Abstract
Erysipelothrix rhusiopathiae, the causative agent of swine erysipelas, is a facultative intracellular Gram-positive bacterium. It has been shown that animals immunized with a filtrate from E. rhusiopathiae cultures are protected against lethal challenge. In this study, we identified and characterized the extracellular proteins of E. rhusiopathiae to search for novel vaccine antigens. A concentrated culture supernatant from the E. rhusiopathiae Fujisawa strain, which has been found to induce protection in mice, was analyzed using two-dimensional electrophoresis. From more than 40 confirmed protein spots, 16 major protein spots were selected and subjected to N-terminal amino acid sequence determination, and 14 protein spots were successfully identified. The identified proteins included housekeeping proteins and other metabolic enzymes. We searched for surface-localized proteins by analyzing the genomes of two E. rhusiopathiae strains: Fujisawa and ATCC 19414. Genome analysis revealed that the ATCC 19414 strain has three putative surface-exposed choline-binding proteins (CBPs): CbpA, CbpB, and CbpC. Each CBP contains a putative choline-binding domain. The CbpC gene is mutated in Fujisawa, becoming a nonfunctional pseudogene. Immunogold electron microscopy confirmed that CbpA and CbpB, as well as the majority of the metabolic enzymes examined, are associated with the cell surface of E. rhusiopathiae Fujisawa. Immunization with recombinant CbpB, but not with other recombinant CBPs or metabolic enzymes, protected mice against lethal challenge. A phagocytosis assay revealed that antiserum against CbpB promoted opsonin-mediated phagocytosis by murine macrophages in vitro. The protective capabilities of CbpB were confirmed in pigs, suggesting that CbpB could be used as a vaccine antigen.
INTRODUCTION
The Gram-positive, non-spore-forming, rod-shaped bacterium Erysipelothrix rhusiopathiae represents a new class, Erysipelotrichia, in the phylum Firmicutes. E. rhusiopathiae has been referred to as the “walled relative” of mycoplasma (1), and E. rhusiopathiae is phylogenetically close to Mollicutes (a class that includes the genus Mycoplasma); the genomic features of this organism represent evolutionary traits of both Firmicutes and Mollicutes (2, 3).
E. rhusiopathiae is ubiquitous in nature and has been isolated from many species of wild and domestic mammals, birds, reptiles, amphibians, and fish (4). E. rhusiopathiae is generally regarded as an opportunistic animal pathogen that causes a variety of diseases in several species of mammals and birds (4). In swine, E. rhusiopathiae can cause erysipelas, which may present as acute septicemia or chronic endocarditis and polyarthritis, resulting in great economic losses to the swine industry (4). Live attenuated vaccines and bacterins have long been used successfully to control swine erysipelas (4). However, the high incidence of swine erysipelas is still a concern for the pork industry worldwide (5–8); therefore, the development of new and more effective vaccines is desired.
In Gram-positive bacteria, secreted and cell surface proteins play a fundamental role in pathogenesis, and they may be good targets for vaccine development (9). In many cases, these surface proteins are covalently anchored to peptidoglycan by the LPXTG motif and interact with components of the host extracellular matrix to adhere to, colonize, and invade cells and tissues (10). In other cases, surface proteins, including Streptococcus pneumoniae choline-binding proteins (CBPs) such as PspA and CbpB, contain tandem repeats beginning with the dipeptide Gly-Trp (GW repeat) at their C termini. S. pneumoniae possesses 10 to 15 different CBPs, and the GW repeat binds to teichoic acid or lipoteichoic acid (LTA) polymers in the cell wall (11–16). There is also a family of cell surface adhesins and invasins of Gram-positive bacteria that do not possess a signal peptide or a peptidoglycan anchor (17, 18). Glycolytic enzymes, including glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and an enolase, which are generally located in the cytosol and do not possess a signal sequence or a peptidoglycan anchor, are located on the surface of Streptococcus pyogenes. These glycolytic enzymes either bind to host components, such as fibronectin and plasminogen, or interact directly with the host cells, thereby enabling pathogens to colonize and invade the host tissue (19, 20). These enzymes are present on the surfaces of nearly all Streptococcus species and other Gram-positive bacteria, indicating that these anchorless housekeeping enzymes can associate with the cell surface after their secretion (17, 18, 21).
Our recent analysis of the genome of E. rhusiopathiae suggested that this bacterium might have an atypical cell wall. E. rhusiopathiae lacks many orthologous genes for the biosynthesis of wall teichoic acids (WTA) and LTA, as well as the dltABCD operon, which is responsible for the incorporation of d-alanine into WTA and LTA (2). Furthermore, recent analysis of the capsular polysaccharide of E. rhusiopathiae indicated that this bacterium indeed has a complex, heterogeneous capsule that is modified by phosphocholine (22). The same study also suggested that there could be some overlap between the biosynthesis pathways of capsular polysaccharides and other cell wall components, including WTA and/or LTA (22). Thus, in addition to its unique phylogenetic position as a member of the Firmicutes, it appears that the cell wall structure of E. rhusiopathiae may also be unique among Gram-positive bacteria.
Irrespective of the above-mentioned findings, the surface display mechanisms of E. rhusiopathiae proteins appear to be similar to those of proteins from other Gram-positive bacteria. The E. rhusiopathiae strain Fujisawa possesses at least 24 surface proteins that contain either an LPXTG motif or a GW repeat (2). Protective antigens are present in E. rhusiopathiae culture supernatants (23–27); however, the nature of these protective antigens is entirely unknown. Thus far, only a few protective antigens have been identified. A 64- to 66-kDa protein found in Triton X-100 extracts of bacterial antigens and whole-cell lysates has been reported to induce partial protection in mice (28). However, surface localization of this protein was not demonstrated. Two surface-localized proteins, surface protective antigen (Spa) (29, 30) and rhusiopathiae surface protein A (RspA) (31), have been shown to function as protective antigens, and these proteins have been detected in supernatants (30, 31).
Although several extracellular proteins produced by E. rhusiopathiae have been characterized, there has been no systematic analysis of the proteins in culture supernatants from this organism. Thus, it is unclear whether the culture supernatant proteins of E. rhusiopathiae are similar to those of other Gram-positive bacteria or whether the supernatants contain other, unknown protective antigens.
