Abstract
Yellow fever virus (YFV) can induce acute, life-threatening disease that is a significant health burden in areas where yellow fever is endemic, but it is preventable through vaccination. The live attenuated 17D YFV strain induces responses characterized by neutralizing antibodies and strong T cell responses. This vaccine provides an excellent model for studying human immunity. While several studies have characterized YFV-specific antibody and CD8+ T cell responses, less is known about YFV-specific CD4+ T cells. Here we characterize the epitope specificity, functional attributes, and dynamics of YFV-specific T cell responses in vaccinated subjects by investigating peripheral blood mononuclear cells by using HLA-DR tetramers. A total of 112 epitopes restricted by seven common HLA-DRB1 alleles were identified. Epitopes were present within all YFV proteins, but the capsid, envelope, NS2a, and NS3 proteins had the highest epitope density. Antibody blocking demonstrated that the majority of YFV-specific T cells were HLA-DR restricted. Therefore, CD4+ T cell responses could be effectively characterized with HLA-DR tetramers. Ex vivo tetramer analysis revealed that YFV-specific T cells persisted at frequencies ranging from 0 to 100 cells per million that are detectable years after vaccination. Longitudinal analysis indicated that YFV-specific CD4+ T cells reached peak frequencies, often exceeding 250 cells per million, approximately 2 weeks after vaccination. As frequencies subsequently declined, YFV-specific cells regained CCR7 expression, indicating a shift from effector to central memory. Cells were typically CXCR3 positive, suggesting Th1 polarization, and produced gamma interferon and other cytokines after reactivation in vitro. Therefore, YFV elicits robust early effector CD4+ T cell responses that contract, forming a detectable memory population.
INTRODUCTION
Members of the family Flaviviridae such as Yellow fever virus (YFV) are important causes of illness both historically and at present, causing a significant health burden in areas where yellow fever is endemic (1). Yellow fever (YF) produces symptoms ranging from a mild flu-like illness to hemorrhagic fever and organ failure, but infection is preventable through vaccination (2, 3). The YF vaccine uses a live attenuated virus (17D) and is safe and extremely effective, generating robust antibody responses that persist for decades (4, 5). The vaccine is known to elicit neutralizing antibodies and strong T cell responses in nearly all recipients (6, 7). Therefore, the YFV 17D vaccine is an excellent and important model for studying human antiviral immunity. Multiple studies have investigated the attributes and dynamics of CD4+ and CD8+ T cell responses following YF vaccination (8–12). By using a variety of readouts, these studies demonstrated that YFV-specific CD8+ T cell responses are polyfunctional, exhibit distinct surface phenotypic markers, and peak approximately 2 to 4 weeks after vaccination. Blom et al. evaluated both CD4+ and CD8+ T cell responses by examining the dynamics and kinetics of CD38 and Ki67 upregulation, noting that CD4 T cell responses precede CD8 T cell responses (12). Polyfunctional CD4+ T cells were also detected after vaccination (11). Previous studies have demonstrated that YF vaccination elicits both Th1 and Th2 YFV-specific CD4+ T cell responses in humans (13). Investigations focused on innate immune responses after YF vaccination have highlighted coordinated efforts of both the innate and adaptive immune systems in mounting strong immune responses against YFV (14–16). However, detailed studies of YFV-specific CD4+ T cells have been hindered by a lack of epitope-specific reagents and a lack of comprehensive epitope knowledge. A recent study attempted to identify HLA class II YFV epitopes, demonstrating that peptides that could elicit T cell responses in peripheral blood mononuclear cells (PBMCs) bound to multiple class II alleles with high affinity (17). However, a direct demonstration that these peptides were CD4+ T cell epitopes was not possible. In general, the epitope specificity, magnitude, and functional phenotype of YFV-specific CD4+ T cell responses remain less characterized than those of CD8+ T cell responses.
In this study, we developed HLA class II tetramers as tools to perform epitope-specific analysis of YFV-specific CD4+ T cells by using in vitro cultures and HLA-DR tetramers to identify epitopes across the YFV genome restricted by seven different HLA-DR types. We then selected a subset of these epitopes and used ex vivo tetramer staining to examine the frequency and phenotype of tetramer-labeled cells. Enzyme-linked immunospot (ELISPOT) and cytokine assays were used to investigate the functional characteristics of YFV-specific CD4+ T cell responses. Our findings provide novel insights into the fine specificity, magnitude, and phenotype of YFV-specific responses and corroborate prior findings on the temporal attributes of YFV-specific CD4+ T cell responses.
MATERIALS AND METHODS
Human subjects and blood samples.
Blood samples were obtained from subjects who had previously received the YF vaccine with informed consent as part of an institutional review board (IRB)-approved protocol. PBMCs were isolated within 18 h by Ficoll underlay. Samples for the longitudinal portion of this study were collected just prior to vaccination and 14, 30, and 60 days after vaccination. All other samples were collected 1 to 5 years after vaccination. Subjects were recruited with informed consent for an IRB-approved study. A total of 36 subjects who had been previously immunized and 6 subjects who were examined longitudinally before and after vaccination were recruited for this study. DNA samples were HLA typed with Dynal Unitray SSP kits (Invitrogen). A summary of the attributes of these subjects is shown in Table 1. Additional subjects who had not received the vaccine were also recruited and used as controls.
Table 1.
Attributes of YFV vaccine recipients
| Subject no.a | Gender | Age (yr)b | Ethnicityc | Vaccination date | Blood draw date | HLA | Ae | Be |
|---|---|---|---|---|---|---|---|---|
| 1 | Female | 61 | Caucasian | 3/5/2005 | 11/16/2009 | DRB1* | 01:01 | 03:01 |
| 2 | Female | 28 | Caucasian | 4/28/2009 | 2/2/2010 | DRB1* | 04:04 | 13:01/13:02d |
| 3 | Female | 46 | Caucasian | 2/5/2008 | 3/25/2010 | DRB1* | 01:01 | 03:01 |
| 4 | Female | 57 | Other | 9/5/2006 | 3/9/2010 | DRB1* | 07:01 | 13:03 |
| 5 | Female | 49 | Caucasian | 3/8/2007 | 9/9/2010 | DRB1* | 15:01 | 15:01 |
| 6 | Female | 48 | Caucasian | 5/1/2007 | 2/4/2010 | DRB1* | 01:03 | 11:01 |
| 8 | Male | 62 | Caucasian | 12/18/2009 | 7/13/2010 | DRB1* | 03:01 | 08:01 |
| 9 | Male | 39 | Caucasian | 12/5/2006 | 6/24/2010 | DRB1* | 07:01 | 11:01 |
| 10 | Female | 44 | Caucasian | 8/7/2009 | 5/4/2010 | DRB1* | 03:01 | 15:01 |
| 11 | Female | 49 | Caucasian | 1/25/2005 | 9/21/2010 | DRB1* | 07:01 | 11:04 |
| 12 | Male | 53 | Caucasian | 10/22/2007 | 5/3/2010 | DRB1* | 03:01 | 03:01 |
| 13 | Male | 53 | Caucasian | 4/17/2006 | 3/24/2010 | DRB1* | 01:03 | 11:01 |
| 14 | Male | 31 | Caucasian | 3/10/2009 | 9/22/2010 | DRB1* | 03:01 | 13:01 |
| 16 | Male | 33 | Caucasian | 12/23/09 | 4/6/2010 | DRB1* | 03:01 | 15:01 |
| 17 | Female | 25 | Caucasian | 1/12/2010 | 9/8/2010 | DRB1* | 07:01 | 15:01 |
| 18 | Female | 29 | Caucasian | 12/30/2008 | 8/23/2010 | DRB1* | 04:04 | 11:01 |
| 19 | Female | 25 | Caucasian | 3/31/2009 | 6/7/2010 | DRB1* | 03:01 | 03:01 |
| 20 | Male | 52 | Caucasian | 3/23/2010 | 9/20/2010 | DRB1* | 04:01 | 14:01 |
| 21 | Female | 23 | Caucasian | 4/29/2008 | 7/12/2010 | DRB1* | 03:01 | 11:01 |
| 22 | Female | 24 | Caucasian | 11/3/2009 | 12/21/2010 | DRB1* | 01:01 | 03:01 |
| 23 | Male | 35 | Asian | 6/1/2005 | 7/16/2010 | DRB1* | 04:03 | 04:04 |
| 24 | Female | 58 | Caucasian | 5/27/2009 | 8/4/2010 | DRB1* | 01:01 | 04:01 |
| 25 | Male | 65 | Caucasian | 8/7/2006 | 8/4/2010 | DRB1* | 03:01 | 13:01/13:02d |
| 26 | Female | 66 | Caucasian | 5/25/2010 | 1/24/2012 | DRB1* | 07:01 | 11:01 |
| 27 | Male | 25 | Caucasian | 1/11/2010 | 9/16/2010 | DRB1* | 01:01 | 13:01 |
| 28 | Female | 51 | Caucasian | 6/8/2010 | 10/19/2010 | DRB1* | 01:01 | 07:01 |
| 29 | Female | 38 | Caucasian | 6/11/2010 | 7/14/2011 | DRB1* | 01:01 | 01:03 |
| 31 | Female | 22 | Caucasian | 6/4/2010 | 7/5/2011 | DRB1* | 03:01 | 15:01 |
| 32 | Male | 29 | Caucasian | 6/19/2008 | 8/3/2010 | DRB1* | 01:01 | 15:01 |
| 33 | Female | 27 | Caucasian | 6/19/2008 | 8/3/2010 | DRB1* | 01:01 | 01:01 |
| 34 | Male | 58 | Caucasian | 7/2/2009 | 10/17/2011 | DRB1* | 01:01 | 15:01 |
| 35 | Male | 54 | Caucasian | 7/13/2010 | 2/22/2011 | DRB1* | 04:01 | 15:01 |
| 38 | Female | 48 | Caucasian | 8/17/2010 | 9/12/2011 | DRB1* | 03:01 | 16:01 |
| 39 | Male | 66 | Caucasian | 8/19/2010 | 1/31/2011 | DRB1* | 08:01 | 15:01 |
| 40 | Female | 76 | Caucasian | 12/20/2010 | 1/11/2011 | DRB1* | 03:01 | 04:04 |
Omitted subject numbers are those of vaccinees who were recruited but had unsuitable HLA types.
