Abstract
Influenza virus is a pathogen that causes morbidity and mortality worldwide. Whereas vaccination is important for prevention of disease, given its limitations, antiviral therapy is at the forefront of treatment and also plays a role in prevention. Currently, two classes of antiviral medications, the adamantanes and the neuraminidase inhibitors, are approved for treatment. Given the resistance patterns of circulating influenza, adamantanes are not recommended. Within the US, two neuraminidase inhibitors are currently approved for both treatment and prevention, while worldwide there are four available. In this review, we will briefly discuss the epidemiology and pathology of influenza and then discuss neuraminidase inhibitors: their mechanism of action, resistance, development, and future applications.
Keywords: influenza, antiviral, neuraminidase, resistance
Introduction
Influenza viruses are ribonucleic acid (RNA) viruses in the family Orthomyxoviridae. There are three subtypes, influenza A, B, and C, with the majority of human infections caused by subtypes A and B. These viruses are responsible for seasonal influenza, causing yearly epidemics in addition to more sporadic, but potentially devastating, pandemics. From the 1970s through 2007, the annual deaths attributable to influenza have ranged from 3,000 to 50,000 in the US.1 This is likely an underestimate, as many persons who die from influenza may die secondary to complications such as bacterial superinfections or cardiovascular events, in addition to a direct result of the virus itself. The majority of those with more severe illness are the elderly, the very young, those with comorbid conditions, and the immunocompromised.2 Influenza causes fevers, chills, myalgias, and upper respiratory symptoms. When severe, it can cause lower respiratory tract infections and can lead to acute respiratory distress syndrome (ARDS). As a result of the high attack rate, extensive resources and health care expenditures are dedicated yearly to the detection, prevention, and treatment of influenza.
Influenza genes and life cycle
Influenza A is a negative sense RNA virus with a low fidelity RNA polymerase. It contains eight gene segments that encode eleven proteins, including hemagglutinin (HA) and neuraminidase (NA) glycoproteins. The influenza A virus uses HA to attach to sialic acid residues on host cells, which then initiates the infection. Once cells are infected, there are direct necrotic effects on the respiratory cell as the virus begins to use the host cell machinery for replication and turns off host cell RNA transcription and translation.3 NA is responsible for virion release and propagation of infection through cleaving sialic acid residues on host cells. Currently, there are 17 known types of HA, with the majority of human disease caused by H1 and H3 and with occasional cases of infection caused by H5, H7, or H9 viruses. There are ten kinds of NA, with N1, N2, and N9 currently circulating in humans. Influenza B is derived from two lineages – the Victoria and Yamagata strains – named for the place of discovery. Influenza B is not responsible for pandemics because it does not have an animal reservoir.
Each year, influenza develops mutations within these genes leading to antigenic drift. When new HA or NA subtypes are introduced via mixing, or within other species (avian or swine), a new subtype is created–this is called antigenic shift and is responsible for pandemics.4 As influenza evolves within populations, this may also lead to antigenic shift. For example, in the current H7N9 outbreak, evolution within the human host could lead to easier transmission between individuals.5 In the twenty-first century, there has been one pandemic: the 2009 swine H1N1 strain.
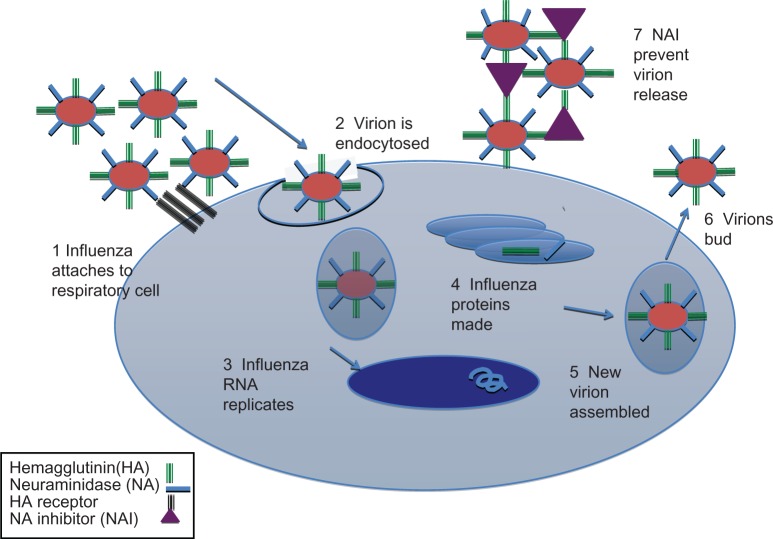
In the replication cycle (Figure 1), influenza first attaches to the host cell by HA, after which the virus is taken into the cell by an endosome. Then the viral RNA replicates and undergoes translation, host genes are suppressed (especially those responsible for immunity), and new virions bud from the surface of the host cell and are released to infect other cells in the host.
Figure 1.
Life cycle of an influenza virus.
Abbreviations: NAI, neuraminidase inhibitor; RNA, ribonucleic acid.
