SUMMARY
Coordinated ribosomal protein (RP) gene expression is crucial for cellular viability, but the transcriptional network controlling this regulon has only been well characterized in the yeast Saccharomyces cerevisiae. We have used whole-genome transcriptional and location profiling to establish that, in Candida albicans, the RP regulon is controlled by the Myb domain protein Tbf1 working in conjunction with Cbf1. These two factors bind both the promoters of RP genes and the rDNA locus; Tbf1 activates transcription at these loci and is essential. Orthologs of Tbf1 bind TTAGGG telomeric repeats in most eukaryotes, and TTAGGG cis-elements are present upstream of RP genes in plants and fungi, suggesting that Tbf1 was involved in both functions in ancestral eukaryotes. In all Hemiascomycetes, Rap1 substituted Tbf1 at telomeres and, in the S. cerevisiae lineage, this substitution also occurred independently at RP genes, illustrating the extreme adaptability and flexibility of transcriptional regulatory networks.
INTRODUCTION
The ribosome is one of the most conserved protein complexes in eukaryotic cells, and its stoichiometric assembly is critical for proper translational activity (Deutschbauer et al., 2005). About 90% of a cell’s transcriptional activity is dedicated to ribosome biogenesis (Rudra and Warner, 2004). This makes the RP subunits and rRNAs among the most abundant biomolecules of the cell. It is therefore critical to coordinate RP expression with nutritional status, and organisms have developed regulatory circuits that both synchronize ribosomal biogenesis with cell growth and coordinate RP and rRNA levels to ensure a constant balance of ribosomal components. Our knowledge of how transcriptional regulation of RP genes is achieved in eukaryotes is limited (Hu and Li, 2007), but recently several transcription factors controlling the RP regulon of S. cerevisiae have been identified (Hall et al., 2006; Jorgensen et al., 2004; Marion et al., 2004; Martin et al., 2004; Rudra et al., 2005; Schawalder et al., 2004; Wade et al., 2004). The most prominent of these are Fhl1, Ifh1, and the Myb domain DNA-binding protein Rap1 that form a regulatory complex at RP gene promoters (Rudra et al., 2005; Wade et al., 2004). Rap1 is a pleiotropic transcriptional activator that also directly binds telomeric repeats and influences telomeric structure and silencing in S. cerevisiae (Lieb et al., 2001).
Within- and between-species phenotypic diversity owes a lot to changes in gene expression driven by the evolution of cisregulatory elements (Wray, 2007). The appearance or disappearance of transcription factor binding motifs in the promoter regions of genes is the most documented mode of transcriptional evolution (Gompel et al., 2005). Broadly, DNA motif turnover can permit related biological processes to either become coregulated or lose their coregulation through addition or subtraction of gene targets from existing regulatory networks. Such reorganization of transcriptional regulation among organisms has been termed rewiring (Scannell and Wolfe, 2004). The prediction of cis-regulatory motifs by bioinformatical analysis of coregulated or functionally related genes is a powerful strategy to detect rewiring (Gasch et al., 2004; Ihmels et al., 2005; Tanay et al., 2005) and has proven useful in unraveling changes of the yeast mating type control (Tsong et al., 2006) and the galactose-utilization regulators (Martchenko et al., 2007).
RP cis-regulatory control is essential for cell viability in S. cerevisiae, and consequently it would appear likely to be under strong selective pressure and to be conserved among related yeast species. However, recent studies have predicted putative cis-elements in various Ascomycetes and suggested that RP gene regulation by Rap1 is unique to Saccharomyces and relatives (Gasch et al., 2004; Tanay et al., 2005). Since RP genes’ transacting regulators remained elusive in other species, the importance of these putative motifs could not be clarified. In this study, using genome-scale molecular methods, we show that the human fungal pathogen C. albicans, as well as most Ascomycetes, regulates RP gene expression through the conserved DNA-binding proteins Tbf1 and Cbf1, proving that co-option of different trans-acting factors occurred in this regulon during the evolution of yeasts.
RESULTS
RP Gene cis-Regulators Have Diverged between Saccharomyces and Candida
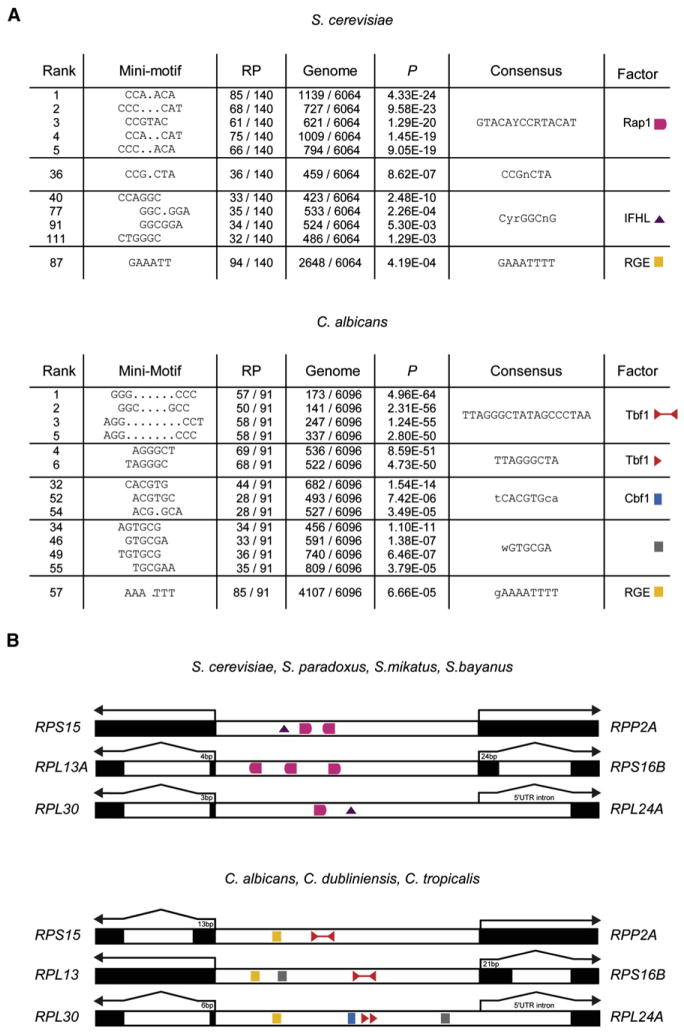
We revisited the detection of putative cis-regulatory elements of RP genes in the fully sequenced and annotated genomes of S. cerevisiae and C. albicans (Braun et al., 2005; Goffeau et al., 1996). Every minimotif composed of two nucleotide triplets spaced by less than 16 base pairs (bp) (XXXn[0–15]XXX) was tested for its enrichment upstream of RP genes. The most statistically significant hits are summarized in Figure 1A, and the complete results are available online (see Tables S1 and S2 available online). As expected, the top-scoring minimotifs for S. cerevisiae correspond to the known Rap1 DNA-binding specificity (Yarragudi et al., 2007) (Figure 1A). IFHL and rapid growth element (RGE) submotifs are also detected in agreement with the role of these cis-elements in the regulation of RP genes of S. cerevisiae (Ihmels et al., 2005; Wade et al., 2004). A more diverse set of motifs was obtained for C. albicans, with the top-scoring ones forming the previously reported palindromic sequence TTAGGGCTATAGCCCTAA (Tanay et al., 2005) (Figure 1A; Table S2). Two distinct clusters with the consensus tCACGTGca and wGTGCGa were also uncovered. Apart from the RGE, no putative motifs common to S. cerevisiae and C. albicans were found, supporting the view that their RP cis-regulatory circuits have significantly diverged (Gasch et al., 2004; Tanay et al., 2005).
Figure 1. Ribosomal Protein Genes’ cis-Regulatory Motifs Have Diverged between S. cerevisiae and C. albicans.
(A) Summary of top-ranked minimotifs (XXXn[0–15]- XXX) detected in both species. Only top representatives of distinct clusters are reported. The graphical representation of enriched motifs is constantly used thereafter.
(B) Conserved cis-regulatory motifs mapping in the promoters of three syntenic RP gene pairs among the Saccharomyces and Candida species.