Age at time of vaccination.
Ethnicity was self-reported by each subject.
The HLA typing method used could not differentiate DRB1*13:01 and DRB1*13:02.
Subjects 5, 12, 19, and 33 are HLA-DRB1 homozygous while all other subjects are heterozygous.
Tetramer-guided epitope mapping.
Biotinylated HLA-DR proteins were purified from the supernatants of transfected insect cell cultures as previously described (18). A total of 546 peptides (17 amino acids in length with an 11-amino-acid overlap) spanning the YFV genome were synthesized (Mimotopes, Clayton, Australia). Pools of five or six peptides each were used to stimulate PBMCs from vaccinated subjects. These same peptide mixtures were loaded into biotinylated HLA-DR proteins that were conjugated with streptavidin-phycoerythrin (PE) to generate tetramers as described previously (19). After 14 days, cultured cells were stained with pooled peptide tetramers and analyzed by flow cytometry. Consistent with previous studies, responses were considered positive when distinct staining greater than 2-fold above the background was observed. Positive wells were stained again with tetramers loaded separately with each of the individual peptides from the positive pool. For each HLA type, mapping experiments were completed for at least three subjects and only epitopes that were detected in two or more subjects were reported as positive.
Epitope density calculations.
Calculations were performed to determine the density of epitopes within the proteins of the YFV genome. Epitope density was calculated as the number of epitopes within each protein adjusted to account for protein length (in amino acids) according to the following formula: Epitope density = (Number of epitopes/Protein length) × 100.
Epitope processing and presentation assays.
To assess whether YFV epitopes could be processed and presented from intact protein, PBMCs were stimulated with whole dialyzed YF vaccine (YF-VAX; Sanofi Pasteur) at 10 μg/ml and replicate wells (48-well plate) were cultured for 14 days with the addition of fresh medium and interleukin-2 (IL-2) as needed starting on day 7, stained with tetramers corresponding to epitopes identified by tetramer-guided epitope mapping, and analyzed by flow cytometry. Again, responses were considered positive when distinct staining greater than 2-fold above the background was observed.
In a separate set of experiments, sorted T cell lines were generated by staining in vitro-expanded T cells with tetramer, sorting gated tetramer-positive CD4+ cells with a FACS Vantage (at single-cell purity), and expanding in a 96-well plate in the presence of 1.0 × 105 irradiated PBMCs and 2 μg/ml phytohemagglutinin (Remel Inc., Lenexa, KS). After expansion, T cells were stained with tetramers to confirm their peptide specificity and then restimulated with two concentrations of whole YF-VAX (2 and 10 μg/ml) and HLA-DR-matched irradiated PBMCs were added as antigen-presenting cells (APC). After 48 h, each well was pulsed for an additional 16 h with 1 μCi of [3H]thymidine (Amersham Biosciences, Piscataway, NJ), cells were collected on a glass filter mat, and uptake was measured with a scintillation counter to assess proliferation.
Ex vivo frequency and phenotypic analysis of YFV-specific CD4+ T cells.
Ex vivo analysis of YFV-specific T cells was carried out as previously described (20, 21). Briefly, 20 to 80 million PBMCs in 200 to 400 μl of T cell culture medium (most commonly, 30 million PBMCs in 200 μl of T cell culture medium) were stained with 20 μg/ml PE-labeled tetramers at room temperature for 100 min. Cells were then stained with anti-CD4 APC (eBioscience), anti-CD14 PerCP (BD Pharmingen), anti-CD19 PerCP (BD Pharmingen), and either anti-CD3–fluorescein isothiocyanate (FITC) (eBioscience) or anti-CD45RA–FITC for 20 min at 4°C. Cells were washed, incubated with anti-PE magnetic beads (Miltenyi Biotec) at 4°C for 20 min, and washed again, and a 1/100 fraction was saved for analysis. The other fraction was passed through a magnetic column (Miltenyi). Bound, PE-labeled cells were flushed and collected. Cells in the bound and precolumn fractions were stained with Via-Probe (BD Bioscience) for 10 min before flow cytometry. Data were analyzed by FlowJo (Tree Star), gating on forward scatter/side scatter and excluding CD14+ and CD19+, and Via-Probe+ (dead) cells and frequencies were calculated on the basis of the number of tetramer-positive cells and the calculated number of CD4+ T cells as previously described (20, 21).
For ex vivo phenotyping studies, PBMCs were stained with tetramer and magnetically isolated with anti-PE beads as described above, except that multiple antibodies, including CD45RA, CXCR3, CCR4, CD38, and CCR7, were added to both fractions after enrichment.
Cytokine analysis of YFV-specific T cell lines.
Short-term YFV-specific T cell lines were generated and analyzed for cytokine production as previously described (22). Briefly, T cells from vaccinated subjects were cultured for 2 weeks with peptide by using adherent CD4− cells as APC and assessed for peptide specificity by tetramer staining. To elicit epitope-specific cytokine secretion, T cell lines were transferred to wells precoated with 0.5 μg of the corresponding HLA/peptide complex and cultured overnight in the presence of soluble CD28/CD49d. Supernatants were harvested and assayed for gamma interferon (IFN-γ), IL-10, IL-13, and IL-5 contents with the Meso Scale Discovery (MSD) Cytokine Multiplex kit and read with the MSD SECTOR Imager 2400.
ELISPOT assays.
For ELISPOT assays, CD8+ T cells were depleted from the PBMCs of vaccinated subjects with CD8 microbeads (Miltenyi). CD4+ T cells were subsequently isolated with a CD4+ T Cell Isolation kit (Miltenyi), and adherent cells from the CD4− fraction were used as APC by plating 1 million cells per well, incubating them for 1 h at 37°C, and then washing them. CD4+ responder cells were seeded at 2.5 million per well, stimulated with YF-VAX at 10 μg/ml for 14 to 17 days, and rested for 3 days before use in ELISPOT assays. The human IFN-γ ELISPOT Ready-SET-Go! kit (eBioscience) was used according to the manufacturer's protocol. Briefly, MultiScreen IP 96-well plates (Millipore) were coated with anti-human IFN-γ capture antibody, washed twice with coating buffer, and blocked with culture medium at room temperature for 2 to 4 h. A total of 1.6 × 105 autologous CD4− cells (frozen on day 0) and 4 × 104 T cells were plated with T cell medium alone, phytohemagglutinin (2 μg/ml), YF-VAX (10 μg/ml), or YF-VAX (10 μg/ml) plus blocking antibodies (10 μg/ml). After ∼40 h of incubation at 37°C in 5% CO2, cells were labeled with biotin-conjugated anti-human IFN-γ detection antibody and avidin-horseradish peroxidase enzyme. Spot-forming colonies (SFCs) were developed with AEC substrate (Vector Laboratories) and stopped after ∼5 min by adding distilled water. Plates were read with an ImmunoSpot S5 Core analyzer (Cellular Technology Ltd., Shaker Heights, OH) after prealignment based on three wells across the plate. SFCs were analyzed with ImmunoCapture 6.2 by the smart-count method. Each condition was analyzed in triplicate; the SFCs for each condition were averaged, and the average background was subtracted to obtain the number of SFCs per condition. For ex vivo ELISPOT assays, the 14- to 17-day culture period was omitted and SFCs were developed and visualized after ∼40 h of incubation.
Statistical analysis.
Single comparisons were done with the Student t test. In cases where an F test indicated significantly different variances, Welch's correction was included. Multiple comparisons were done with the Kruskal-Wallis test, followed by Dunn's multiple-comparison test.