Influenza prevention
Current prevention strategies include both vaccination and prophylaxis with antivirals. In the event of novel influenza viruses (such as the current circulating H7N9) or the potential for a new pandemic strain, development of an effective vaccine may be too slow to be effective against spread of infection. Additionally, for some patients who are immunocompromised, vaccination, while beneficial, may not be entirely protective.6 This is also true for the elderly and infants, who do not mount as aggressive an immune response to vaccination and who are at increased risk for more severe disease. Therefore, other preventive measures and treatment must be effective, widely available, and well tolerated.
Vaccination currently targets the HA glycoprotein. Vaccines are designed to match major HA epitopes within the predicted circulating strains of influenza. Given that these epitope sequences may change yearly, vaccination is only effective for the current circulating strains. In years where the predicted strains do not match the actual circulating strains, or during a pandemic where the HA is novel, vaccination may not be as effective or readily available. Other targets for vaccination are currently being examined, including some sites within the HA that are highly conserved and would therefore lead to the creation of antibodies that can neutralize HA from a wide variety of subtypes.7 The matrix (M) protein, which affects virion packaging and release, is highly conserved within influenza subtypes and is therefore an attractive target for future influenza vaccine development. Because it would introduce only T cell immunity, it would likely not replace current antibody-inducing vaccines.8
Epidemiology
During the 2012–2013 influenza season in the US, influenza B (>60% Yamagata, >30% Victoria lineages) and influenza A (H3N2 and 2009 H1N1) strains were the major causes of infection.9 According to the US Centers for Disease Control and Prevention (CDC), over 12,000 patients were hospitalized with laboratory-confirmed influenza during 2012–2013.10 That represents approximately only 9% of all cases/hospitalizations within the US, as these numbers are derived from only 15 states. Thus, the estimate for the 2012–2013 season of hospitalized patients is closer to 137,000.10 Of the samples tested by CDC for the entire season, only two H1N1 and two H3N2 samples demonstrated resistance to the neuraminidase inhibitors (NAIs) oseltamivir and zanamivir.10 In addition, from January to September of 2012, there were approximately 307 human influenza cases with a swine variant H3N2 (H3N2v) in the US; this strain did not demonstrate sustained transmission among humans, and the majority of affected persons had prolonged exposure to pigs.10
Within the US Department of Veterans Affairs (VA) population, 27,972 respiratory samples were tested during the 2012–2013 influenza season; of these, 5,110 (18%) were positive: 76% for influenza A and 23% for influenza B. Of these, 261strains (H3N2, H1N1, and B) underwent NA gene sequencing, and none were found to have NAI resistance mutations (Mark Holodniy, personal observation 2013).
In addition to the current seasonal strains, there have been sporadic human cases of avian H5N1 influenza, and most recently, an H7N9 outbreak in the People’s Republic of China. H5N1 is a highly pathogenic avian influenza that causes high mortality in birds11 and has a case fatality rate of approximately 60% in humans.12 Fortunately, H5N1 has not yet developed the ability for efficient person-to-person transmission. Human infection with H7N9 was first recognized in Eastern China13 and has recently caused significant morbidity. As of June 1, 2013, 131 patients were documented to have H7N9 infection, with the majority of patients developing severe pneumonia.13–15 The Chinese Center for Disease Control and Prevention has instituted the Chinese National Influenza-Like Illness Surveillance Network, which as of April 28, 2013 had collected 20,739 samples and tested them for H7N9.16 Among those, six were positive for H7N9.16 The majority of patients were male, and the time from onset of symptoms to presentation was around 13 days.17 At the time of this writing, there have been 32 deaths attributed to H7N9.13,18 No cases of H7N9 human-to-human transmission have been documented, although there have been over a thousand close contacts tested.17 Some patients reported exposure to poultry markets, and in Shanghai some birds tested positive for H7N9.19
Current antiviral medications
There are two classes of drugs used to treat influenza: adamantanes and NAIs. The adamantanes are only effective against influenza A viruses, as they inhibit the M2 protein, which is not coded by influenza B. Adamantanes prevent hydrogen ion influx and prevent the virus from uncoating.20 There are two drugs that are licensed (amantadine and rimantadine), but given the high degree of resistance among influenza A strains to these drugs, they are not approved for use and are ineffective against influenza B (which lacks the M2 protein).4,21–24 In the US, there are currently two licensed NAIs – oseltamivir (Tamiflu™; Genentech Inc, South San Francisco, CA, USA) and zanamivir (Relenza™; GlaxoSmithKline, Research Triangle Park, NC, USA). In addition, laninamivir (Inavir®; Daiichi Sankyo, Tokyo, Japan), which is currently licensed in Japan, is undergoing study in the US.25 Peramivir, which is licensed in Japan (Rapiacta®; Shionogi, Tokyo, Japan) and South Korea (PeramiFlu®; Green Cross, Yongin-Si, South Korea) has also been studied in the US, but is not currently approved. There are other promising targets. The influenza RNA polymerase has multiple sites of enzymatic activity; these could be exploited for future drug development.26–28 Additionally, the nucleoprotein (NP), which among other functions wraps the influenza RNA molecules into viral particles, could also be exploited for prevention; drugs using this strategy are currently being investigated.26 Other potential targets include the nonstructural proteins (NS1 and NS2) that suppress the host immune response.26
Neuraminidase glycoprotein and inhibition
The NA protein is a homotetrameric glycoprotein with a stalk region and an enzymatically active head.29 The NA active site cleaves sialic acid at the glycosidic bond on the host cell as well as in respiratory mucus, leading to spread of the virus.30 NAIs act to prevent viral progeny from being released and thus slow the spread of infection.31
