Abstract
Bacteriophages are the largest reservoir of genetic diversity. Here we describe the novel phage ϕJM-2012. This natural isolate from marine Vibrio cyclitrophicus possesses very few gene contents relevant to other well-studied marine Vibrio phages. To better understand its evolutionary history, we built a mathematical model of pairwise relationships among 1,221 phage genomes, in which the genomes (nodes) are linked by edges representing the normalized number of shared orthologous protein families. This weighted network revealed that ϕJM-2012 was connected to only five members of the Pseudomonas ϕKZ-like phage family in an isolated network, strongly indicating that it belongs to this phage group. However, comparative genomic analyses highlighted an almost complete loss of colinearity with the ϕKZ-related genomes and little conservation of gene order, probably reflecting the action of distinct evolutionary forces on the genome of ϕJM-2012. In this phage, typical conserved core genes, including six RNA polymerase genes, were frequently displaced and the hyperplastic regions were rich in both unique genes and predicted unidirectional promoters with highly correlated orientations. Further, analysis of the ϕJM-2012 genome showed that segments of the conserved N-terminal parts of ϕKZ tail fiber paralogs exhibited evidence of combinatorial assortment, having switched transcriptional orientation, and there was recruitment and/or structural changes among phage endolysins and tail spike protein. Thus, this naturally occurring phage appears to have branched from a common ancestor of the ϕKZ-related groups, showing a distinct genomic architecture and unique genes that most likely reflect adaptation to its chosen host and environment.
INTRODUCTION
Bacteriophages, which are the most abundant biological entities on Earth, typically outnumber their bacterial hosts by about 10-fold (1, 2). In the ocean, phages act as major regulators of host populations by lysing between 10 and 50% of the bacteria each day (3). They have key effects in shaping viral and host genomes through horizontal gene transfers (HGTs), with an estimated infection frequency of up to 1015 times/s (4), and help select for infection-resistant bacteria (5). Over a wide range of spaces and a time scale exceeding 3 billion years, phages have greatly impacted microbial population dynamics, genomic diversity, and evolution (6, 7). Metagenomic studies of the oceanic viral fraction found only a few sequences homologous to known viruses (8, 9), indicating that a huge amount of viral diversity still remains to be explored.
The study of marine phage genomes is relatively new (10), with only about 35 marine phages completely sequenced to date (11). Nevertheless, these genomes have revealed some interesting features. For example, several marine Vibrio phages, including ϕVpV262 with T7-like features (12), T4-like ϕKVP40 (13), and the recently identified phage ϕSIO-2 (11), exhibit diverse morphologies, different patterns of host specificity, and/or unusual genomic architectures, reflecting their unique ecological and evolutionary properties. Furthermore, the presence of genes that share robust structural or sequence similarities with those found in unrelated bacteria and phages of nonmarine origin (14) enables us to infer the potential spread of these phages in the ocean and study the distribution of mobile genetic elements.
However, phage genomes are typically characterized by independent gene contents reflecting numerous HGT events (15), lack of universal genes (16), and frequent disconnects between genetic relatedness and morphological distinctions (17). Thus, classical taxonomic classification has proven difficult, often limiting our understanding of the genomic diversity and evolution of phages (18, 19). As a promising approach, recent studies have proposed reticulated network modeling, in which pairwise relationships of phage genomes are represented as weighted phage-phage similarities in terms of gene/protein contents (15, 20). Because the resulting phylogenomic network includes the evolutionary reconstruction process for individual gene phylogeny through normalization/weighting of the average number of shared genes, it is generally accepted as more adequate for tracing true evolutionary history (21, 22).
Here we report the discovery and characterization of a marine Vibrio phage, ϕJM-2012. This phage exhibits a myoviral lineage and possesses very few homologs with other well-known marine Vibrio phages. A mathematical model of reticulate networks based on ortholog clustering (15, 20) and subsequent comparative analysis provides strong molecular support for the notion that ϕJM-2012 diverged from a common ancestor of the ϕKZ-related groups (23–27), most likely because of the imposition of distinct evolutionary forces upon its genome. Thus, given that ϕKZ-like phages have limited genetic diversity and a narrow host range (23–29), these data provide novel insights into the diversity and evolution of ϕKZ-related groups at the genomic and amino acid sequence levels.
MATERIALS AND METHODS
Bacterial strain and its bacteriophage.
The bacterial host we used, Vibrio cyclitrophicus, was obtained from the body fluid of an ascidian with soft-tunic syndrome (30). Bacteriophage ϕJM-2012 was isolated from single plaques on host bacteria by using 0.2-μm-filtered and autoclaved seawater supplemented with 1% Bacto tryptone and 0.5% yeast extract (Gibco, Gaithersburg, MD). After incubation for 24 h at 15°C, a single plaque was picked from the lawn of host cells, eluted in 0.2-μm-filtered autoclaved seawater, and combined with a host culture in a plaque assay. Following several rounds of plaque isolation, the phage particles were purified by CsCl density gradient equilibrium centrifugation as previously described (31).
Morphological characterization by transmission electron microscopy (TEM).
A solution (8 μl) of purified bacteriophage was dropped onto 400-mesh Formvar carbon-coated copper grids (Ted Pella Inc., Redding, CA). After 2 min, the bacteriophage solution was removed with filter paper and the grids were stained with 8 μl of 1% aqueous uranyl acetate (Merck, Darmstadt, Germany). The grids were examined with a Philips TECNAI F12 FEI transmission electron microscope (FEI, Hillsboro, OR) at an accelerating voltage of 120 kV. Bacteriophages were morphologically classified according to the International Committee on Taxonomy of Viruses (ICTV; http://www.ictvdb.org/) classification scheme.
Genome sequencing and bioinformatic analysis.
Bacteriophage DNA was prepared for sequencing as previously described (31). The genomic sequence of the bacteriophage was determined with an FLX Titanium genome sequencer (Roche, Mannheim, Germany) according to the manufacturer's standard procedures. All reads were assembled with Newbler Assembler (version 2.3; 454 Life Sciences, Branford, CT) and the CLC Genomics Workbench software program (version 4.5.1; CLC Bio, Aarhus, Denmark). Sequencing and assembly were performed by ChunLab Inc. (Seoul, South Korea), and the bacteriophage was designated ϕJM-2012. The potential gene products of the predicted open reading frames (ORFs) were compared with the proteins in the nonredundant GenBank databases by using BLASTp or PSI-BLAST (http://www.ncbi.nlm.nih.gov/blast/) (32) with E values of <10−4 (as of December 2012). The functional assignment of each ORF was predicted by local alignment algorithm-based database searches with Pfam (version 27.0; http://pfam.sanger.ac.uk/) (33), HHpred (version 2.0; http://toolkit.tuebingen.mpg.de/hhpred) (34), and “pdb70_1Dec12.” Protein structures were predicted with PSIPRED (version 3.0; http://bioinf.cs.ucl.ac.uk/) (35) and/or Phyre2 (version 2.0; http://www.sbg.bio.ic.ac.uk/phyre2/) (36). Multiple-sequence alignment of the protein sequences of interest was performed with Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) (37). Phylogenetic analyses of the protein sequences of interest were performed with MEGA5 (version 5.2.1; http://www.megasoftware.net/) (38). To estimate the robustness of the trees, we used the maximum-likelihood algorithm provided with bootstrap support (n = 1,000 replicates). To search for tRNA genes, we used tRNAscan-SE (version 1.2.1; http://lowelab.ucsc.edu/tRNAscan-SE/) (39). The PHIRE program (version 1.00; http://www.agr.kuleuven.ac.be/logt/PHIRE.htm) (40) was used to identify phage-specific promoters by using the following default parameters: string length, 20; degeneracy, 4; dominanNum, 4; window size, 20. Sequence logos were calculated by WebLogo (version 2.8.2; http://weblogo.berkeley.edu/) (41). The ARNold program (http://rna.igmors.u-psud.fr/toolbox/arnold/) (42) was used to predict the rho-independent transcription terminators.
Protein family clustering and comparative analysis.
To represent the evolutionary relationship of ϕJM-2012 with other bacteriophages as a network, we built a similarity network by using the A CLAssification of Mobile genetic Elements (ACLAME) database (version 0.4; http://aclame.ulb.ac.be) as previously described (20), with some modifications. Briefly, to cluster protein sequences into orthologous families, we performed pairwise similarity comparisons of each predicted protein of the ϕJM-2012 genome (n = 173) with BLAST in ACLAME; we used a Markov clustering (MCL) algorithm and the database of “viruses and prophages” (as of November 2012), within an E value threshold of 0.0001. Fifty-eight potential gene products of ϕJM-2012 were found to be associated with protein families in ACLAME, while the remaining proteins were defined as “dummy” families, e.g., the unclassified protein families 1 to 115, and considered to assess the actual similarity between ϕJM-2012 and those in the ACLAME database. Because the ACLAME database is not currently updated and does not include the most recently identified bacteriophages, including ϕKZ-like bacteriophages, ϕPA3 (23), 201ϕ2-1 (25), and ϕOBP (27), we further examined protein families from a number of completely sequenced bacteriophages. We downloaded a total of 1,652 predicted protein sequences representing the genomes of ϕKZ (GenBank accession number NC_004629), ϕPA3 (HQ630627), 201ϕ2-1 (NC_010821), ϕEL (NC_007623), and ϕOBP (NC_016571) from the GenBank database (http://www.ncbi.nlm.nih.gov/GenBank/index.html). Of these, 1,145 predicted protein sequences retrieved from ϕPA3, 201ϕ2-1, and ϕOBP were analyzed with the ACLAME database as described for ϕJM-2012. The protein sequences that were not assigned to any protein families in the ACLAME database were manually subjected to all-to-all BLASTp searches against a total set of 1,825 predicted protein sequences from the six phages with a cutoff E value of 10−4. A sequence was added to a cluster if it shared a reciprocal best BLAST hit relationship with at least one of the sequences in the cluster; it was thereafter considered the unclassified protein family when we assessed phage-to-phage similarity. In addition, dot plots were generated on the basis of the all-to-all BLASTp search results.