RESULTS
Identification of CD4+ T cell epitopes across the YFV genome.
The tetramer-guided epitope-mapping approach was used to identify CD4+ T cell epitopes across the YFV genome as described in Materials and Methods. Detailed results identifying epitopes within one representative YFV protein for a representative DRB1*03:01 subject are shown in Fig. 1. Similar experiments were performed for all YFV proteins (a total of 108 peptide pools), examining responses in at least three vaccinated subjects by using each of the seven HLA types studied. Tetramer staining results were qualitatively similar for each of the HLA-DR types examined, revealing distinct sets of epitopes for each HLA restriction. For a list summarizing all of the epitopes identified (including the number of subjects tested and the number of positive responses), see Table S1 in the supplemental material. As noted in that table, only epitopes that elicited detectable responses in at least two subjects were reported as positive. A total of 112 epitopes restricted by seven common HLA-DRB1 alleles, 01:01, 03:01, 04:01, 04:04, 07:01, 11:01, and 15:01, were observed in subjects with the corresponding haplotype. As summarized in Table 2, epitopes were present within every YFV protein. However, only the envelope and NS3 proteins contained epitopes for each of the HLA-DRB1 alleles tested. The envelope, NS3, and NS5 proteins contained the highest number of epitopes, but these were also the largest proteins. On the basis of epitope density calculations (as described in Materials and Methods), the capsid, envelope, NS2a, and NS3 proteins had the highest epitope densities. Therefore, these proteins appear to be important targets of CD4+ T cell responses.
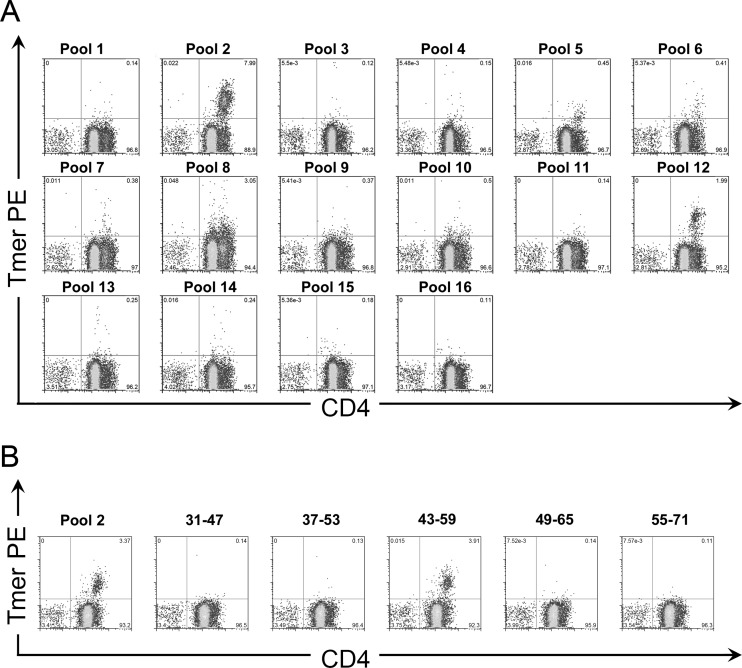
Fig 1.
Identification of CD4+ T cell epitopes within YFV proteins. (A) Staining of in vitro-stimulated cells from an HLA-DRB1*03:01 vaccinee with tetramers (Tmer) loaded with peptide pools spanning the YFV envelope protein. Peptide pools 2 and 12 contained DRB1*03:01-restricted epitopes. (B) Identification of the antigenic peptide within envelope pool 2 by staining of in vitro-stimulated cells with tetramers loaded with single peptides. The pool 2 epitope was identified within the Env 43-59 peptide.
Table 2.
Densities of YFV protein epitopes among seven common HLA DRB1 allelesa
| HLA-DRB1 (no. of subjects) | C (101 aa)b | PrM (164 aa) | Env (493 aa) | NS1 (352 aa) | NS2a (224 aa) | NS2b (130 aa) | NS3 (623 aa) | NS4a (126 aa) | NS4b (250 aa) | NS5 (905 aa) | Total (3,368 aa) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 01:01 (4) | 2 | 2 | 4 | 2 | 2 | 1 | 5 | 1 | 2 | 4 | 25 |
| 03:01 (6) | 0 | 0 | 2 | 4 | 1 | 0 | 4 | 0 | 0 | 0 | 11 |
| 04:01 (3) | 0 | 1 | 8 | 1 | 0 | 0 | 3 | 0 | 2 | 5 | 20 |
| 04:04 (3) | 0 | 0 | 3 | 1 | 1 | 0 | 4 | 0 | 1 | 2 | 12 |
| 07:01 (4) | 1 | 1 | 1 | 3 | 3 | 0 | 2 | 0 | 1 | 4 | 16 |
| 11:01 (5) | 3 | 0 | 6 | 0 | 1 | 0 | 2 | 1 | 1 | 8 | 22 |
| 15:01 (5) | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 3 | 6 |
| Total | 6 | 4 | 25 | 11 | 8 | 2 | 21 | 2 | 7 | 26 | 112 |
| Epitope density | 5.9 | 2.4 | 5.1 | 3.1 | 3.4 | 1.5 | 3.4 | 1.6 | 2.8 | 2.9 | 3.4 |
Values for YFV proteins containing the highest number of epitopes or the highest epitope density are in boldface.
aa, amino acids.
Selection of naturally processed YFV epitopes for ex vivo analysis.
Although the 112 YFV peptides identified as epitopes elicited positive responses in vitro, some could be cryptic epitopes that are not normally processed and presented in vivo. Others could be subdominant epitopes that comprise only a minor component of the CD4+ T cell response. We investigated the processing and presentation of peptide epitopes from intact protein by two assay methods, tetramer analysis of vaccine-stimulated PBMCs and measurement of the proliferation of tetramer-positive T cell lines in response to whole, dialyzed vaccine. Our initial experiments focused on DRB1*03:01 and DRB1*15:01, two of the most common HLA-DRB1 types in our study cohort (DRB1*01:01 was also common in the subjects recruited for epitope identification studies, but some of these subjects were not available for further study). Epitopes that gave a positive response in either assay were verified as being processed and presented. As summarized in Table 3, 8 of the 11 peptides that elicited DRB1*03:01-restricted responses and 6 out of 6 peptides that elicited DRB1*15:01-restricted responses were shown to be naturally processed (14 out of 17). The remaining three peptides appeared to contain cryptic epitopes that are not presented from intact antigen, a supposition that is supported by the observation that T cells recognizing these peptides were naive (Table 3). Since it was not feasible (because of blood volume limits) to perform ex vivo staining for every possible epitope in every subject, we next sought to identify the most useful tetramers for detecting YFV-specific CD4+ T cells directly ex vivo. For example, we assessed the ability of the 11 DRB1*03:01 tetramers and 6 DRB1*15:01 tetramers that correspond to naturally processed epitopes to detect YFV-specific CD4+ T cells directly ex vivo. Only a subset of these tetramers consistently labeled a clear population of YFV-specific CD4+ T cells (representative data shown in Fig. 2). The tetramer specificities that were less effective included epitopes that were not naturally processed, low-frequency epitopes that may be subdominant, and one tetramer that had a high background. Similar experiments were completed to select effective tetramers for other HLA types, which are in boldface in Table S1 in the supplemental material. In total, we identified 34 tetramers that are recommended for ex vivo analysis. However, additional subdominant epitopes that are not in boldface, such as DRB1*15:01/NS2b 19-35, could still be informative if a larger sample were used for analysis.
Table 3.