To illustrate the reorganization of the cis-regulatory elements of RP genes, we examined the promoter regions of three syntenic RP gene pairs (Figure 1B). For each, we mapped the conserved and thus likely functional instances of the motifs identified above in four closely related Saccharomyces species and three Candida species (Figure 1B). In all three intergenic regions, Saccharomyces has at least one conserved Rap1-binding site, which was shown experimentally to be bound by Rap1 (Lieb et al., 2001). In C. albicans, the same RP gene promoters have a conserved TTAGGGCTATAGCCCTAA palindromic elements or a TTAGGG direct repeat (Figure 1B).
The palindrome TAGGGCTATAGCCCTA with at most three mismatches is found 70 times in the entire genome of C. albicans: 66 are located upstream of RP genes (Table S3). This palindrome was also screened in the genome of the closely related species Candida tropicalis, and, although it is conserved upstream of the same RP genes, the central six nucleotides show more variability (Table S4). It was also noticed that TTAGGG direct repeats occur upstream of many C. albicans RP genes. This suggests that the functional part of the palindrome is limited to TTAGGG, a motif associated with telomeric repeats in humans and many other species (Brown, 1989) and known to be the target of Tbf1, a subtelomeric Myb DNA-binding protein of S. cerevisiae (Brigati et al., 1993). Altogether, TTAGGG elements in a palindrome or as direct repeats are found upstream of 82% of C. albicans RP genes. The second minimotif cluster with RP enrichment in C. albicans is tCACGTGca, the binding target of S. cerevisiae Cbf1, a regulator of sulfur metabolism genes and a key player in the maintenance of centromeres (Maerkl and Quake, 2007) (Figure 1A). Interestingly, all direct repeats of TTAGGG at RP genes and almost half of the palindromic elements are flanked by a Cbf1 motif within 15 bp (Figure 1B; Table S5). The presence in C. albicans of orthologs of Tbf1 (orf19.801) and Cbf1 (orf19.2876) with very high conservation of their DNA-binding domains led us to further investigate these factors.
Tbf1 and Cbf1 Are trans-Factors Associated with RP Gene Promoters in C. albicans
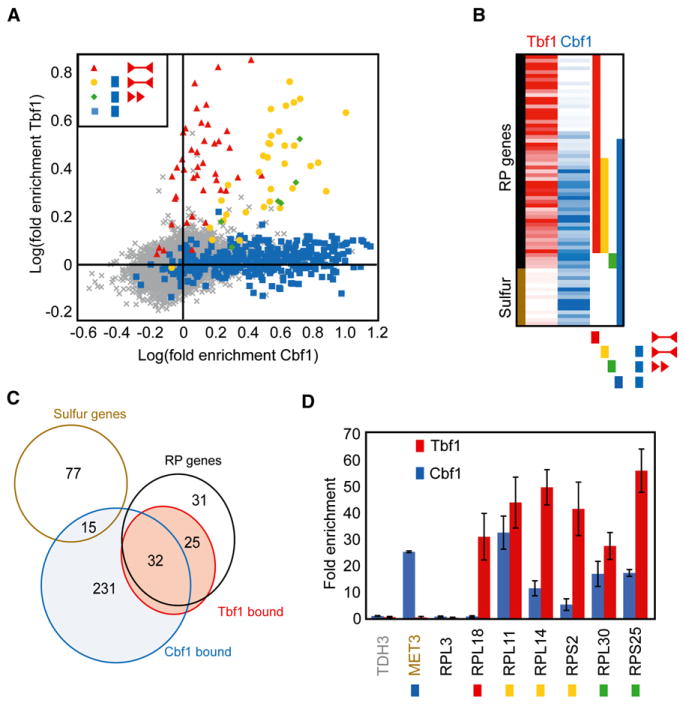
We first validated the in vivo physical interaction of Tbf1 with RP gene promoters. We performed whole-genome location profiling by chromatin immunoprecipitation (ChIP) coupled with microarray analysis (ChIP-CHIP) using an HA-tagged version of this transcription factor expressed in C. albicans (Guillemette et al., 2005). Using a cutoff of two standard deviations above the mean of log ratios (giving a 1.4-fold enrichment cutoff), Tbf1 was associated with 82 of the 11,817 probes in our microarray layout; 68 of them were located within 300 bp of a palindromic or directly repeated TTAGGG element (Figure 2A; Table S6). These probes span 64 intergenic regions, 57 of which are upstream of RP genes (Figures 2B and 2C). Tbf1 function seems therefore restricted to the control of the RP regulon as predicted by our bioinformatics analysis of RP cis-regulatory elements. Some of the intergenic probes on the arrays were more than 300 bp from putative Tbf1-binding sites, and it is possible that these elements were bound by Tbf1 but not detected by our experimental setup.
Figure 2. Whole-Genome Location Profiling by ChIP-CHIP Reveals that Tbf1 and Cbf1 Are Associated with RP Gene Promoters.
(A) Tbf1 and Cbf1 location data overlap the presence of the TTAGGG and CACGTG cis-regulatory sequences within 300 bp of probes.
(B) Tbf1 binds many RP genes, Tbf1 and Cbf1 signals overlap on a subset of RP genes, and Cbf1 alone binds promoters of the sulfur starvation regulon. Color saturation follows fold enrichment values. The different combinations of DNA elements were coded with colored blocks used in subsequent figures. Tbf1 palindrome alone, red; Cbf1, blue; Tbf1 palindrome + Cbf1, yellow; Tbf1 direct repeats + Cbf1, green.
(C) Cbf1 is a pleiotropic trans-acting factor, while Tbf1 is restricted to RP gene promoters.
(D) Validation of genome-wide binding data by ChIP of in vivo TAP-tagged Tbf1 and Cbf1 coupled with qPCR for a subset of nine target genes. Error bars reflect one standard deviation from the mean of three biological replicates.
We also determined the binding sites of Cbf1 using the same method. The Cbf1 signal was detected by 365 probes (in this case, the two standard deviation cutoff gave a 2.0-fold enrichment cutoff), of which 242 are found within 300 bp of a CACGTG element (Figure 2A). In contrast to Tbf1, Cbf1 was bound to promoters of genes with a wide variety of functions arguing for a more general regulatory role. Of 285 bound intergenic regions, 35 are upstream of RP genes (Figures 2B and 2C). In addition to the RP regulon, C. albicans Cbf1 binds 15 genes in the sulfur starvation response, in agreement with its conserved involvement in this function (Biswas et al., 2003b).
Our genome-wide location data confirm that Tbf1 and Cbf1 overlap on 32 RP gene promoters, all of which contain the predicted motifs (Figures 2B and 2C). We can infer a binding mode for C. albicans Tbf1 and Cbf1 based on the symmetry of bound DNA elements. It appears that Tbf1 only binds to palindromic elements or alternatively to a direct repeat of TTAGGG flanked by a 5′ Cbf1-binding site with a 7 bp spacing. In our experiments, Tbf1 does not bind isolated or direct repeats of the TTAGGG motif without a flanking Cbf1 site (Figure S1). These findings suggest a functional relationship between the DNA-binding proteins Tbf1 and Cbf1.
We further validated our findings for a panel of nine intergenic regions presenting all different combinations of the Tbf1 and Cbf1 elements. Site-specific primers were used to test the binding by ChIP of Tbf1-TAP and Cbf1-TAP followed by quantitative PCR (qPCR). These experiments confirmed the direct binding of Tbf1 to RPL18, RPL11, RPL14, RPS2, RPL30, and RPS25 and of Cbf1 to MET3, RPL11, RPL14, RPS2, RPL30, and RPS25 (Figure 2D). TDH3 and RPL3, lacking either Tbf1 or Cbf1 sites, showed no signal by qPCR (Figure 2D).