Network construction.
The resulting output was parsed in the form of a matrix in which the rows represented mobile genetic elements (MGEs) and the columns represented the protein families. We then determined the similarities of ϕJM-2012, ϕPA3, ϕOBP, and 201ϕ2-1 to the related MGEs as previously described (15). From this, we obtained P values representing the probability of finding a common number of protein families between each pair of MGE vehicles on the basis of the following hypergeometric equation:
| (1) |
where c is the number of protein families in common; a is the number of protein families of MGEs in ACLAME hits; b is the number of protein families in ϕJM-2012, ϕPA3, ϕOBP, and 201ϕ2-1, respectively; and n is the total number of protein families in the ACLAME database. The significance (Sig) value was obtained with the following equation Sig = −log(E value) = −log(P value × T), where the P value was obtained from the equation above and T = 1,221 × 1,220. Afterwards, the MGEs with Sig values of >0 were used to construct a new network of the bacteriophage genome-encoded protein-sharing relationships among ϕJM-2012, ϕPA3, ϕOBP, and 201ϕ2-1. This was visualized with the Cytoscape software (version 1.1; http://cytoscape.org/) (43) by using an edge-weighted spring-embedded model where MGEs that shared more protein families are located closer together in the network.
Nucleotide sequence accession numbers.
The sequences of the bacterial strain used here and its bacteriophage, ϕJM-2012, have been deposited in GenBank and assigned accession numbers KF488567 and JQ340088, respectively.
RESULTS
Morphology of phage ϕJM-2012.
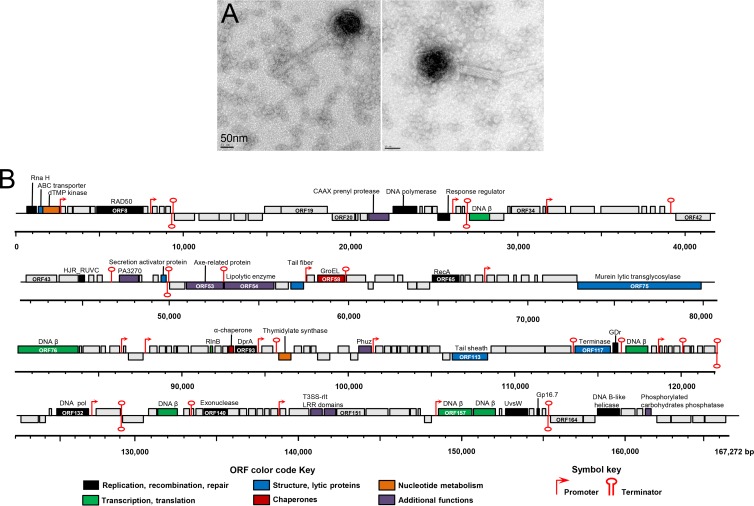
TEM examination of purified ϕJM-2012 particles revealed that each particle comprised an isometric head, a contractile tail, and kinked tail fibers that were probably associated with the tail (Fig. 1A). The diameter of the DNA-filled phage heads was 124 ± 4 nm. The noncontracted phage tails, which consisted of a neck, a tail sheath, and a central tube, were 260 ± 5 nm in length and 29 ± 2 nm in width, and the contracted tail was 131 ± 11 nm long. On the basis of its morphological traits, ϕJM-2012 was presumptively identified as a member of the Myoviridae family.
Fig 1.
Morphology and genome map of bacteriophage ϕJM-2012. (A) Electron micrographs of negatively stained purified viral particles. (B) The 167,292-bp genome of ϕJM-2012 represented in four tiers, with markers spaced at 1-kbp intervals. The ORFs are shown as boxes on either the positive (upper line) or negative (lower line) DNA strand, depending on whether they are transcribed rightward or leftward, respectively. The ORF numbers are shown within boxes, and each box is colored according to its putative function, which is shown above the ORFs. Other sequences, including the predicted promoters and terminators, are indicated. The numbers of phage proteins detected per ORF are provided in Table S1 in the supplemental material. LRR, leucine-rich repeat.
Genome overview and annotation.
The entire genome sequence of ϕJM-2012, which was determined with the Roche Genome Sequencer system, was found to be a 167,292-bp-long double-stranded DNA comprising a total of 173 predicted ORFs. Of these ORFs, 38 were transcribed leftward and the remaining 135 were transcribed rightward (Fig. 1B). The average G+C content was 35.40%; this parameter did not show any large-scale variations across the sequences, but eight ORFs (ORF14, ORF36, ORF106, ORF110, ORF113, ORF133, ORF142, and ORF164, mostly related to hypothetical proteins) had G+C contents of >40%. The ORF density was about 92.82%, there were 118 inter-ORF sequences of >15 bp and only eight sequences longer than 250 bp, and 20 sequences overlapped (data not shown). No tRNA sequence was identified by the tRNAscan-SE program. Prediction of initiation codon usage identified 160 ORFs with AUG start codons, while eight and five predicted ORFs used the UUG and GUG start codons, respectively. We also identified a total of 14 putative phage-specific promoters with a consensus sequence of WTTTAYAYCTATATATTATA (see Table S1 in the supplemental material), and these motifs were predicted to be unidirectional (Fig. 1B). In addition, we found 21 potential rho-independent transcription terminators (see Table S1).
Annotation of the 173 ORFs with BLASTp, PSI-BLAST, and HHpred revealed that 65 of them were homologous to sequences of phage or microbial origin in the NCBI nonredundant protein database (E values of <10−4), and 37 ORFs were predicted to have structural similarities to known proteins (see Table S1). Because the HHpred probability score is the most relevant statistical measure (34) and a score exceeding 90% generally indicates a true positive hit (34, 44), we used this as the threshold. Among the 37 assignable ORFs, 11 (ORF1, ORF8, ORF24, ORF45, ORF65, ORF93, ORF118, ORF132, ORF140, ORF160, and ORF166) were predicted to have functions in DNA replication, recombination, and repair; 6 (ORF31, ORF76, ORF89, ORF119, ORF157, and ORF158) were putatively associated with translation or transcription; 2 (ORF3 and ORF95) were putatively assigned to nucleotide metabolism; and 6 (ORF2, ORF51, ORF56, ORF75, ORF113, and ORF117) appeared to represent phage structural proteins (Fig. 1B).
Further examination of the ϕJM-2012 genome revealed very few genes with homologs in the known marine Vibrio phages. The gene content of ϕJM-2012 was highly related to those of various other phages, especially a number of Pseudomonas ϕKZ-like phages (see Table S1 in the supplemental material). In order to estimate the mosaicism of the phage genome that is mostly attributable to HGT events (15), we analyzed the pairwise relationships of the gene contents of ϕJM-2012 by using the ACLAME database (20).
Pairwise comparison of ϕJM-2012 genome.
ACLAME (version 0.4, released in August 2009), one of the most effective methods of identifying homologs/orthologs among the rapidly diverging MGE sequences of plasmids, bacteriophages, and transposons, implements the procedure of clustering of individual proteins to the protein families that can have functions in common (20). Hence, the member proteins belonging to identical protein families in the ACLAME database indicate the similarity of their phylogenetic profiles (45).
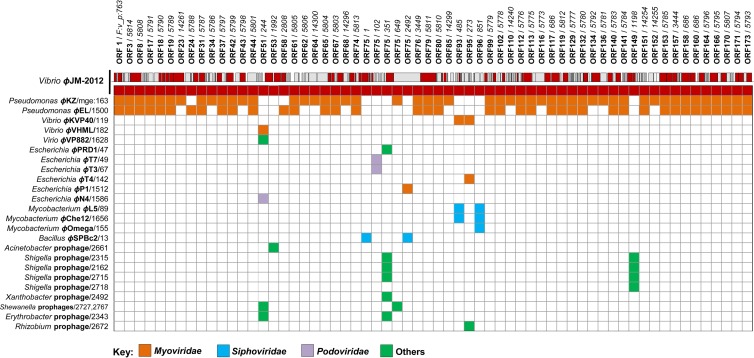
A pairwise comparison of the ϕJM-2012 genome by using the ACLAME database with an E value threshold of 0.0001 and a database of “viruses and phages” containing 16,057 protein families clustered from 54,218 predicted proteins encoded by 1,217 virus and prophage/phage genomes (45) allowed us to assign 58 ORFs to 56 protein families found in 83 phages or prophages (see Table S2 in the supplemental material). Although these ORFs occupied a relatively small portion (about 33%) of the ϕJM-2012 genome, this could suggest mosaicism. We then constructed a matrix in which “Family:vir_proph (F:v_p)” and “mge” were represented as a protein family identifier and a phage identifier, respectively (Fig. 2). Of the 83 MGEs identified, ϕJM-2012 shared more genetic elements with phages belonging to the Myoviridae family than with Siphoviridae, Podoviridae, or as-yet uncharacterized viruses or prophages, suggesting a possible myoviral lineage (Fig. 2). At the nucleotide level, none of the ORFs tested was related to any phage or prophage gene in the ACLAME database.
Fig 2.