The NA active site is highly conserved across both influenza A and B32 and has many charged amino acids that act to bind the sialic acid residues.28,32–34 The majority of these bonds are either hydrogen bonds or charge-charge interactions.34 The degree of polarity within the active site has an impact on the oral bioavailability of NAIs, because NAIs that have a polar side chain, such as zanamivir, are not orally bioavailable.34 The active site contains eight functional residues in addition to eleven framework residues.30,32 Within the different NA subtypes, there are further differences and similarities, and thus they have been grouped into two families: group 1 contains subtypes NA1, NA4, NA5, and NA8; group 2 contains NA2, NA3, NA6, NA7, and NA9.35 Most NAs within group 1 contain a region (the “150 loop/cavity”) that rearranges when bound by a drug or sialic acid to open a cavity that could be exploited for development of new agents.26,33 Influenza B NA appears to have segregated from influenza A relatively early in its evolution into group II and thus is distinct from influenza A NA groups.36
In general, rearrangements necessary for binding of NAIs to influenza B-NA require greater conformational changes than those for influenza A-NA, and this may play a role in the generally better efficacy of NAIs in the treatment of influenza A compared with influenza B.34 It has also been observed that some mutations leading to substitutions within the NA active site that cause oseltamivir resistance, are only active for group 1 NA.33
Current neuraminidase inhibitors
As mentioned previously, there are two NAIs currently licensed in the US and two other NAIs that are licensed in Japan; we will discuss them individually in more depth.
Zanamivir
Zanamivir was the first NAI to be developed37 and was licensed in 1999. It exhibits higher affinity to the NA binding site than does the native sialic acid.21 Given its polarity, it is not well absorbed and is thus delivered as an inhaled agent.21,31 After inhalation, 15% of the drug deposits within the lower respiratory tract.31 Therefore, it can precipitate bronchospasm in patients with pulmonary disease and cannot be used in patients who require mechanical ventilation.31 It is also available in an intravenous (IV) form for compassionate use and is currently undergoing clinical trials to determine optimal dosing.28 For influenza treatment, a 10 mg dose is inhaled twice daily (approved for patients older than 7 years) for 5 days, whereas prophylaxis is given once daily for 10 days (although it can be administered for up to 28 days) in patients older than 5 years of age.28,38 Zanamivir also appears to have higher activity against influenza B and H1N1 strains than oseltamivir, but slightly lower activity against H3N2 compared with oseltamivir.28 It remains active against influenza strains that contain the H275Y (or H274Y) neuraminidase mutation.31 The H275Y mutation is associated with resistance to oseltamivir and peramivir; however, zanamivir, given its more closely retained homology to the natural substrate, is unaffected.4 In immunocompetent patients, no resistance to zanamivir therapy has been demonstrated, though the N294S (N295S) neuraminidase mutation causes decreased sensitivity to zanamivir.21
Oseltamivir
Oseltamivir was developed based on the structure of the active site of zanamivir and is administered as a prodrug that is converted by the liver into its active form.39 Like zanamivir, it is licensed in the US for both treatment and prophylaxis.28 Unlike zanamivir, treatment and prophylaxis dosages are based both on weight and renal function. It has been approved for children as young as 1 year. It is relatively well tolerated, with the most common side effect being gastrointestinal upset, which is mitigated by ingestion along with food; however, there have been reports of neurological side effects in children, noted mostly in Japan.22,40 It is also available for intravenous administration in patients who cannot tolerate oral dosing.28 Resistance to oseltamivir has been documented in patients who have been treated with the drug, but it also has been found in patients without exposure to NA inhibitors.21
Peramivir
Peramivir is offered only as an intravenous formulation because of its low oral bioavailability.28,33 In initial clinical trials, oral peramivir efficacy was no better than placebo, as its absorption was low.33 However, it achieves very high concentrations in the bloodstream.41 It has a slightly lower activity against influenza than either zanamivir or oseltamivir.31 The half-maximal inhibitory concentration (IC50) for influenza B is higher than that for influenza A, but is lower than that for both oseltamivir and zanamivir.42 In addition to gastrointestinal side effects, peramivir has been linked to neutropenia and occasionally to electrocardiographic (ECG) abnormalities.31 It is delivered at 600 mg intravenously twice daily for 5 days in adults, and by weight-based dosing for pediatric patients.28 Like oseltamivir, peramivir resistance is mediated by the H275Y NA mutation.31
Laninamivir
Laninamivir is another inhaled NAI, but is currently licensed only in Japan.28,31 It is a prodrug, like oseltamivir, and is converted to the active form in the respiratory tract.43 It requires only one dose because of its long half-life and is highly concentrated within tissues.28 It is very active against both influenza A and B and has good activity against strains that contain the H275Y NA mutation.28
Mutations in neuraminidase associated with NAI resistance
Numerous excellent reviews have discussed NAI resistance.4,21,28,44,45 NAI resistance is mediated by mutations that change the conformation of the NA active site leading to decreased binding of the NAI to the NA, or by changes in amino acids that change the contact/interaction with the drug.28 Depending on the drug-NA interaction, different changes can lead to different drug susceptibility or resistance. Though ideally the NAI should be as similar to the natural substrate as possible, some differences are needed to ensure that the drug is absorbed and retained.26 In general, the majority of resistance has been seen with oseltamivir; relatively few strains have been found to have zanamivir resistance.44 This may be because zanamivir is more closely related to the natural substrate; however, oseltamivir is also used much more frequently.44 No laninamivir-resistant strains have yet been described.28 Please see Table 1 for a partial list of resistance mutations.