Tbf1 Is Essential and Activates RP Gene Expression
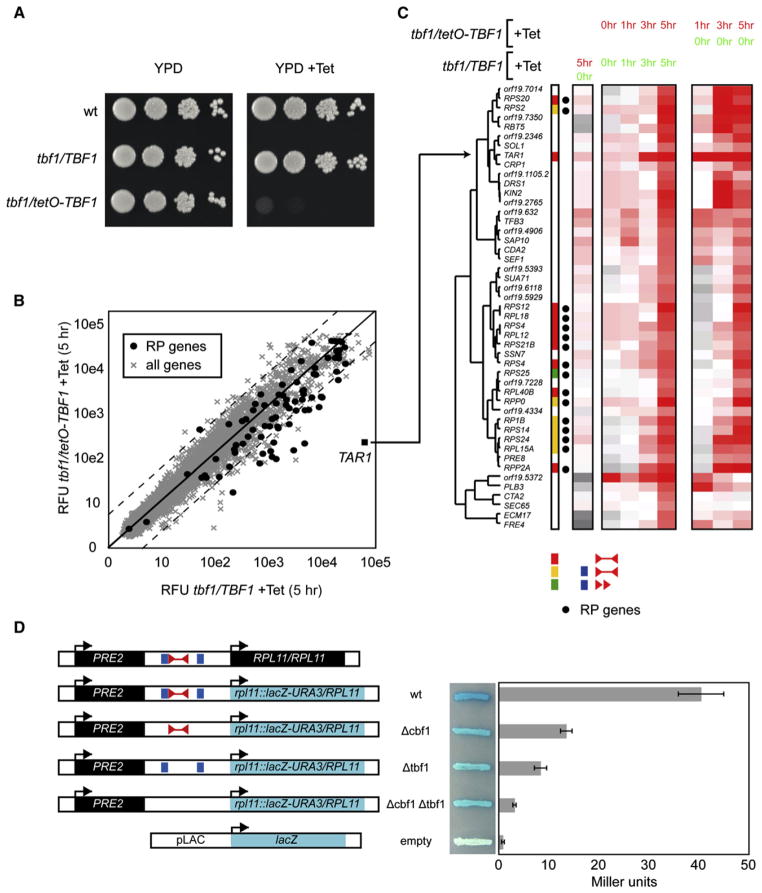
To confirm the role of Tbf1 in RP gene regulation, a conditional mutant was generated with a tetracycline-regulatable cassette (tetO-TBF1) in a heterozygous background (tbf1/TBF1) (Roemer et al., 2003). We observed a specific 400-fold downregulation of TBF1 in the tbf1/tetO-TBF1 strain after tetracycline addition (Figure S2A). Turning off Tbf1 expression with tetracycline causes a severe growth defect in the tbf1/tetO-TBF1 strain (Figure 3A). Microarray expression profiling of this strain shows that RP gene expression is specifically decreased upon addition of tetracycline to the medium (Figure 3B; Table S7). Forty-seven genes are downregulated 2-fold after a 5 hr Tbf1 shutoff (Figures 3B and 3C). Sixteen of these are RP genes that possess a Tbf1 element in their promoter region (p = 4.26 × 10−18) (Figures 3B and 3C). Time course experiments comparing the heterozygous tbf1/TBF1 and the tbf1/tetO-TBF1 strains show that RP gene downregulation is highly specific to the shutoff strain and is not caused by a nonspecific effect of tetracycline addition to the medium (Figure 3C). In addition, all but two RP genes cluster together based on their expression pattern (Figure 3C). Our expression profiling data were confirmed by reverse transcription and qPCR (RT-qPCR) analysis with four target RP genes (RPL18, RPL11, RPS20, and RPP2A); all four start to show downregulation after 2 hr of tetracycline treatment (Figures S2B–S2E).
Figure 3. Tbf1 Is an Essential Transcriptional Activator of RP Genes and of the TAR1 Gene.
(A) The tbf1/tetO-TBF1 conditional mutant is growth defective. Ten-fold dilutions of the indicated strains were spotted on YPD with or without 100 μg/ml of tetracycline.
(B) Expression profiling of the tbf1/tetO-TBF1 conditional mutant shows that it specifically downregulates RP genes and TAR1 after Tbf1 shutoff (relative fluorescence units, RFU). Dashed lines represent the 2-fold limits.
(C) Hierarchical clustering of the 47 genes downregulated more than 2-fold in the tbf1/tetO-TBF1 strain treated with tetracycline shows that RP genes cluster together and all contain a Tbf1 element. Experimental and reference samples are identified by the time of tetracycline treatment in red and green, respectively.
(D) LacZ reporter assays demonstrate that transcription at the RPL11 locus is driven by the Tbf1- and Cbf1-binding elements. Error bars represent one standard deviation from the mean of three biological replicates.
We also used a lacZ reporter assay to validate the role of the Tbf1- and Cbf1-binding elements in RP transcriptional regulation. We deleted the Tbf1- and the Cbf1-binding motifs in the intergenic region upstream of RPL11 and replaced the RPL11 ORF by lacZ. This reporter’s activity is reduced by at least 4- and 2.5-fold in the absence of the Tbf1 and Cbf1-binding sites respectively, while the deletion of both sites causes an even more dramatic decrease in expression (Figure 3D).
Tbf1 Regulates the rDNA Locus
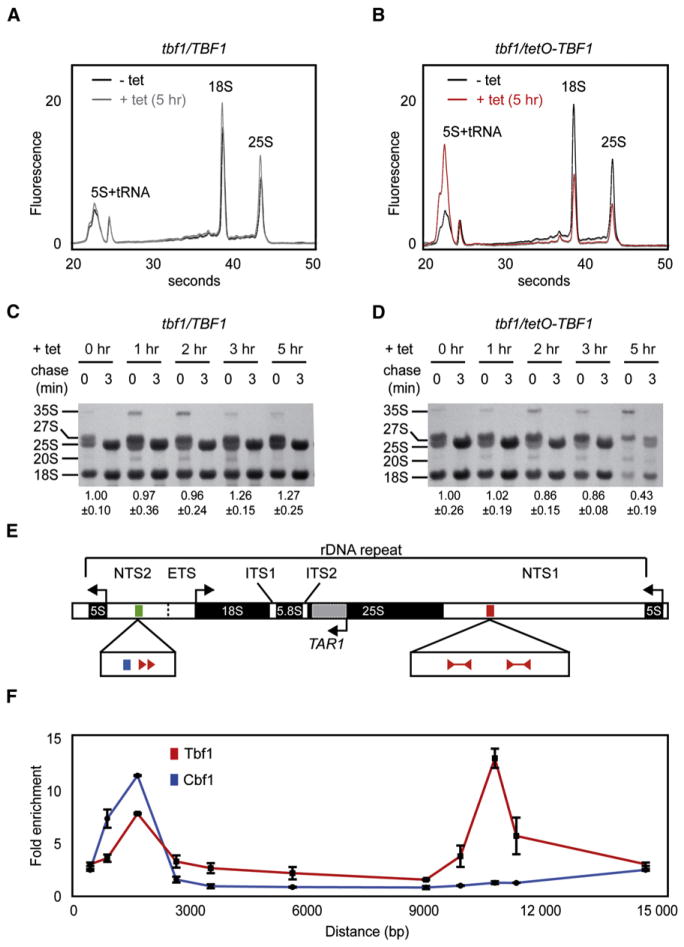
In our Tbf1 shutoff profiling data, the most rapidly downregulated gene was TAR1, a mitochondrial protein encoded on the anti-sense strand of the 25S rDNA (Coelho et al., 2002). We observed a 20-fold decrease in TAR1 expression after a 5 hr treatment with tetracycline, and this downregulation was visible even 1 hr after tetracycline addition (Figures 3B and 3C). Also, in the course of RNA quality control for microarray analysis, we had observed that 18S and 28S rRNA abundance relative to small RNAs was decreased in the tbf1/tetO-TBF1 strain treated with tetracycline (Figures 4A and 4B). We also performed time course [C3H3]-methionine pulse-chase labeling to evaluate the rate of synthesis and processing of rRNA after tetracycline addition. The amount of CH3 incorporated in the rRNA of the conditional TBF1 mutant during a 3 min pulse is greatly reduced after 3 and 5 hr of tetracycline treatment while the heterozygous tbf1/TBF1 strain is not affected (Figures 4C and 4D), implying that shutting off TBF1 causes a decrease in rRNA synthesis. In addition, the processing of the 27S and 20S rRNA into 25S and 18S subunits, respectively, appears slowed down when TBF1 expression is low. In fact, a 27S rRNA band is still visible after a 3 min pulse followed by a 3 min chase at 5 hr of tetracycline treatment, while it is absent earlier in the time course (Figures 4C and 4D). Therefore, rRNA is synthesized and processed slowly in the TBF1 conditional mutant. This is reminiscent of the phenotype of S. cerevisiae fhl1 and fhl1/ifh1 mutants (Rudra et al., 2005). We therefore hypothesized that Tbf1 shutoff has either a direct or an indirect effect on rRNA synthesis. To test the direct effect hypothesis, we looked for conserved Tbf1 and Cbf1 motifs in the rDNA locus of C. albicans and found two occurrences of the palindromic Tbf1-binding site in the terminator NTS1 region of the 35S rRNA in addition to a TTAGGG direct repeat flanked by a Cbf1 element in the NTS2 region (Figure 4E). We used ChIP and qPCR to map Tbf1 and Cbf1 occupancy at the rDNA locus of C. albicans. Both regulators are bound upstream of the 5S and 35S rRNA genes, while only Tbf1 is present in the NTS1 region (Figure 4F). The predicted DNA motifs are thus occupied, suggesting a direct involvement of Tbf1 and Cbf1 in rRNA regulation.
