterostrophus | 5 | TCAAGTGGTCTCCTCTATT/AATGTGCTGTCTCAATCC | 54.7 | ||
| Tr106272 | PKS9 | 2,612 | 54 | 0.0 | AAR92209 | Uncharacterized PKS2 from Gibberella moniliformis | 5 | CCGTATCTCTGCTGTATC/ATCGTCTGTGATGAAGTG | 54.7 | ||
| Tr73618 | Singlet, PKS2S | 2,567 | 41/40 | 0.0 | BAC20566/AAD34559 | MlcB from P. citrinum and LovF from A. terreus synthesize diketide portion of lovastatin and citrinin, respectively | 31, 34 | TACCATTACACAGACTTG/AGCAATCACAACATCATA | 50.2 | ||
| Tr59482 | PKS5 | RD clade III, T-toxin | 2,205 | Uncharacterized T-toxin-like synthase | 34 | 0.0 | ABB76806 | PKS1 and PKS2 of C. heterostrophus required for synthesis of T-toxin | 35 | TCTCATTGATGCGTGGTA/GCTTGGACTCTCATTCATATC | 57 |
| Tr65116 | PKS7 | RD clade IV, fumonisins | 2,434 | Uncharacterized fumonisin-like synthase | 40 | 0.0 | ELQ64206 | Mycocerosic acid synthase from M. oryzae | 33 | AAGAAGATGTCCGCAACT/AAGCACTCATACACAACCT | NA |
According to the JGI genome portal (http://genome.jgi-psf.org/Trire2/Trire2.home.html).
Based on the PKS grouping of Baker et al. (6).
NA, not available.
Somatic incompatibility.
The somatic compatibility of Δpks4 mutants and the parental strain was tested in confrontation assays on 2.4% potato dextrose agar (PDA; BD Difco, Germany) at 28°C for 7 days in darkness. All three strains were cultivated together on multiple plates, which were then micro- and macroscopically screened for flat zones, barrage zones, and anastomoses, which indicate somatic incompatibility between the opponents.
Morphological observations.
The parental strain QM 6a and both deletion mutants were cultivated in darkness and a 12-h light cycle on 2.4% PDA (BD Difco, Germany) at 28°C for 7 days. The spore density was measured quantitatively per cm2 of the developed colony. For this purpose, three 6.2-cm2 agar fragments were cut from cultures pregrown at 28°C for 7 days and were rinsed separately in 15 ml of water containing 0.1% Tween 80 until visually all conidia were washed out. The optical density at 590 nm (OD590) of the suspension was measured in a Biolog (Hayward, CA) turbidimeter calibrated to the Biolog standard for filamentous fungi. The final concentration value was calculated based on the calibration curve inferred from serial dilutions of the standard suspension. Furthermore, conidial size was assessed by measuring the length of 40 conidia per each strain under 400× magnification with a light microscope.
Mycelial growth rate and carbon source utilization.
Growth rates and carbon utilization profiles of the strains were analyzed using a phenotype microarray system with Biolog FF microplates for filamentous fungi (Biolog Inc., Hayward, CA) as described by Druzhinina et al. (23) and Atanasova et al. (24). Briefly, the strains were cultivated on 2.4% PDA for 5 days in darkness, and conidial inocula were prepared by rolling a sterile, wetted cotton swab over conidiating areas of the plates. The conidia were then suspended in sterile Biolog FF inoculating fluid (0.25% phytagel, 0.03% Tween 40), gently mixed, and adjusted to a transmission of 75% at 590 nm (by using a Biolog standard turbidimeter; 4 × 107 spores in 10 ml of phytagel). A total of 90 μl of the conidial suspension was dispensed into each of the wells of the Biolog FF microplates and incubated at 28°C in darkness and with a 12-h ligh-dark cycle. The OD750 (for detection of mycelial growth) was measured after 18, 24, 42, 48, 66, 72, 90, and 96 h using a microplate reader. The growth rate of each strain was assessed based on the averaged mycelial density, measured on all 95 carbon sources after 0, 24, 48, and 72 h of incubation in darkness. Statistical analyses were performed using the Statistica software package (version 6.1; StatSoft Inc., Tulsa, OK).
Response to illumination.
All three strains were inoculated in the Biolog FF phenotype microarray plates containing 95 carbon sources and water as described above, incubated in light (20-cm distance to a Master TLD 15-W/840 lamp) or in darkness at 28°C for 5 days. Mycelial density was measured at 750 nm. The growth rates and carbon utilization patterns were compared, and the data were statistically analyzed.
UV sensitivity.
The tolerance to UV irradiation of the pigment-deficient spores was tested at 254 nm by using four HNS 15-W OFR UV lamps. The conidia collected after 5 days of growth on PDA plates were filtered through sterile glass wool to remove hyphal fragments. The volume of the spore suspension that contained 4 × 107 (75% turbidity) was determined using a Biolog turbidimeter at 590 nm. The suspensions were then diluted several times to obtain ca. 20 spores per 100 μl, which were then plated on four PDA plates per sample (QM 6a and both Δpks4 mutants) and four plates for each sample control. Open plates were then exposed to UV illumination for 5, 7, and 10 min, whereas the control was protected from UV light. Finally, the plates were incubated for 48 h at 28°C, the single-spore colonies observed under 10× magnification were counted, and the percentage of germinated spores was normalized to the spore numbers obtained from control plates. Statistical analyses were performed using the Statistica software package (version 6.1; StatSoft Inc., Tulsa, OK).
Mechanical stability of T. reesei conidial cell walls.
Spores of the mutants and the parental strain collected from 5-day-old cultures on PDA plates were transferred to a carbon supporter and were coated with a 4-nm Au-Pd layer under vacuum conditions in a high-vacuum evaporation unit (Sputter Coater Quorum Q150T S; Quorum Technologies, Germany). Spores of QM 6a and both mutants were then separately studied, observing 10 ± 2 different fields for each strain under a FEI Quanta 200 field emission scanning electron microscope (FEGSEM) with 5 kV and high pressure under 25,000× magnification.
Mating tests.
In vitro matings were carried out on PDA medium (BD Difco, Germany) at room temperature and cycling daylight in dual confrontation assays of QM 6a or pks4 deletion mutants (MAT1-2) against T. reesei CBS 999.97 (MAT1-1). Plates were incubated for 10 to 20 days, until stromata formed. Single ascospore progeny were recovered from mature perithecia, and the obtained F1 strains were then purified and tested for somatic incompatibility as described above. Strains that expressed the somatic incompatibility reaction were considered unique genets and thus were used further in this study. The obtained F1 generations of the MAT1-1 and MAT1-2 pks4 deletion progeny were further crossed as described above.
PCR verification of MAT loci and pks4 inheritance.