In this study, we systematically analyzed the culture supernatant proteins of E. rhusiopathiae. To identify protein antigens in the culture supernatant, we employed a proteomic approach using two-dimensional (2-D) gel electrophoresis and N-terminal amino acid sequencing. We also examined the putative CBPs identified by genome analysis of E. rhusiopathiae strain Fujisawa (serotype 1a) and E. rhusiopathiae strain ATCC 19414 (serotype 2). We examined the surface association of the extracellular proteins by immunogold electron microscopy and further analyzed the protective capacities of the proteins in mice and pigs.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The wild-type E. rhusiopathiae strain Fujisawa (serotype 1a), originally isolated from a septicemic pig, and the E. rhusiopathiae type strain, ATCC 19414 (serotype 2), were used. The bacteria were grown either in brain heart infusion (BHI; Difco Laboratories, Detroit, MI) containing 0.1% Tween 80 (pH 7.6) (BHI-T80) or in the dialyzable components of BHI-T80 medium (see below). Escherichia coli M15 (Qiagen GmbH, Hilden, Germany) was grown in Luria-Bertani medium. When appropriate, the medium was supplemented with ampicillin (100 μg/ml), kanamycin (50 μg/ml), or isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM).
Identification of secreted proteins. (i) Preparation of supernatant samples.
To exclude macromolecules derived from the medium and Tween 80 micelles, E. rhusiopathiae strain Fujisawa was grown at 37°C for 16 h in the dialyzable components of BHI-T80 medium, from which macromolecules with molecular masses of more than 14,000 Da and Tween 80 micelles were filtered out using a cellulose membrane (molecular weight cutoff [MWCO], 14,000) as previously described (32). After centrifugation of the overnight culture, the supernatant was passed through 0.45-μm-pore-size membrane filters and dialyzed against phosphate-buffered saline (PBS) at 4°C. The supernatant was concentrated using a Vivaspin 20 device (MWCO, 10,000; Sartorius Stedim Biotech, Göttingen, Germany), and the proteins were purified with a cleanup kit (Bio-Rad Laboratories, Hercules, CA). The purified proteins were dissolved in a rehydration buffer and used for 2-D gel electrophoresis. As a control, the dialyzable components of BHI-T80 were concentrated, and the proteins were purified with the cleanup kit. The protein concentration was determined with a bicinchoninic acid (BCA) protein assay kit (Thermo Scientific, Rockford, IL).
(ii) 2-D gel electrophoresis analysis.
The protein sample (185 μl, 200 μg) and an equivalent amount of control sample were applied to a rehydration equilibration tray (Bio-Rad, Hercules, CA) onto which an IPG strip (11 cm, pI 4 to 7) was laid. The tray was fixed on a Protean IEF cell (Bio-Rad), and isoelectric focusing (IEF) gel electrophoresis was performed. The strips were rehydrated overnight and focused with a program of 250 V for 15 min followed by 8,000 V for 35,000 V-h, with a current limit of 50 μA. After IEF gel electrophoresis, the strips were reduced and equilibrated with equilibration buffers I and II (Bio-Rad), which contained dithiothreitol and iodoacetamide, respectively, for 15 min. The strips were then subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with 4% stacking and 12.5% resolving Tris-HCl gels (Bio-Rad). The proteins were transferred to polyvinylidene difluoride membranes (Immobilon-P; Millipore, Bedford, MA), which were then stained with Coomassie brilliant blue (CBB) R-250 (Nacalai Tesque, Kyoto, Japan) and destained with water containing 25% methanol and 7% acetic acid. The protein spots were excised from the membrane and subjected to N-terminal amino acid sequence analysis.
(iii) N-terminal amino acid sequence analysis.
The N-terminal amino acid sequence was analyzed with a Procise 491 HT automated protein sequencer (Applied Biosystems, Foster City, CA), and the five N-terminal amino acids were sequenced using the Edman degradation method. The protein was first incubated with phenylisothiocyanate. After cleavage of each successive N-terminal amino acid, the anilinothiazolinone derivative formed was converted to the more stable phenylthiohydantoin (PTH) derivative. The PTH derivatives were separated by online reverse-phase high-performance liquid chromatography using a C18 column with solvents supplied by the manufacturer and were identified by comparison with the PTH-amino acid standards. After the N-terminal amino acid sequences were obtained, the corresponding putative genes in the E. rhusiopathiae Fujisawa genome (accession no. AP012027) (2) were identified using in silico Molecular Cloning Genomics Edition software (33) (In Silico Biology, Yokohama, Japan), and their nucleotide sequences were determined.
Identification of putative CBPs by E. rhusiopathiae genome analysis.
The genome sequence of the E. rhusiopathiae type strain, i.e., ATCC 19414 (serotype 2), which consists of four large contigs (accession no. ACLK 00000000.2), was obtained from the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov/genomes/MICROBES/microbial_taxtree.html) and compared to the sequence of Fujisawa (accession no. AP012027) (2). ATCC 19414 strain-specific coding sequences (CDS) with a putative choline-binding domain (the GW module) at the C terminus were identified.
Cloning, expression, and purification of recombinant proteins.
Recombinant proteins were expressed and purified as previously described (30). Briefly, genomic DNA from the Fujisawa strain was PCR amplified with the KOD FX polymerase (Toyobo, Osaka, Japan) and the primers shown in Table 1. The PCR products were digested with restriction enzymes and cloned into a pQE-30 expression vector (Qiagen Inc., Santa Clarita, CA) that had been predigested with the same enzymes (Table 1). The constructs were transformed into E. coli M15 cells by electroporation, and positive clones were confirmed by sequencing. Histidine (His)-tagged recombinant protein was purified with a QIAexpress kit (Qiagen Inc., Valencia, CA) according to the manufacturer's protocol. The fractions containing recombinant protein were pooled, dialyzed with PBS at 4°C, and subsequently used for immunization. The purified protein concentration was determined using a Pierce BCA protein assay kit.
Table 1.