Figure 4. Tbf1 Controls Transcription of the rDNA Locus.
(A and B) Tbf1 shutoff by tetracycline in the tbf1/tetO-TBF1 strain causes a rapid decrease in 18S and 28S rRNA abundance, as observed on a total RNA electropherogram.
(C and D) Tbf1 shutoff by tetracycline slows down rRNA transcription and processing as measured by pulse-chase labeling with [C3H3]-methionine. Values underneath the autoradiograms are the mean intensities in arbitrary units/μg of RNA ± standard deviation of three pulse-chase experiments.
(E) Physical map of the C. albicans rDNA locus (Venema and Tollervey, 1999) highlighting TAR1 and rRNA genes as well as Tbf1 and Cbf1 cis-regulatory motifs.
(F) Tbf1 and Cbf1 are detected at the rDNA locus by ChIP followed by qPCR, and the enrichment signals overlap their respective DNA-binding motifs. Error bars reflect one standard deviation from the mean of three biological replicates.
Tbf1 and Cbf1 Were Substituted by Rap1 in Saccharomyces and Relatives
To better characterize the evolution of the Tbf1 and Cbf1 RP cis-elements, we established a phylogeny of 15 fungi whose genomes have been completely sequenced by using previously published sequence alignments (James et al., 2006). We then assessed enrichment of the different regulatory motifs in orthologous RP gene sets of each species. We validated our method with other gene groups and the known regulatory elements Mbp1 and RGE and recapitulated previous findings (Gasch et al., 2004; Ihmels et al., 2005) (Figure 5A).
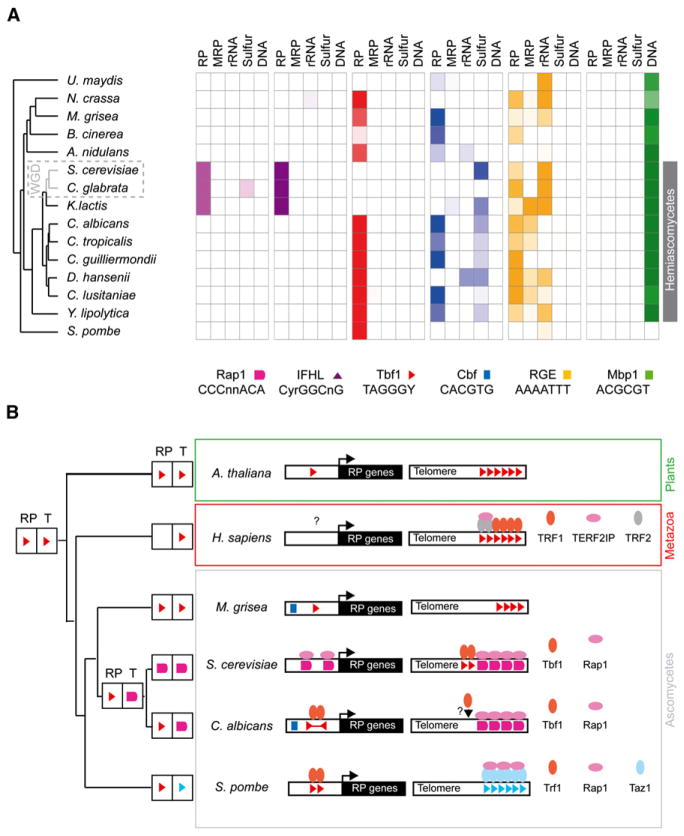
Figure 5. Tbf1 and Cbf1 Elements Are Enriched Upstream of RP Genes in Many Distantly Related Fungal Species and Are Absent in Saccharomyces and Relatives.
(A) Orthologous gene sets of cytosolic ribosomal protein genes (RP), mitochondrial ribosomal protein genes (MRP), nucleolar genes (rRNA), sulfur metabolism genes (sulfur), and DNA replication genes (DNA) from 15 completely sequenced fungal genomes were tested for enrichment of the Rap1, IFHL, Tbf1, Cbf1, RGE, and Mbp1 motifs. Color saturation follows enrichment p values (Table S8).
(B) Tbf1 and Rap1 or their orthologs exchanged function as DNA-binding proteins of the telomeres (T) and RP gene promoters during the evolution of eukaryotes. Blue triangles, Taz1-binding motif (GGTTACA).
Tbf1 elements are highly enriched in RP gene intergenic regions of most Ascomycetes except in S. cerevisiae and relatives (Figure 5A). Even S. pombe, the earliest diverging species of our Ascomycetes tree, has highly enriched Tbf1 direct repeats at RP genes, corresponding to the previously published HomolE element (Gross and Kaufer, 1998; Tanay et al., 2005). These Ascomycetes’ Tbf1 elements have adopted strikingly different symmetries (Figure 5B), probably arising from the flexibility of the Tbf1 dimer, given that its human ortholog, TRF1, binds various orientations of the TTAGGG motif (Bianchi et al., 1999).
The Cbf1 element (CACGTG) is enriched in the sulfur starvation response only in Hemiascomycetes, while it is present upstream of RP genes in most Ascomycetes alongside Tbf1 except in Botrytis cinerea, Debaryomyces hansenii, and S. pombe, where Cbf1 or Tbf1 motifs have been lost selectively (Figure 5A), suggesting that these elements can function independently. The Cbf1 and Tbf1 elements are absent upstream RP genes of S. cerevisiae and related species where Rap1 and IFHL sites are exclusively found (Figure 5A). This phylogenetic distribution of the Rap1 consensus was previously reported with a larger spectrum of fungi (Gasch et al., 2004; Tanay et al., 2005). Our results challenge the suggested model that the HomolE/Tbf1 element evolved gradually from the IFHL motif bound by the Fhl1/Ifh1 heterodimer (Tanay et al., 2005). Instead, our findings suggest that the ancestral Ascomycetes RP cisregulatory circuit contained Tbf1 and Cbf1, but not Rap1 or IFHL, motifs and that a transcription factor substitution occurred in the phylum Hemiascomycetes prior to K. lactis speciation and the whole-genome duplication (WGD) of S. cerevisiae (Scannell et al., 2006) (Figure 5A).
DISCUSSION
The ribosome is the protein production unit of the cell. Given the essentiality and the conservation of genes within this regulon, its cis-regulatory circuitry could be expected to be under constant selective pressure blocking genotypic/phenotypic exploration. It is therefore remarkable that Rap1, the key transcriptional activator of RP genes in S. cerevisiae, does not play this role in C. albicans. Instead we have shown that Tbf1 is the main regulator of RP genes in C. albicans.