Mycelia were harvested after 2 to 4 days of growth on PDA (Difco, Germany) at 28°C, and genomic DNA was isolated by using a DNeasy plant minikit (Qiagen, Germany) following the manufacturer's protocol. The mating type of the progeny strains was determined by PCR amplification of mat1-1-3 and mat1-2-1 as described by Druzhinina et al. (25). Furthermore, the progeny were screened for inheritance of the pks4 gene by using the PCR primers PKS4-2fw (TCATTATACACGGACTTT) and PKS4-1rev (TATAAGCCTGACTGTAGT) under the following conditions: 1 min of an initial denaturation step at 94°C, followed by 30 cycles of 1 min of denaturation at 94°C, 1 min of annealing at 50°C, and 90 s of extension at 72°C. Final extension was carried out at 72°C for 7 min. The results were verified by qPCR with the primer pair for the pks4 gene listed in Table 1 under the conditions described above. The cycle thresholds obtained for pks4 were compared to that for the tef1 housekeeping gene.
Mycoparasitic potential.
The mycoparasitic abilities of T. reesei QM 6a and the two pks4 deletion mutants were assessed in dual confrontation tests against the plant pathogenic fungi Rhizoctonia solani, Sclerotinia sclerotiorum, and Alternaria alternata (the “hosts”). Agar plugs with fresh cultures of Trichoderma and the host fungus were each placed on opposite sides of a PDA plate, 1 cm from the edge. The cultures were incubated at 28°C for 10 days under a 12-h light-dark cycle. The antagonistic potential was quantified as the percent reduction of the host's growth, corrected for its growth when confronted with itself (the growth of antagonist against itself was set as the zero inhibition rate).
Production of VOCs.
The impact of pks4 gene on volatile organic compounds (VOCs) production was tested in “sandwich” culture assays. T. reesei strains were first cultivated on a PDA plate for 2 days at 28°C, and then they were sealed together with a plate containing a fresh plug of a host fungus, so that the host fungus was facing the T. reesei culture from the top. No hyphal contact was established between the two confronted fungi. The plates were cocultivated under the same conditions for 4 days. The colony radii of the host fungi were measured every 24 h.
Production of water-soluble compounds.
Water-soluble compounds (WSCs) were assessed by growing Trichoderma strains on the PDA plates covered by cellophane, which was removed together with Trichoderma mycelium after 60 h. Agar plugs of the host fungi were then put in the middle of the plates and were cultivated for a further 4 days under 12-h light-dark cycling at 28°C. The colony radii of the fungi were measured every 24 h. Additionally, the same experiment was performed when T. reesei was pregrown under the same conditions as described above under the influence of volatile compounds of the host fungus. The latter was plated on a fresh PDA plate and was sealed on top of the T. reesei plate. The plate with the host fungus was then removed simultaneously with Trichoderma and the cellophane. The same host fungus was then plated on this plate, and its growth was observed for 4 days.
Gene expression analysis.
Confrontation assays for T. reesei QM 6a and both Δpks4 strains against Rhizoctonia solani were carried out on potato dextrose agar plates covered with cellophane at 28°C and with a 12-h illumination cycle. To compensate for the effects caused by nutrient depletion, self-confrontation experiments were included as controls. Peripheral hyphal zones from each confrontation stage were sampled and shock-frozen in liquid nitrogen. Mycelia of five replicate plates were harvested and pooled together before the RNA extraction when (i) the hyphae were ca. 20 mm apart (before the contact) and (ii) at contact of Trichoderma and host hyphae. The biomass was ground to a fine powder under liquid nitrogen, and total RNA was isolated by using the RNeasy extraction kit (Qiagen, Germany). For cDNA synthesis, RNA (5 μg) that was DNase treated (DNase I, RNase-free; Fermentas, Germany) was reverse transcribed by using the RevertAid first-strand cDNA kit (Fermentas, Germany) according to the manufacturer's protocol with a combination of the provided oligo(dT) and random hexamer primers.
All real-time PCR experiments were performed in an iCycler IQ (Bio-Rad, Germany). For the reaction mixture, IQ SYBR green supermix (Bio-Rad, Germany) was prepared for 25-μl assay mixtures with the standard MgCl2 concentration (3 mM) and final primer concentrations of 100 nM each. The primers used are given in Table 1. The amplification protocol consisted of an initial denaturation step (3 min at 95°C) followed by 40 cycles of denaturation (15 s at 95°C), annealing (20 s at the temperature given in Table 1 for each primer), and extension (72°C for 15 s). The tef1 (elongation factor 1α-encoding) gene, whose expression remained constant under all conditions tested (±20 relative percent), was used as a reference. Determination of the PCR efficiency was performed using triplicate reaction mixtures from a dilution series of cDNA (0.1, 10−2, 10−3, and 10−2). Amplification efficiency was then calculated from the given slopes in the IQ5 optical system software v2.0. The qPCR were performed with the cDNA of 5 pooled biological replicates for each species and condition separately. Expression ratios were calculated by the Pfaffl test model implemented in the relative expression software tool (REST) (26). The expression of pks genes in the mutants under nonantagonistic conditions (confrontation to itself) was measured by using QM 6a as a control. The mathematical model used for the expression analysis was based on correction for exact PCR efficiencies and the mean crossing-point deviation between the sample group and the control group (26).
RESULTS
Generation of pks4 knockout strains of T. reesei QM 6a.
To generate pks4 knockout strains, a linear DNA construct was designed to replace the reading frame of pks4 with a hygromycin B (hyg) selection marker under the T. reesei pki1 promoter via homologous recombination (21). Transformants with the expected albino phenotype were picked from plates, and the deletion of pks4 was verified by PCR and by qPCR as described in Materials and Methods. Two verified deletants, Δpks4-1 and Δpks4-2, were selected for further studies.
Biolog phenotype microarray analyses verified that the phenotypes of both candidate strains were consistent and similar to the parental strain (see Fig. S1 in the supplemental material). Furthermore, the genetic identity of the mutants and the influence of the pks4 knockout on the recognition of the QM 6a genotype were tested in a plate confrontation assay. Both mutants expressed somatic incompatibility reactions (a flat zone) to the parental QM 6a strain but not to each other, and no anastomoses were observed in contact with QM 6a. These findings suggested that the two deletion strains are genetically identical. In contrast, the parental strain recognized the mutants as nonself and thus reacted antagonistically, which eventually led to the partial overgrowth of the mutants.
Δpks4 mutants are devoid of conidial pigmentation.