Table 1.
Neuraminidase inhibitor resistance mutations
| Influenza type | Mutation | Low | Medium | High | Sensitive |
|---|---|---|---|---|---|
| H1N1 | H274Y | O, P | Z | ||
| Q136K | Z | O | |||
| N70S | Z | O | |||
| Y155H | O | Z, P | |||
| 1222V/M | O | P, Z | |||
| pdm09H1N1 | H274Y | O, P | Z | ||
| I222V | O | Z | |||
| I222R | P, Z | O | |||
| E119G | P | Z | O | ||
| E119V | O, P | Z | |||
| H5N1 | V116A | P | O, Z | ||
| H274Y | O, P | Z | |||
| E119G | P, Z | O | |||
| D198G | O, Z | P | |||
| N294S | O | P, Z | |||
| S246N | O | Z | |||
| H252Y | O | P, Z | |||
| I222L | O | Z | |||
| I222V | O | P, Z | |||
| I222M/T | O | P, Z | |||
| H3N2 | N294S | O | Z | ||
| R292K | Z | O, P | |||
| E119A/D | O, P, Z | ||||
| E119G | P | Z | O | ||
| E119I | Z | O, P | |||
| E119V | O | P, Z | |||
| Q136K | Z | O | |||
| R371K | O, Z | ||||
| D151A/D | Z | O | |||
| Influenza B | E119A/D | O, Z, P | |||
| E119G | O, Z | P | |||
| E119V | O, P | Z | |||
| R371K | Z | O | |||
| D198N | O, Z | P | |||
| D198E | Z | O, P | |||
| H274Y | O, P | Z | |||
| I222T | O | Z | |||
| R292K | O, Z | P | |||
| N294S | O |
Notes: S, Sensitive; L, Low <10 fold change compared to wildtype; M, Medium 10–100 fold change; H, High >100 fold change; O, oseltamivir; Z, zanamivir; P, peramivir. Adapted with permission from McKimm-Breschkin JL. Influenza neuraminidase inhibitors: antiviral action and mechanisms of resistance. Influenza Other Respi Viruses. John Wiley & Sons Ltd21 and Antiviral Res, 98(2), Samson M, Pizzorno A, Abed Y, Boivin G. Influenza virus resistance to neuraminidase inhibitors, 174–185, Copyright 2013, with permission from Elsevier.28
As an example of the mechanism whereby resistance develops, when oseltamivir binds to the NA target, the NA undergoes a conformational change.21,28 The H274Y (H275Y) and N294S mutations lead to decreased affinity for oseltamivir because they prevent some of the conformational changes from taking place, whereas the R292K lowers affinity for the drug.28,46 However, only in N1 is the H274Y mutation significant, whereas the other two mutations are important for N2 viruses.21,47 H274Y is not located in the active site; however, it causes repositioning in a side chain and decreased binding to oseltamivir and greater binding to sialic acid.26 H274Y does not interfere with binding to zanamivir.26 Also, although oseltamivir binding is inhibited, it appears that sialic acid binding also occurs at a decreased rate compared with the wild-type virus; however, there are compensatory mutations such as R222Q and D344N that allow increased binding of sialic acid.26,48,49 The mutations E119V and R292K also confer resistance to oseltamivir: the E119 mutations via decreasing side chain interaction with the drug; and R292 by inhibiting the rotation of the E276 residue.50 Additionally, R292K decreases affinity to the drug because of decreased interaction with the carboxylate group.50 It could be inferred that other NA subtypes with H274Y mutation (group 1) would be resistant to oseltamivir and that those within group 2 with the E119V and R292K mutations would be resistant as well. The I222V mutation has also been shown to decrease susceptibility to oseltamivir within N1 influenza strains in vitro and appears to have an added effect when paired with H274Y.4,21,51 The McKimm-Breschkin article21 offers a useful summary of the mechanisms of resistance to NAIs that includes a discussion of structural issues.