Matrix view showing pairwise relationships of the phage ϕJM-2012 proteins with the 1,217 prophage/phage genomes in the ACLAME database. The matrix shown was input into the MCL algorithm to produce a list of clustered proteins (i.e., families) based on the ACLAME database with a threshold E value of 0.0001. In the matrix view, the columns indicate the protein families that contain ϕJM-2012-encoded proteins. The first row represents ϕJM-2012, while the succeeding rows correspond to closely related phages (MGEs). A shared cell indicates that an MGE protein in that row is found in the same protein family as a ϕJM-2012 protein (above a certain threshold) in the same column, and the Myoviridae (orange), Siphoviridae (blue), Podoviridae (purple), and as-yet-uncharacterized viruses or prophages (green) are colored according to ICTV classes.
Notably, the ϕJM-2012 genome showed biased pairwise relationships with two Pseudomonas ϕKZ-like phages; of the 56 protein families identified, 48 were shared with ϕKZ (24) while 42 were shared with ϕEL (26). However, ϕJM-2012 shared only a few protein families with the marine Vibrio phages and other phages. In addition, ORF75 clustered with eight protein families mostly related to putative lytic transglycosylases (LTs) and phage tail tape measure proteins (TMPs) (see Table S2 in the supplemental material); this suggested that ORF75 (2,273 amino acids [aa]) could be a multidomain protein that shares one or more domains with various proteins (46). Of the protein families common to 42 prophages, that encoded by ORF149 was functionally categorized as being relevant to type III secretion system (T3SS)-related proteins of Shigella prophages.
Because of the poorly defined functions in the current ACLAME database, arising from the recently characterized ϕKZ-like phages (28), many ϕKZ-specific protein families shared with ϕJM-2012 were functionally unassigned. However, by HHpred analysis, we were able to identify seven protein families, including member proteins of phage-encoded morphogenetic proteases (47), tubulin-like protein PhuZ (48), phage-encoded chaperonin (GroEL) (49), a tail sheath protease-resistant fragment (50), a cell-piercing protein (51), a DnaB family replicative helicase, and a fragment similar to a CAAX protease 1 homolog (this study) (see Table S2 in the supplemental material).
Protein-sharing network for ϕJM-2012.
To establish the evolutionary relationship of ϕJM-2012 with other phages and prophages, we built a mathematical model of protein-sharing networks (15). In parallel, because of the lack of sequence information in the ACLAME database, we also generated a new reticulate classification for ϕPA3 (23), 201ϕ2-1 (25), and the recently characterized phage ϕOBP (27); these three members of the ϕKZ-like groups have completely sequenced genomes and show high relatedness in their gene contents (see Table S1 in the supplemental material). Inferring a gene content-based phylogenomic network requires proper normalization or weighting to counter the possibility of larger genomes sharing more genes (21, 52). Accordingly, we estimated the phage-phage similarity, which we defined as the significance (Sig) score (15), by considering the uncharacterized protein families (see Materials and Methods). Our results revealed that ϕJM-2012 shared 48, 49, 49, 42, and 45 protein families with ϕPA3, ϕKZ, 201ϕ2-1, ϕEL, and ϕOBP, respectively (see Table S3 in the supplemental material). We also determined the number of families shared across the five species (data not shown).
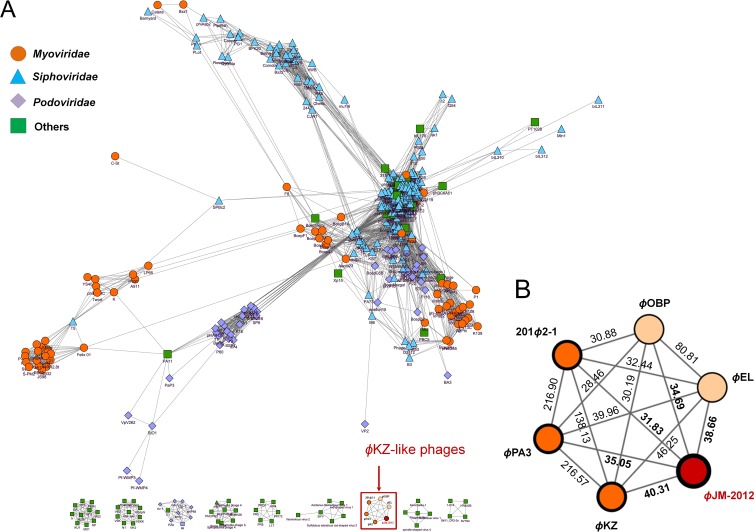
Next, we assessed all of the statistical relationships against a total of 16,758 protein families clustered from 55,536 predicted proteins of the 1,221 virus and prophage/phage genomes and generated Sig scores for each phage. Of the MGEs comprising the entire reticulated network, 796 that did not show any similarity to the ϕJM-2012 genome (Sig scores of <0) were excluded for clarity; the excluded MGEs were mostly prophage genomes. The resulting network consisted of 425 nodes indicating viruses and prophage/phages as MGEs and 7,256 edges representing the significant relationships between phages (Fig. 3), in which each MGE was placed with respect to the weight of its phage-phage connection. Sixty-four of the MGEs were placed into nine small interconnected components (Fig. 3A); within the largest component, the phages belonging to Myoviridae, Siphoviridae, Podoviridae, or uncharacterized and other phages were grouped into several highly interconnected regions that had some groups or phages interspaced among them.
Fig 3.
Protein-sharing network for ϕJM-2012. (A) A network representation for ϕJM-2012 was produced with Cytoscape in ACLAME version 0.4. Nodes represent viruses and/or phages, and edges represent their statistically weighted pairwise similarities. Viruses and phages with significance (Sig) scores of >0 are shown. For clarity, only 7,256 edges from 425 viruses and phages (including ϕJM-2012 and five representative ϕKZ-like phages) are shown; each node is depicted as a different color and shape, representing viruses and/or phages belonging to Myoviridae, Siphoviridae, and Podoviridae of a given ICTV class or uncharacterized and other phages. (B) Enlargement of the red box in panel A. Values are the significance (Sig) scores for similarities estimated with the hypergeometric equation shown in Materials and Methods. The graph shows the relationships of ϕJM-2012 to five ϕKZ-like phages, i.e., ϕKZ, ϕPA3, and 201ϕ2-1 of the ϕKZ-like viruses (dark orange) and ϕEL and ϕOBP of the ϕEL-like viruses (light orange).
Interestingly, Vibrio phage ϕJM-2012 was restricted to a single isolated network comprising five ϕKZ-like species that were linked to ϕJM-2012 with Sig scores ranging from 31.83 to 40.31 (Fig. 3B). Since we normalized the number of shared protein families with respect to the weighted similarities (15), none of the 81 MGEs that had only one or two shared protein families (see Table S2 in the supplemental material) could be reliably placed in the network. Our results therefore suggest that ϕJM-2012 shares its closest evolutionary linkage with the ϕKZ-like phages. Additionally, two other remarkable features emerged from this analysis. First, the small network distribution of the five ϕKZ-like phages is unusual; they failed to link with any component within the reticulate representation (Fig. 3A), possibly because they form an evolutionarily distinct branch of the Myoviridae family (28). Second, although the ϕKZ-like phages were densely interconnected, ϕKZ, ϕPA3, and 201ϕ2-1 were more closely related (Sig score range, 138.13 to 216.90), suggesting two possible groups in this phage family, the other group being ϕEL and ϕOBP (Sig score, 80.81) (Fig. 3B). This supports the recent taxonomic classification of the ϕKZ-like phages as ϕKZ-like and ϕEL-like viruses (25, 27), whereas the similarity values of ϕJM-2012 suggest that it is probably an emerging member of the ϕKZ-related group.
Comparative analysis of genomic organization.
Given the gene content-based relationships described above, we next examined the potential conservation of gene order in the ϕJM-2012 genome with respect to that of the ϕKZ-like phages. The lack of appreciable DNA homology among the ϕKZ-related genomes (27) was also reflected in the ϕJM-2012 genome, as described in its pairwise comparison. At the genome-wide predicted-protein level, the alignments and pairwise similarities of the six phages are shown as a dot matrix plot in which each protein sequence is shown as the best hit shared by two phages (E value cutoff, 10−4) in the BLASTp search. To clarify the shared core genes and non-ϕKZ-related ORFs (unique genes), we screened the preliminary ACLAME database clustering for the six phages. We found that most of the unique genes were functionally unassigned (E values of >10−4 in PSI-/BLASTp or ACLAME, probability score of <90% in HHpred).
In the ϕJM-2012 genome, we observed a considerable disruption of gene order compared to that of the ϕKZ- and ϕEL-like viruses (Fig. 4A, see the nearly continuous dotted line along the diagonal). To further evaluate the extent of disruption between ϕJM-2012 and the other two genera, we measured the neighborhood disruption frequency (NDF). This is the number of measured breakpoints between gene neighbors divided by the number of genes shared by the genomes; an NDF of 0 indicates that there is absolute conservation of gene order, while an NDF of 1 indicates absolute shuffling (53). As shown in the dot matrix, the ϕJM-2012 genome exhibited a higher degree of NDFs (close to 1) than those found between and within the two genera, reflecting extensive genome rearrangement in ϕJM-2012 (53, 54).
Fig 4.