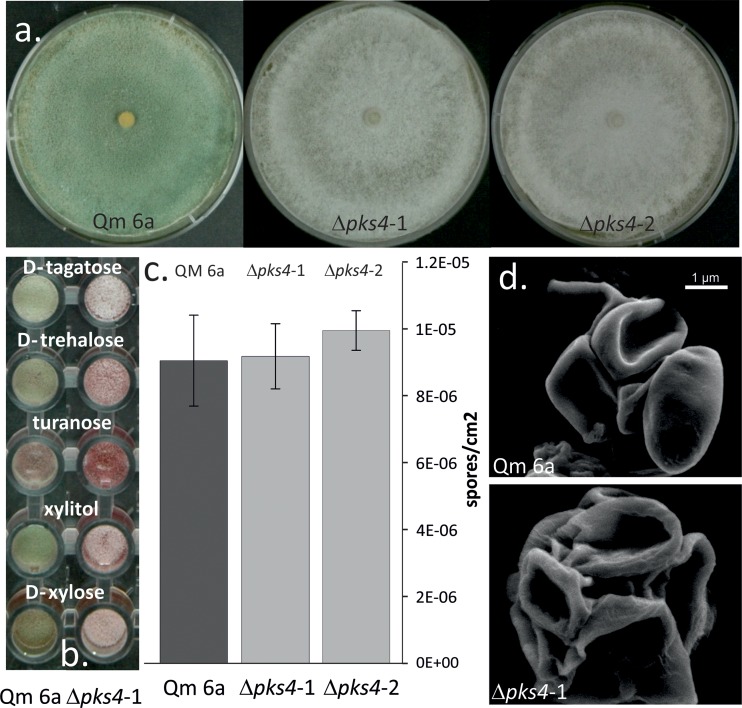
The default hypothesis of this work was that PKS4 is involved in pigment formation. This was confirmed: morphological examination on PDA plates revealed that both pks4 deletion mutants lost their green conidial pigmentation (Fig. 1a), while formation of the yellow pigment and its secretion into the medium, a characteristic of T. reesei, remained unaffected. No green conidia were formed under any of the cultivation conditions in either light or darkness, and the color was not recovered by cultivation on any of 95 carbon sources of the Biolog phenotype microarrays, 5 of which are shown in Fig. 1b.
Fig 1.
Conidiation morphology of QM 6a and the pks4 deletion mutants. (a) Plate macromorphology and conidial pigmentation of QM 6a and Δpks4 cultures. (b) Conidial pigmentation of QM 6a and Δpks4 mutants on selected carbon sources of the Biolog FF microplate phenotype microarrays. (c) Conidial density of QM 6a and Δpks4 mutants' spores produced after 7 days of cultivation PDA. No statistically significant difference in the intensity of conidia production was observed (ANOVA, P > 0.05). (d) Conidial mechanical stability observed under a FEI Quanta 200 FEGSEM after application of vacuum conditions and high pressure, under 25,000× magnification.
The Δpks4 strains did not show any statistically significant difference in the intensity of conidia production on PDA: after 7 days, QM 6a, Δpks4-1, and Δpks4-2 produced on average 9.03 × 106, 9.16 × 106, and 9.94 × 106 spores per cm2, respectively (analysis of variance [ANOVA], P > 0.05) (Fig. 1c). Spore size remained unchanged (ANOVA, P > 0.05) (data not shown); however, the conidia of the deletion mutants showed less mechanical stability against reduced air pressure (Fig. 1d).
In order to test whether pks4 is solely responsible for conidial pigmentation, we crossed the Δpks4 mutants (which, due to their QM 6a background, possess MAT1-2 [27]) with the T. reesei MAT1-1 strain CBS 999.97. In total, 34 pure single-spore strains were isolated from mature ascospores. A total of 21, 10, and 3 cultures contained purely white, green, or yellowish conidia, respectively. All strains from the first-generation progeny (F1) were screened by PCR for mating types, and there was a nearly equal distribution (15 MAT1-2 and 18 MAT1-1) that was independent of the phenotype (see Fig. S2 in the supplemental material). Specific pks4 primers were designed (Table 1) and used to test the F1 progeny for the presence of the pks4 gene. We found that all the strains with green conidia had indeed inherited the wild-type gene, whereas the yellow and albino conidia phenotype conidia did not contain the pks4 gene (see Fig. S2). This proved that the loss of the conidial green pigmentation is directly caused by the pks4 deletion and that this gene is involved in its production.
Loss of green pigmentation caused reduced resistance to UV.
The lack of green conidia pigmentation was also reflected in an increased sensitivity to UV light: after 7 min of UV exposure (see Materials and Methods for details) sixty-four percent of green spores survived, whereas only 8 to 20% of white conidia were able to germinate (Table 2). Prolongation of exposure time (to 10 min) led to a ca. 92% reduction of germination for both Δpks4 mutants (Table 2), while germination of QM 6a was decreased by only 60%.
Table 2.
Spore germination after exposure to UV light for 7 and 10 min
| T. reesei strain | % germinated spores (mean ± SD) after UV exposure for: |
|
|---|---|---|
| 7 min | 10 min | |
| QM 6a | 63.6 ± 10.1 | 39.4 ± 6.4 |
| Δpks4-1 | 19.7 ± 6.5 | 5.9 ± 8.4 |
| Δpks4-2 | 7.7 ± 6.5 | 7.7 ± 6.5 |
pks4 contributes to the development of the reddish-brown color of stroma.
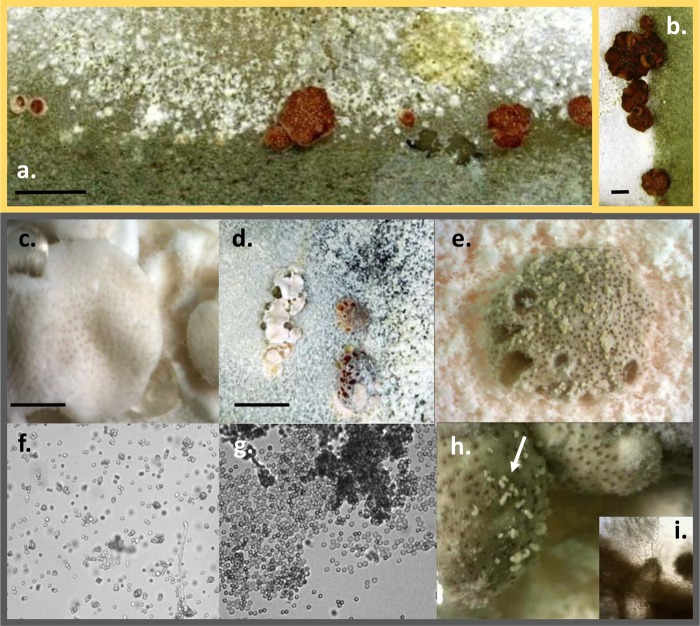
The wild-type morphology of the T. reesei teleomorph is conspicuous, with perithecial openings that appear as dark brown dots against the reddish-brown to brown background of stroma (Fig. 2a and b) (36). Crossing of the two Δpks4 mutants with the MAT1-1 tester strain CBS 999.79, as described above, produced stroma with fruiting bodies that were normally pigmented (see Fig. S3 in the supplemental material) and which ejected viable and fertile ascospores. Crossings between the albino strains from the F1 progeny described above showed that the pigmentation of both the stroma surface and perithecial openings was altered (Fig. 2c to e, h, and i). Thus, the young stromata appeared to be white with slightly brownish dots of perithecial openings (Fig. 2c and d). However, the mature and overmature teleomorphs developed brown pigmentation of both the stroma surface and perithecial walls and openings (Fig. 2d, e, and h). Importantly, the surfaces of mature stromata were covered by whitish cirri that originated from perithecial openings (Fig. 2e and h). Microscopic investigation showed that these structures were composed entirely of mature ascospores, as some of them started to germinate (Fig. 2g), indicating possible defects in the mechanism of ascospore discharge. The morphology of the asci was normal.