Zanamivir and laninamivir are the drugs most similar to the natural target, and therefore resistance is seen less frequently, although it does occur.21,52 It is expected that resistance to these two drugs would cause more harm to the virus as it would lead to greater changes and potentially decreased fitness.21 Peramivir has similarities to both oseltamivir and zanamivir, and thus many of the mutations that confer resistance to either of these drugs lead to cross-resistance with peramivir.21
The N294S mutation leads to decreased susceptibility to both zanamivir and oseltamivir and is seen primarily in N1 subtypes.21 I223R and K mutations can cause resistance to zanamivir.4,53,54 The I222 mutations (I222V, I222R, and I222T) have been seen across N subtypes (and also within influenza B) and result primarily in decreased susceptibility to oseltamivir.21 E119 mutants (E119V, E119I in vivo, E119A/D/G in vitro) can cause decreased susceptibility to different NAIs depending on the substitution21, and viral fitness can increase when accompanied by the I222V mutation, as demonstrated in H3N2 isolates.55
The majority of mutations to NAIs are caused by mutations within the NA gene itself. Changes to the HA gene and product can also lead to decreased susceptibility to NAIs. These changes in turn decrease the need for NA activity.28,31
Transmission and the effect of NAI resistance
In general, influenza is highly infectious and is transmitted via aerosolized respiratory secretions and large droplets and has an incubation period of 1 to 4 days.2 Persons who are immunocompromised can shed virus for several weeks.56,57
For the 2012–2013 season, oseltamivir resistance within the US was reported to be approximately 1% for H3N2 and 0% for 2009 H1N1 and influenza B strains in circulation.10 However, in 2008–2009, H1N1 oseltamivir resistance had been quite high and widespread in the US, Europe, and other countries.44,58,59 This was not the case in 2009 pandemic H1N1. Additionally, it was once thought that oseltamivir resistance mediated by the H274Y/H275Y mutation led to the decreased viral fitness and transmissibility previously seen in in vitro phenotypic models. This was true from 1999 through 2006. However, during the 2007 season, H1N1 containing the H274Y mutation increased and eventually became the dominant strain in circulation.47,60 What was even more alarming was that resistance in these strains arose without any drug pressure or exposure, suggesting that mutated strain was more fit than the nonresistant strains.47 It has been shown that decreased viral fitness associated with H274Y is mediated by V234M and R22Q NA mutations.49 However, many H274Y strains have unimpaired transmission.61 Within the H3N2 strains, the majority of resistance is mediated by E119V and R292K NA mutations; to date, these appear to result in decreased viral fitness and require higher titers in experimental models to cause infection in ferrets.47
During the 2009 pandemic, the novel H1N1 virus demonstrated susceptibility to oseltamivir. NAI-resistant strains were reported, the majority of those strains were found in immunocompromised hosts and were thought to have arisen secondarily to increased exposure to oseltamivir and prolonged shedding. Studies demonstrated that transmission of oseltamivir-resistant strains may be slightly less efficient than that of the wild-type virus, but resistant strain viral titers tend to be comparable to that of the wild-type virus in vitro.47 As discussed in Govorkova’s excellent review62, in some animal models, transmissibility of resistant strains is comparable to that in the wild type, though this is somewhat controversial as other studies have found less efficient transmission of resistant strains.63
For avian H5N1, experimental models have shown that mutations like H274Y and N294S within NA can lead to decreased enzymatic function, but viral fitness appears to be similar to that of wild-type influenza, which is of concern given the high pathogenicity in humans of this avian influenza subtype.47 For the emergent H7N9 subtype, there has not been documentation of transmission between persons.21 However, NAI resistance appears to arise relatively quickly following treatment with oseltamivir.64 The NA gene resistance mutation was documented as R292L and was associated with patients who deteriorated quickly, maintained high viral loads, and required extracorporeal membrane oxygenation (ECMO) support.64
Diagnosis of NAI-resistant influenza mutants
Despite knowledge of mutations associated with resistance to NAIs, there are currently no rapid diagnostics available for clinical laboratories for the detection of strains; the majority of resistance testing is performed at surveillance laboratories. The two main types of diagnostic tests include genotypic and phenotypic assays.
Phenotypic assays rely on culture and are sometimes inaccurate. The sialic acid structures within the cell types used do not bind HA uniformly; some have even lower HA binding in the presence of NAIs, which decreases the utility of NA activity as a measure of antiviral activity.4,47 Therefore, other phenotypic assays that measure NAI enzyme inhibitory activity are more accurate.65 However, though NAI inhibition may be measured accurately, it does not necessarily correlate with viral replication. Currently, most of these tests utilize either chemiluminescent or fluorescent read-out, and both require that the virus be cultured first.4 Chemiluminescent assay is less expensive but requires a higher viral titer for improved accuracy, whereas the fluorescent assay is generally thought to be more accurate.65 Although there are some limitations to phenotypic assays, they are able to detect the effect of both known and unknown resistance mutations and provide a sensitivity profile for the antiviral drug because they measure inhibitory concentration of the drug.65,66 Kits are commercially available and contain reagents, but still require culture of the virus.67 Finally, some studies have combined phenotypic assays with sequencing.68
Genotypic resistance testing is rapid and can be done without viral culture. Current methodologies used for analyzing the influenza NA gene include the polymerase chain reaction (PCR) with Sanger sequencing, next generation sequencing (NGS), real-time PCR (RT PCR), probe hybridization technologies, and single nucleotide polymorphism analysis, among others.4,65 The disadvantages include cost (although the methodologies mentioned are significantly less expensive than phenotypic assays) and the potential that genotypic resistance does not always correlate with phenotypic resistance.65 PCR and population sequencing are used most frequently, although they can be time consuming depending on whether population-based sequencing or clonal analysis is performed. Given that novel mutations can be discovered through whole gene sequencing, assays using PCR with sequencing are attractive applications.65 RT PCR is faster and is used in clinical laboratories for diagnosis of influenza as well as for subtyping. Detection of resistance mutations can be done utilizing a number of methods, such as allele-specific PCR for specific single nucleotide polymorphisms or probe hybridization.65,69,70
NGS analysis of influenza sequences has generally been performed using pyrosequencing.65,71 We have utilized another NGS technique using targeted resequencing to demonstrate low frequency mutations associated with NAI resistance.72 In our study, we sequenced the NA gene from nine clinical samples and were able to detect oseltamivir resistance (H275Y) at 0.18% in one of the nine clinical samples.72 Presently, it is unclear what the clinical significance of low-level frequency mutants may be.