Natural processing of YFV epitopes
| HLA | Epitope | Processed | CD45RA |
|---|---|---|---|
| DRB1*03:01 | Env 43-59 | Yes | Negative |
| DRB1*03:01 | Env 331-347 | Yes | Negative |
| DRB1*03:01 | NS1 7-29 | No | Positive |
| DRB1*03:01 | NS1 85-101 | Yes | Negative |
| DRB1*03:01 | NS1 121-143 | Yes | Negative |
| DRB1*03:01 | NS1 163-179 | Yes | Negative |
| DRB1*03:01 | NS2a 13-29 | Yes | Negative |
| DRB1*03:01 | NS3 151-173 | No | Positive |
| DRB1*03:01 | NS3 229-251 | No | Positive |
| DRB1*03:01 | NS3 283-299 | Yes | Negative |
| DRB1*03:01 | NS3 355-371 | Yes | Negative |
| DRB1*15:01 | Env 457-473 | Yes | Negative |
| DRB1*15:01 | NS2b 19-35 | Yes | Negative |
| DRB1*15:01 | NS3 145-161 | Yes | Negative |
| DRB1*15:01 | NS5 325-341 | Yes | Negative |
| DRB1*15:01 | NS5 367-389 | Yes | Negative |
| DRB1*15:01 | NS5 547-563 | Yes | Negative |
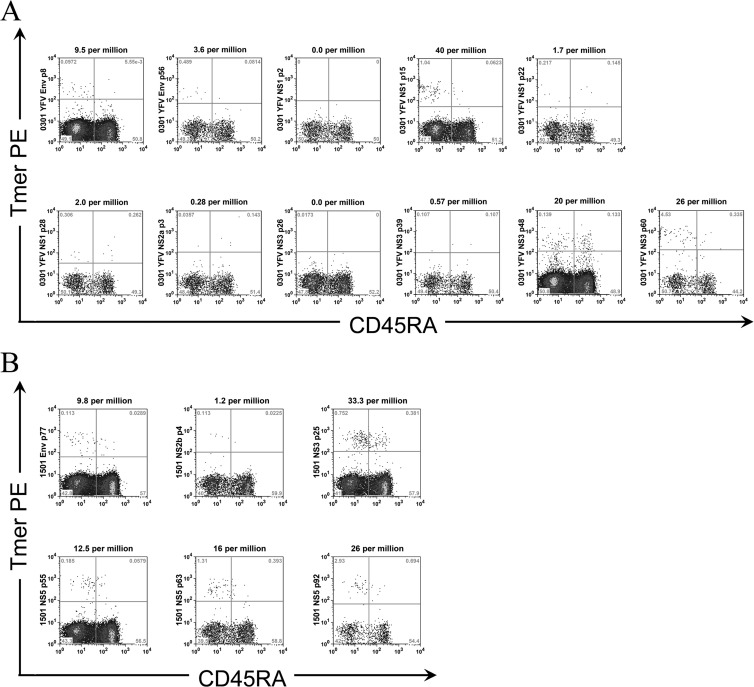
Fig 2.
Selection of YFV epitopes for ex vivo analysis. (A) Frequencies of YFV-specific T cells in a vaccinated DRB1*03:01 subject. The frequencies of T cells per million CD4+ T cells are as indicated. Env p8 (43-59), Env p56 (331-347), NS1 p15 (85-101), and NS3 p60 (355-371) effectively labeled YFV-specific CD4+ T cells. NS3 p48 (283-299) had a high background, while NS1 p2 (7-23), NS1 p22 (127-143), NS1 p28 (163-179), NS2a p3 (13-29), NS3 p26 (151-167), and NS3 p39 (229-245) did not label a distinct population. (B) Frequencies of YFV-specific T cells per million CD4+ T cells in a vaccinated DRB1*15:01 subject. Env p77 (457-473), NS3 p25 (145-161), NS5 p55 (325-341), NS5 p63 (367-383), and NS5 p92 (547-563) effectively labeled YFV-specific CD4+ T cells, while NS2b p4 (19-35) did not label a distinct population.
Ex vivo frequencies of YFV-specific CD4+ T cells.
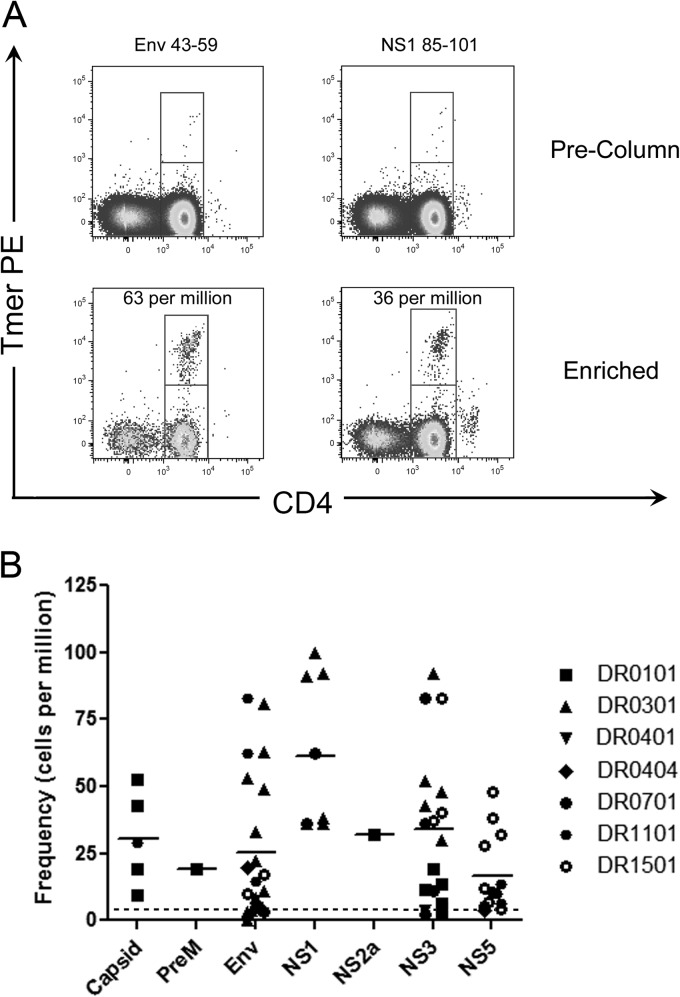
We next used anti-PE magnetic bead enrichment to detect YFV-specific T cells without in vitro expansion. This approach enabled us to directly characterize the frequency of YFV-specific T cells in the peripheral blood of vaccinated subjects. For these experiments, we focused on 16 subjects who were readily available for these experiments by using representative epitopes shown to perform well for ex vivo analysis (see Table S1 in the supplemental material). Representative staining results for a previously vaccinated DRB1*03:01 subject are shown in Fig. 3A. As summarized in Fig. 3B, we analyzed ex vivo frequencies across seven HLA types with multiple tetramers per subject. We observed diverse frequencies that ranged from 0 to 100 cells per million CD4+ T cells. Analysis of variance indicated no difference in the frequency of YFV-specific T cells with respect to HLA type (P = 0.14). Among all of the subjects examined, T cells specific for the NS1 protein had the highest frequencies, followed by NS3, capsid, envelope, and NS5 (the mean ± the standard error of the mean [SEM] frequencies were 61 ± 10, 34 ± 7, 31 ± 8, 23 ± 6, and 17 ± 4 cells per million, respectively), but only the differences between NS1 and the envelope protein and between NS1 and NS5 were statistically significant (Bonferroni's multiple-comparison test, P < 0.05). There were insufficient data available for the PrM and NS2a proteins for inclusion in this analysis. In these 16 subjects, frequencies were examined 4 to 67 months after vaccination but there was no correlation between T cell frequency and the elapsed time since vaccination (r2 = 0.001, P = 0.76). At earlier time points after vaccination, T cell frequencies varied markedly with time (see longitudinal analysis of YFV-specific CD4+ T cell responses below) but frequencies were apparently more stable >4 months after vaccination.
Fig 3.
Frequencies of YFV-specific T cells. (A) Frequencies of YFV-specific T cells in a vaccinated DRB1*03:01 subject. The frequencies of T cells per million CD4+ T cells are as indicated. (B) Frequencies of YFV-specific T cells in 16 vaccinated subjects with DRB1*01:01, DRB1*03:01, DRB1*04:01, DRB1*04:04, DRB1*07:01, DRB1*11:01, or DRB1*15:01 HLA-DR haplotypes. Each data point represents the frequency of T cells specific for a single YFV protein epitope in a single subject. The dotted reference line indicates the average number of CD45RA− T cells from naive, unvaccinated subjects.
Ex vivo surface phenotyping of YFV-specific CD4+ T cells.