Fig 2.
Teleomorph morphology of T. reesei QM 6a wild type and Δpks4 mutants. (a) Stromata of wild-type T. reesei strains Qm 6a and CBS 999.97, mated in vitro. (b) Fruiting bodies of strains Δpks4-1 and Qm 6a mated in vitro. (c) Young stromata and mature, partially melanized brownish fruiting bodies of F1 Δpks4 strains mated in vitro. Bar, 2 mm. (d) Partially melanized overmature fruiting structures of F1 Δpks4 strains with dark perithecia openings. Bar, 10 mm. (e) Magnified young white stromata of F1 Δpks4 strains with melanized openings. (f) Conidia of F1 Δpks4 strains. (g and h) Ascospores from the cirri of F1 Δpks4 attached to overmature perithecia, indicated by the arrows in panel h. Magnification (panels f and g), ×200. (i) Pigmented perithecium from the stroma shown in panel h. Magnification, ×200.
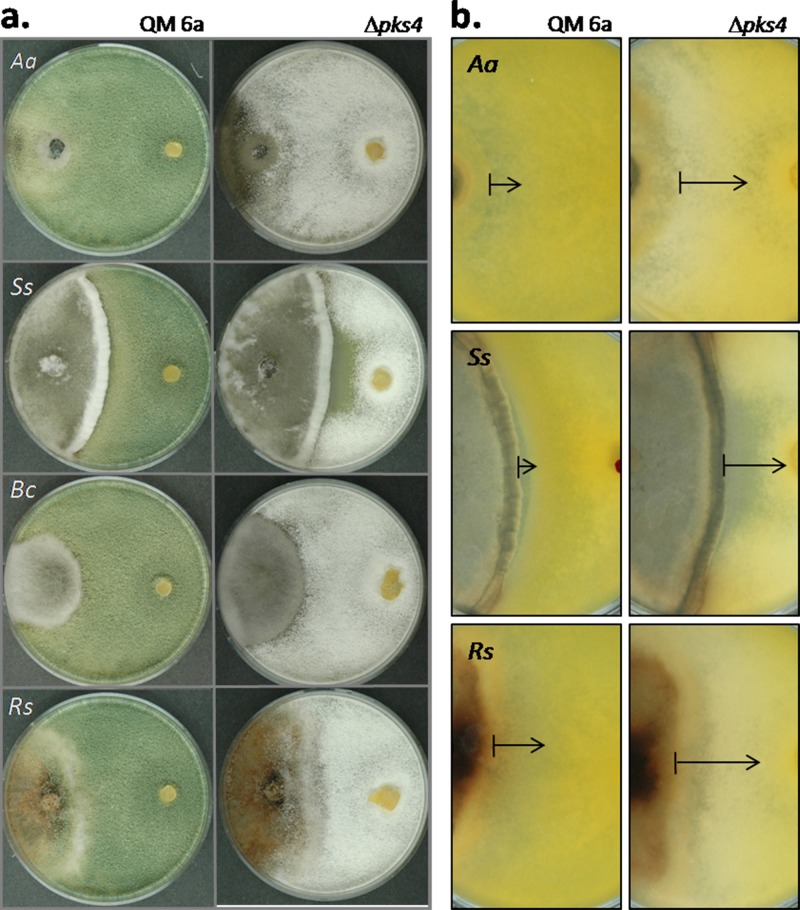
Loss of pks4 slightly reduces the mycoparasitic potential of T. reesei and decreases the defense against other fungi.
Dual confrontation assays with Rhizoctonia solani, Sclerotinia sclerotiorum, and Alternaria alternata showed that the ability of T. reesei QM 6a to attack and inhibit growth of these fungi was reduced in the Δpks4 mutants (Fig. 3a) by 37%, 13%, and 40%, respectively, for each host fungus. Interestingly, there were no significant differences in the abilities of QM 6a and the pks4 mutants to overgrow their opponents, as can be observed for A. alternata and R. solani in Fig. 3a, and thus the mycoparasitic capacity was not affected. However, a difference was detected with respect to the ability to protect against the host's metabolites: in confrontations of QM 6a with A. alternata, S. sclerotiorum, or R. solani, a narrow antibiosis zone was observed, which is indicative of secretion of metabolites toxic for T. reesei (Fig. 3b). In the case of QM 6a, this zone was clear and thin, while it was considerably enlarged and displayed diffuse borders in the confrontations with the Δpks4 mutants, suggesting that metabolites secreted by the other fungi had penetrated the colonies of the deletion mutants.
Fig 3.
Antagonistic potentials of QM 6a and the pks4-1 deletion mutant. Confrontations panels are shown only for Δpks4-1, but both mutants revealed identical mycoparasitic patterns. (a) Front sides of confrontation plates, with plant-pathogenic fungi always on the left side and Trichoderma QM 6a or the mutant always on the right. (b) Back sides of confrontation plates, with plant-pathogenic fungi always on the left side and T. reesei QM 6a or the mutant strain always on the right. Antibiosis zones for Δpks4 strains are indicated by the arrows. Aa, Ss, Bs, and Rs mark Alternaria alternata, Sclerotinia sclerotiorum, Botrytis cinerea, and Rhizoctonia solani, respectively.
pks4 affects the production of water-soluble and volatile inhibitory compounds.
In order to test whether the loss of pks4 is also reflected in an alteration of water-soluble and volatile metabolites produced by T. reesei, strain QM 6a and the two Δpks4 strains were grown in sealed sandwich cultures (see Materials and Methods) with R. solani, S. sclerotiorum, and A. alternata so that the host fungi were facing Trichoderma cultures from the top. No hyphal contact was established between the two fungi. Compared to the effect caused by both of the pks4 deletion mutants versus that of QM 6a, the growth of all four host fungi was strongly reduced by VOCs (Fig. 4; R. solani is shown as one example; see also Table S1 in the supplemental material), indicating that the pks4 deletion mutants displayed enhanced production of VOCs compared to QM 6a.
Fig 4.

Effects of VOCs and WSCs from pks4 deletion mutants and QM 6a on growth of R. solani. (a) Reduction of R. solani growth by the VOCs secreted by T. reesei QM 6a and Δpks4 mutants after 4 days of incubation. Both mutants consistently reduced the growth of R. solani. (b) R. solani growth on PDA medium containing WSCs of T. reesei and the pks4 deletion mutants. (c) Growth of R. solani on medium with WSCs secreted by the pks4 mutants and QM 6a in the presence of VOCs from R. solani during growth of the T. reesei strains.
In contrast, the formation of fungicidal WSCs (see Materials and Methods) by T. reesei was reduced in the Δpks4 strains (Fig. 4b). Interestingly, the secretion of WSCs by T. reesei QM 6a and the Δpks4 mutants was inhibited by the presence R. solani VOCs (no hyphal contact), and this effect was even enhanced in the Δpks4 strains (Fig. 4c; see also Table S2 in the supplemental material).