Dot plot of protein sequence comparisons and locations of the RNAP genes on the genomes. (A) A total of 1,825 protein sequences from six phage genomes (ϕJM-2012, ϕKZ, ϕPA3, 201ϕ2-1, ϕEL, and ϕOBP) were aligned and manually compared by using BLASTp searches with a cutoff E value of 10−4. A sequence was represented as a dot if it shared a reciprocal best BLASTp hit relationship. (B) The positions of the RNAP genes are shown as vertical arrows on the genomes. The arrow box for each ORF indicates its transcriptional orientation. Within boxes, the ORF numbers and predicted amino acid sequences are shown and each box is colored according to its protein family clustered by the ACLAME database. From top to bottom, sets of ORFs from ϕJM-2012, ϕPA3, ϕKZ, 201ϕ2-1, ϕEL, and ϕOBP are listed, respectively.
With respect to the 55 shared core genes, the rearrangement patterns were considered by using reciprocal best BLAST hit, because ϕJM-2012 did not appear to have a biased relationship to either the ϕKZ- or ϕEL-like viruses. The results of this analysis suggested that gene rearrangement events may have potentially inverted two or three consecutive ORFs in a number of places (e.g., ORF17 to ORF19, ORF42 and ORF43, ORF64 and ORF65, ORF79 and ORF80, ORF112 and ORF113, and ORF116 and ORF117), disrupted 19 ORFs with gaps, and repositioned 16 ORFs to different locations in the genome (see Table S3 in the supplemental material). The interruption of gene order by frequent inversions, insertions, and transpositions resulted in ϕJM-2012 sharing only a few gene sets with the counterpart gene products in the ϕKZ- or ϕEL-like viruses, even though most of these genes are conserved in colinear (synteny) blocks within the ϕKZ-related genomes (25, 27). In particular, on the basis of ACLAME clustering, we observed gene order disruption among the putative genes for six RNA polymerase (RNAP) beta and beta′ subunit genes of ϕJM-2012 compared to those in the other ϕKZ-like phages (Fig. 4B). This group of multisubunit RNAPs is widely conserved among the ϕKZ-like phages (23).
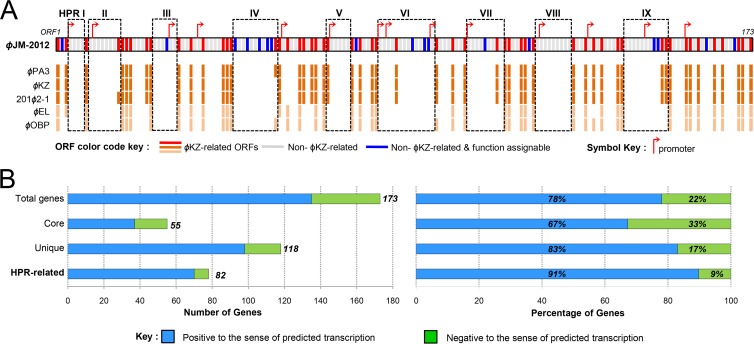
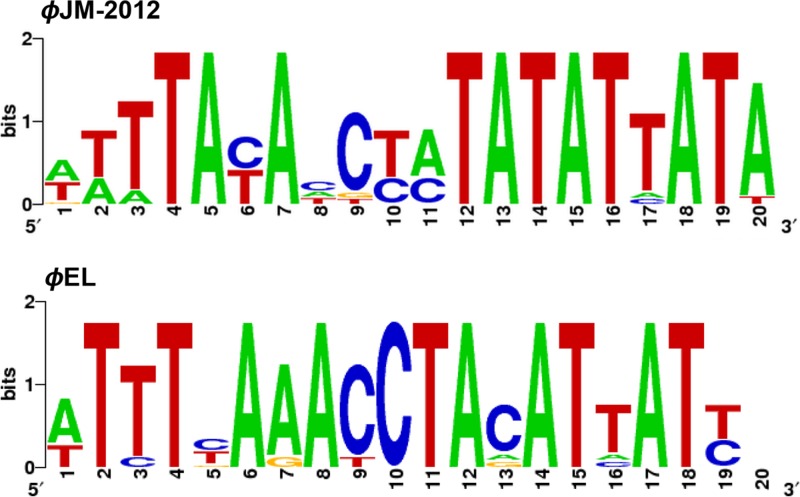
As expected, the large-scale genomic rearrangement in the ϕJM-2012 genome facilitated the positional relocation of shared core genes, resulting in extensive disruption of synteny conservation and hyperplastic regions (HPRs) typically exhibited by ϕKZ-related genomes (27). Nonetheless, we found specific regions with dense grouping of unique genes and a few accessory genes. Further, most of these genes are not annotated and are also substantially smaller than the remaining genes (∼506 versus 1,057 bp), which shows HPR characteristics (55, 56). Hence, we assumed that these regions are ϕJM-2012-specific HPRs and were designated HPRs I to IX (ORF4 to ORF7, ORF9 to ORF16, ORF25 to ORF30, ORF45 to ORF55, ORF68 to ORF73, ORF81 to ORF98, ORF103 to ORF111, ORF120 to ORF128, and ORF142 to ORF152, respectively) (Fig. 5A). In addition, of the 14 putative phage-specific promoters, 10 were found in eight HPRs, all of them immediately preceding unique genes. Except for one, the remaining sequences are located in the upstream regions of unique genes not localized in the HPRs. In addition, the transcriptional orientations of HPR gene sequences were more highly correlated with their predicted unidirectional promoters than were the orientations of core genes (Fig. 5B). Most of the HPR-related genes (about 91%) were positive for sense-oriented transcription. Notably, a consensus sequence of the promoter motifs is highly similar to those of ϕEL (26) (Fig. 6).
Fig 5.
Schematic representation of the HPRs and regional relevance of the sense and antisense genes in the ϕJM-2012 genome. (A) Genes were classified into the following three categories: core, present in all ϕKZ-related groups; accessory, present in one or more phages; unique, present only in ϕJM-2012. Red cells correspond to core or accessory genes of ϕJM-2012. Unique genes are further divided into those that were functionally assignable in HHpred searches (blue) or not assignable (gray). Purple arrows indicate predicted promoters. Regions with more hyperplastic characteristics (HPRs I to IX) are boxed. Dark and light orange cells represent gene products shared with the ϕKZ- and ϕEL-like viruses, respectively. (B) Comparisons of genes that are positive (blue) or negative (green) with respect to the sense of their predicted transcriptional orientations in the ϕJM-2012 genome.
Fig 6.
Logo representation of phage-specific promoters of ϕJM-2012 in comparison to a motif of phage ϕEL. The motifs that contain similar base characteristics indicate that these promoters may be functionally interchangeable.
Unique genes and potential hot spot in HPRs of ϕJM-2012.
The high level of sequence divergence among phage proteins and the large number of novel viruses often lead to a lack of sequence similarity (27, 57). Since structural similarity is considered an ideal measure for predicting viral protein function and evolutionary links (57), in the absence of significant sequence similarity, we examined the unique genes by using the structure-predicting tools HHpred, PSIPRED, and Phyre2.
Among the unique HPR genes, the most potentially informative match appeared to be ORF45 in HPR IV. Our initial HHpred analysis revealed that ORF45 had strong similarities to the Holliday junction resolvases (HJRs) of Escherichia coli RuvC (see Table S1 in the supplemental material). HJR, a site-specific recombinase belonging to the tyrosine recombinase family, catalyzes DNA rearrangement via viral DNA integration into and excision from the host genomes (58). The overall tertiary structure of ORF45 (determined by Phyre2) was similar to that of E. coli RuvC (data not shown). Their C termini differed; this is likely to reflect distinct dimerization interactions, as seen for E. coli RuvC and Thermus thermophilus RuvC (58).
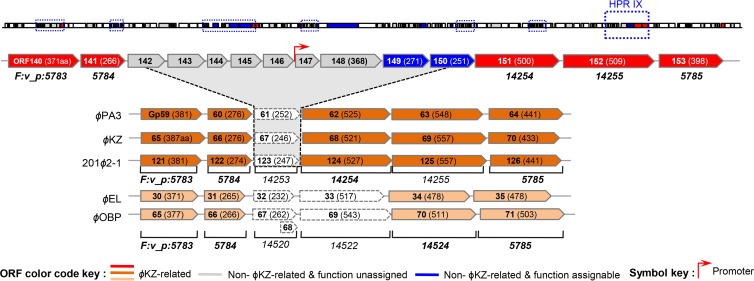
In HPR IX, an apparent insertion of nine extra genes was found between ORF141 and ORF151 (Fig. 7). Interestingly, the regions harboring the flanking gene products of the identical protein families in the ϕKZ- and ϕEL-like viruses resemble the HPRs of ϕJM-2012. The functions of the newly acquired genes are mostly unknown. However, the ACLAME database assigned ORF149 to the prophage-encoded invasion plasmid antigen (IpaH) of Shigella flexneri (see Table S2 in the supplemental material). Its secondary structure (predicted by HHpred) was found to be highly similar to that of a partial leucine-rich repeat domain at the N terminus of IpaH3 (59) (see Table S1). ORF150 was also a candidate for the Salmonella enterica serovar Typhimurium virulence factor SspH2. We noticed that these two novel genes together with the Salmonella prophage Gifsy-1-carrying T3SS substrate GogB moron (60, 61), were grouped into the same protein family in the ACLAME database; this family was functionally annotated as “gene ontology (GO) interaction with host via protein secreted by T3SS.” Such prophage-carrying morons frequently encode proven or suspected virulence factors. More importantly, we identified a putative promoter positioned between ORF146 and ORF147 (Fig. 7); such an arrangement is typically observed in transcriptionally autonomous moron units (60, 62).
Fig 7.