Primers used in this study
| Protein target | Primer sequence (5′–3′)a |
|---|---|
| GroEL | cgcggatccATGGCTAAAGATGTACGTTAT |
| cccaagcttTTAATACATTGGCGGCATTTC | |
| FBA | cgcggatccATGGCATTAGTATCAGCTAAA |
| aactgcagTTAAGCTTGGTTAGCTGAACCC | |
| TPI | cgcggatccATGAGAAAACCGATTATCTTT |
| cccaagcttTTATTTAATTGCATCGATGAT | |
| PGP | cgcggatccATGAAAGTTTTAATTACAGGT |
| acgcgtcgacTTAGCTGATTGCGCCACCTT | |
| GAPDH | cgcggatccATGACAGTTAAAGTAGCAATT |
| cccaagcttTTAGAATTTTGAAGCAACGTA | |
| IMPDH | cgcggatccATGATGAATGGTAAAGTAATT |
| acgcgtcgacTTATTTGCCATGATAATTTG | |
| LysA | cgcggatccATGAATCTGAAAGAACTGTGT |
| cccaagcttTTATTTCTCCTTAACTGAATT | |
| CbpA | cgggatccTTTGAAGAACCGGATGCATCAT |
| cccaagcttTTAAAGCATACGACCTGAACG | |
| CbpB | cccgagctcGAGGAGCGAAAACCGAGAGAA |
| cccaagcttCTATTTTTTTAGTGCTCCTTG | |
| CbpC | cgggatccCAAGAACTAAAACAAGCAGAAC |
| aactgcagTTATAACCAGACCCCACTCTGA |
Lowercase letters indicate the linker containing a restriction enzyme site for cloning.
ELISA.
The antibody response in the animals was examined by an enzyme-linked immunosorbent assay (ELISA) with recombinant proteins as described previously (34). Briefly, recombinant proteins were diluted to 10 μg/ml in carbonate buffer (pH 9.6) and used to coat the wells of polystyrene plates (100 μl/well). Mouse and pig sera were diluted 1:100 in PBS, and the bound antibodies were detected with horseradish peroxidase-conjugated goat anti-mouse or rabbit anti-pig IgG (Zymed Laboratories). Sera from animals immunized with PBS containing adjuvant were used as negative controls.
Confirmation of surface expression.
For immunogold electron microscopy, samples were collected 6 h and 16 h after culture initiation. The bacteria were placed on Formvar-coated nickel grids (400 mesh; Nisshin, Tokyo, Japan) and washed with PBS. The grids were then incubated with antisera (diluted 1:100 in PBS), followed by incubation with 10-nm colloidal gold-conjugated goat anti-mouse immunoglobulins (IgG, IgM, and IgA) (BB International, Cardiff, United Kingdom). The grids were washed with PBS, incubated with 1% ammonium acetate solution for 10 min, and air dried. The dried grids were observed with a transmission electron microscope (model H-7500; Hitachi, Japan). Grids that had been incubated with PBS and with antiserum against recombinant SpaA were included as negative and positive controls, respectively.
Animal experiments.
All animal experiments in this study were performed according to the regulations and guidelines of the Animal Ethics Committee of the National Institute of Animal Health, Tsukuba, Ibaraki, Japan.
To examine the immunogenicity of the recombinant proteins (seven supernatant proteins and three putative CBPs), 10 groups of eight 6- to 8-week-old female BALB/c mice (Japan SLC, Inc., Hamamatsu, Japan) were immunized with each recombinant protein (see Fig. S1 in the supplemental material). Each mouse was injected twice (once subcutaneously [s.c.] and once intramuscularly [i.m.]) with 100 μl (3.6 μg) of each recombinant protein sample, containing 10% Polygen adjuvant (MVP Laboratories, Inc., Omaha, NE). Eight mice were injected with PBS containing the adjuvant as a control. After 2 weeks, the mice were boosted at the same injection sites with the same dose of the same protein sample. One week after boosting, blood was collected from each mouse, and the pooled sera were assayed for an antibody response by Western blotting (see Fig. S1). After 3 days, for a preliminary vaccine potency trial, these mice were challenged s.c. with 0.1 ml of a bacterial suspension containing 103 CFU (approximately 100 times the 50% lethal dose [LD50]) of Fujisawa and observed for death (see Table S1).
To examine the vaccine efficacy of CbpB, which has been found to exhibit a potential protective capability (see Table S1 in the supplemental material), a group of 10 6- to 8-week-old female BALB/c mice were immunized i.m. with approximately 10 times the previous dose of recombinant CbpB protein (30 μg) emulsified in Freund's complete adjuvant. After 2 weeks, the mice received an i.m. booster of the same protein (30 μg) emulsified in Freund's incomplete adjuvant. As negative controls, two groups of 10 mice each were inoculated with PBS or CbpA, both of which were emulsified in Freund's adjuvant. Seven days later, blood was collected from each mouse. Then, after 3 days, the mice were challenged s.c. with 3.5 × 102 CFU of the Fujisawa strain and observed for 17 days for clinical symptoms or death. The collected blood samples were subjected to antibody response analysis via ELISA, using the recombinant proteins as antigens.
The vaccine efficacy of CbpB was further examined in germfree piglets obtained through caesarian section. Seven 2-week-old piglets were immunized i.m. with 1 mg of recombinant CbpB protein emulsified in Freund's complete adjuvant. As a control, three piglets were inoculated with PBS emulsified in Freund's complete adjuvant. After 2 weeks, the piglets were given an i.m. booster of 1 mg of the same protein or PBS, both of which were emulsified in Freund's incomplete adjuvant. Seven days later, the piglets were challenged i.m. with 3.8 × 107 CFU of the Fujisawa strain and were observed for 10 days for death or clinical symptoms.
Phagocytosis assay.
A phagocytosis assay was performed essentially as described previously (35), except that the murine macrophage-like J774.1 cell line was used. Briefly, 90-μl bacterial suspensions containing 107 CFU of Fujisawa in PBS were sensitized for 10 min by incubation with 10 μl normal serum or immune serum from mice immunized with recombinant CbpB. Subsequently, the suspensions were incubated with rotation at 37°C for 60 min with murine macrophage-like J774.1 cells (106). Phagocytosis was stopped by shaking the tubes in ice water; uningested bacteria were removed by washing four times with cold PBS. The cells were resuspended in cold PBS, and cytospin smears were prepared, Gram stained, and examined by light microscopy under oil immersion. The experiment was repeated three times, and the results are expressed as the phagocytosis index, which is defined as the percentage of bacterium-ingesting macrophages times the average number of bacteria in a macrophage times 100.
Statistical analysis.
Statistical significance was determined by Student's unpaired t test. For the vaccine studies, the mortality/survival profiles for each infected group were plotted and analyzed via the log rank test, using GraphPad Prism 5 software.
RESULTS
Identification of E. rhusiopathiae extracellular proteins. (i) Culture supernatant proteins.