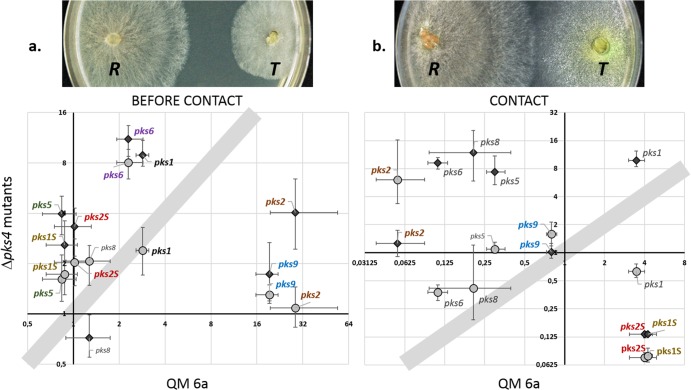
pks4 influences the expression of other PKS-encoding genes during confrontation with other fungi.
Because of the impact of pks4 on the production of components that inhibit fungal growth, we were interested in whether the loss of PKS4 would impact other PKS-encoding genes in T. reesei. To this end, we assessed their expression in QM 6a and the two Δpks4 mutants when confronted with R. solani. Expression of pks genes was inspected both prior to contact and at contact of the hyphae (Fig. 5). The expression of two pks genes, pks3 (Trire2:105804) and pks7 (Trire2:65116), was not detectable under the conditions of our experiments, which also included confrontations of T. reesei QM 6a and the Δpks4 strains with themselves.
Fig 5.
Expression of pks genes in Δpks4 mutants compared to QM 6a before contact (a) and at contact (b) with Rhizoctonia solani. The gray circles and black diamonds represent data for the Δpks4-1 and Δpks4-2 mutants, respectively. Shadowed areas indicate that there was no difference in regulation of the pks4 deletion mutants compared to QM 6a. Protein numbers of pks genes in the genome of T. reesei are as follows (in parentheses): pks1 (Trire2:65172), pks2 (Trire2:65891), pks5 (Trire2:59482), pks6 (Trire2:60118), pks8 (Trire2:81964), pks9 (Trire2:106272), pks1S (Trire2:73621), pks2S (Trire2:73618). Those genes whose names are shown in color exhibited a consistent expression trend for both Δpks4 mutants. Vertical and horizontal bars indicate standard deviations for the wild type and the mutants, respectively. The images above the plots show the sampling stage of the T. reesei (T) strains under each condition of confrontation with R. solani (R).
Expression analysis of the remaining eight pks genes in the Δpks4 mutants and QM 6a prior to and at the time of contact, respectively, with R. solani revealed striking changes in the patterns of transcript formation that depended on the stage of the interaction. Before contact, both QM 6a and the mutants upregulated four pks genes from lovastatin/citrinin reducing clade I (pks1, pks2, pks6, and pks9; for protein ID numbers, see Table 1 and Fig. 5). Yet, contrary to pks9 (Trire2:106272) and pks2 (Trire2:65891), which were much highly expressed in QM 6a than in the mutants (Fig. 5a), pks6 and pks1 were strongly upregulated in the two mutant strains (Fig. 5a). The remaining two reducing genes, pks5 (Trire2:59482) from the clade of fumonisin-like synthases and the singlet pks2S (Trire2:73618), as well as the nonreducing pks1S (Trire2:73621), were not influenced in QM 6a prior to contact with R. solani but showed upregulation in both mutants (Fig. 5a). At contact with R. solani, the pattern was different: QM 6a downregulated the majority of its pks genes, including pks9 and pks2, which were strongly upregulated before the contact with R. solani (Fig. 5). Interestingly, the two singlet pks genes (pks1S and pks2S) were differentially downregulated at the contact between the pks4 mutants and R. solani (Fig. 5b), whereas they were strongly upregulated in QM 6a at this stage.
Most of the eight pks genes were downregulated in the mutants, whereas two singlet genes, pks2S, a member of the lovastatin/citrinin clade, and pks1S, which belongs to nonreducing clade III, were upregulated (see Fig. S4 in the supplemental material).
DISCUSSION
In this study, we functionally characterized the role of the polyketide synthase PKS4 in T. reesei. pks4 belongs to the nonreducing clade I of fungal pks-encoding genes, which includes genes associated with pigment production, such as aurofusarin (7–9) and bikaverin (10, 11), but also DHN melanin (12–19). While the former comprise substances of relatively low molecular weights, melanins—the dark to black pigments—are of high molecular mass that derive from oxidative polymerization of phenolic compounds (20, 37). Melanins are pigments that occur in all biological kingdoms and serve many functions, such as defense against environmental stresses including UV light, oxidizing agents, and ionizing radiation, and they also contribute to fungal pathogenesis (38).The chemical structures of the conidial pigments of T. reesei, and of Trichoderma spp. in general, have not been elucidated yet, but due to their green and sometimes yellowish color they appear not to be melanins. Yellow pigments from Aspergillus niger have been shown to be dimeric linear naphtho-y-pyrones (4, 7, 13). However, Benitez et al. (39) preliminarily characterized the conidial pigment of Trichoderma spp. (T. viride at that time) as a nonindolic melanin-like polyphenol. Consistent with these data, Csiktusnádi Kiss et al. (40) identified the main pigment fractions of T. harzianum as oxidation polymers originating from monomer molecules containing polar substructures and double bonds in the alkyl chain. Here, we showed that the final dark brown component of the fruiting body and the stroma color, which likely represents melanin, are independent of PKS4, but the colorization is delayed in the early stages of fruiting body development. In the absence of pks4, the young stroma of T. reesei are colorless (white) with slightly darkened openings of perithecia, indicating some retained pigmentation. Mature and overmature stromata, however, showed some dark brown coloration on the surface and perithecia walls, indicating that melanin was still synthesized. The question of whether the pigments synthesized by PKS4 are melanins is important, because our data showed that PKS4 is involved in antagonism and defense against other fungi and in the mechanical stability of the conidium. A role in antagonism is also supported by earlier findings indicating that pks4 is upregulated during antagonism and mycoparasitic contact of T. reesei with R. solani (41). A role in defense, stress resistance, virulence, and mechanical stability has so far been shown for melanins but not for the low-molecular-weight pigments formed by PKS4 orthologues mentioned above. In addition, some polyketides have been shown to be involved in sexual development (42–46) but none of them is a PKS4 orthologue, and the mechanism of involvement is still only poorly understood. In most cases, this may be related to cell wall stabilization (46). T. reesei PKS4 therefore exhibits a biological function otherwise typical for melanin-synthesizing PKSs: in human pathogenic fungi, such as Cryptococcus neoformans (47–59), Sporothrix schenckii (51, 52), Paracoccidioides brasiliensis (53), Histoplasma capsulatum (54), and the opportunistic pathogen Aspergillus fumigatus (20), melanin is involved in virulence, probably because of resistance against oxidative stress. Also, it contributes to resistance against antifungal drugs in H. capsulatum (55). Melanins have also been demonstrated to play crucial roles in plant pathogenic fungi: in Magnaporthe grisea, it accumulates between the plasma membrane and the cell wall of the appressorium and creates the turgor pressure needed for penetration (56). In addition, expression of an A. alternata melanin biosynthetic PKS in the insect pathogen Metarhizium anisopliae resulted in increased virulence (57). In wood decaying Basidiomycota, such as Phellinus weirii (58) and Pleurotus ostreatus (I. S. Druzhinina, unpublished data), melanin is crucially important in reactions of somatic incompatibility when physical borders between genetically unique individuals are marked by thick melanized walls (barrage reaction) impermeable for competitive fungi, which could also be the reason for the effect of PKS4 on vegetative compatibility for T. reesei.