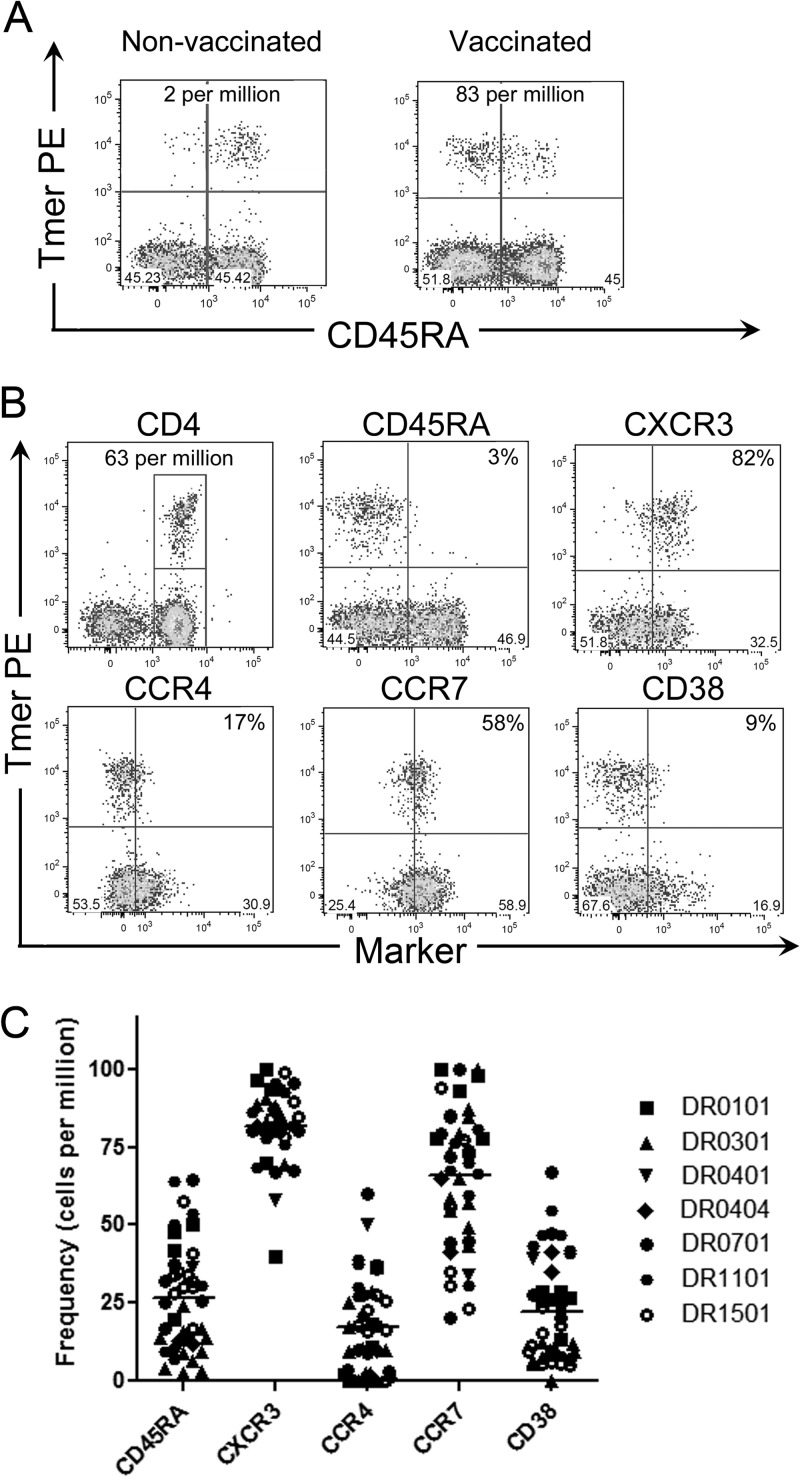
As described in other recent CD4+ T cell phenotyping studies, we combined ex vivo tetramer enrichment and antibody staining for cell surface markers of interest (20, 21, 23). To validate this approach, we first compared CD45RA expression for YFV-specific T cells in vaccinated and unvaccinated subjects. Representative results depicting frequency and CD45RA expression for a pair of vaccinated and unvaccinated subjects are shown in Fig. 4A. In this particular experiment, extra PBMCs from the unvaccinated subject (20 million versus 80 million) were examined to produce a similar number of enriched events. As expected, YFV-specific T cells were infrequent (mean ± SEM frequency = 4 ± 2 cells per million) and had a predominantly CD45RA+ phenotype in unvaccinated subjects. In contrast, YFV-specific T cells occurred at higher frequencies (mean ± SEM frequency = 36 ± 5 cells per million, P < 0.0001) and had a predominantly CD45RA− phenotype in vaccinated subjects. We then used additional markers, including CXCR3, CCR4, CCR7, and CD38, to characterize the phenotype of YFV-specific T cells in vaccinated subjects. Detailed staining results for one representative subject are shown in Fig. 4B, while results for 17 subjects of various HLA types (each analyzed with tetramers specific for one to four different epitopes) are summarized in Fig. 4C. YFV-specific T cells in vaccinated subjects were predominantly CXCR3 positive and correspondingly had lower expression of CCR4. YFV-specific cells had variable expression of CCR7, and the majority expressed only low levels of CD38, indicating that they had not been recently activated. These results indicate that YFV-specific T cells in vaccinated subjects have a predominantly Th1-like memory phenotype, with various proportions of effector memory-like and central-memory-like cells.
Fig 4.
Phenotype of YFV-specific T cells. (A) Ex vivo analysis of 80 million PBMCs from an unvaccinated DRB1*15:01 subject (left) and 20 million PBMCs from a YF-vaccinated DRB1*15:01 subject (right) stained with PE-labeled NS3 145-161 tetramer (Tmer) and CD45RA antibody. The frequencies of T cells per million CD4+ T cells are indicated. (B) Ex vivo analysis of a representative vaccinated DRB1*03:01 subject stained with PE-labeled Env 43-59 tetramer and CD45RA, CXCR3, CCR4, CCR7, and CD38 antibodies. The value in the upper right quadrant of each panel is the percentage of epitope-specific cells that express that marker. (C) Summary of ex vivo T cell phenotypes for vaccinated subjects with DRB1*01:01, DRB1*03:01, DRB1*04:01, DRB1*04:04, DRB1*07:01, DRB1*11:01, or DRB1*15:01 HLA-DR haplotypes. T cells specific for single YFV protein epitopes were stained with PE-labeled tetramers and CD45RA, CXCR3, CCR4, CCR7, and CD38 antibodies. Each data point represents the expression of the corresponding surface marker by T cells from a single vaccinated subject.
Cytokine profiles of YFV-specific T cells.
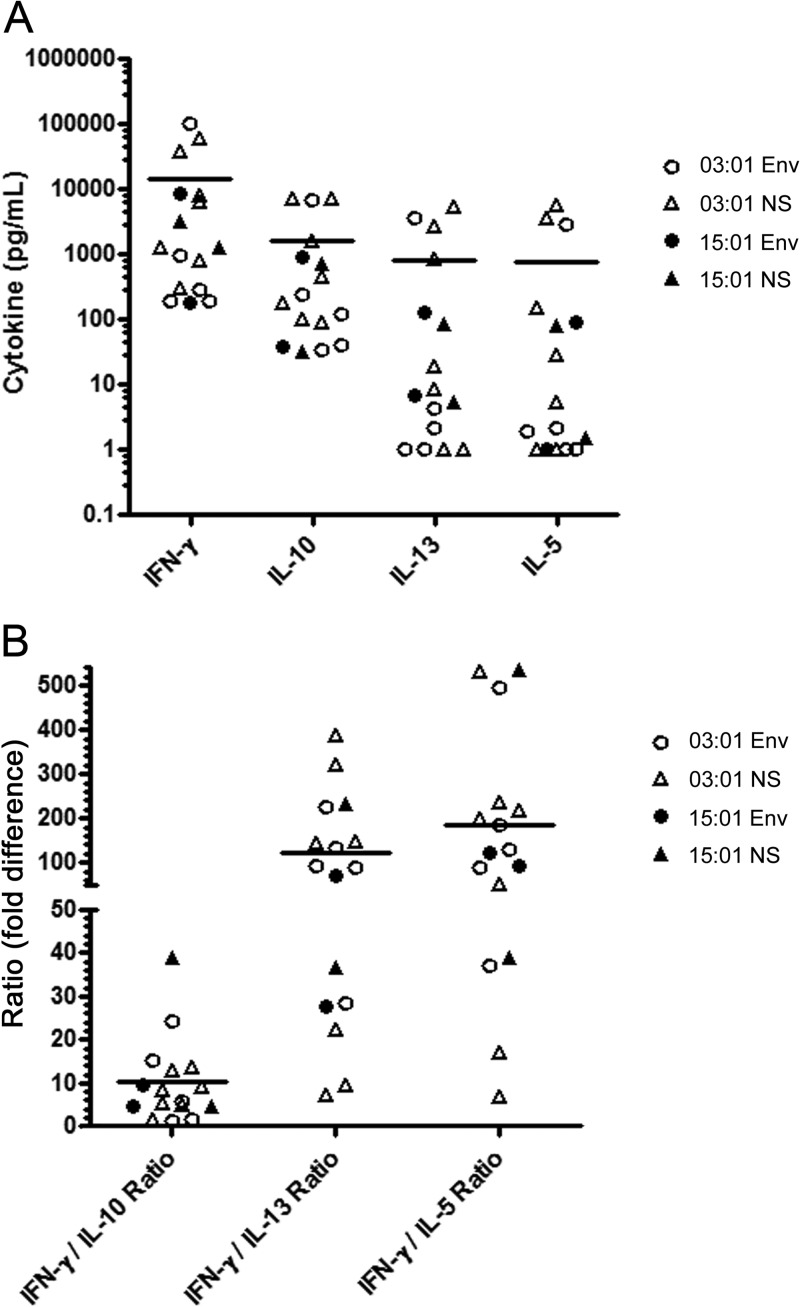
Ex vivo phenotyping indicated that the majority of YFV-specific T cells in vaccinated subjects expressed CXCR3, implying a Th1 phenotype. To further assess T cell polarization, we examined the cytokine content of supernatants from peptide/HLA-activated YFV-specific T cell lines (generated as described in Materials and Methods) with the MSD multiplex kit. A summary of the results of these assays is shown in Fig. 5A. Various cell lines secreted low (0 to 1,000 pg/ml), moderate (1,000 to 10,000 pg/ml), or high (>10,000 pg/ml) levels of IFN-γ and either low (0 to 1000 pg/ml) or moderate (1,000 to 10,000 pg/ml) levels of IL-10, IL-13, and IL-5. In all cases, IFN-γ was the predominant cytokine. As shown in Fig. 5B, levels of IFN-γ were an average of 10-fold higher than IL-10 levels, 100-fold higher than IL-13 levels, and 200-fold higher than IL-5 levels. Despite the wide range of cytokine levels observed, the level of IFN-γ secretion did not vary significantly by HLA DRB1 restriction (03:01 versus 15:01, P = 0.15) or epitope specificity (envelope versus nonstructural, P = 0.86). There was a similar lack of correlation between HLA restriction/epitope specificity and levels of IL-10, IL-13, and IL-5. In total, these data indicated that YFV-specific CD4+ T cells in vaccinated subjects were predominantly CXCR3+ and capable of secreting IFN-γ (Th1-like cells), but some also secreted lower levels of other cytokines.