Reorganization and Convergence of the RP Genes and rDNA cis-Regulatory Network
RP genes’ trans-acting factor specificity is achieved differently in S. cerevisiae and C. albicans. Specificity is obtained by protein complex formation in S. cerevisiae, where the essential cofactors Fhl1, Ifh1, and Hmo1 always associate with the pleiotropic regulator Rap1 at RP genes and provide it with transcriptional activation potential (Lieb et al., 2001; Rudra et al., 2005). Experimental evidence suggests that the assembly of this complex at RP gene promoters depends mostly on the Rap1-binding motif and possibly on a second, less obvious, DNA element (Rudra et al., 2005; Schawalder et al., 2004; Wade et al., 2004). In contrast, the unique presence of the Tbf1 cis-element at RP gene promoters provides the transcription factor specificity in C. albicans. The presence of Cbf1-binding motifs alongside only half of Tbf1 sites indicates that Cbf1 is not required for RP specialization of Tbf1. Instead, based on our data, Cbf1 probably facilitates access of Tbf1 to its targets, or, alternatively, Tbf1 and Cbf1 may bind synergistically to their adjacent elements.
Beyond this significant difference, the organization of the cis-regulatory network of RP genes and rDNA in both organisms is remarkably similar. First, the coverage of the RP regulon by Rap1 and Tbf1 is comparable, reaching more than 80% of RP genes (Lieb et al., 2001). Second, trans-factors of RP genes also act at rDNA in both species. In S. cerevisiae, Hmo1 binds rDNA repeats, and the disruption of either Hmo1 or Fhl1 produces a shortage of rRNA (Gadal et al., 2002; Rudra et al., 2005). Interestingly, C. albicans Tbf1 shutoff causes a similar effect on rRNA levels, suggesting that it could act as a RNA polymerase I and II transcriptional activator. Since they are bound by Tbf1 and Cbf1, the rDNA-regulatory regions were also reconnected during the transcription factor substitution of the RP regulon. Third, the key RP regulators of both species are essential. Tbf1, Cbf1, and Rap1 have orthologs in C. albicans and S. cerevisiae with conserved DNA-binding domains and motif specificity (Biswas et al., 2003a, 2003b) (Figure S3). Deletion of C. albicans Rap1 has no detrimental effect, although its removal or the inactivation of one of its cofactors is lethal in S. cerevisiae (Biswas et al., 2003a; Uemura et al., 2004). Conversely, in C. albicans, Cbf1 loss causes severe growth defects, while its deletion in S. cerevisiae is less significant (Biswas et al., 2003b). Interestingly, Tbf1 is essential in both species; in S. cerevisiae, given that it plays no role in RP regulation, Tbf1 essentiality is likely related to its subtelomeric function (Berthiau et al., 2006; Brigati et al., 1993). This role is supported by the localization of the Tbf1 direct repeat motif (TTAGGGn[0–6]TTAGGG), which is found 23 times in S. cerevisiae genome, 20 of which are located within 360 bp of the chromosome ends (Table S9; Figure 5B). Unfortunately, the lack of reliably assembled chromosome ends for the C. albicans genome precludes the detection of subtelomeric-associated motifs in this species.
Ultimately, both circuits converged to one crucial DNA-binding trans-factor, perhaps sacrificing robustness of the RP cis-regulatory network to the simplicity of regulation. However, during the transition period, the ancestor of Saccharomyces and related species most likely had both regulatory circuits, and thus redundancy could have allowed loss of the no-longer-essential Tbf1/Cbf1 cis-regulatory elements in this branch. The emergence of similar network features and the convergent co-option of the new transacting factor Rap1 by most RP genes were most likely driven by the constant selective pressure to maintain stoichiometric assembly of RPs and rRNAs. The wholesale substitution of Tbf1 by Rap1 as the principal regulator of RP gene expression occurred prior to the WGD of the yeast lineage, suggesting that dramatic transcriptional rewiring of essential functions may occur in the absence of extensive duplication of regulatory circuits.
Implications of Transcription Factor Substitution for the Study of Biological Systems
There are fundamental differences between the trans-factor substitution characterized here and the previously identified RP transcriptional rewiring (Ihmels et al., 2005). The latter involved a change in the coregulation of mitochondrial ribosomal protein (MRP) and RP regulons between S. cerevisiae and C. albicans. This divergence was detected from analysis of a compendium of expression profiles from both species and involved the selective loss of the RGE element in the MRP gene set of S. cerevisiae (Ihmels et al., 2005). The RGE element is an example of a pleiotropic cis-regulator that was used to change the transcriptional wiring between two related biological processes by the gain or loss of cis-regulatory elements. This type of evolutionary change involves a single trans-factor that is differentially recruited to gene sets (functions) in two organisms. In comparison, the substitution of Tbf1 by Rap1 implies the complete replacement of the key essential RP trans-acting factor and the mutation of the corresponding cis-regulatory elements within a single biological function.
Since the coregulation of RP genes is maintained after the substitution in both S. cerevisiae and C. albicans, this rewiring is mostly cryptic if only coordinate expression of the RP genes is considered. This change would therefore appear to have no effect on the organismal phenotype. However, since transcription is the endpoint of signal transduction pathways, the substitution of a trans-factor may allow the cell to respond differently to external or internal signals. In S. cerevisiae, the assembly of the essential factors Rap1, Ifh1, and Fhl1 is dictated by TOR, PKA, and Sch9 signaling (Jorgensen et al., 2004; Martin et al., 2004; Schawalder et al., 2004; Wade et al., 2004). Under conditions of rapid proliferation, Rap1 and Fhl1 recruit Ifh1 to RP gene promoters, where it activates transcription to maximal levels. In turn, Ifh1 is released from RP genes under conditions of starvation or stress (Martin et al., 2004; Schawalder et al., 2004; Wade et al., 2004). This raises the question of how the signals upstream of Tbf1 in C. albicans and Rap1 in S. cerevisiae have rewired such that RP gene transcription responds to the correct growth stimuli; what could have changed is the nature of the environmental cues that affect transcription of RP genes, and not their coregulation. Further analyses of the signals that regulate Tbf1 in C. albicans and Rap1 in S. cerevisiae would help us understand other aspects of this rewiring event and the significance of transcription factor substitution for yeast adaptability.
Rap1 and Tbf1 Are Myb Domain Proteins Involved at Telomeres in Many Eukaryotes
It seems unlikely that the substitution of the transcriptional circuit controlling a biological process as extensive and critical as the ribosome occurred through random mutations alone. Since the substitution to accommodate the new trans-acting factor involved the convergence of new cis-elements upstream of so many genes, a more realistic scenario may be that some form of interaction between Tbf1 and Rap1 facilitated the transition.
Repeats of the TTAGGG motif are present at telomeres in all branches of the eukaryotic phylogeny including those of humans, plants, and many fungal species such as U. maydis, N. crassa, M. grisea, and A. nidulans (Li et al., 2000). C. albicans and S. cerevisiae Tbf1 proteins are orthologous to human TRF1, a Myb domain protein that binds directly to the human telomeric repeats (Broccoli et al., 1997). However, in Hemiascomycetes, telomeric repeats are composed of the Rap1-binding motif, and in S. cerevisiae, Tbf1 only binds TTAGGG in subtelomeric regions, where it inhibits silencing by the telomeric Rap-Sir complex (Berthiau et al., 2006; Cohn et al., 1998; Conrad et al., 1990; Fourel et al., 2001; Lieb et al., 2001; McEachern and Blackburn, 1994) (Figure 5B). In humans and S. pombe, Rap1 does not bind DNA directly but is tethered to telomeres by protein-protein interactions with TRF2 and Taz1, respectively (Li et al., 2000; Park et al., 2002) (Figure 5B). Thus, Tbf1 and Rap1 are universally found at eukaryotic telomeres, but their role as the telomeric repeat-binding factors was exchanged in Hemiascomycetes. Subsequently, before speciation of K. lactis, a second substitution of Tbf1 by Rap1 occurred at the RP regulon in Saccharomyces and relatives (Figures 5A and 5B).