An interesting consequence of pks4 loss of function that has not been reported for any other PKS was its effect on the expression of the other eight pks genes of T. reesei. During normal growth in the absence of a competing fungus, all but two of these eight genes were significantly downregulated in the pks4 mutants. Since growth of the mutant strains and the wild type occurred at the same rate, these differences are not the consequence of variable rates of nutrient uptake. Furthermore, an antagonistic interaction with R. solani revealed that loss of pks4 function influenced the expression of the other pks genes in different ways, with some being upregulated, some downregulated, and some not changed. From these data, we conclude that PKS4—or rather the function of its product in, for example, protection and defense against stress—is an important signal for the expression of these pks genes. This is definitely an area that requires further investigations.
Loss of function of pks4 also led to a decreased synthesis of water-soluble inhibitory components by T. reesei, and it is tempting to speculate that they are formed by one or more of the affected PKSs. Since their expression was tested in the absence of other fungi and they were all found to be downregulated (see Fig. S4 in the supplemental material), it is, however, not possible to predict which ones are the responsible producers. In addition, these components could also be products of other enzymes, such as nonribosomal peptide synthases, of which T. reesei produces 10 (59).
Finally, it was interesting that deletion of pks4 increased the production of VOCs by T. reesei. The chemical natures of the VOCs from T. reesei have not been identified yet, but their diversity compared to those from other Trichoderma spp. showed that they are composed mainly of long aliphatic acids and that they have alcohols and esters (60, 61), which are usually products of fatty acid catabolism (62). Biosynthesis of these compounds may be favored by the lack of PKS activity, which results in an increased access to the cellular pools of acetyl- and malonyl-CoA.
The functions of PKSs have so far mainly been investigated with respect to the role of their products in human or plant pathogenesis. Our data showed that PKS4 also influences several biological functions in T. reesei that are not only related to the interaction with other organisms. Transcriptomic analyses of T. reesei have recently shown that many pks genes are maximally expressed during rapid vegetative growth (63–65), which is not a pattern that would be expected for genes whose functions are traditionally viewed as unrelated to growth (i.e., secondary metabolites). It will thus be worthwhile to perform a deeper investigation of the regulation and role of pks genes in fungal physiology.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by a grant of the Austrian Science Fund (FWF) to C.P.K. (P 21266). S.E.B. and B.P.K. were supported by funding from the U.S. DOE Biomass Program.
We thank Aldin Saracevic and Yuan Zhi-Lin for laboratory assistance and Bernhard Pummer for the SEM images.
Footnotes
Published ahead of print 13 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00103-13.
REFERENCES
- 1. Druzhinina IS, Seidl-Seiboth V, Herrera-Estrella A, Horwitz BA, Kenerley CM, Monte E, Mukherjee PK, Zeilinger S, Grigoriev IV, Kubicek CP. 2011. Trichoderma: the genomics of opportunistic success. Nat. Rev. Microbiol. 16:749–759 [DOI] [PubMed] [Google Scholar]
- 2. Druzhinina IS, Shelest E, Kubicek CP. 2012. Novel traits of Trichoderma predicted through the analysis of its secretome. FEMS Microbiol. Lett. 337:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Druzhinina IS, Kubicek CP. 2013. Ecological genomics of Trichoderma. In Martin F. (ed), Ecological genomics of fungi. Wiley-Blackwell, Oxford, United Kingdom [Google Scholar]
- 4. Mukherjee PK, Horwitz BA, Kenerley CM. 2012. Secondary metabolism in Trichoderma: a genomic perspective. Microbiology 158:35–45 [DOI] [PubMed] [Google Scholar]
- 5. Kroken S, Glass NL, Taylor JW, Yoder OC, Turgeon BG. 2003. Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc. Natl. Acad. Sci. U. S. A. 100:15670–15675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baker SE, Perrone G, Richardson NM, Gallo A, Kubicek CP. 2012. Phylogenomic analysis of polyketide synthase-encoding genes in Trichoderma. Microbiology 158:147–154 [DOI] [PubMed] [Google Scholar]
- 7. Frandsen RJ, Schütt C, Lund BW, Staerk D, Nielsen J, Olsson S, Giese H. 2011. Two novel classes of enzymes are required for the biosynthesis of aurofusarin in Fusarium graminearum. J. Biol. Chem. 286:10419–10428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim JE, Han KH, Jin J, Kim H, Kim JC, Yun SH, Lee YW. 2005. Putative polyketide synthase and laccase genes for biosynthesis of aurofusarin in Gibberella zeae. Appl. Environ. Microbiol. 71:1701–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Malz S, Grell MN, Thrane C, Maier FJ, Rosager P, Felk A, Albertsen KS, Salomon S, Bohn L, Schafer W, Giese H. 2005. Identification of a gene cluster responsible for the biosynthesis of aurofusarin in the Fusarium graminearum species complex. Fungal Genet. Biol. 42:420–433 [DOI] [PubMed] [Google Scholar]
- 10. Linnemannstöns P, Schulte J, del Mar Prado M, Proctor RH, Avalos J, Tudzynski B. 2002. The polyketide synthase gene pks4 from Gibberella fujikuroi encodes a key enzyme in the biosynthesis of the red pigment bikaverin. Fungal Genet. Biol. 37:134–148 [DOI] [PubMed] [Google Scholar]
- 11. Wiemann P, Willmann A, Straeten M, Kleigrewe K, Beyer M, Humpf HU, Tudzynski B. 2009. Biosynthesis of the red pigment bikaverin in Fusarium fujikuroi: genes, their function and regulation. Mol. Microbiol. 72:931–946 [DOI] [PubMed] [Google Scholar]
- 12. Baker SE. 2008. Aspergillus genomics and DHN-melanin conidial pigmentation, p 73–85 In Varga J, Samson RA. (ed),Aspergillus in the genomic era. Wageningen Academic Publishers, Wageningen, The Netherlands [Google Scholar]