Combination therapy
Given that there are currently only two NAIs licensed in the US, and that there is increasing concern for resistance, the use of combination therapy is promising as it raises the potential of decreasing emerging resistance and increasing efficacy of therapy.73,74 In addition, combination therapy may help in instances in which treatment is started later or in hosts with compromised immune systems.75 Some initial combinations that have been evaluated have been combinations of NAIs themselves, whereas others have investigated combinations of novel NAIs and novel therapies.76,77
In a placebo-controlled study, patients presenting with influenza during the 2008–2009 influenza season were randomized to receive oseltamivir monotherapy with an inhaled placebo (176 patients), oseltamivir/zanamivir combination therapy (192 patients), or a zanamivir plus an oral placebo pill (173 patients).78 The outcome was defined as influenza viral copy number at 48 hours after treatment. The dual therapy was associated with higher viral copy numbers than oseltamivir monotherapy, potentially because zanamivir caused decreased oseltamivir binding.78 In another small study, dual therapy with oseltamivir and zanamivir was compared to oseltamivir alone during the 2009 pandemic; there did not appear to be a difference in efficacy as measured by influenza viral load and symptoms (though the study was underpowered, with only 24 patients).79
Other therapies that have been considered for combination therapy include the NAIs plus adamantanes and ribavirin. Ribavirin is an RNA polymerase inhibitor and is more commonly used for the treatment of respiratory syncytial virus and hepatitis C virus infections. It is available for inhalation therapy, as tablets for oral administration, and, outside the US, intravenously. However, it is teratogenic, can cause hemolytic anemia, neutropenia, and thrombocytopenia, and for those reasons carries a black box warning.
In an in vitro study, Hoopes et al looked at a mathematical and experimental model of emerging resistance with triple combination antiviral drug therapy (TCAD).80 Using oseltamivir, amantadine, and ribavirin, and serial passaging of influenza virus in culture, resistance emerged rapidly when single agents were used. Those authors also demonstrated that although resistance mutations to oseltamivir did occur in the TCAD-treated cells, overall there was a very high barrier to resistance given that multiple mutations were needed to overcome treatment efficacy of the combination, and that in combination amantadine remained active, likely secondary to the effect of ribavirin. Nguyen et al looked at TCAD with amantadine, oseltamivir, and ribavirin using a murine influenza infection model and found that there was a greater than 90% survival in mice treated with TCAD compared with monotherapy (0% amantadine, 20% oseltamivir, 0% ribavirin alone) or dual therapy (50% oseltamivir/amantadine, 60% oseltamivir/ribavirin).75
Kim et al looked at the efficacy of TCAD therapy in critically ill influenza patients admitted to their hospital in Korea using a retrospective analysis.81 One hundred twenty-seven patients were studied, of whom 24 received TCAD and the others received oseltamivir alone. At 14 days, there was a statistically significant trend toward survival in the TCAD group, with 17% mortality for TCAD versus 35% for oseltamivir alone. However, there was no significant difference in survival at 90 days.
Within the US, two clinical trials have been completed that compared TCAD to oseltamivir. Both studies (NCT00867139 and NCT00979251)82,83 looked at TCAD versus oseltamivir monotherapy in immunocompromised hosts. Seo et al published their results of two investigations (Trial NCT00867139)84,86: one on the pharmacokinetics in healthy individuals and another looking at efficacy within immunocompromised hosts. There were two substudies for the TCAD therapy in immunocompromised hosts; one randomized patients older than 7 years to TCAD (oseltamivir, amantadine, ribavirin) or oseltamivir alone, and the other substudy was open label for patients who had more severe disease and who were excluded from randomization. All patients had either undergone chemotherapy or hematopoietic stem cell transplant within the last 2 years and needed to have a positive test for influenza. Three patients were randomized and four participated in the open label study. Given lack of funding, the trial was stopped early. The patients receiving TCAD did demonstrate a decrease in viral load, although one patient progressed to ARDS. A third study, NCT01227967, is examining the efficacy and safety of TCAD and is currently recruiting patients.85
Future neuraminidase inhibitor development
Given the limitations of current drug therapy (for instance, modality of administration and resistance), new therapeutics are needed. In addition to NAIs, other agents are being investigated that have other viral targets, as well as recent studies of other combination therapies.26,28 Quantitative structural relationship models have been used to model new potential NAIs.34
Zanamivir analogs, including laninamivir, have been synthesized.87 With the discovery of the 150 loop/cavity, other compounds are currently being evaluated that would lead to tighter binding of the NA and thus to greater efficacy.33 Given that NA exists on the surface of virions in tetramers, there is increasing interest in finding compounds that would link multiple NAs on the surface of virions, ie, multivalent preparations that may exhibit greater binding affinity and potentially improved pharmacokinetics, in addition to increased potency.33 There has also been interest in developing derivatives of peramivir.33 In addition, benzoic acid is a relatively inexpensive chemical compound that could be utilized for future NAI synthesis.88 Because current NAIs are expensive to produce and require long manufacturing times, synthesis of lower cost benzoic acid derivatives having anti-influenza activity could be quite useful in pandemics.33
NA binds to its substrate by ionic bonds, but recently Kim et al have demonstrated transient covalent bonding.89 This finding may also lead to new drug development. The group has been investigating 2,3 difluorosialic acid compounds as potential therapeutics; these compounds are attractive because they have relatively high affinity and slow dissociation from the NA, leading to very long half lives and prolonged inactivation of NA.89 In their review, Chamni and De-Eknamkul discuss the current patents and the chemical structures of NAIs.90 Additionally, other compounds that interfere with the influenza binding of sialic acid residues, such as DAS181 which cleaves sialic acid residues on host cells, preventing binding by NA, are in clinical trial.91 Other compounds like favipiravir (T-705) are also being developed and have applications for viral infections beyond influenza.27,92,93 In the future, medications such as these, in addition to newer NAIs, will be increasingly important as influenza resistance to current therapies increases.