Fig 5.
Cytokine analysis of supernatants from YFV-specific T cell lines. (A) Supernatants from a total of 16 peptide/MHC-activated YFV-specific T cell lines specific for epitopes from the envelope protein or nonstructural proteins isolated from vaccinated subjects with a DRB1*15:01 or a DRB1*03:01 haplotype were analyzed for IFN-γ, IL-10, IL-13, and IL-5 content with the MSD Cytokine Multiplex kit. Each symbol indicates the level of secreted cytokine for a single cell line. The y axis uses a logarithmic scale to depict the wide range of cytokine levels observed (0 to 100,000 pg/ml). (B) Cytokine ratios for IFN-γ production versus IL-10, IL-13, and IL-5 production by the same 16 T cell lines specific for epitopes from the envelope protein (circles) or nonstructural proteins isolated from vaccinated subjects with a DRB1*15:01 or a DRB1*03:01 haplotype. Each symbol indicates the ratio of secreted cytokines for a single cell line.
ELISPOT analysis of YFV-specific CD4+ T cells.
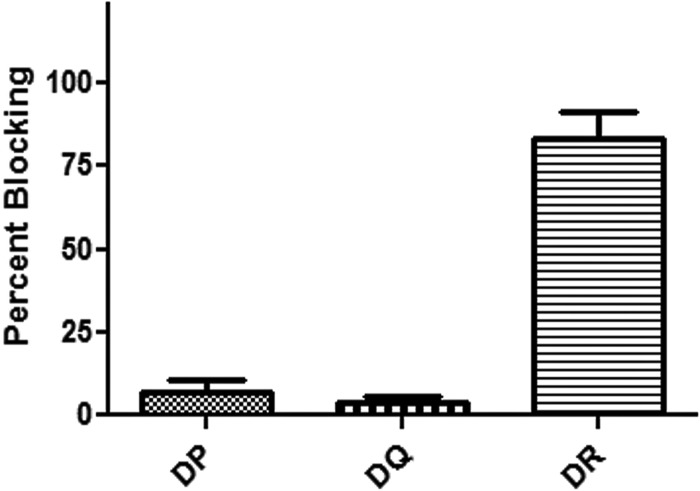
To verify the frequencies of YFV-specific T cells observed with tetramers, ex vivo ELISPOT assays (with no in vitro expansion) were performed with samples from two subjects with a total of three epitopes. The frequencies of IFN-γ-secreting T cells measured by ELISPOT assay were not significantly different from the frequencies measured in samples from the same subjects with tetramers (see Table S2 in the supplemental material, P = 0.51). In vitro ELISPOT assays were then performed in the presence or absence of HLA-DP, HLA-DQ, or HLA-DR blocking antibodies to determine the relative contributions of DP-, DQ-, and DR-restricted T cells to the overall response to YFV. As depicted in Fig. 6, HLA-DR antibody addition blocked approximately 83% of the response while HLA-DP and HLA-DQ antibodies blocked 4 and 7%, respectively. These results indicate that the majority of the YFV-specific response is HLA-DR restricted.
Fig 6.
HLA blocking of YFV-specific CD4+ T cell responses. Each bar indicates the average percent blocking observed after the addition of HLA-DP-, HLA-DQ-, or HLA-DR-blocking antibodies (triplicate SFCs enumerated by ELISPOT assay compared with an unblocked control) measured in 10 different vaccinated subjects (error bars indicated SEM).
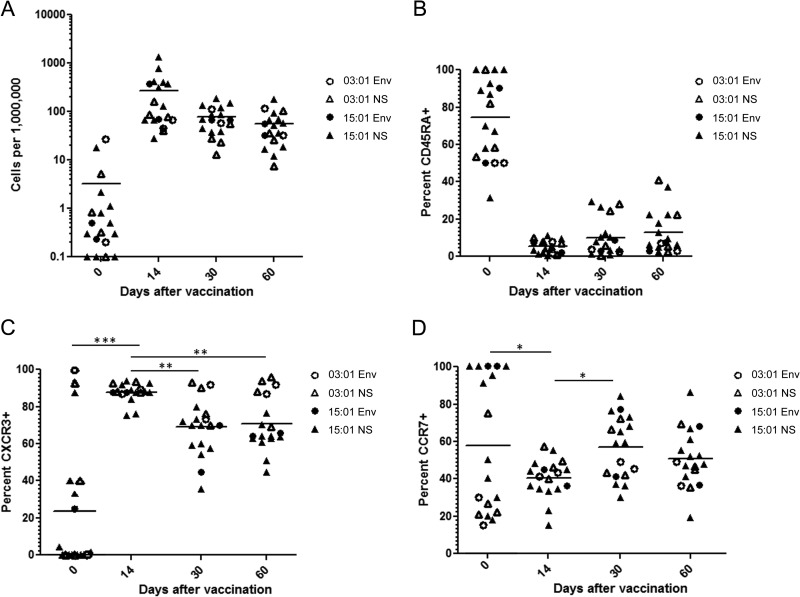
Longitudinal analysis of YFV-specific CD4+ T cell responses.
To investigate the temporal dynamics of CD4+ T cell responses to YFV, we monitored the ex vivo frequencies and phenotypes of YFV-specific T cells in samples drawn just prior to vaccination and 14, 30, and 60 days after vaccination. Figure 7A depicts longitudinal T cell frequencies measured in samples from two subjects with DR03:01 haplotypes (analyzed with Env 43-59, NS1 85-101, and NS3 355-371 tetramers) and four subjects with DR15:01 haplotypes (analyzed with two to four of the following: Env 457-473, NS3 145-161, NS5 367-383, and NS5 547-563 [depending on the blood volume that was available for the prevaccination draw]). As seen before in unvaccinated subjects, YFV-specific T cells were predominantly CD45RA+ prior to vaccination (Fig. 7B) and occurred at relatively low frequencies, averaging around three cells per million (Fig. 7A). The frequency of YFV-specific T cells increased dramatically within 14 days after vaccination, reaching an average of approximately 250 cells per million, and subsequently declined, reaching a range similar to that observed in long-term vaccinees. Figure 7B, C, and D summarize the longitudinal changes observed in the cell surface expression of CD45RA, CXCR3, and CCR7, respectively, following vaccination. The phenotype of the response after vaccination was predominantly CD45RA− and CXCR3+ at all time points. As expected, YFV-specific CD4+ T cells were predominantly CD45RA+CCR7+ (naive) before vaccination and were equivalently CD45RA− at all times after vaccination. There was a significant upregulation of CXCR3 on day 14 that was reduced by day 30. There was a corresponding transient reduction in CCR7 expression on day 14, consistent with an effector response, which was no longer observed by day 30. These results indicate that YFV-specific T cells undergo robust expansion during the first 2 weeks after vaccination and rapidly acquire a Th1-like memory phenotype. YFV-specific T cells then decline in frequency and transition from an effector-like phenotype to a more central-memory-like phenotype within the first month after vaccination. Beyond 1 month, YFV-specific CD4+ T cell responses reach frequencies and display phenotypic markers that are similar to those observed in the subjects who were examined years after vaccination.
Fig 7.
Longitudinal changes in the frequency and phenotype of YFV-specific CD4+ T cells. (A) Ex vivo frequencies of YFV-specific T cells in six vaccinated subjects with a DRB1*15:01 or a DRB1*03:01 haplotype measured before and 14, 30, and 60 days after vaccination with two to four different tetramers loaded with epitopes from the envelope (Env) protein or nonstructural (NS) proteins. Each data point represents the frequency of T cells specific for a single YFV epitope in a single subject. (B) Changes in cell surface expression of CD45RA for YFV-specific T cells in the same six vaccinated subjects measured before and 14, 30, and 60 days after vaccination with tetramers corresponding to the same epitopes. (C) Changes in cell surface expression of CXCR3 for YFV-specific T cells in the same six vaccinated subjects measured before and 14, 30, and 60 days after vaccination. CXCR3 expression was significantly increased 14 days following vaccination (P < 0.05) and subsequently declined (P < 0.05). (D) Changes in cell surface expression of CCR7 for YFV-specific T cells in the same six vaccinated subjects measured before and 14, 30, and 60 days after vaccination. CCR7 expression was significantly decreased 14 days following vaccination (P < 0.05) but recovered by 30 days following vaccination (P < 0.05).