- 13. Chiang YM, Meyer KM, Praseuth M, Baker SE, Bruno KS, Wang CC. 2011. Characterization of a polyketide synthase in Aspergillus niger whose product is a precursor for both dihydroxynaphthalene (DHN) melanin and naphtho-c-pyrone. Fungal Genet. Biol. 48:430–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jørgensen TR, Park J, Arentshorst M, van Welzen AM, Lamers G, Vankuyk PA, Damveld RA, van den Hondel CA, Nielsen KF, Frisvad JC, Ram AF. 2011. The molecular and genetic basis of conidial pigmentation in Aspergillus niger. Fungal Genet. Biol. 48:544–553 [DOI] [PubMed] [Google Scholar]
- 15. Langfelder K, Jahn B, Gehringer H, Schmidt A, Wanner G, Brakhage AA. 1998. Identification of a polyketide synthase gene (pksP) of Aspergillus fumigatus involved in conidial pigment biosynthesis and virulence. Med. Microbiol. Immunol. (Berl.) 187:79–89 [DOI] [PubMed] [Google Scholar]
- 16. Tsai HF, Chang YC, Washburn RG, Wheeler MH, Kwon-Chung KJ. 1998. The developmentally regulated alb1 gene of Aspergillus fumigatus: its role in modulation of conidial morphology and virulence. J. Bacteriol. 180:3031–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tsai HF, Fujii I, Watanabe A, Wheeler MH, Chang YC, Yasuoka Y, Ebizuka Y, Kwon-Chung KJ. 2001. Pentaketide melanin biosynthesis in Aspergillus fumigatus requires chain length shortening of a heptaketide precursor. J. Biol. Chem. 276:29292–29298 [DOI] [PubMed] [Google Scholar]
- 18. Watanabe A, Fujii I, Sankawa U, Mayorga ME, Timberlake WE, Ebizuka Y. 1999. Re-identification of Aspergillus nidulans wA gene to code for a polyketidesynthase of naphthopyrone. Tetrahedron Lett. 40:91–94 [Google Scholar]
- 19. Watanabe A, Fujii I, Tsai H, Chang YC, Kwon-Chung KJ, Ebizuka Y. 2000. Aspergillus fumigatus alb1 encodes naphthopyrone synthase when expressed in Aspergillus oryzae. FEMS Microbiol. Lett. 192:39–44 [DOI] [PubMed] [Google Scholar]
- 20. Heinekamp T, Thywißen A, Macheleidt J, Keller S, Valiante V, Brakhage AA. 2012. Aspergillus fumigatus melanins: interference with the host endocytosis pathway and impact on virulence. Front. Microbiol. 3:440. 10.3389/fmicb.2012.00440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yu JH, Hamari Z, Han KH, Seo JA, Reyes-Domínguez Y, Scazzocchio C. 2004. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41:973–981 [DOI] [PubMed] [Google Scholar]
- 22. Penttila M, Nevalainen H, Ratto M, Salminen E, Knowles J. 1987. A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei. Gene 61:155–164 [DOI] [PubMed] [Google Scholar]
- 23. Druzhinina IS, Schmoll M, Seiboth B, Kubicek CP. 2006. Global carbon utilization profiles of wild-type, mutant, and transformant strains of Hypocrea jecorina. Appl. Environ. Microbiol. 72:2126–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Atanasova L, Druzhinina IS. 2010. Global nutrient profiling by phenotype microarrays: a tool complementing genomic and proteomic studies in conidial fungi. J. Zhejiang Univ. Sci. B 11:151–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Druzhinina IS, Komoń-Zelazowska M, Atanasova L, Seidl V, Kubicek CP. 2010. Evolution and ecophysiology of the industrial producer Hypocrea jecorina (anamorph Trichoderma reesei) and a new sympatric agamospecies related to it. PLoS One 5(2):e9191. 10.1371/journal.pone.0009191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pfaffl MW, Horgan GW, Dempfle L. 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30:e36. 10.1093/nar/30.9.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Seidl V, Seibel C, Kubicek CP, Schmoll M. 2009. Sexual development in the industrial workhorse Trichoderma reesei. Proc. Natl. Acad. Sci. U. S. A. 106:13909–13914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pihet M, Vandeputte P, Tronchin G, Renier G, Saulnier P, Georgeault S, Mallet R, Chabasse D, Symoens F, Bouchara JP. 2009. Melanin is an essential component for the integrity of the cell wall of Aspergillus fumigatus conidia. BMC Microbiol. 9:177. 10.1186/1471-2180-9-177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li Y, Chooi YH, Sheng Y, Valentine JS, Tang Y. 2011. Comparative characterization of fungal anthracenone and naphthacenedione biosynthetic pathways reveals an α-hydroxylation-dependent Claisen-like cyclization catalyzed by a dimanganesethioesterase. J. Am. Chem. Soc. 133:15773–15785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chooi YH, Cacho R, Tang Y. 2010. Identification of the viridicatumtoxin and griseofulvin gene clusters from Penicillium aethiopicum. Chem. Biol. 17:483–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Abe Y, Suzuki T, Mizvno T, Ono C, Iwamoto K, Hosobuchi M, Yoshikawa H. 2002. Molecular cloning and characterization of an ML-236B (compactin) biosynthetic gene cluster in Penicillium citrinum. Mol. Genet. Genomics 267:636–646 [DOI] [PubMed] [Google Scholar]
- 32. O'Callaghan J, Caddick MX, Dobson AD. 2003. A polyketide synthase gene required for ochratoxin A biosynthesis in Aspergillus ochraceus. Microbiology 149:3485–3491 [DOI] [PubMed] [Google Scholar]
- 33. Xue M, Yang J, Li Z, Hu S, Yao N, Dean RA, Zhao W, Shen M, Zhang H, Li C, Liu L, Cao L, Xu X, Xing Y, Hsiang T, Zhang Z, Xu JR, Peng YL. 2012. Comparative analysis of the genomes of two field isolates of the rice blast fungus Magnaporthe oryzae. PLoS Genet. 8(8):e1002869. 10.1371/journal.pgen.1002869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hendrickson L, Davis CR, Roach C, Nguyen DK, Aldrich T, McAda PC, Reeves CD. 1999. Lovastatin biosynthesis in Aspergillus terreus: characterization of blocked mutants, enzyme activities and a multifunctional polyketide synthase gene. Chem. Biol. 6:429–439 [DOI] [PubMed] [Google Scholar]
- 35. Baker SE. 2006. Aspergillus niger genomics: past, present and into the future. Med. Mycol. 1:S17–S21 [DOI] [PubMed] [Google Scholar]
- 36. Samuels GJ, Chaverri P, Farr DF, McCray EB. 2010, last update Trichoderma online. Systematic Mycology and Microbiology Laboratory, ARS, USDA: http://nt.ars-grin.gov/taxadescriptions/keys/FrameListAllTaxa.cfm?gen=Trichoderma [Google Scholar]
- 37. Riley PA. 1997. Melanin. Int. J. Biochem. Cell Biol. 29:1235–1239 [DOI] [PubMed] [Google Scholar]
- 38. Eisenman HC, Casadevall A. 2012. Synthesis and assembly of fungal melanin. Appl. Microbiol. Biotechnol. 93:931–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Benítez T, Villa TG, García Acha I. 1976. Some chemical and structural features of the conidial wall of Trichoderma viride. Can. J. Microbiol. 22:318–321 [DOI] [PubMed] [Google Scholar]