It has been shown that a protective antigen(s) is present in the culture filtrate from the attenuated E. rhusiopathiae strain Koganei 65-0.15 (serotype 1a) (23). We confirmed that a 10-fold-concentrated culture supernatant from the Fujisawa strain could confer partial protection in mice (unpublished result). In a preliminary experiment, we determined that all control mice (n = 3) inoculated with PBS with Freund's adjuvant died within 3 days of challenge, whereas all mice (n = 8) immunized with the 30-fold-concentrated culture supernatant from the Fujisawa strain with Freund's adjuvant survived for 14 days after lethal challenge with 2.6 × 103 CFU of the homologous strain (P < 0.01) (data not shown). The protective antigens SpaA and RspA have been detected in supernatants of the Fujisawa strain (30, 31); however, it is unknown whether additional protective antigens are present in the supernatant.
To search for supernatant proteins with potential protective capabilities, E. rhusiopathiae Fujisawa was grown to stationary phase, and a 2-D protein profile of the culture supernatant was analyzed. No protein spots were observed for the control, which consisted of concentrated dialyzable components of BHI-T80 (data not shown). In the culture supernatant from E. rhusiopathiae Fujisawa, approximately 40 protein spots were detected (Fig. 1); among these, 16 major protein spots were selected, and their N-terminal amino acid sequences were determined. Two pairs of protein spots (spots 2 and 25 and spots 11 and 12) were revealed to have the same sequences and similar masses but different isoelectric points in the 2-D gel, suggesting that they were charge variants. The N-terminal amino acid sequence data were used to query the Fujisawa genome database. Fourteen of the 16 amino acid sequences were 100% identical to the inferred sequences of open reading frames (ORFs) present in the Fujisawa genome sequence (Table 2). Amino acid sequence analysis revealed that the supernatant included several housekeeping glycolytic enzymes, including class II fructose-1,6-bisphosphate aldolase (FBA), triosephosphate isomerase (TPI), and GAPDH. The supernatant also contained other metabolic enzymes, including the pyruvate dehydrogenase complex E1 component α-subunit (PDHα), which is involved in the conversion of pyruvate to acetyl-coenzyme A (acetyl-CoA), as well as adenylate kinase (AK) and inosine-5′-monophosphate dehydrogenase (IMPDH), both of which are involved in purine metabolism. Other proteins identified in the supernatant included the chaperonin GroEL, ribosomal recycling factor, diaminopimelate decarboxylase (LysA), and type I pyroglutamyl-peptidase (PGP).
Fig 1.
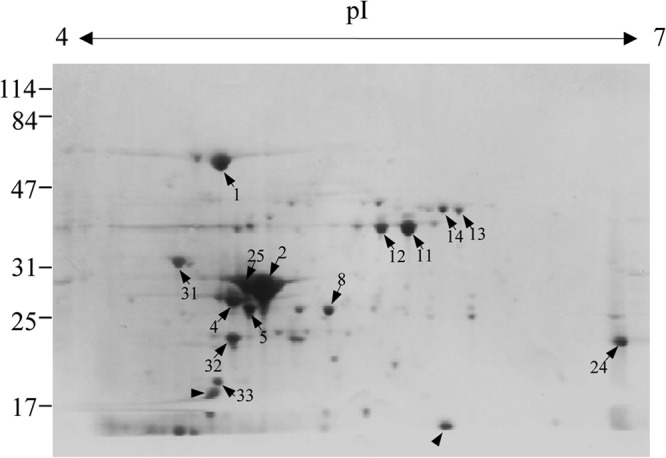
Two-dimensional gel electrophoresis profile of the secreted proteins of E. rhusiopathiae. The sample was prepared from a supernatant of the E. rhusiopathiae Fujisawa strain that was cultured at 37°C for 16 h in dialyzable components of BHI-T80 medium. The spots labeled with Arabic numerals were selected for N-terminal amino acid sequencing. Arrowheads indicate the spots whose amino-terminal sequences were not determined by Edman degradation. The positions of the protein molecular mass standards (kDa) are shown to the left.
Table 2.
Proteins identified in the culture supernatant of E. rhusiopathiae Fujisawa
| Spot no. | N-terminal amino acid sequence | Locus tag | Identical or homologous protein | pI/molecular mass (kDa) |
|---|---|---|---|---|
| 1 | AKDVR | ERH_1267 | Chaperonin GroEL | 4.56/58.4 |
| 2 | ALVSA | ERH_1632 | Fructose-1,6-bisphosphate aldolase, class II (FBA) | 4.73/30.3 |
| 4 | MRKPI | ERH_1335 | Triosephosphate isomerase (TPI) | 4.59/26.9 |
| 5 | MNLLI | ERH_1152 | Adenylate kinases (AK) | 4.70/24.4 |
| 8 | MKVLI | ERH_0382 | Type I pyroglutamyl-peptidase (PGP) | 5.00/23.1 |
| 11 | TVKVA | ERH_1534 | Glyceraldehyde-3-phosphate dehydrogenase, type I (GAPDH) | 5.32/35.5 |
| 12 | TVKVA | ERH_1534 | Glyceraldehyde-3-phosphate dehydrogenase, type I (GAPDH) | 5.32/35.5 |
| 13 | MMNGK | ERH_1297 | Inosine-5′-monophosphate dehydrogenase (IMPDH) | 5.52/39.5 |
| 14 | AQVKK | ERH_0438 | Pyruvate dehydrogenase complex E1 component, α-subunit (PDHα) | 5.34/41.6 |
| 24 | MNLKE | ERH_0207 | Diaminopimelate decarboxylase (LysA) | 5.87/44.5a |
| 25 | ALVSA | ERH_1632 | Fructose-1,6-bisphosphate aldolase, class II (FBA) | 4.73/30.3 |
| 31 | DANVD | ERH_1662 | 3D domain-containing hypothetical protein | 4.49/28.7a |
| 32 | MQSII | ERH_0644 | Ribosome recycling factor | 4.68/20.2 |
| 33 | LTLRA | ERH_0928 | Hypothetical protein | 5.12b/19.6 |
The molecular mass of the protein observed in the 2-D gel was lower than the predicted value shown in the table.
The isoelectric point of the protein observed in the 2-D gel was higher than the predicted value shown in the table.
(ii) Putative choline-binding proteins.
We searched for surface-localized proteins and proteins containing the GW module encoded in the genome sequences of E. rhusiopathiae Fujisawa (serotype 1a) and ATCC 19414 (serotype 2). In the Fujisawa strain, we identified CbpA (ERH_0407), which contains a GW module at the N and C termini, and CbpB (ERH_0768), which contains a GW module at the C terminus (Fig. 2), in addition to SpaA (ERH_0094), which is the major protective antigen of the organism (29, 30, 36) and also contains a GW module at the C terminus. In ATCC 19414, an additional protein (designated CbpC) was identified. CbpC contains a GW module at its C terminus and was truncated in Fujisawa (ERH_0738) due to a frameshift mutation. PCR and sequencing analyses confirmed that the CbpC gene was also functional in the Koganei 65-0.15 strain (serotype 1a), which is the vaccine strain in Japan (data not shown).