Analyses of unique genes in the HPRs and potential “hot spots.” HPRs of ϕJM-2012, ϕKZ, ϕPA3, 201ϕ2-1, ϕEL, and ϕOBP. The arrow box of each ORF indicates its transcriptional orientation. The genes between ORF140 and ORF153 are shown for the five ϕKZ-like phages. The protein family of each gene based on the ACLAME database is shown below the box. Red, dark orange, light orange, and gray boxes represent the core genes of ϕJM-2012, ϕKZ-like viruses, and ϕEL-like viruses and unique genes of ϕJM-2012 having no functional assignment, respectively. Blue boxes (ORF149 and ORF150) represent two moron-like elements related to the virulence genes of T3SS family members. Bioinformatically predicted promoters are indicated.
Unique genes of structural and infectivity traits.
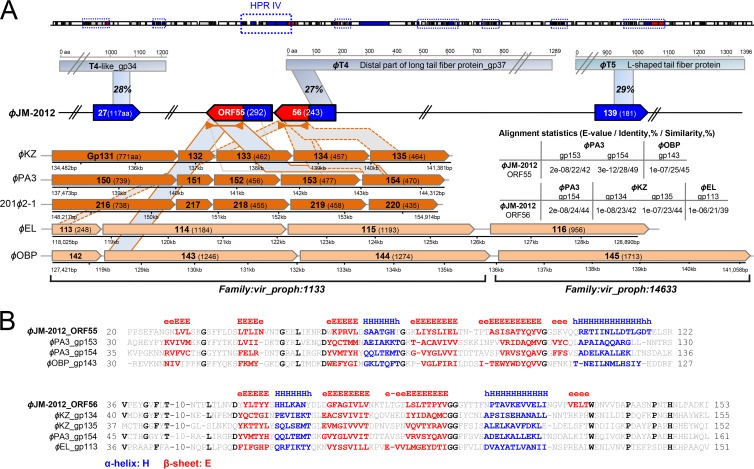
Regarding structural components, we saw striking interspecies differences in the tail fiber-related genes. The extensive swapping of tail fiber genes between loci complicates the detection of sequence homology (7, 63), so we used local alignment algorithm-based searches (e.g., HHpred or pfam) in conjunction with the secondary-structure prediction tool PSRPRED. HHpred analysis initially identified a single tail fiber-related gene immediately adjacent to HPR IV (ORF56) (see Table S1 in the supplemental material). The C-terminal residues of ORF56 were predicted to encode an N-terminal portion of the distal-end subunit of bacteriophage T4 gene product 37 (gp37) (Fig. 8A, top), which is responsible for host recognition and attachment (64). Regarding the N terminus of this ORF, residues 1 to 146 showed notable similarities in sequence (Fig. 8A, right panel) and predicted secondary structure (Fig. 8A, bottom, and B) to the N-terminal portions of ϕKZ gp134 and gp135 and ϕPA3 gp154, which all diverged from ϕKZ gp131. gp131 and its orthologs/paralogs, which appear to confer tail fiber-associated substrate binding, have conserved N-terminal features, such as predominant β-strands and sequence similarity patterns (65). The N-terminal part of ORF56 (aa 35 to 153) also shared secondary-structure similarity with ϕEL gp113 (aa 35 to 151) of the ϕOBP ortholog (27). A comparable structural similarity pattern was observed in the N terminus of the preceding ORF, ORF55 (Fig. 8A, bottom, and B). In addition, given the extraordinary variability of phage tail fiber genes and the typical lack of sequence similarity among virus structural proteins (7), we further sought to identify additional relevant genes by using a broad E value cutoff of 1 in the ACLAME database. We identified two candidates that matched members of the tail fiber protein families, ORF27, which matched the gp34 proximal tail fiber subunit of a T4-like A. salmonicida phage, and ORF139, which matched the L-shaped tail fiber protein of a T5-like E. coli phage (Fig. 8A, top). This suggests that, unlike its evolutionary neighbors, ϕJM-2012 shows mosaicism among its tail fiber-related genes (at least for the few ORFs identified to date).
Fig 8.
Comparison of the tail fiber genes of five ϕKZ-like phages and ϕJM-2012. (A) At the top are the tail fiber genes encoding the T4-like, T4, and T5 proteins and the corresponding regions of the ϕJM-2012 ORFs. The shading shows the regions with sequence or structural similarities. Because of space constraints, T4-like gp34, T4 gp37, the T5 tail fiber gene, and the ϕJM-2012 genome are truncated as indicated. At the bottom are ϕKZ tail fiber orthologs and paralogs grouped into two protein families based on the ACLAME database. The number of each gene product and its protein sequence length are shown within arrow boxes that also indicate their transcriptional orientations. The shading indicates the regions that show robust structural similarities. On the right are the alignment statistics for our comparison of the tail fiber genes. (B) Amino acid sequence alignment and secondary-structure comparison of ϕJM-2012 ORF55 and ORF56 and ϕKZ tail fiber proteins. Secondary-structure elements are shown in the top part of the alignment. The beta-strand residues (E) are red, and alpha-helix residues are blue. Conserved residues are indicated by bold black letters.
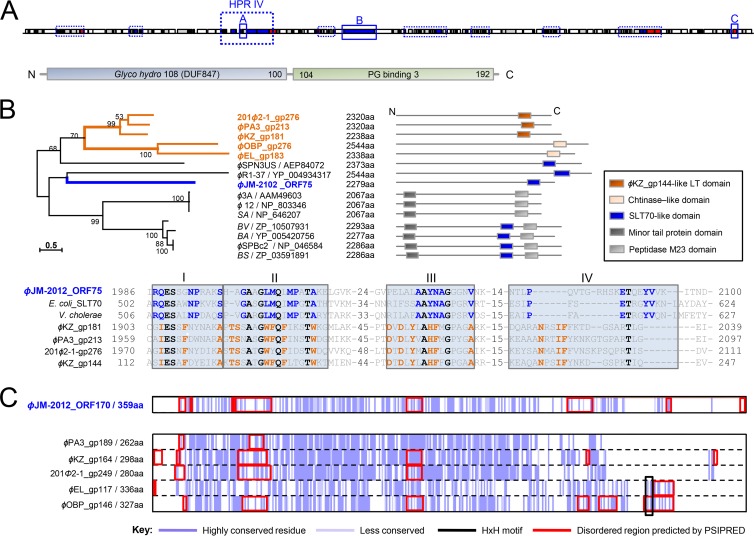
Our HHpred- and ACLAME-based analyses revealed that ϕJM-2012 harbors two identifiable lysozyme-like proteins, represented by ORF51 and ORF75. ORF51, which is located in HPR IV, encodes two domains, one from N-terminal glycoside hydrolase family 108 (PF05838; 7.6e-06) and a C-terminal peptidoglycan (PG)-binding domain (PF09374; 4.3e-08), connected via a short linker (Fig. 9A). This modular structure is characteristic of a family of phage muraminidases responsible for mediating phage release (66, 67), which is different from the lysis-related PG hydrolases of the ϕKZ-related group (68). The second identified ORF, ORF75, was preliminarily predicted to be a potential multidomain protein orthologous to members of the LT or TMP family, the latter of which includes ϕKZ gp181 (Fig. 2). The size of ORF75 and the presence of a small C-terminal lysozyme-like domain, both of which are characteristic of the TMPs (69), seem to support this alignment. However, the phylogenetic position of ORF75 among the other TMP family members (all >2,000 aa) indicated that the features of ORF75 differed from those of ϕKZ gp181 or its orthologs (Fig. 9B). Sequence analysis of the functional domains showed that ORF75 is more similar to soluble LT 70 (SLT70). More specifically, within motif IV, which serves as a signature for distinguishing the individual subfamilies of LTs (70), Pro2080, Glu2089, Thr2090, Tyr2093, and Val2094 were conserved between ORF75 and SLT70 family members (Fig. 9B).
Fig 9.
Sequence features of infection-related structural genes. (A) Modular structure of ϕJM-2012 ORF51, as predicted by Pfam analysis. The predicted glycoside hydrolase family 108 domain and PG-binding domain 3 are light blue and green, respectively. (B) Upper left, maximum-likelihood tree of ϕJM-2012 ORF75 and TMPs. Abbreviations: SA, Staphylococcus aureus subsp. aureus; BV, Bacillus vallismortis; BA, Bacillus amyloliquefaciens subsp. plantarum; BS, Bacillus subtilis subsp. subtilis. GenBank accession numbers are included. Upper right, schematic representations of the modular structures of the proteins. Each box represents the functional domain. Bottom, the repeated consensus sequences (consensus motifs I to IV) found in the C-terminal LT domains of ϕJM-2012 ORF75, E. coli SLT70 (GenBank accession no. NP_418809), V. cholerae SLT70 (WP_000197919), ϕPA3 gp213, ϕKZ gp181, 201ϕ2-1 gp276, and ϕKZ gp144. (C) Sequence conservation histogram based on Clustal Omega alignment, where dark purple represents the most highly conserved residues. The HxH motifs and disordered regions predicted by HHpred and PSIPRED are boxed in black and red, respectively.
With respect to the cell-puncturing devices used by phages to penetrate the bacterial cell envelope during infection, the ACLAME database initially categorized the candidate member proteins of ϕJM-2012, ϕPA3, ϕKZ, and 201ϕ2-1 as potentially corresponding to two known orthologs of EL-like viruses that are similar to the injection needle protein of myovirus P2 (P2-Gpv) (27) (see Table S3 in the supplemental material); however, their C-terminal regions were found to be quite different. Multiple-sequence alignment revealed that the double-histidine iron-binding motif (HxH) typically conserved in ϕP2-Gpv (51) was lacking in all three corresponding proteins of ϕKZ-like viruses, as well as in ORF170 of ϕJM-2012 (Fig. 9C). ORF170 failed to yield any reliable bioinformatic matches, and PSIPRED identified variations in the length and number of disordered residues in the C-terminal part that are critical for conformational changes in the tail tube complexes, including the tail spike (57, 71). In addition, the extreme C terminus of ORF75 did not show any clear sequence or structural similarity to the C-terminal distal end of ϕKZ gp181 and its orthologs that have been proposed to act as membrane-puncturing needles (68, 72; data not shown).