DISCUSSION
The YF vaccine has been approved for decades and remains one of the most effective vaccines known. While neutralizing antibody responses to viral structural proteins are considered to be the predominant protective effect elicited by vaccination, limited published data imply that T cell help may play some role in promoting protection (24, 25). Activated CD4+ T cells have been shown to appear early after YF vaccination and are known to produce cytokines that support the activation and differentiation of B cells (26). A higher frequency of YFV CD4+ T cells at early time points after vaccination was also found to be correlated with a higher antibody titer (11). Furthermore, it has been demonstrated that CD4+ T cells are present in the hepatic lesions of YFV-infected persons, suggesting a possible role in virus control and/or clearance (27). Previous studies have demonstrated increases in the total number and activation status of CD4+ T cells following vaccination (11, 12, 14). However, the epitope specificity of the CD4+ T cell response elicited by the YF vaccine had remained relatively uncharacterized and the frequency and phenotype of YFV epitope-specific CD4+ T cells had never been examined directly. In this study, we developed and used HLA class II tetramers to identify CD4+ T cell epitopes restricted by seven common HLA-DR types across the entire YFV genome. YFV is known to be highly immunogenic, activating dendritic cell subsets via multiple Toll-like receptors and mobilizing a potent adaptive response (15). Therefore, it was not altogether unexpected that CD4+ T cells recognize epitopes within all of the structural and nonstructural proteins of the YFV genome (see Table S1 in the supplemental material). While a broad array of epitopes were recognized by CD4+ T cells, only the envelope and NS3 proteins were recognized by every subject in our study, each containing an epitope for all of the HLA-DRB1 alleles that we tested. These results agree with an earlier study in which responses in immunized mice focused on the envelope and NS3 proteins (28). However, NS1 epitopes were recognized by the highest frequencies of T cells. While the envelope, NS3, and NS5 proteins contained the highest overall numbers of epitopes, the smaller capsid protein also had a high epitope density, suggesting that these proteins are important targets of the CD4+ T cell response. On the basis of homology, these proteins are likely to be important in responses to other flaviviruses.
Since our epitope identification experiments relied on in vitro cultures stimulated with synthetic peptides, these results must be interpreted with the caveat that some peptides may not be naturally processed and presented and/or elicit responses that originate from the naive T cell compartment. While it was not feasible to examine this issue exhaustively, we addressed this by investigating the processing and presentation of DRB1*03:01−- and DRB1*15:01-restricted epitopes. Our results indicated that all of the DR15:01-restricted epitopes and 8 of 11 DR03:01-restricted epitopes could be processed and presented from intact protein. Correspondingly, T cells that were labeled by DRB1*03:01 tetramers loaded with peptides that were not processed and presented from intact protein were shown to be predominantly CD45RA+ CCR7+ in vaccinated subjects, suggesting a naive phenotype. These results indicate that approximately 80% of the peptide epitopes identified are naturally processed and recognized by YFV-specific memory T cells. While the vaccinees in our study had diverse HLA-DR types, they were primarily of Caucasian descent. Therefore, it is possible that our results are skewed because of this lack of diversity in our study population.
Our work used primarily tetramers to detect YFV-specific T cells. Because tetramers preferentially detect high-affinity T cells, it could be argued that tetramers may underestimate T cell frequencies by failing to detect low-affinity cells. However, a limited comparison indicated that frequencies measured by ELISPOT assays (which more readily detect low-affinity T cells) were similar to frequencies determined with tetramers. Because our study included only HLA-DR tetramers, we were not able to perform a detailed characterization of HLA-DQ- and HLA-DP-restricted T cells. However, ELISPOT assays indicated that the vast majority of the YFV-specific T cell response is HLA-DR restricted.
The HLA-restricted T cell epitopes defined in our work and the corresponding tetramer reagents provided the means to measure the frequency of YFV-specific CD4+ T cells directly ex vivo. This analysis revealed that YFV-specific memory T cells were readily detectable in subjects examined years after vaccination with a wide range of frequencies, ranging from 0 to 100 cells per million CD4+ T cells. T cells specific for these same epitopes were much less frequent and had a predominantly naive phenotype in unvaccinated subjects. The observation that moderate frequencies of YFV-specific T cells were detected in all vaccinated subjects, even though some were vaccinated up to 5 years earlier, suggests that YFV-specific T cell responses persist at detectable levels several years after vaccination. Comparing T cell frequencies for various YFV proteins, we observed that T cells specific for NS1 and NS3 epitopes were the most frequent. This raises the possibility that key internal viral proteins are more immunogenic than structural proteins, perhaps because their sequences are more functionally constrained. The induction of high frequencies of T cells specific for nonstructural proteins almost certainly reflects the fact that the YFV vaccine consists of a live, attenuated virus strain that is capable of sufficient replication to allow the presentation of all viral antigens in subcutaneous tissues.
The combined use of ex vivo tetramer staining and cytokine profile analysis allowed unambiguous characterization of the functional phenotype of YFV-specific T cells. As expected, YFV-specific cells were CD45RA negative in vaccinated subjects, implying a memory phenotype. Additional staining indicated that these cells were also CD45RO positive. While previous studies have suggested that YF vaccination elicits both Th1 and Th2 YFV-specific CD4+ T cell responses (13), we observed Th1-like responses. Longitudinal analysis of YFV-specific CD4+ T cell responses demonstrated that peak frequencies occurred approximately 2 weeks following vaccination. These kinetics are somewhat different from the observations of Akondy et al. (10), who observed that YFV tetramer-specific CD8+ T cell responses peak at day 30. However, earlier activation of CD4+ than CD8+ T cells after YFV vaccination has been observed by examining either DR+ (26) or CD38+ Ki67+ (12) CD4+ and CD8+ T cells.
In their detailed analyses of YFV-specific CD8+ T cells, both Blom et al. and Akondy et al. observed a predominantly CD45RA− CCR7− phenotype during the peak response and gradual reexpression of CD45RA but not CCR7. For YFV-specific CD4+ T cells, we observed a similar loss of CD45RA and CCR7 expression coinciding with peak frequencies. However, we observed that CD45RA was typically not reexpressed and CCR7 expression was gradually regained on CD4+ T cells as responses contracted. YFV-specific CD4+ T cells in long-term vaccinees had some diversity in their surface expression of CCR7, implying various proportions of effector-like and central-memory-like cells. This variation may partially explain the diverse magnitudes of cytokine secretion observed in various YFV-specific T cell lines. The YFV-specific CD4+ T cell response elicited by vaccination is apparently a Th1-like memory response, as exhibited by the predominant production of IFN-γ but also lesser amounts of IL-5, IL-10, and IL-13 that would be expected to support B cell and CD8+ T cell responses. These characteristics are consistent with the observation that the majority of YFV-specific T cells expressed CXCR3, a Th1-associated marker. These observations are consistent with a recent vaccine study reporting tumor necrosis factor alpha and IFN-γ responses that emerged 15 days after vaccination (29). Likewise, YFV-specific CD8+ T cells were also observed to be polyfunctional, expressing CD107a and secreting IFN-γ (12). Cumulatively, these results suggest differences in expansion kinetics and phenotypic behavior of YFV-specific CD4+ and CD8+ T cells.
In total, we report the identification of 112 HLA-DR-restricted YFV-specific T cell epitopes, a subset of which is well suited for ex vivo T cell analysis. Our results also indicate that a limited panel of HLA-DR epitopes could be used to characterize YFV-specific CD4+ T cells and to monitor longitudinal changes in the magnitude and phenotype of YFV-specific CD4+ T cell responses in the context of vaccination or infection. Given the homology between YFV and other members of the family Flaviviridae, the approaches used in this study should be applicable to the West Nile and dengue viruses, for which there are currently no approved vaccines.
Supplementary Material
ACKNOWLEDGMENTS
We thank Gladys Doronio and Susan Holt for subject recruitment and Diana Sorus for assistance in preparing the manuscript.
This work was supported by National Institutes of Health contract HHSN272200900043C.
Footnotes
Published ahead of print 18 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01160-13.