The fact that Tbf1 and Rap1 orthologs physically interact at the eukaryotic telomeres and that both are Myb domain proteins with similar GC-rich DNA specificities may have facilitated these evolutionary transitions. It is also possible that, once recruited at telomeric repeats in Hemiascomycetes, Rap1 acquired molecular properties, rendering it competent for RP gene transcriptional regulation. Considering that both RP gene expression and telomere maintenance require the ability to alter chromatin in order to ensure a high yield of RP mRNAs or telomere stability (Fourel et al., 2002; Moretti et al., 1994; Moretti and Shore, 2001; Morse, 2000; Reid et al., 2000; Rohde and Cardenas, 2003), it is possible that there is selective pressure to connect these functions to the same highly competent DNA-binding protein. This view is supported by the fact that, in plants (Oriza sativa and Arabidopsis thaliana) and in many Ascomycetes, the telomeric repeats as well as a major RP cis-regulatory element are composed of TTAGGG (Li et al., 2005; Richards and Ausubel, 1988; Tremousaygue et al., 1999; Yu et al., 2000). The O. sativa telomeric repeat-binding protein is a Tbf1/TRF1 ortholog (Yu et al., 2000), and thus it would be of interest to establish if telomere-binding proteins of the TRF1 family are also involved in RP gene and/or rDNA transcriptional regulation in plants. This would support a model in which RP gene regulation and telomeric maintenance were Tbf1 dependent in ancestral eukaryotes. Thus, in addition to illustrating the ability of the cell to rewire essential regulatory functions to different conserved trans-acting factors, our work establishes a Tbf1-mediated control of RP transcriptional regulation that applies to most Ascomycetes and possibly plants and proves that the well-established Rap1 circuit is limited to S. cerevisiae and its close relatives.
EXPERIMENTAL PROCEDURES
Strains, Media, and Plasmids
ChIP-CHIP experiments were conducted in the BWP17 strain background, and the tetracycline titrable allele of TBF1 was generated in the CAI4 background. Cell growth, transformation, and DNA preparation were carried out using standard procedures. Cells were grown at 30°C. Synthetic medium was SC -Ura-Cys-Met (0.67% yeast nitrogen base, 2% glucose, amino acid dropout), and rich medium was YPD (1% yeast extract, 2%peptone, 2% dextrose). When stated, tetracycline was added to a concentration of 100 μg/ml for the indicated time (Roemer et al., 2003).
A 3XHA epitope was added by PCR to the C terminus of the CBF1 and TBF1 ORFs. PCR products were cloned in the vector pCaEXP (Care et al., 1999). CBF1-HA was inserted between BamHI and BspEI sites, and TBF1-HA was inserted into a BspEI site. The resulting expression vectors pCBF1-HA and pTBF1-HA were integrated at the RPS1 locus of Candida albicans BWP17 (Care et al., 1999). TBF1 and CBF1 were also TAP tagged in vivo with a TAP-URA3 PCR product containing 100 bp homology upstream and downstream of each ORF (H.L., unpublished data). Correct integration of the TAP tag was verified by PCR and sequencing.
To generate rpl11::lacZ-URA3 reporters, the WT or mutated RPL11 promoter was cloned upstream of a lacZ-URA3 cassette (H.L., unpublished data). The resulting constructs were then PCR amplified with oligos that have 100 bp of homology upstream and downstream of the RPL11 ORF and transformed in C. albicans strain BWP17. Transformants were selected on −ura plates, and correct integration was checked by PCR and sequencing.
Whole-Genome Location Profiling by ChIP-CHIP
ChIP experiments were performed as described previously with some modifications (Guillemette et al., 2005). Briefly, cells were grown to an optical density at 600 nm of 0.6 in 50 ml of SD-Cys-Met. We followed the ChIP protocol available at http://www.ircm.qc.ca/microsites/francoisrobert/en/317.html with the following exceptions: chromatin was sonicated to an average 300 bp, and 700 μl of whole-cell extracts (WCE) were incubated with monoclonal 12CA5 anti-HA supernatant immobilized on protein-L-agarose beads (Sigma). Immunoprecipitated DNA was used for either whole-genome location profiling or gene-specific real-time qPCR analysis. For whole-genome location profiling, tagged ChIPs were labeled with Cy5 dye, and untagged (mock) ChIPs were labeled with Cy3 dye.
Microarrays containing 11,817 70-mer oligonucleotide probes were cohybridized with tagged immunoprecipitated (Cy5-labeled) and mock immunoprecipitated (untagged strain; Cy3-labeled) DNA samples. Our C. albicans whole-genome microarrays contained single spots of 5423 intergenic 70-mer oligonucleotide probes combined with 6394 intragenic 70-mer oligonucleotide probes already in use in our C. albicans ORF microarray. Microarray hybridization, washing, scanning, and normalization were performed as described (Martchenko et al., 2007). The significance cutoff was determined using the distribution of log ratios for each factor. It was set at two standard deviations from the mean of log-transformed fold enrichments (1.4- and 2.0- fold for Tbf1 and Cbf1 data sets, respectively). Values shown are an average of two biological replicates derived from independently isolated transformants of tagged and mock constructs.
For qPCR validations, in vivo TAP-tagged Tbf1 and Cbf1 were ChIPed as described above except that IgG Sepharose (GE Healthcare) was used.
Quantitative PCR
Quantitative real-time PCR was performed using the Corbett Rotor-Gene RG-3000A (Corbett Research, Sydney, Australia) with SYBR Green fluorescence (QIAGEN). Real-time PCR was performed using 1 ng Tbf1-TAP or Cbf1-TAP ChIPed DNA or total genomic DNA extracted from WCE. Cycling was for 15 min at 95°C, followed by 45 cycles of 95°C, 10 s;, 56°C, 15 s; and 72°C, 15 s. All samples were tested in triplicate, and means were used for further calculations. Fold enrichments of tested promoter sequences were estimated by using the coding sequence of the C. albicans ACT1 ORF as a reference. Oligonucleotide sequences are given in Table S10.
Gene Expression Profiling, RT-qPCR, and β-Galactosidase Assays
Cells were grown without tetracycline to an OD600 of 0.2 in YPD. Tetracycline was then added to the appropriate samples at 100 μg/ml for the indicated times. Total RNA was extracted by the hot phenol extraction method (Carlson and Botstein, 1982) with the difference that glass beads (Sigma) were added to samples. Total RNA was subsequently cleaned up on RNeasy columns (QIAGEN). For labeling, 20 μg of total RNA was reverse transcribed using oligo(dT)21 in the presence of Cy3 or Cy5-dCTP (Perkin-Elmer-Cetus/NEN) and Superscript II reverse transcriptase (Invitrogen). Thereafter, template RNA was degraded by adding 2.5 units RNase H (USB) and 1 μg RNase A (Pharmacia) followed by incubation for 15 min at 37°C. The labeled cDNAs were purified with QIAquick PCR Purification Kit (QIAGEN). Microarrays hybridization, washing, scanning, and normalization were performed as described (Martchenko et al., 2007). Hierarchical clustering and heat map display of the expression profiling data (Figure 3C) were done with the Cluster and Treeview programs (http://rana.lbl.gov/EisenSoftware.htm).
For RT-qPCR, RNA samples were converted to cDNAs by reverse transcription (RT) with SuperScript III (Invitrogen) with oligodT and random octamers following the manufacturer’s protocol. qPCR was performed as above, and all target genes validated were normalized to the C. albicans ACT1 ORF. Oligonucleotides used for RT-qPCR are listed in Table S10. Expression ratios were calculated by using the ΔΔCt method.
Solid and liquid β-galactosidase assays were performed as described (Martchenko et al., 2007).
RNA Species Distributions and [C3H3]-Methionine Labeling
Electropherograms of RNA species distributions were obtained by capillary electrophoresis with fluorescence detection on an Agilent Bioanalyser 2100 (Agilent Technologies). Bioanalyzer RNA 6000 Nano Chips (Agilent Technologies) were loaded with 250 ng of total RNA before and after treatment with tetracycline by following the manufacturer’s protocol.
[C3H3]-methionine pulse-chase labeling following tetracycline treatment was performed as described (Rudra et al., 2005), except that 3 min pulse and chase were used.