DISCUSSION
We describe here the isolation and genomic characterization of a novel marine V. cyclitrophicus bacteriophage that we designated ϕJM-2012. Our mathematical estimation of the pairwise relationships between its gene content and those of ϕKZ-related genomes and our subsequent comparative analysis provide strong molecular support for the notion that ϕJM-2012 underwent adaptive reconstruction because of the imposition of distinct evolutionary forces upon its genome.
A phylogenomic network is a model that can be used to display and quantify the effect of non-tree-like reticulate processes on the evolutionary history of an organism (21, 22). Networks of shared genes are reconstructed from the presence/absence pattern of all protein families that are a set of homologous proteins (i.e., proteins with a common origin) found in diverse species. Here we used a protein-sharing network that is specific for the taxonomic classification of phages and plasmids and mathematically weights the similarity between phage genomes in terms of shared gene contents (15, 20). The considerable genome size variation of phage species ranging from 17 kbp to 0.5 Mbp (73) requires a proper normalization or weighting to counter the possibility that larger genomes share more genes (21, 52). Because a weighted edge signifies the strength of the connection, such mathematical models are better able to trace the true evolutionary linkage than are unweighted networks (21). Our network representation further considered probable close relatives that were absent from the ACLAME database. Remarkably, ϕJM-2012 was found to interconnect solely with the five ϕKZ-like phages, strongly indicating that ϕJM-2012 and the ϕKZ-related groups evolved from a common ancestor. In addition, the results of our network approach support the recent taxonomic subdivision of the ϕKZ-related groups into the ϕKZ- and ϕEL-like viruses (25, 27).
As an emerging new genus, the Pseudomonas ϕKZ-like phages show limited genetic diversity and a narrow host range, having been isolated only from Pseudomonas species to date (27). The ϕKZ-like phages also lack similarity to other phages at the DNA and protein levels (29) and thus form a distinct evolutionary branch within the Myoviridae family. These apparently unique evolutionary constraints raise two obvious questions, i.e., (i) how did this natural phage evolve to interact with an altered (not just expanded) host range within a common bacterial species, and (ii) what factors facilitate its niche-adaptive processes?
We therefore investigated the genomic organization of ϕJM-2012 and the ϕKZ-related groups, as genomic evolution can be traced by patterns of colinear (synteny) blocks (i.e., chromosomal regions that share a conserved order of genes within close relatives) (54, 74) and HPRs (55, 56). Surprisingly, our comparative analysis of the six phage genomes showed that ϕJM-2012 had undergone extensive genome rearrangement and showed little lineage-specific synteny with the ϕKZ-related genomes. Given that synteny blocks can be a strong indicator of conserved gene function (54, 60, 75), this suggests that distinct evolutionary forces acting on the genome may have altered its functional relatedness to the ϕKZ-like phages. In addition, a potential reengineering of the regulatory system of ϕJM-2012 can be evidenced by the loss of the conserved positional patterns of notable orthologs, such as RNAPs (54, 76), that are highly associated with the evolution of transcriptional regulation among the ϕKZ-like phages (23).
With respect to the HPRs specific to ϕJM-2012, our identification of regional features has two intriguing implications. First, the presence of unidirectional promoters that showed a consensus sequence similar to that of ϕEL and their preponderance in most of the HPRs may reflect the fact that ϕJM-2012 retained this particular feature of the ϕKZ-related hyperplastic portion (27), despite the extensive disruption of its genomic architecture. Second, relative to the other regions, the higher correlation of putative promoters and the transcriptional orientations of the predicted gene sequences in the HPRs suggest that the HPR-related genes may be functional (77). Thus, these variable regions could have specific niche-defining roles in the adaptation of ϕJM-2012 to its particular host and environment (78).
We further examined the gene contents and clustering of the HPRs. In HPR IV, ϕJM-2012 was found to harbor a site-specific recombinase (HJR)-related gene. Although future studies are warranted to functionally characterize this gene, its presence in or absence from the six phage genomes indicated that HJR, which is known to be associated with a temperate life cycle (79), could play an important role in the rearrangement of the ϕJM-2012 genome (53). Another notable feature that we identified was the localization of prophage-related genes (with the ACLAME database) in HPRs IV and IX (the latter of which is likely to be a hot spot, as it contains two moron-like elements). Prophage-carrying morons encode proven or suspected virulence factors (e.g., extracellular toxins, outer membrane proteins, and enzymes) and thus provide selective benefits for both phages and (via selection) their bacterial hosts (60). Their location is not random, and many phage-encoding virulence factors have been found near the tail fiber regions (80). By further analysis with HHpred, we found that the prophage-related genes in HPR IV of ϕJM-2012, which are adjacent to the tail fiber-related genes, were similar to those of the S. enterica serovar Typhi phage Vi type I tail spike-carrying acetylxylan esterase domain (81), and this portion of HPR IV, which includes the HJR, phage muramidase, prophage-related, and tail fiber-related genes, may be similar to the pathogenicity islet in S. enterica serovar Typhi that is critical for toxin secretion (67). Although it is unclear whether their positional relationships within HPRs are strategic, this finding prompted us to speculate on some potential mechanisms underlying the movement of transferable elements.
Essentially, there are two current models to explain genome mosaicism: homologous/nonhomologous recombinations and illegitimate moron accumulation (60, 62). Curiously, we failed to detect any specific linker sequences around the HPRs of ϕJM-2012. However, this does not necessarily indicate the presence of nonhomologous or illegitimate recombination, since it could be explained by the presence of relatively short and little-known conserved sequences at the gene boundaries (19, 60) and the lack of information on intermediate-sequence similarities (60). Thus, it is difficult to explicitly determine the genetic mechanisms of this recombination (19, 60). Nevertheless, on the basis of the presence of relevant genes, we propose that all types of recombination (illegitimate, homologous, and site specific) are likely to be responsible for the mosaic architecture of the ϕJM-2012 genome.
Our assessment of candidate infection-related structural proteins in ϕJM-2012 and its closest relatives revealed considerable changes in conserved motifs, alterations in structural elements, and genetic exchanges. This may explain the shift of ϕJM-2012 in host tropism, because structural proteins are involved in host recognition, attachment, penetration, and lysis (57, 64, 68). The modular structures and sequence features of the functional domains of the two putative structural lysins of ϕJM-2012 were different from those of the lysis-related PG hydrolases and infection-related structural lysins of ϕKZ-like phages, respectively (27, 68, 72). With respect to injection needle-like proteins, the lack of an HxH motif and the functional importance of the disordered region allow us to hypothesize that ORF170 reflects a distinctive ability of ϕJM-2012 to puncture the cell membrane, or (more likely) this phage harbors yet-to-be-identified components that participate in this process. For example, ORF53 and ORF54, which carry an acetyl esterase domain, were located immediately adjacent to the tail fiber-related genes, in an arrangement similar to that of the tail spike proteins in S. enterica serovar Typhi phage Vi type I (81). In particular, tail fiber-related genes evolve much more rapidly than other phage genes, since they allow shuffling of sequences between otherwise unrelated genes (63). A classical case is phage Mu, which contains a recombinase capable of reversing the orientations of receptor-interacting genes, leading to new receptor recognition specificity (82, 83). In addition, the similarities among the tail fiber genes of coliphages and those of other phage families indicate the presence of illegitimate recombination-driven domain exchanges (84). In the present study, we found that ϕJM-2012 harbored two obvious fragments of ORF55 and ORF56, providing evidence of an illegitimate-recombination-like process in the conserved structural parts of ϕKZ tail fiber orthologs or paralogs. This could provide some important insights into how the evolutionary process operates in diverged members of the ϕKZ tail fiber gene family (65). Notably, the orientations of ORF55 and ORF56 were reversed compared to those in the other ϕKZ tail fiber orthologs or paralogs (which undergo sense-positioned transcription). Given the absence of predicted reverse promoters in ϕJM-2012, it can be inferred that these structural genes have nonessential functions (27, 77). In contrast, ORF27 and ORF139, which harbor new relevant genes, appear to be functional. Given that tail fiber proteins are generally indispensable to phage survival, such changes may be related to its ability to infect a different host.
Finally, comparative genomic analysis revealed a highly disrupted genome of ϕJM-2012. Compared to small-scale changes in gene order and inversion processes that are often involved in the genetic divergence of ϕKZ-related genomes with a common bacterial host, Pseudomonas species, our observations clearly suggest that genomic rearrangement is a key component of its high level of divergence. This dramatic change in gene order most likely resulted from natural selection and/or adaptation to a heterogeneous environment, although it is difficult to detect the fragile sites (synteny breakpoints) in the ϕJM-2012 genome. In addition, relative to the shared core genes, the higher proportion of unique genes that are well preceded by promoters on the transcriptional strand may have facilitated its evolution. Collectively, our results yield novel insights into the diversity and evolution of ϕKZ-related phages at the genomic and amino acid sequence levels.
Supplementary Material
ACKNOWLEDGMENTS
We thank Gipsi Lima-Mendez (Laboratoire de Bioinformatique des Génomes et des Réseaux, Université Libre de Bruxelles) for her expert help with our reticulate analysis and Gye-Min Lee (Department of Information and Statistics, Gyeongsang National University) for help with the statistical equation.