Fig 2.
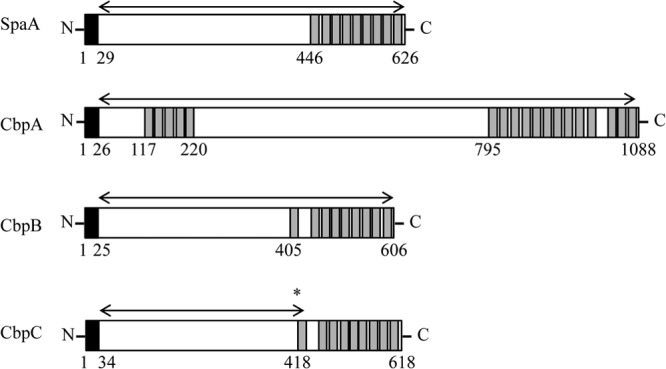
Schematic representation of the domain structures of CBPs of E. rhusiopathiae Fujisawa. The signal sequence and choline-binding domain are indicated by black and gray boxes, respectively. Arrows indicate the regions of the recombinant proteins constructed. CbpC of Fujisawa is truncated by a frameshift mutation from a single nucleotide deletion (asterisk) in cbpC, and therefore, the schematic of CbpC is depicted as if it has no mutation. Numbers indicate the positions of amino acids.
Recombinant extracellular proteins and their immunogenicity in mice.
A His tag expression system was used to express the recombinant extracellular proteins of E. rhusiopathiae in E. coli. We selected nine secreted proteins that were abundant in the supernatant, as well as three CBPs, and cloned the corresponding genes into E. coli. Of the nine secreted proteins, we confirmed the expression of seven proteins; however, the expression of the other two proteins (PDHα and a hypothetical protein [ERH_0928]) was unsuccessful. The seven recombinant supernatant proteins and the three putative CBPs were purified and separated by SDS-PAGE (see Fig. S1 in the supplemental material). To examine the immunogenicity of the extracellular proteins, we immunized mice with two 3.6-μg doses of the recombinant proteins, given 2 weeks apart, and the antibody responses to the proteins were analyzed by Western blotting. With the exception of GroEL, all the recombinant culture supernatant proteins and CBPs were confirmed to be immunogenic in mice (see Fig. S1).
Surface expression analysis by immunogold electron microscopy and dot immunoblotting.
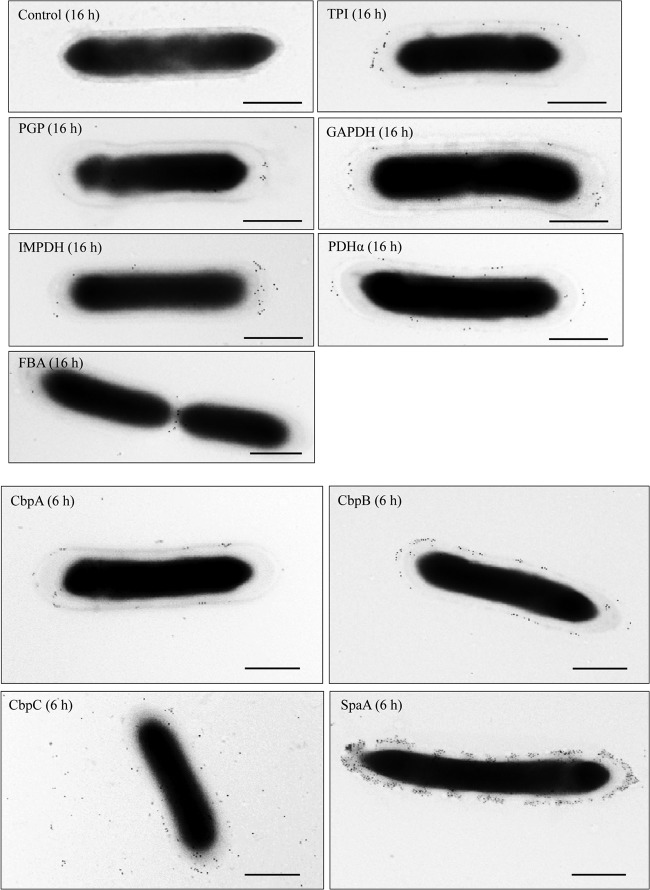
In Gram-positive bacteria, glycolytic enzymes secreted into the supernatant can reassociate with the cell surface after secretion (17, 18, 21). Immunogold electron microscopy confirmed that a majority of the supernatant proteins, with the exception of FBA, were indeed localized on the cell surface of the Fujisawa strain (Fig. 3). For FBA, which was detected as a major spot in the 2-D protein profile (Fig. 1), gold particles were often observed at the cell division site; however, peripheral localization was not demonstrated (Fig. 3). Furthermore, CbpA and CbpB were located on the cell surface of Fujisawa, as was SpaA. CbpC was dispersed, although some gold particles remained on the cell surface (Fig. 3). Using dot immunoblotting, we observed the presence of CbpB on the cell surface of ATCC 19414 and Fujisawa cells, and we found that the CbpB signal was much stronger in ATCC 19414 than in Fujisawa (data not shown). CbpA was detected in both strains. As expected, CbpC, which is truncated in Fujisawa due to a frameshift mutation, was barely detected in that strain (data not shown).
Fig 3.
Immunogold electron microscopy analysis of the surface association of extracellular proteins of E. rhusiopathiae Fujisawa. E. rhusiopathiae Fujisawa was collected 6 h and 16 h after culture initiation. The bacteria were incubated with antisera, followed by incubation with 10-nm-diameter colloidal gold-conjugated goat anti-mouse immunoglobulin. Bacteria incubated with antiserum against recombinant SpaA and with PBS were included as positive and negative controls, respectively. Numbers in parentheses indicate the time of collection after culture initiation. Bars = 500 nm.
Protection experiments in animals.
We found that the mice immunized with either the culture supernatant proteins, CbpA, or truncated CbpC died within 5 days, as did the control mice. The mean time to death was significantly prolonged only for the mice immunized with CbpB (see Table S1 in the supplemental material).