Informatics Procedures
The upstream DNA sequence of every gene was extracted up to −1500 bp or to the next ORF. To uncover enriched putative RP cis-regulatory elements, every minimotif composed of two nucleotide triplets separated by less than 16 bp (XXXn[0–15]XXX) was tested for its overrepresentation upstream of RP genes compared to its occurrence in the upstream region of all genes in the genome considered. Genes possessing the minimotif were defined as those with at least one instance of the motif or its reverse complement in their upstream region. The enrichments were calculated using a hypergeometric distribution. The significance threshold after Bonferroni correction was set at p < 10−2 (Figure 1A, Tables S1 and S2).
The maximum likelihood Ascomycetes phylogeny was produced using the PHYLIP package (Felsenstein, 2004). The concatenated DNA multiple sequence alignment used is already published (James et al., 2006). The columns containing gaps were dropped, producing an alignment of 2119 bp.
The various gene sets used in Figure 5A were defined using Gene Ontology (GO) annotations of C. albicans (obtained from the Candida Genome Database, www.candidagenome.org). These sets were used as queries to produce orthologous gene lists of high-scoring (e < 1e-40) BLAST hits in every species (Altschul et al., 1997). GO terms used are GO:0005830 (RP), GO:0005761 (MRP), GO:0005730 (rRNA), GO:0006790 (sulfur), and GO:0000067 (DNA). The motifs in Figure 5A were scored for enrichment in the different gene sets using the above-mentioned procedure for motif detection. Considering the plasticity of the TTAGGG symmetries, four motif variants of Tbf1 were tested, and the highest score was reported (Table S8). The color saturation in the heat map display of Figure 5A is linear and follows the log of the p value starting at p = 10−2 and reaching full saturation at p = 10−10.
Supplementary Material
Acknowledgments
We are grateful to Sadri Znaidi for help with optimizing the ChIP-CHIP protocol and to Jean-Sébastien Deneault and the Biotechnology Research Institute (BRI) microarray facility members for technical help. We want to thank the Broad Institute (http://www.broad.mit.edu/annotation/fgi), Genolevures (http://cbi.labri.fr/Genolevures/), and the Sanger Center (http://www.sanger.ac.uk/Projects/Fungi) for rendering their sequence data available. We are grateful to the Assembling the Fungal Tree of Life project (http://aftol.org/) for making fungal alignments available. This work was supported by grants from the Canadian Institute for Health Research (CIHR). H.L. was supported by a CIHR fellowship. H.L. also acknowledges partial support from a Conseil National de Recherches Canada (CNRC) fellowship. A.S. was supported by a National Institutes of Health (NIH) postdoctoral fellowship. This is National Research Council (NRC) publication 49549. H.H. was responsible for all bioinformatics procedures. H.L. conducted all experiments, with the help of A.S. and M.M. on mutant microarray analysis. T.R. provided the Tbf1 conditional mutant. E.P., A.N., and M.W. oversaw the work. The authors declare no competing financial interest.
Footnotes
ACCESSION NUMBERS
Microarray data have been deposited into the National Center for Biological Information (NCBI)’s Gene Expression Omnibus (GEO) under the following ID numbers: platforms, GPL6474 and GPL6475; experiments, GSM264698, GSM264703, GSM264705, GSM264707, GSM265362, GSM265364, GSM265365, GSM265367, GSM265368, GSM265370, GSM265373, GSM265375, GSM265376, GSM265377, GSM265378, and GSM265379; and series, GSE10622, GSE10458, and GSE10499.
Supplemental Data include three figures and ten tables and can be found with this article online at http://www.molecule.org/cgi/content/full/29/5/552/DC1/.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthiau AS, Yankulov K, Bah A, Revardel E, Luciano P, Wellinger RJ, Geli V, Gilson E. Subtelomeric proteins negatively regulate telomere elongation in budding yeast. EMBO J. 2006;25:846–856. doi: 10.1038/sj.emboj.7600975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A, Stansel RM, Fairall L, Griffith JD, Rhodes D, de Lange T. TRF1 binds a bipartite telomeric site with extreme spatial flexibility. EMBO J. 1999;18:5735–5744. doi: 10.1093/emboj/18.20.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas K, Rieger KJ, Morschhauser J. Functional analysis of CaRAP1, encoding the Repressor/activator protein 1 of Candida albicans. Gene. 2003a;307:151–158. doi: 10.1016/s0378-1119(03)00456-6. [DOI] [PubMed] [Google Scholar]
- Biswas K, Rieger KJ, Morschhauser J. Functional characterization of CaCBF1, the Candida albicans homolog of centromere binding factor 1. Gene. 2003b;323:43–55. doi: 10.1016/j.gene.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Braun BR, van Het Hoog M, d’Enfert C, Martchenko M, Dungan J, Kuo A, Inglis DO, Uhl MA, Hogues H, Berriman M, et al. A human-curated annotation of the Candida albicans genome. PLoS Genet. 2005;1:36–57. doi: 10.1371/journal.pgen.0010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigati C, Kurtz S, Balderes D, Vidali G, Shore D. An essential yeast gene encoding a TTAGGG repeat-binding protein. Mol Cell Biol. 1993;13:1306–1314. doi: 10.1128/mcb.13.2.1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broccoli D, Smogorzewska A, Chong L, de Lange T. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat Genet. 1997;17:231–235. doi: 10.1038/ng1097-231. [DOI] [PubMed] [Google Scholar]
- Brown WR. Molecular cloning of human telomeres in yeast. Nature. 1989;338:774–776. doi: 10.1038/338774a0. [DOI] [PubMed] [Google Scholar]
- Care RS, Trevethick J, Binley KM, Sudbery PE. The MET3 promoter: a new tool for Candida albicans molecular genetics. Mol Microbiol. 1999;34:792–798. doi: 10.1046/j.1365-2958.1999.01641.x. [DOI] [PubMed] [Google Scholar]
- Carlson M, Botstein D. Two differentially regulated mRNAs with different 5′ ends encode secreted with intracellular forms of yeast invertase. Cell. 1982;28:145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Coelho PS, Bryan AC, Kumar A, Shadel GS, Snyder M. A novel mitochondrial protein, Tar1p, is encoded on the antisense strand of the nuclear 25S rDNA. Genes Dev. 2002;16:2755–2760. doi: 10.1101/gad.1035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn M, McEachern MJ, Blackburn EH. Telomeric sequence diversity within the genus Saccharomyces. Curr Genet. 1998;33:83–91. doi: 10.1007/s002940050312. [DOI] [PubMed] [Google Scholar]
- Conrad MN, Wright JH, Wolf AJ, Zakian VA. RAP1 protein interacts with yeast telomeres in vivo: overproduction alters telomere structure and decreases chromosome stability. Cell. 1990;63:739–750. doi: 10.1016/0092-8674(90)90140-a. [DOI] [PubMed] [Google Scholar]
- Deutschbauer AM, Jaramillo DF, Proctor M, Kumm J, Hillenmeyer ME, Davis RW, Nislow C, Giaever G. Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics. 2005;169:1915–1925. doi: 10.1534/genetics.104.036871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package), version 3.6. Seattle: Department of Genome Sciences, University of Washington; 2004. [Google Scholar]
- Fourel G, Boscheron C, Revardel E, Lebrun E, Hu YF, Simmen KC, Muller K, Li R, Mermod N, Gilson E. An activation-independent role of transcription factors in insulator function. EMBO Rep. 2001;2:124–132. doi: 10.1093/embo-reports/kve024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourel G, Miyake T, Defossez PA, Li R, Gilson E. General regulatory factors (GRFs) as genome partitioners. J Biol Chem. 2002;277:41736–41743. doi: 10.1074/jbc.M202578200. [DOI] [PubMed] [Google Scholar]
- Gadal O, Labarre S, Boschiero C, Thuriaux P. Hmo1, an HMGbox protein, belongs to the yeast ribosomal DNA transcription system. EMBO J. 2002;21:5498–5507. doi: 10.1093/emboj/cdf539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Moses AM, Chiang DY, Fraser HB, Berardini M, Eisen MB. Conservation and evolution of cis-regulatory systems in ascomycete fungi. PLoS Biol. 2004;2:e398. doi: 10.1371/journal.pbio.0020398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, et al. Life with 6000 genes. Science. 1996;274:563–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- Gompel N, Prud’homme B, Wittkopp PJ, Kassner VA, Carroll SB. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature. 2005;433:481–487. doi: 10.1038/nature03235. [DOI] [PubMed] [Google Scholar]