REFERENCES
- 1.Barrett AD, Higgs S. 2007. Yellow fever: a disease that has yet to be conquered. Annu. Rev. Entomol. 52:209–229 [DOI] [PubMed] [Google Scholar]
- 2.Monath TP. 2005. Yellow fever vaccine. Expert Rev. Vaccines 4:553–574 [DOI] [PubMed] [Google Scholar]
- 3.Monath TP. 2012. Review of the risks and benefits of yellow fever vaccination including some new analyses. Expert Rev. Vaccines 11:427–448 [DOI] [PubMed] [Google Scholar]
- 4.Theiler M, Smith HH. 1937. The use of yellow fever virus modified by in vitro cultivation for human immunization. J. Exp. Med. 65:787–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poland JD, Calisher CH, Monath TP, Downs WG, Murphy K. 1981. Persistence of neutralizing antibody 30-35 years after immunization with 17D yellow fever vaccine. Bull. World Health Organ. 59:895–900 [PMC free article] [PubMed] [Google Scholar]
- 6.Niedrig M, Lademann M, Emmerich P, Lafrenz M. 1999. Assessment of IgG antibodies against yellow fever virus after vaccination with 17D by different assays: neutralization test, haemagglutination inhibition test, immunofluorescence assay and ELISA. Trop. Med. Int. Health 4:867–871 [DOI] [PubMed] [Google Scholar]
- 7.Co MD, Terajima M, Cruz J, Ennis FA, Rothman AL. 2002. Human cytotoxic T lymphocyte responses to live attenuated 17D yellow fever vaccine: identification of HLA-B35-restricted CTL epitopes on nonstructural proteins NS1, NS2b, NS3, and the structural protein E. Virology 293:151–163 [DOI] [PubMed] [Google Scholar]
- 8.Santos AP, Bertho AL, Dias DC, Santos JR, Marcovistz R. 2005. Lymphocyte subset analyses in healthy adults vaccinated with yellow fever 17DD virus. Mem. Inst. Oswaldo Cruz 100:331–337 [DOI] [PubMed] [Google Scholar]
- 9.Miller JD, van der Most RG, Akondy RS, Glidewell JT, Albott S, Masopust D, Murali-Krishna K, Mahar PL, Edupuganti S, Lalor S, Germon S, Del Rio C, Mulligan MJ, Staprans SI, Altman JD, Feinberg MB, Ahmed R. 2008. Human effector and memory CD8+ T cell responses to smallpox and yellow fever vaccines. Immunity 28:710–722 [DOI] [PubMed] [Google Scholar]
- 10.Akondy RS, Monson ND, Miller JD, Edupuganti S, Teuwen D, Wu H, Quyyumi F, Garg S, Altman JD, Del Rio C, Keyserling HL, Ploss A, Rice CM, Orenstein WA, Mulligan MJ, Ahmed R. 2009. The yellow fever virus vaccine induces a broad and polyfunctional human memory CD8+ T cell response. J. Immunol. 183:7919–7930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kohler S, Bethke N, Böthe M, Sommerick S, Frentsch M, Romagnani C, Niedrig M, Thiel A. 2012. The early cellular signatures of protective immunity induced by live viral vaccination. Eur. J. Immunol. 42:2363–2373 [DOI] [PubMed] [Google Scholar]
- 12.Blom K, Braun M, Ivarsson MA, Gonzalez VD, Falconer K, Moll M, Ljunggren HG, Michaëlsson J, Sandberg JK. 2013. Temporal dynamics of the primary human T cell response to yellow fever virus 17D as it matures from an effector- to a memory-type response. J. Immunol. 190:2150–2158 [DOI] [PubMed] [Google Scholar]
- 13.Santos AP, Matos DC, Bertho AL, Mendonça SC, Marcovistz R. 2008. Detection of Th1/Th2 cytokine signatures in yellow fever 17DD first-time vaccinees through ELISpot assay. Cytokine 42:152–155 [DOI] [PubMed] [Google Scholar]
- 14.Gaucher D, Therrien R, Kettaf N, Angermann BR, Boucher G, Filali-Mouhim A, Moser JM, Mehta RS, Drake DR, III, Castro E, Akondy R, Rinfret A, Yassine-Diab B, Said EA, Chouikh Y, Cameron MJ, Clum R, Kelvin D, Somogyi R, Greller LD, Balderas RS, Wilkinson P, Pantaleo G, Tartaglia J, Haddad EK, Sékaly RP. 2008. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J. Exp. Med. 205:3119–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Querec T, Bennouna S, Alkan S, Laouar Y, Gorden K, Flavell R, Akira S, Ahmed R, Pulendran B. 2006. Yellow fever vaccine YF-17D activates multiple dendritic cell subsets via TLR2, 7, 8, and 9 to stimulate polyvalent immunity. J. Exp. Med. 203:413–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Querec TD, Akondy RS, Lee EK, Cao W, Nakaya HI, Teuwen D, Pirani A, Gernert K, Deng J, Marzolf B, Kennedy K, Wu H, Bennouna S, Oluoch H, Miller J, Vencio RZ, Mulligan M, Aderem A, Ahmed R, Pulendran B. 2009. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat. Immunol. 10:116–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Melo AB, Nascimento EJ, Braga-Neto U, Dhalia R, Silva AM, Oelke M, Schneck JP, Sidney J, Sette A, Montenegro SM, Marques ET. 2013. T-cell memory responses elicited by yellow fever vaccine are targeted to overlapping epitopes containing multiple HLA-I and -II binding motifs. PLoS Negl. Trop. Dis. 7:e1938. 10.1371/journal.pntd.0001938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Novak EJ, Liu AW, Nepom GT, Kwok WW. 1999. MHC class II tetramers identify peptide-specific human CD4(+) T cells proliferating in response to influenza A antigen. J. Clin. Invest. 104:R63–R67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Novak EJ, Liu AW, Gebe JA, Falk BA, Nepom GT, Koelle DM, Kwok WW. 2001. Tetramer-guided epitope mapping: rapid identification and characterization of immunodominant CD4+ T cell epitopes from complex antigens. J. Immunol. 166:6665–6670 [DOI] [PubMed] [Google Scholar]
- 20.Kwok WW, Roti M, Delong JH, Tan V, Wambre E, James EA, Robinson D. 2010. Direct ex vivo analysis of allergen-specific CD4+ T cells. J. Allergy Clin. Immunol. 125:1407–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wambre E, Delong JH, James EA, LaFond RE, Robinson D, Kwok WW. 2012. Differentiation stage determines pathologic and protective allergen-specific CD4+ T-cell outcomes during specific immunotherapy. J. Allergy Clin. Immunol. 129:544–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roti M, Yang J, Berger D, Huston L, James EA, Kwok WW. 2008. Healthy human subjects have CD4+ T cells directed against H5N1 influenza virus. J. Immunol. 180:1758–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwok WW, Tan V, Gillette L, Littell CT, Soltis MA, LaFond RB, Yang J, James EA, DeLong JH. 2012. Frequency of epitope-specific naive CD4(+) T cells correlates with immunodominance in the human memory repertoire. J. Immunol. 188:2537–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brandriss MW, Schlesinger JJ, Walsh EE, Briselli M. 1986. Lethal 17D yellow fever encephalitis in mice. I. Passive protection by monoclonal antibodies to the envelope proteins of 17D yellow fever and dengue 2 viruses. J. Gen. Virol. 67:229–234 [DOI] [PubMed] [Google Scholar]
- 25.Wrammert J, Miller J, Akondy R, Ahmed R. 2009. Human immune memory to yellow fever and smallpox vaccination. J. Clin. Immunol. 29:151–157 [DOI] [PubMed] [Google Scholar]
- 26.Martins MA, Silva ML, Marciano AP, Peruhype-Magalhães V, Eloi-Santos SM, Ribeiro JG, Correa-Oliveira R, Homma A, Kroon EG, Teixeira-Carvalho A, Martins-Filho OA. 2007. Activation/modulation of adaptive immunity emerges simultaneously after 17DD yellow fever first-time vaccination: is this the key to prevent severe adverse reactions following immunization? Clin. Exp. Immunol. 148:90–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quaresma JA, Barros VL, Pagliari C, Fernandes ER, Andrade HF, Jr, Vasconcelos PF, Duarte MI. 2007. Hepatocyte lesions and cellular immune response in yellow fever infection. Trans. R. Soc. Trop. Med. Hyg. 101:161–168 [DOI] [PubMed] [Google Scholar]
- 28.van der Most RG, Harrington LE, Giuggio V, Mahar PL, Ahmed R. 2002. Yellow fever virus 17D envelope and NS3 proteins are major targets of the antiviral T cell response in mice. Virology 296:117–124 [DOI] [PubMed] [Google Scholar]
- 29.Silva ML, Martins MA, Espírito-Santo LR, Campi-Azevedo AC, Silveira-Lemos D, Ribeiro JG, Homma A, Kroon EG, Teixeira-Carvalho A, Elói-Santos SM, Martins-Filho OA. 2011. Characterization of main cytokine sources from the innate and adaptive immune responses following primary 17DD yellow fever vaccination in adults. Vaccine 29:583–592 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.