Guidelines for prophylaxis
Several guidelines have been developed establishing who should receive zanamivir or oseltamivir for influenza prophylaxis. Jackson et al offer a systematic review that details studies looking at the efficacy of prophylaxis, either postexposure or seasonal and outbreak prevention.94 Generally, the adamantanes are not recommended, given the high level of established resistance among influenza strains, but doses of 100 mg orally twice daily can be used if the strain is thought to be susceptible.31 For close household contacts of persons with influenza who have not received the vaccine and who have comorbidities that could lead to complications if they were to become infected, either zanamivir or oseltamivir is effective.95 Other persons who should receive prophylaxis include health care workers who had not practiced proper precautions, and persons who live in nursing homes or other long-term care facilities.
In general, dosing for children is weight based. For household or other close contact postexposure prophylaxis within the US, oseltamivir is approved for adults and children older than 13 years and is dosed at 75 mg orally twice daily for 10 days. Zanamivir is approved for prophylaxis in adults and children older than 5 years, and is dosed at 10 mg inhaled daily for 10 days. Laninamivir has only been licensed for treatment at this time, although given its long half-life it has potential for influenza prophylaxis.28 Patients who receive postexposure prophylaxis should also be vaccinated.95
For persons at risk for influenza complications, such as long-term care facility residents or persons who cannot receive influenza vaccination, prophylaxis is approved for duration of up to 6 weeks, generally during the time of potential exposure.2,95,96 Long-term administration is safe and relatively well tolerated in adults, and in a recent open label study has been found to be safe for long durations in children.97 However, although safe and well tolerated with proven efficacy2, long-term prophylaxis does raise the concern for promotion of NAI resistance.98 In general, prophylaxis with oseltamivir is approximately 68% to 90% effective, and zanamivir is approximately 68% to 80% effective in preventing influenza infection.31,99
Conclusion
In summary, influenza is a respiratory pathogen that causes significant morbidity and mortality globally. There have been four pandemics in the last 100 years, and the threat of a new pandemic is ever present. Vaccination remains the most important cornerstone of prevention. However, depending on vaccination rates, vaccine effectiveness (based on vaccine composition and the population vaccinated), the amount of protection afforded varies considerably. Therefore, antiviral medications such as the NAIs are increasingly important both for treatment and for prophylaxis of vulnerable populations.
Although oseltamivir resistance among circulating influenza strains was low during the 2012–2013 influenza season, given the history of natural variants that circulate and can be transmitted with resistance, there remains a pressing need for other antiviral agents that are active against influenza. Zanamivir continues to demonstrate efficacy and low levels of resistance, but its use is limited in patients who have significant respiratory comorbidities. Laninamivir will be a useful addition if approved in the US, as well as other agents that are currently in the very early stages of drug development. The detection of resistance will also need to be streamlined, allowing for better treatment and prophylaxis decisions to be made in real time.
Acknowledgment
This study was funded by intramural funds from the Department of Veterans Affairs. The views expressed in this article are those of the authors and do not necessarily reflect the position or policy of the Departments of Veterans Affairs or the United States Government.
Footnotes
Disclosure
Both authors report no conflicts of interest in this work.