- Gross T, Kaufer NF. Cytoplasmic ribosomal protein genes of the fission yeast Schizosaccharomyces pombe display a unique promoter type: a suggestion for nomenclature of cytoplasmic ribosomal proteins in databases. Nucleic Acids Res. 1998;26:3319–3322. doi: 10.1093/nar/26.14.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemette B, Bataille AR, Gevry N, Adam M, Blanchette M, Robert F, Gaudreau L. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 2005;3:e384. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DB, Wade JT, Struhl K. An HMG protein, Hmo1, associates with promoters of many ribosomal protein genes and throughout the rRNA gene locus in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:3672–3679. doi: 10.1128/MCB.26.9.3672-3679.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Li X. Transcriptional regulation in eukaryotic ribosomal protein genes. Genomics. 2007;90:421–423. doi: 10.1016/j.ygeno.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Ihmels J, Bergmann S, Gerami-Nejad M, Yanai I, McClellan M, Berman J, Barkai N. Rewiring of the yeast transcriptional network through the evolution of motif usage. Science. 2005;309:938–940. doi: 10.1126/science.1113833. [DOI] [PubMed] [Google Scholar]
- James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ, Celio G, Gueidan C, Fraker E, Miadlikowska J, et al. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature. 2006;443:818–822. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, Tyers M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004;18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Oestreich S, de Lange T. Identification of human Rap1: implications for telomere evolution. Cell. 2000;101:471–483. doi: 10.1016/s0092-8674(00)80858-2. [DOI] [PubMed] [Google Scholar]
- Li X, Zhong S, Wong WH. Reliable prediction of transcription factor binding sites by phylogenetic verification. Proc Natl Acad Sci USA. 2005;102:16945–16950. doi: 10.1073/pnas.0504201102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb JD, Liu X, Botstein D, Brown PO. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat Genet. 2001;28:327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- Maerkl SJ, Quake SR. A systems approach to measuring the binding energy landscapes of transcription factors. Science. 2007;315:233–237. doi: 10.1126/science.1131007. [DOI] [PubMed] [Google Scholar]
- Marion RM, Regev A, Segal E, Barash Y, Koller D, Friedman N, O’Shea EK. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc Natl Acad Sci USA. 2004;101:14315–14322. doi: 10.1073/pnas.0405353101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martchenko M, Levitin A, Hogues H, Nantel A, Whiteway M. Transcriptional rewiring of fungal galactose-metabolism circuitry. Curr Biol. 2007;17:1007–1013. doi: 10.1016/j.cub.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DE, Soulard A, Hall MN. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell. 2004;119:969–979. doi: 10.1016/j.cell.2004.11.047. [DOI] [PubMed] [Google Scholar]
- McEachern MJ, Blackburn EH. A conserved sequence motif within the exceptionally diverse telomeric sequences of budding yeasts. Proc Natl Acad Sci USA. 1994;91:3453–3457. doi: 10.1073/pnas.91.8.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti P, Shore D. Multiple interactions in Sir protein recruitment by Rap1p at silencers and telomeres in yeast. Mol Cell Biol. 2001;21:8082–8094. doi: 10.1128/MCB.21.23.8082-8094.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti P, Freeman K, Coodly L, Shore D. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- Morse RH. RAP, RAP, open up! New wrinkles for RAP1 in yeast. Trends Genet. 2000;16:51–53. doi: 10.1016/s0168-9525(99)01936-8. [DOI] [PubMed] [Google Scholar]
- Park MJ, Jang YK, Choi ES, Kim HS, Park SD. Fission yeast Rap1 homolog is a telomere-specific silencing factor and interacts with Taz1p. Mol Cells. 2002;13:327–333. [PubMed] [Google Scholar]
- Reid JL, Iyer VR, Brown PO, Struhl K. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol Cell. 2000;6:1297–1307. doi: 10.1016/s1097-2765(00)00128-3. [DOI] [PubMed] [Google Scholar]
- Richards EJ, Ausubel FM. Isolation of a higher eukaryotic telomere from Arabidopsis thaliana. Cell. 1988;53:127–136. doi: 10.1016/0092-8674(88)90494-1. [DOI] [PubMed] [Google Scholar]
- Roemer T, Jiang B, Davison J, Ketela T, Veillette K, Breton A, Tandia F, Linteau A, Sillaots S, Marta C, et al. Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol Microbiol. 2003;50:167–181. doi: 10.1046/j.1365-2958.2003.03697.x. [DOI] [PubMed] [Google Scholar]
- Rohde JR, Cardenas ME. The tor pathway regulates gene expression by linking nutrient sensing to histone acetylation. Mol Cell Biol. 2003;23:629–635. doi: 10.1128/MCB.23.2.629-635.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudra D, Warner JR. What better measure than ribosome synthesis? Genes Dev. 2004;18:2431–2436. doi: 10.1101/gad.1256704. [DOI] [PubMed] [Google Scholar]
- Rudra D, Zhao Y, Warner JR. Central role of Ifh1p-Fhl1p interaction in the synthesis of yeast ribosomal proteins. EMBO J. 2005;24:533–542. doi: 10.1038/sj.emboj.7600553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell DR, Wolfe K. Rewiring the transcriptional regulatory circuits of cells. Genome Biol. 2004;5:206. doi: 10.1186/gb-2004-5-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell DR, Byrne KP, Gordon JL, Wong S, Wolfe KH. Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature. 2006;440:341–345. doi: 10.1038/nature04562. [DOI] [PubMed] [Google Scholar]
- Schawalder SB, Kabani M, Howald I, Choudhury U, Werner M, Shore D. Growth-regulated recruitment of the essential yeast ribosomal protein gene activator Ifh1. Nature. 2004;432:1058–1061. doi: 10.1038/nature03200. [DOI] [PubMed] [Google Scholar]
- Tanay A, Regev A, Shamir R. Conservation and evolvability in regulatory networks: the evolution of ribosomal regulation in yeast. Proc Natl Acad Sci USA. 2005;102:7203–7208. doi: 10.1073/pnas.0502521102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremousaygue D, Manevski A, Bardet C, Lescure N, Lescure B. Plant interstitial telomere motifs participate in the control of gene expression in root meristems. Plant J. 1999;20:553–561. doi: 10.1046/j.1365-313x.1999.00627.x. [DOI] [PubMed] [Google Scholar]
- Tsong AE, Tuch BB, Li H, Johnson AD. Evolution of alternative transcriptional circuits with identical logic. Nature. 2006;443:415–420. doi: 10.1038/nature05099. [DOI] [PubMed] [Google Scholar]
- Uemura H, Watanabe-Yoshida M, Ishii N, Shinzato T, Haw R, Aoki Y. Isolation and characterization of Candida albicans homologue of RAP1, a repressor and activator protein gene in Saccharomyces cerevisiae. Yeast. 2004;21:1–10. doi: 10.1002/yea.1048. [DOI] [PubMed] [Google Scholar]
- Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu Rev Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- Wade JT, Hall DB, Struhl K. The transcription factor Ifh1 is a key regulator of yeast ribosomal protein genes. Nature. 2004;432:1054–1058. doi: 10.1038/nature03175. [DOI] [PubMed] [Google Scholar]
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- Yarragudi A, Parfrey LW, Morse RH. Genome-wide analysis of transcriptional dependence and probable target sites for Abf1 and Rap1 in Saccharomyces cerevisiae. Nucleic Acids Res. 2007;35:193–202. doi: 10.1093/nar/gkl1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu EY, Kim SE, Kim JH, Ko JH, Cho MH, Chung IK. Sequence-specific DNA recognition by the Myb-like domain of plant telomeric protein RTBP1. J Biol Chem. 2000;275:24208–24214. doi: 10.1074/jbc.M003250200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.