This work was supported by a grant from the World Class University Program (R32-10253) funded by the Ministry of Education, Science and Technology of South Korea.
Footnotes
Published ahead of print 25 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02656-13.
REFERENCES
- 1.Wommack KE, Colwell RR. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64:69–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aalto AP, Bitto D, Ravantti JJ, Bamford DH, Huiskonen JT, Oksanen HM. 2012. Snapshot of virus evolution in hypersaline environments from the characterization of a membrane-containing Salisaeta icosahedral phage 1. Proc. Natl. Acad. Sci. U. S. A. 109:7079–7084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suttle CA. 2007. Marine viruses—major players in the global ecosystem. Nat. Rev. Microbiol. 5:801–812 [DOI] [PubMed] [Google Scholar]
- 4.Duhaime MB, Wichels A, Waldmann J, Teeling H, Glöckner FO. 2011. Ecogenomics and genome landscapes of marine Pseudoalteromonas phage H105/1. ISME J. 5:107–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stoddard LI, Martiny JB, Marston MF. 2007. Selection and characterization of cyanophage resistance in marine Synechococcus strains. Appl. Environ. Microbiol. 73:5516–5522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canchaya C, Fournous G, Chibani-Chennoufi S, Dillmann ML, Brüssow H. 2003. Phage as agents of lateral gene transfer. Curr. Opin. Microbiol. 6:417–424 [DOI] [PubMed] [Google Scholar]
- 7.Seguritan V, Alves N, Jr, Arnoult M, Raymond A, Lorimer D, Burgin AB, Jr, Salamon P, Segall AM. 2012. Artificial neural networks trained to detect viral and phage structural proteins. PLoS Comput. Biol. 8:e1002657. 10.1371/journal.pcbi.1002657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Angly FE, Felts B, Breitbart M, Salamon P, Edwards RA, Carlson C, Chan AM, Haynes M, Kelley S, Liu H, Mahaffy JM, Mueller JE, Nulton J, Olson R, Parsons R, Rayhawk S, Suttle CA, Rohwer F. 2006. The marine viromes of four oceanic regions. PLoS Biol. 4:e368. 10.1371/journal.pbio.0040368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bench SR, Hanson TE, Williamson KE, Ghosh D, Radosovich M, Wang K, Wommack KE. 2007. Metagenomic characterization of Chesapeake Bay virioplankton. Appl. Environ. Microbiol. 73:7629–7641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mann NH. 2005. The third age of phage. PLoS Biol. 3:e182. 10.1371/journal.pbio.0030182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baudoux AC, Hendrix RW, Lander GC, Bailly X, Podell S, Paillard C, Johnson JE, Potter CS, Carragher B, Azam F. 2012. Genomic and functional analysis of Vibrio phage SIO-2 reveals novel insights into ecology and evolution of marine siphoviruses. Environ. Microbiol. 14:2071–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardies SC, Comeau AM, Serwer P, Suttle CA. 2003. The complete sequence of marine bacteriophage VpV262 infecting Vibrio parahaemolyticus indicates that an ancestral component of a T7 viral supergroup is widespread in the marine environment. Virology 310:359–371 [DOI] [PubMed] [Google Scholar]
- 13.Miller ES, Heidelberg JF, Eisen JA, Nelson WC, Durkin AS, Ciecko A, Feldblyum TV, White O, Paulsen IT, Nierman WC, Lee J, Szczypinski B, Fraser CM. 2003. Complete genome sequence of the broad-host-range vibriophage KVP40: comparative genomics of a T4-related bacteriophage. J. Bacteriol. 185:5220–5233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rohwer F, Segall A, Steward G, Seguritan V, Breitbart M, Wolven F, Azam F. 2000. The complete genomic sequence of the marine phage Roseophage SIO1 shares homology with nonmarine phages. Limnol. Oceanogr. 45:408–418 [Google Scholar]
- 15.Lima-Mendez G, Van Helden J, Toussaint A, Leplae R. 2008. Reticulate representation of evolutionary and functional relationships between phage genomes. Mol. Biol. Evol. 25:762–777 [DOI] [PubMed] [Google Scholar]
- 16.Edwards RA, Rohwer F. 2005. Viral metagenomics. Nat. Rev. Microbiol. 3:504–510 [DOI] [PubMed] [Google Scholar]
- 17.Sabehi G, Shaulov L, Silver DH, Yanai I, Harel A, Lindell D. 2012. A novel lineage of myoviruses infecting cyanobacteria is widespread in the oceans. Proc. Natl. Acad. Sci. U. S. A. 109:2037–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rohwer F, Edwards R. 2002. The phage proteomic tree: a genome-based taxonomy for phage. J. Bacteriol. 184:4529–4535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatfull GF, Hendrix RW. 2011. Bacteriophages and their genomes. Curr. Opin. Virol. 1:298–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leplae R, Lima-Mendez G, Toussaint A. 2010. ACLAME: a CLAssification of Mobile genetic Elements, update 2010. Nucleic Acids Res. 38:D57–D61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dagan T. 2011. Phylogenomic networks. Trends Microbiol. 19:483–491 [DOI] [PubMed] [Google Scholar]
- 22.Bapteste E, van Iersel L, Janke A, Kelchner S, Kelk S, McInerney JO, Morrison DA, Nakhleh L, Steel M, Stougie L, Whitfield J. 2013. Networks: expanding evolutionary thinking. Trends Genet. 29:439–441 [DOI] [PubMed] [Google Scholar]
- 23.Monson R, Foulds I, Foweraker J, Welch M, Salmond GP. 2011. The Pseudomonas aeruginosa generalized transducing phage ϕPA3 is a new member of the ϕKZ-like group of ‘jumbo' phages, and infects model laboratory strains and clinical isolates from cystic fibrosis patients. Microbiology 157:859–867 [DOI] [PubMed] [Google Scholar]
- 24.Mesyanzhinov VV, Robben J, Grymonprez B, Kostyuchenko VA, Bourkaltseva MV, Sykilinda NN, Krylov VN, Volckaert G. 2002. The genome of bacteriophage ϕKZ of Pseudomonas aeruginosa. J. Mol. Biol. 317:1–19 [DOI] [PubMed] [Google Scholar]
- 25.Thomas JA, Rolando MR, Carroll CA, Shen PS, Belnap DM, Weintraub ST, Serwer P, Hardies SC. 2008. Characterization of Pseudomonas chlororaphis myovirus 201ϕ2-1 via genomic sequencing, mass spectrometry, and electron microscopy. Virology 376:330–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hertveldt K, Lavigne R, Pleteneva E, Sernova N, Kurochkina L, Korchevskii R, Robben J, Mesyanzhinov V, Krylov VN, Volckaert G. 2005. Genome comparison of Pseudomonas aeruginosa large phages. J. Mol. Biol. 354:536–545 [DOI] [PubMed] [Google Scholar]
- 27.Cornelissen A, Hardies SC, Shaburova OV, Krylov VN, Mattheus W, Kropinski AM, Lavigne R. 2012. Complete genome sequence of the giant virus OBP and comparative genome analysis of the diverse ϕKZ-related phages. J. Virol. 86:1844–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krylov VN, Dela Cruz DM, Hertveldt K, Ackermann H-W. 2007. “phiKZ-like viruses,” a proposed new genus of myovirus bacteriophages. Arch. Virol. 152:1955–1959 [DOI] [PubMed] [Google Scholar]
- 29.Krylov V, Pleteneva E, Bourkaltseva M, Shaburova O, Volckaert G, Sykilinda N, Kurochkina L, Mesyanzhinov V. 2003. Myoviridae bacteriophages of Pseudomonas aeruginosa: a long and complex evolutionary pathway. Res. Microbiol. 154:269–275 [DOI] [PubMed] [Google Scholar]
- 30.Jang HB, Kim YK, Del Castillo CS, Nho SW, Cha IS, Park SB, Ha MA, Hikima JI, Hong SJ, Aoki T, Jung TS. 2012. RNA-seq-based metatranscriptomic and microscopic investigation reveals novel metalloproteases of Neobodo sp. as potential virulence factors for soft tunic syndrome in Halocynthia roretzi. PLoS One 7:e52379. 10.1371/journal.pone.0052379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Green MR, Sambrook J. 2012. Molecular cloning: a laboratory manual, 4th ed, vol 1 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 32.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sonnhammer EL, Eddy SR, Durbin R. 1997. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins 28:405–420 [DOI] [PubMed] [Google Scholar]
- 34.Söding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33:W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGuffin LJ, Bryson K, Jones DT. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404–405 [DOI] [PubMed] [Google Scholar]
- 36.Kelley LA, Sternberg MJE. 2009. Protein structure prediction on the web: a case study using the Phyre server. Nat. Protoc. 4:363–371 [DOI] [PubMed] [Google Scholar]
- 37.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lavigne R, Sun WD, Volckaert G. 2004. PHIRE, a deterministic approach to reveal regulatory elements in bacteriophage genomes. Bioinformatics 20:629–635 [DOI] [PubMed] [Google Scholar]
- 41.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naville M, Ghuillot-Gaudeffroy A, Marchais A, Gautheret D. 2011. ARNold: a web tool for the prediction of Rho-independent transcription terminators. RNA Biol. 8:11–13 [DOI] [PubMed] [Google Scholar]
- 43.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lopes A, Amarir-Bouhram J, Faure G, Petit MA, Guerois R. 2010. Detection of novel recombinases in bacteriophage genomes unveils Rad52, Rad51 and Gp2.5 remote homologs. Nucleic Acids Res. 38:3952–3962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lima-Mendez G, Toussaint A, Leplae R. 2011. A modular view of the bacteriophage genomic space: identification of host and lifestyle marker modules. Res. Microbiol. 162:737–746 [DOI] [PubMed] [Google Scholar]