- 40. Csiktusnádi Kiss G, Forgács E, Cserháti T, Vizcaino JA. 2000. Colour pigments of Trichoderma harzianum. Preliminary investigations with thin-layer chromatography-Fourier transform infrared spectroscopy and high-performance liquid chromatography with diode array and mass spectrometric detection. J. Chromatogr. A 896:61–68 [DOI] [PubMed] [Google Scholar]
- 41. Atanasova L, Crom S, Gruber S, Coulpier F, Seidl-Seiboth V, Kubicek CP, Druzhinina IS. 2013. Comparative transcriptomics reveals different strategies of Trichoderma mycoparasitism. BMC Genomics 14:121. 10.1186/1471-2164-14-121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wolf JC, Mirocha CJ. 1973. Regulation of sexual reproduction in Gibberella zeae (Fusarium roseum ‘Graminearum') by F-2 (zearalenone). Can. J. Microbiol. 19:725–734 [DOI] [PubMed] [Google Scholar]
- 43. Zimmerman WC, Blanchette RA, Burnes TA, Farrell RL. 1995. Melanin and perithecial development in Ophiostoma piliferum. Mycologia 87:857–863 [Google Scholar]
- 44. Graziani S, Vasnier C, Daboussi MJ. 2004. Novel polyketide synthase from Nectria haematococca. Appl. Environ. Microbiol. 70:2984–2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Engh I, Nowrousian M, Kück U. 2007. Regulation of melanin biosynthesis via the dihydroxynaphthalene pathway is dependent on sexual development in the ascomycete Sordaria macrospora. FEMS Microbiol. Lett. 275:62–70 [DOI] [PubMed] [Google Scholar]
- 46. Nowrousian M. 2009. A novel polyketide biosynthesis gene cluster is involved in fruiting body morphogenesis in the filamentous fungi Sordaria macrospora and Neurospora crassa. Curr. Genet. 55:185–198 [DOI] [PubMed] [Google Scholar]
- 47. Nosanchuk JD, Rosas AL, Lee SC, Casadevall A. 2000. Melanisation of Cryptococcus neoformans in human brain tissue. Lancet 355:2049–2050 [DOI] [PubMed] [Google Scholar]
- 48. Rosas AL, Nosanchuk JD, Feldmesser M, Cox GM, McDade HC, Casadevall A. 2000. Synthesis of polymerized melanin by Cryptococcus neoformans in infected rodents. Infect. Immun. 68:2845–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rosas AL, Nosanchuk JD, Gómez BL, Edens WA, Henson JM, Casadevall A. 2000. Isolation and serological analyses of fungal melanins. J. Immunol. Methods 244:69–80 [DOI] [PubMed] [Google Scholar]
- 50. Casadevall A, Rosas AL, Nosanchuk JD. 2000. Melanin and virulence in Cryptococcus neoformans. Curr. Opin. Microbiol. 3:354–358 [DOI] [PubMed] [Google Scholar]
- 51. Romero-Martinez R, Wheeler M, Guerrero-Plata A, Rico G, Torres-Guerrero H. 2000. Biosynthesis and functions of melanin in Sporothrix schenckii. Infect. Immun. 68:3696–3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Morris-Jones R, Youngchim S, Gómez BL, Aisen P, Hay RJ, Nosanchuk JD, Casadevall A, Hamilton AJ. 2003. Synthesis of melanin-like pigments by Sporothrix schenckii in vitro and during mammalian infection. Infect. Immun. 71:4026–4033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gómez BL, Nosanchuk JD, Díez S, Youngchim S, Aisen P, Cano LE, Restrepo A, Casadevall A, Hamilton AJ. 2001. Detection of melanin-like pigments in the dimorphic fungal pathogen Paracoccidioides brasiliensis in vitro and during infection. Infect. Immun. 69:5760–5767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nosanchuk JD, Gómez BL, Youngchim S, Díez S, Aisen P, Zancopé-Oliveira RM, Restrepo A, Casadevall A, Hamilton AJ. 2002. Histoplasma capsulatum synthesizes melanin-like pigments in vitro and during mammalian infection. Infect. Immun. 70:5124–5131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hamilton AJ, Holdom MD. 1999. Antioxidant systems in the pathogenic fungi of man and their role in virulence. Med. Mycol. 37:375–389 [DOI] [PubMed] [Google Scholar]
- 56. Howard RJ, Valent B. 1996. Breaking and entering: host penetration by the fungal rice blast pathogen Magnaporthe grisea. Annu. Rev. Microbiol. 50:491–512 [DOI] [PubMed] [Google Scholar]
- 57. Tseng MN, Chung PC, Tzean SS. 2011. Enhancing the stress tolerance and virulence of an entomopathogen by metabolic engineering of dihydroxynaphthalene melanin biosynthesis genes. Appl. Environ. Microbiol. 77:4508–4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li CY. 1981. Phenoloxidase and peroxidase activities in zone lines of Phellinus weirii. Mycologia 73:811–821 [Google Scholar]
- 59. Kubicek CP, Herrera-Estrella A, Seidl-Seiboth V, Martinez DA, Druzhinina IS, Thon M, Zeilinger S, Casas-Flores S, Horwitz BA, Mukherjee PK, Mukherjee M, Kredics L, Alcaraz LD, Aerts A, Antal Z, Atanasova L, Cervantes-Badillo MG, Challacombe J, Chertkov O, et al. 2011. Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma. Genome Biol. 12:R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nemcovic M, Jakubíková L, Víden I, Farkas V. 2008. Induction of conidiation by endogenous volatile compounds in Trichoderma spp. FEMS Microbiol. Lett. 284:231–236 [DOI] [PubMed] [Google Scholar]
- 61. Siddiquee S, Cheong BE, Taslima K, Kausar H, Hasan MM. 2012. Separation and identification of volatile compounds from liquid cultures of Trichoderma harzianum by GC-MS using three different capillary columns. J. Chromatogr. Sci. 50:358–367 [DOI] [PubMed] [Google Scholar]
- 62. Savage TJ, Hristova MK, Croteau R. 1996. Evidence for an elongation/reduction/C1-elimination pathway in the biosynthesis of n-heptane in xylem of Jeffrey pine. Plant Physiol. 111:1263–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Metz B, Seidl-Seiboth V, Haarmann T, Kopchinskiy A, Lorenz P, Seiboth B, Kubicek CP. 2011. Expression of biomass-degrading enzymes is a major event during conidium development in Trichoderma reesei. Eukaryot. Cell 10:1527–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Portnoy T, Margeot A, Le Crom S, Linke R, Atanasova L, Fekete E, Sándor E, Karaffa L, Druzhinina IS, Seiboth B, Kubicek CP. 2011. The CRE1 carbon catabolite repressor of the fungus Trichoderma reesei: a master regulator of carbon assimilation. BMC Genomics 12:269. 10.1186/1471-2164-12-269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Karimi-Aghcheh R, Bok JW, Phatale PA, Smith KM, Baker SE, Lichius A, Omann M, Zeilinger S, Seiboth B, Rhee C, Keller NP, Freitag M, Kubicek CP. 2013. Functional analyses of Trichoderma reesei LAE1 reveal conserved and contrasting roles of this regulator. G3 3:369–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.