To further examine the potential of CbpB as a vaccine antigen, mouse protection assays were performed. Ten mice were immunized with two 30-μg doses of recombinant CbpB each, and two additional groups of 10 mice were immunized with recombinant CbpA or PBS as negative controls. The mice immunized with CbpB developed IgG antibodies against CbpB; however, the IgG responses to CbpA were low and variable (Fig. 4). After a challenge infection with 3.5 × 102 CFU (approximately 10 times the 50% lethal dose) of the Fujisawa strain, two mice immunized with CbpB showed clinical symptoms, such as depression, and died. However, the other 8 mice survived, with no clinical symptoms. Mice immunized with CbpA or PBS died within 4 days after the challenge infection (P < 0.01 compared with CbpB group) (Fig. 5). Sera obtained from the CbpB-immunized mice promoted the phagocytosis of E. rhusiopathiae by murine macrophages (Fig. 6). This result strongly suggests that the anti-CbpB antibodies act as opsonins, similar to the antibodies against SpaA, which is a major protective antigen of E. rhusiopathiae (30, 36).
Fig 4.
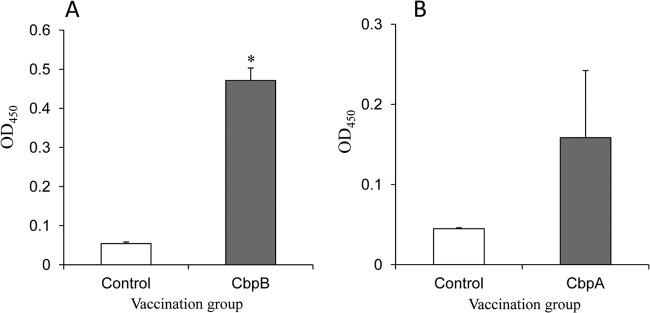
ELISA detection of anti-CBP IgG antibodies in the sera of mice inoculated with recombinant CbpB (A) and CbpA (B). The results are expressed as means with standard deviations for the control (n = 10) and CBP-vaccinated (n = 10) mice. *, P < 0.01. OD450, optical density at 450 nm.
Fig 5.
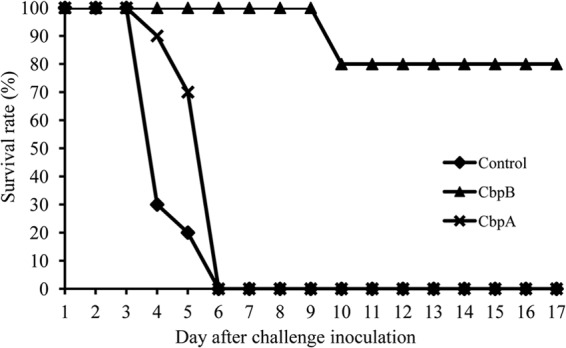
Survival of CBP-immunized mice after challenge with E. rhusiopathiae Fujisawa. Mice were immunized with two doses of CbpA, CbpB, or PBS and were challenged with 10 LD50 of Fujisawa 1 week after the final vaccination. Results are plotted as percent survival for each vaccination group (n = 10 mice per group).
Fig 6.
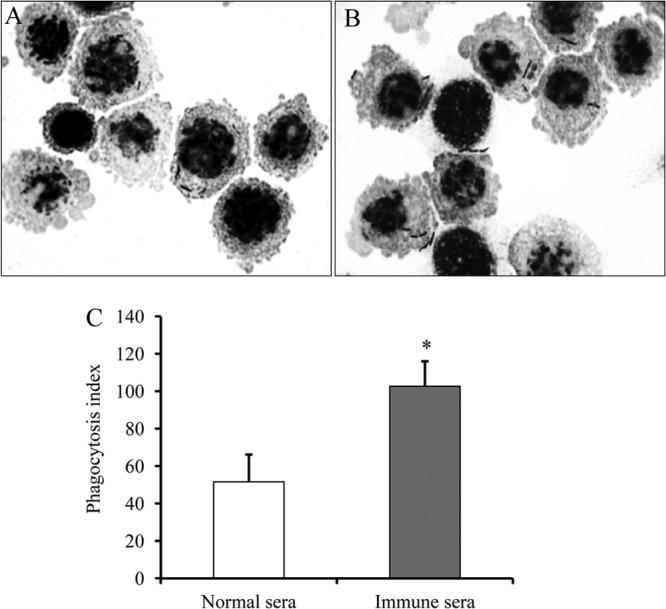
Phagocytosis by murine macrophage-like J774.1 cells of E. rhusiopathiae treated with normal sera (A) or antisera against CbpB (B). A suspension of J774.1 cells (106) was incubated with strain Fujisawa (107 CFU) under rotation at 37°C for 60 min. After removing uningested bacteria by washes, cytospin smears were prepared, Gram stained, and examined by light microscopy. (C) Results of phagocytosis assays. Results are expressed as mean phagocytic indexes (for three independent experiments) with standard deviations. *, P < 0.01.
The efficacy of CbpB as a vaccine antigen was further confirmed in pigs. All piglets in the control group (n = 3) died within 2 days following the challenge inoculation. In the immunized group (n = 7), one piglet died on day 3 after the challenge inoculation; however, the remaining piglets, including one piglet that developed systemic urticarial lesions, survived (P < 0.01). We confirmed that IgG antibodies were produced in the CbpB-vaccinated piglets at the time of the challenge inoculation (Fig. 7). We detected high levels of anti-CbpB IgG antibodies in convalescent-phase serum samples from nonvaccinated pigs that were naturally infected with E. rhusiopathiae (see Fig. S2 in the supplemental material). Taken together, these results suggest that CbpB is an immunodominant antigen generated by E. rhusiopathiae during the course of infection in pigs.
Fig 7.