- 46.Lima-Mendez G, Toussaint A, Leplae R. 2007. Analysis of the phage sequence space: the benefit of structured information. Virology 365:241–249 [DOI] [PubMed] [Google Scholar]
- 47.Thomas JA, Weintraub ST, Wu W, Winkler DC, Cheng N, Steven AC, Black LW. 2012. Extensive proteolysis of head and inner body proteins by a morphogenetic protease in the giant Pseudomonas aeruginosa phage ϕKZ. Mol. Microbiol. 84:324–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kraemer JA, Erb ML, Waddling CA, Montabana EA, Zehr EA, Wang H, Nguyen K, Pham DS, Agard DA, Pogliano J. 2012. A phage tubulin assembles dynamic filaments by an atypical mechanism to center viral DNA within the host cell. Cell 149:1488–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kurochkina LP, Semenyuk PI, Orlov VN, Robben J, Sykilinda NN, Mesyanzhinov VV. 2012. Expression and functional characterization of the first bacteriophage-encoded chaperonin. J. Virol. 86:10103–10111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aksyuk AA, Kurochkina LP, Fokine A, Forouhar F, Mesyanzhinov VV, Tong L, Rossmann MG. 2011. Structural conservation of the myoviridae phage tail sheath protein fold. Structure 19:1885–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Browning C, Shneider MM, Bowman VD, Schwarzer D, Leiman PG. 2012. Phage pierces the host cell membrane with the iron-loaded spike. Structure 20:326–339 [DOI] [PubMed] [Google Scholar]
- 52.Dutilh BE, Huynen MA, Bruno WJ, Snel B. 2004. The consistent phylogenetic signal in genome trees revealed by reducing the impact of noise. J. Mol. Evol. 58:527–539 [DOI] [PubMed] [Google Scholar]
- 53.Suyama M, Bork P. 2001. Evolution of prokaryotic gene order: genome rearrangements in closely related species. Trends Genet. 17:10–13 [DOI] [PubMed] [Google Scholar]
- 54.Springman R, Badgett MR, Molineux IJ, Bull JJ. 2005. Gene order constrains adaptation in bacteriophage T7. Virology 341:141–152 [DOI] [PubMed] [Google Scholar]
- 55.Millard AD, Zwirglmaier K, Downey MJ, Mann NH, Scanlan DJ. 2009. Comparative genomics of marine cyanomyoviruses reveals the widespread occurrence of Synechococcus host genes localized to a hyperplastic region: implications for mechanisms of cyanophage evolution. Environ. Microbiol. 11:2370–2387 [DOI] [PubMed] [Google Scholar]
- 56.Arbiol C, Comeau AM, Kutateladze M, Adamia R, Krisch HM. 2010. Mobile regulatory cassettes mediate modular shuffling in T4-type phage genomes. Genome Biol. Evol. 2:140–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pell LG, Kanelis V, Donaldson LW, Howell PL, Davidson AR. 2009. The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc. Natl. Acad. Sci. U. S. A. 106:4160–4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen L, Shi K, Yin Z, Aihara H. 2013. Structural asymmetry in the Thermus thermophilus RuvC dimer suggests a basis for sequential strand cleavages during Holliday junction resolution. Nucleic Acids Res. 41:648–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu Y, Li H, Hu L, Wang J, Zhou Y, Pang Z, Liu L, Shao F. 2008. Structure of a Shigella effector reveals a new class of ubiquitin ligases. Nat. Struct. Mol. Biol. 15:1302–1308 [DOI] [PubMed] [Google Scholar]
- 60.Brüssow H, Canchaya C, Hardt WD. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68:560–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Coombes BK, Wickham ME, Brown NF, Lemire S, Bossi L, Hsiao WW, Brinkman FS, Finlay BB. 2005. Genetic and molecular analysis of GogB, a phage-encoded type III-secreted substrate in Salmonella enterica serovar typhimurium with autonomous expression from its associated phage. J. Mol. Biol. 348:817–830 [DOI] [PubMed] [Google Scholar]
- 62.Hendrix RW, Lawrence JG, Hatfull GF, Casjens S. 2000. The origins and ongoing evolution of viruses. Trends Microbiol. 8:504–508 [DOI] [PubMed] [Google Scholar]
- 63.Sullivan MB, Coleman ML, Weigele P, Rohwer F, Chisholm SW. 2005. Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol. 3:e144. 10.1371/journal.pbio.0030144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bartual SG, Otero JM, Garcia-Doval C, Llamas-Saiz AL, Kahn R, Fox GC, van Raaij MJ. 2010. Structure of the bacteriophage T4 long tail fiber receptor-binding tip. Proc. Natl. Acad. Sci. U. S. A. 107:20287–20292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sycheva LV, Shneider MM, Sykilinda NN, Ivanova MA, Miroshnikov KA, Leiman PG. 2012. Crystal structure and location of gp131 in the bacteriophage phiKZ virion. Virology 434:257–264 [DOI] [PubMed] [Google Scholar]
- 66.Pei J, Grishin NV. 2005. COG3926 and COG5526: a tale of two new lysozyme-like protein families. Protein Sci. 14:2574–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hodak H, Galán JE. 2013. A Salmonella Typhi homologue of bacteriophage muramidases controls typhoid toxin secretion. EMBO Rep. 14:95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fokine A, Miroshnikov KA, Shneider MM, Mesyanzhinov VV, Rossmann MG. 2008. Structure of the bacteriophage phiKZ lytic transglycosylase gp144. J. Biol. Chem. 283:7242–7250 [DOI] [PubMed] [Google Scholar]
- 69.Katsura I. 1987. Determination of bacteriophage lambda tail length by a protein ruler. Nature 327:73–75 [DOI] [PubMed] [Google Scholar]
- 70.Blackburn NT, Clarke AJ. 2001. Identification of four families of peptidoglycan lytic transglycosylases. J. Mol. Evol. 52:78–84 [DOI] [PubMed] [Google Scholar]
- 71.Leiman PG, Basler M, Ramagopal UA, Bonanno JB, Sauder JM, Pukatzki S, Burley SK, Almo SC, Mekalanos JJ. 2009. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. U. S. A. 106:4154–4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fokine A, Battisti AJ, Bowman VD, Efimov AV, Kurochkina LP, Chipman PR, Mesyanzhinov VV, Rossmann MG. 2007. Cryo-EM study of the Pseudomonas bacteriophage phiKZ. Structure 15:1099–1104 [DOI] [PubMed] [Google Scholar]
- 73.Oliveira H, Melo LD, Santos SB, Nóbrega FL, Ferreira EC, Cerca N, Azeredo J, Kluskens LD. 2013. Molecular aspects and comparative genomics of bacteriophage endolysins. J. Virol. 87:4558–4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Comeau AM, Arbiol C, Krisch HM. 2010. Gene network visualization and quantitative synteny analysis of more than 300 marine T4-like phage scaffolds from the GOS metagenome. Mol. Biol. Evol. 27:1935–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grote J, Thrash JC, Huggett MJ, Landry ZC, Carini P, Giovannoni SJ, Rappé MS. 2012. Streamlining and core genome conservation among highly divergent members of the SAR11 clade. mBio 3:e00252–12. 10.1128/mBio.00252-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bull JJ, Springman R, Molineux IJ. 2007. Compensatory evolution in response to a novel RNA polymerase: orthologous replacement of a central network gene. Mol. Biol. Evol. 24:900–908 [DOI] [PubMed] [Google Scholar]
- 77.Sullivan MB, Krastins B, Hughes JL, Kelly L, Chase M, Sarracino D, Chisholm SW. 2009. The genome and structural proteome of an ocean siphovirus: a new window into the cyanobacterial ‘mobilome'. Environ. Microbiol. 11:2935–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Comeau AM, Bertrand C, Letarov A, Tétart F, Krisch HM. 2007. Modular architecture of the T4 phage superfamily: a conserved core genome and a plastic periphery. Virology 362:384–396 [DOI] [PubMed] [Google Scholar]
- 79.Schmuki MM, Erne D, Loessner MJ, Klumpp J. 2012. Bacteriophage P70: unique morphology and unrelatedness to other Listeria bacteriophages. J. Virol. 86:13099–13102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Boyd EF, Brüssow H. 2002. Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. Trends Microbiol. 10:521–529 [DOI] [PubMed] [Google Scholar]
- 81.Pickard D, Toribio AL, Petty NK, van Tonder A, Yu L, Goulding D, Barrell B, Rance R, Harris D, Wetter M, Wain J, Choudhary J, Thomson N, Dougan G. 2010. A conserved acetyl esterase domain targets diverse bacteriophages to the Vi capsular receptor of Salmonella enterica serovar Typhi. J. Bacteriol. 192:5746–5754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Giphart-Gassler M, Plasterk RH, van de Putte P. 1982. G inversion in bacteriophage Mu: a novel way of gene splicing. Nature 297:339–342 [DOI] [PubMed] [Google Scholar]
- 83.Grundy FJ, Howe MM. 1984. Involvement of the invertible G segment in bacteriophage Mu tail fiber biosynthesis. Virology 134:296–317 [DOI] [PubMed] [Google Scholar]
- 84.Haggård-Ljungquist E, Halling C, Calendar R. 1992. DNA sequences of the tail fiber genes of bacteriophage P2: evidence for horizontal transfer of tail fiber genes among unrelated bacteriophages. J. Bacteriol. 174:1462–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.