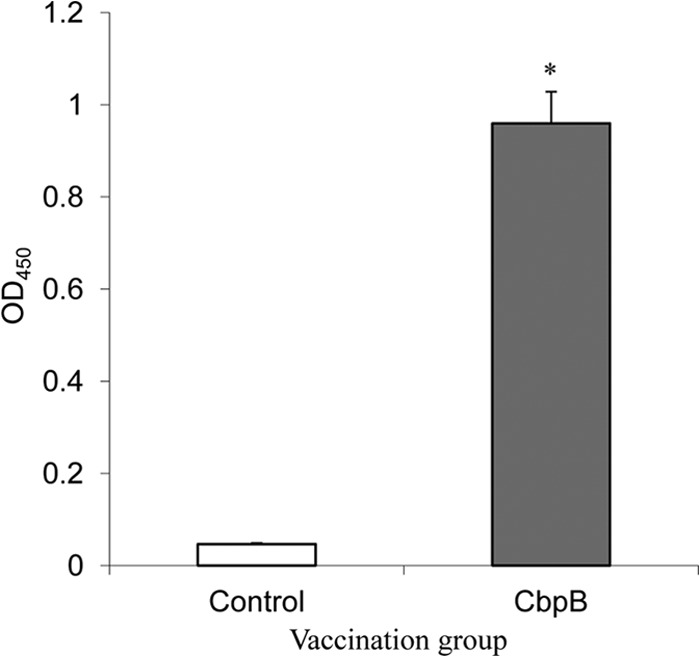
ELISA detection of anti-CbpB IgG antibodies in the sera of piglets inoculated with recombinant CbpB. The results are expressed as means with standard deviations for the control (n = 3) and CbpB-vaccinated (n = 7) piglets. *, P < 0.01. OD450, optical density at 450 nm.
DISCUSSION
In the present work, we investigated the immunogenicity, protective capability, and surface association of the extracellular proteins of E. rhusiopathiae.
The major protective antigen of E. rhusiopathiae is SpaA, which is a putative CBP (29, 30). Its precise function and its role in virulence have yet to be clarified; however, the importance of SpaA as an opsonin-inducing protective antigen is well known (30, 36). In this study, we found that an additional CBP, designated CbpB, is a novel protective antigen of E. rhusiopathiae. A phagocytosis assay confirmed that CbpB is also an opsonin-inducing protective antigen.
It has been shown that animals immunized with a culture filtrate from E. rhusiopathiae are protected against homologous and heterologous strains (23–25). As observed in other Firmicutes bacteria, anchorless housekeeping glycolytic enzymes, including TPI, FBA, and GAPDH, were present in the supernatant of the Fujisawa strain. We also found that GroEL, which is a highly conserved heat shock protein essential to all living organisms, was also present in the supernatant. Other proteins identified in the supernatant included type I PGP, a member of the C15 family of cysteine peptidases, and LysA, an enzyme that catalyzes the final step in lysine biosynthesis (37). PDHα and AK were also identified in the supernatant. In this study, we found that the majority of the culture supernatant proteins were immunogenic; however, we could not confirm their protective capabilities. It is possible that unexamined supernatant proteins, including AK and PDHα, might display protective properties. Alternatively, culture supernatant proteins with protective capabilities may have been degraded by proteases released following cell lysis, effectively preventing their detection. In this study, we confirmed the presence of CbpB in Fujisawa culture supernatants through Western blotting with serum from a germfree pig that was immunized with the CbpB protein (see Fig. S3 in the supplemental material). This result indicates that protective antigens that occur naturally on the cell surface of E. rhusiopathiae, including SpaA (30, 32) and RspA (31), are also present in the supernatant.
In summary, we demonstrated that a novel CBP, designated CbpB, elicits protective immunity by inducing opsonic antibodies, suggesting that CbpB could be used as a vaccine antigen to control erysipelas in animals. As observed for other Firmicutes bacteria, several metabolic enzymes, including glycolytic enzymes, are present in the supernatant of E. rhusiopathiae. However, we could not confirm the protective capabilities of these supernatant proteins. Our results suggest that CBPs are likely to be involved in the observed protection conferred by immunization with the supernatant of E. rhusiopathiae.
Supplementary Material
ACKNOWLEDGMENTS
We thank K. Shiraiwa for technical assistance.
This work was supported in part by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (to Y.S.).
Footnotes
Published ahead of print 9 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00549-13.
REFERENCES
- 1.Weisburg WG, Tully JG, Rose DL, Petzel JP, Oyaizu H, Yang D, Mandelco L, Sechrest J, Lawrence TG, Van Etten J. 1989. A phylogenetic analysis of the mycoplasmas: basis for their classification. J. Bacteriol. 171:6455–6467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ogawa Y, Ooka T, Shi F, Ogura Y, Nakayama K, Hayashi T, Shimoji Y. 2011. The genome of Erysipelothrix rhusiopathiae, the causative agent of swine erysipelas, reveals new insights into the evolution of Firmicutes and the organism's intracellular adaptations. J. Bacteriol. 193:2959–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis JJ, Xia F, Overbeek RA, Olsen GJ. 2013. Genomes of the class Erysipelotrichia clarify the firmicute origin of the class Mollicutes. Int. J. Syst. Evol. Microbiol. 63:2727–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wood RL. 1992. Erysipelas, p 475–486 In Leman AD, Straw BE, Mengeling WL, D'Allaire S, Taylor DJ. (ed), Diseases of swine, 7th ed. Iowa State University Press, Ames, IA [Google Scholar]
- 5.Bender JS, Shen HG, Irwin CK, Schwartz KJ, Opriessnig T. 2010. Characterization of Erysipelothrix species isolates from clinically affected pigs, environmental samples, and vaccine strains from six recent swine erysipelas outbreaks in the United States. Clin. Vaccine Immunol. 17:1605–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coutinho TA, Imada Y, Barcellos DE, Oliveira SJ, Moreno AM. 2011. Phenotypic and molecular characterization of recent and archived Erysipelothrix spp. isolated from Brazilian swine. Diagn. Microbiol. Infect. Dis. 69:123–129 [DOI] [PubMed] [Google Scholar]
- 7.Eamens GJ, Forbes WA, Djordjevic SP. 2006. Characterisation of Erysipelothrix rhusiopathiae isolates from pigs associated with vaccine breakdowns. Vet. Microbiol. 115:329–338 [DOI] [PubMed] [Google Scholar]
- 8.Nagai S, To H, Kanda A. 2008. Differentiation of Erysipelothrix rhusiopathiae strains by nucleotide sequence analysis of a hypervariable region in the spaA gene: discrimination of a live vaccine strain from field isolates. J. Vet. Diagn. Invest. 20:336–342 [DOI] [PubMed] [Google Scholar]
- 9.Fischetti VA. 2006. Surface proteins on gram-positive bacteria, p 12–25 In Fischetti VA, Novick RP, Ferretti JJ, Portnoy DA, Rood JI. (ed), Gram-positive pathogens, 2nd ed. ASM Press, Washington, DC [Google Scholar]
- 10.Navarre WW, Schneewind O. 1999. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63:174–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bergmann S, Hammerschmidt S. 2006. Versatility of pneumococcal surface proteins. Microbiology 152:295–303 [DOI] [PubMed] [Google Scholar]
- 12.Frolet C, Beniazza M, Roux L, Gallet B, Noirclerc-Savoye M, Vernet T, Di Guilmi AM. 2010. New adhesin functions of surface-exposed pneumococcal proteins. BMC Microbiol. 10:190. 10.1186/1471-2180-10-190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hakenbeck R, Madhour A, Denapaite D, Brückner R. 2009. Versatility of choline metabolism and choline-binding proteins in Streptococcus pneumoniae and commensal streptococci. FEMS Microbiol. Rev. 33:572–586 [DOI] [PubMed] [Google Scholar]
- 14.Jedrzejas MJ. 2001. Pneumococcal virulence factors: structure and function. Microbiol. Mol. Biol. Rev. 65:187–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenow C, Ryan P, Weiser JN, Johnson S, Fontan P, Ortqvist A, Masure HR. 1997. Contribution of novel choline-binding proteins to adherence, colonization and immunogenicity of Streptococcus pneumoniae. Mol. Microbiol. 25:819–829 [DOI] [PubMed] [Google Scholar]
- 16.Yother J, White JM. 1994. Novel surface attachment mechanism of the Streptococcus pneumoniae protein PspA. J. Bacteriol. 176:2976–2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chhatwal GS. 2002. Anchorless adhesins and invasins of Gram-positive bacteria: a new class of virulence factors. Trends Microbiol. 10:205–208 [DOI] [PubMed] [Google Scholar]
- 18.Pancholi V, Chhatwal GS. 2003. Housekeeping enzymes as virulence factors for pathogens. Int. J. Med. Microbiol. 293:391–401 [DOI] [PubMed] [Google Scholar]
- 19.Pancholi V, Fischetti VA. 1998. Alpha-enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J. Biol. Chem. 273:14503–14515 [DOI] [PubMed] [Google Scholar]
- 20.Pancholi V, Fischetti VA. 1992. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J. Exp. Med. 176:415–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boël G, Jin H, Pancholi V. 2005. Inhibition of cell surface export of group A streptococcal anchorless surface dehydrogenase affects bacterial adherence and antiphagocytic properties. Infect. Immun. 73:6237–6248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi F, Harada T, Ogawa Y, Ono H, Ohnishi-Kameyama M, Miyamoto T, Eguchi M, Shimoji Y. 2012. Capsular polysaccharide of Erysipelothrix rhusiopathiae, the causative agent of swine erysipelas, and its modification with phosphorylcholine. Infect. Immun. 80:3993–4003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi S, Sato H, Hirose K, Saito H. 1992. Immunological characterization of protective antigens prepared by alkaline treatment of whole cells and from the culture filtrate of Erysipelothrix rhusiopathiae. Vet. Microbiol. 30:73–85 [DOI] [PubMed] [Google Scholar]
- 24.Sato H, Hirose K, Saito H. 1995. Protective activity and antigenic analysis of fractions of culture filtrates of Erysipelothrix rhusiopathiae. Vet. Microbiol. 43:173–182 [DOI] [PubMed] [Google Scholar]
- 25.Sawada T, Takahashi T. 1987. Cross protection of mice and swine inoculated with culture filtrate of attenuated Erysipelothrix rhusiopathiae and challenge exposed to strains of various serovars. Am. J. Vet. Res. 48:239–242 [PubMed] [Google Scholar]
- 26.White RR, Verwey WF. 1970. Isolation and characterization of a protective antigen-containing particle from culture supernatant fluids of Erysipelothrix rhusiopathiae. Infect. Immun. 1:380–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White RR, Verwey WF. 1970. Solubilization and characterization of a protective antigen of Erysipelothrix rhusiopathiae. Infect. Immun. 1:387–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galán JE, Timoney JF. 1990. Cloning and expression in Escherichia coli of a protective antigen of Erysipelothrix rhusiopathiae. Infect. Immun. 58:3116–3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Makino S, Yamamoto K, Murakami S, Shirahata T, Uemura K, Sawada T, Wakamoto H, Morita H, Morita Y. 1998. Properties of repeat domain found in a novel protective antigen, SpaA, of Erysipelothrix rhusiopathiae. Microb. Pathog. 25:101–109 [DOI] [PubMed] [Google Scholar]
- 30.Shimoji Y, Mori Y, Fischetti VA. 1999. Immunological characterization of a protective antigen of Erysipelothrix rhusiopathiae: identification of the region responsible for protective immunity. Infect. Immun. 67:1646–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimoji Y, Ogawa Y, Osaki M, Kabeya H, Maruyama S, Mikami T, Sekizaki T. 2003. Adhesive surface proteins of Erysipelothrix rhusiopathiae bind to polystyrene, fibronectin, and type I and IV collagens. J. Bacteriol. 185:2739–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogawa Y, Minagawa Y, Shi F, Eguchi M, Muneta Y, Shimoji Y. 2012. Immunostimulatory effects of recombinant Erysipelothrix rhusiopathiae expressing porcine interleukin-18 in mice and pigs. Clin. Vaccine Immunol. 19:1393–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohyama A, Kurokawa K, Enai K, Saitoh H, Kanaya S, Altaf-Ul-Amin M, Ogasawara N. 2006. Bioinformatics tool for genomic era: a step towards the in silico experiments—focused on molecular cloning. J. Comp. Aid Chem. 7:102–115 [Google Scholar]
- 34.Shimoji Y, Oishi E, Kitajima T, Muneta Y, Shimizu S, Mori Y. 2002. Erysipelothrix rhusiopathiae YS-1 as a live vaccine vehicle for heterologous protein expression and intranasal immunization of pigs. Infect. Immun. 70:226–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimoji Y, Yokomizo Y, Sekizaki T, Mori Y, Kubo M. 1994. Presence of a capsule in Erysipelothrix rhusiopathiae and its relationship to virulence for mice. Infect. Immun. 62:2806–2810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Imada Y, Goji N, Ishikawa H, Kishima M, Sekizaki T. 1999. Truncated surface protective antigen (SpaA) of Erysipelothrix rhusiopathiae serotype 1a elicits protection against challenge with serotypes 1a and 2b in pigs. Infect. Immun. 67:4376–4382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scapin G, Blanchard JS. 1998. Enzymology of bacterial lysine biosynthesis. Adv. Enzymol. Relat. Areas Mol. Biol. 72:279–324 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.