References
- 1.Estimates of deaths associated with seasonal influenza – United States, 1976–2007. MMWR Morb Mortal Wkly Rep. 2010 Aug 27;59(33):105–1062. [PubMed] [Google Scholar]
- 2.Williams JM. 2009 update in prevention, evaluation, and outpatient treatment of influenza. Curr Med Res Opin. 2009 Apr;25(4):817–828. doi: 10.1185/03007990802708244. [DOI] [PubMed] [Google Scholar]
- 3.Treaner JJ. Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases. 7th ed. Philadelphia: Churchill Livingstone; 2010. Avian influenza; pp. 2270–2271. [Google Scholar]
- 4.Cheng VC, To KK, Tse H, Hung IF, Yuen KY. Two years after pandemic influenza A/2009/H1N1: what have we learned? Clin Microbiol Rev. 2012 Apr;25(2):223–263. doi: 10.1128/CMR.05012-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bao CJ, Cui LB, Zhou MH, Hong L, Gao GF, Wang H. Live-animal markets and influenza A (H7N9) virus infection. N Engl J Med. 2013;368(24):2337–2339. doi: 10.1056/NEJMc1306100. [DOI] [PubMed] [Google Scholar]
- 6.Beck CR, McKenzie BC, Hashim AB, Harris RC, Nguyen-Van-Tam JS, University of Nottingham Influenza and the ImmunoCompromised (UNIIC) Study Group Influenza vaccination for immunocompromised patients: systematic review and meta-analysis by etiology. J Infect Dis. 2012;206(8):1250–1259. doi: 10.1093/infdis/jis487. [DOI] [PubMed] [Google Scholar]
- 7.Corti D, Voss J, Gamblin SJ, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333(6044):850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 8.Lambe T, Spencer AJ, Mullarkey CE, et al. T-cell responses in children to internal influenza antigens, 1 year after immunization with pandemic H1N1 influenza vaccine, and response to revaccination with seasonal trivalent-inactivated influenza vaccine. Pediatr Infect Dis J. 2012 Jun;31(6):e86–e91. doi: 10.1097/INF.0b013e318255e443. [DOI] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention National and Regional Level Outpatient Illness and Viral Surveillance 2013Available from: http://gis.cdc.gov/grasp/fluview/fluportaldashboard.htmlAccessed August 7, 2013
- 10.Centers for Disease Control and Prevention Seasonal Influenza (Flu) Situation update: summary of weekly FluView Available from http://www.cdcgov/flu/weekly/summary.htmAccessed August 7, 2013
- 11.Centers for Disease Control and Prevention Seasonal Influenza (Flu) Highly Pathogenic Avian Influenza A (H5N1) Virus Available from http://www.cdc.gov/flu/avianflu/h5n1-virus.htmAccessed August 7, 2013
- 12.Centers for Disease Control and Prevention Seasonal Influenza (Flu) Highly Pathogenic Avian Influenza A (H5N1) in People Available from http://www.cdc.gov/flu/avianflu/h5n1-people.htmAccessed August 7, 2013
- 13.World Health Organization Influenza Frequently Asked Questions on human infection caused by the avian influenza A (H7N9) virus 2013Available from http://www.who.int/influenza/human_animal_interface/faq_H7N9/en/index.htmlAccessed August 7, 2013
- 14.Ke Y, Wang Y, Zhang W, Huang L, Chen Z. Deaths associated with avian influenza A(H7N9) virus in China. Ann Intern Med. 2013;159(2):159–160. doi: 10.7326/0003-4819-159-2-201307160-00669. [DOI] [PubMed] [Google Scholar]
- 15.Gao HN, Lu HZ, Cao B, et al. Clinical findings in 111 cases of influenza A (H7N9) virus infection. N Engl J Med. 2013 Jun 13;368(24):2277–2285. doi: 10.1056/NEJMoa1305584. [DOI] [PubMed] [Google Scholar]
- 16.Xu C, Havers F, Wang L, et al. Monitoring avian influenza A(H7N9) virus through national influenza-like illness surveillance, China. Emerg Infect Dis. 2013;19(8):1289–1292. doi: 10.3201/eid1908.130662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ke Y, Wang Y, Liu S, et al. High severity and fatality of human infections with avian influenza A (H7N9) infection in China. Clin Infect Dis. 2013 Aug 7; doi: 10.1093/cid/cit371. Epub. [DOI] [PubMed] [Google Scholar]
- 18.Gao R, Cao B, Hu Y, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368(20):1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 19.Lee SS, Wong NS, Leung CC. Exposure to avian influenza H7N9 in farms and wet markets. Lancet. 2013;381(9880):1815. doi: 10.1016/S0140-6736(13)60949-6. [DOI] [PubMed] [Google Scholar]
- 20.Takeda M, Pekosz A, Shuck K, Pinto LH, Lamb RA. Influenza a virus M2 ion channel activity is essential for efficient replication in tissue culture. J Virol. 2002;76(3):1391–1399. doi: 10.1128/JVI.76.3.1391-1399.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKimm-Breschkin JL. Influenza neuraminidase inhibitors: antiviral action and mechanisms of resistance. Influenza Other Respi Viruses. 2013;7(Suppl 1):25–36. doi: 10.1111/irv.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schirmer P, Holodniy M. Oseltamivir for treatment and prophylaxis of influenza infection. Expert Opin Drug Saf. 2009;8(3):357–371. doi: 10.1517/14740330902840519. [DOI] [PubMed] [Google Scholar]
- 23.Deyde VM, Xu X, Bright RA, et al. Surveillance of resistance to adamantanes among influenza A(H3N2) and A(H1N1) viruses isolated worldwide. The Journal of Infectious Diseases. 2007;196(2):249–257. doi: 10.1086/518936. [DOI] [PubMed] [Google Scholar]
- 24.Bright RA, Medina MJ, Xu X, et al. Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: a cause for concern. Lancet. 2005;366(9492):1175–1181. doi: 10.1016/S0140-6736(05)67338-2. [DOI] [PubMed] [Google Scholar]
- 25.Biota Safety, tolerability and pharmacokinetic study of multiple doses of CS-8958 Available from: http://clinicaltrials.gov/ct2/results?term=laninamivirAccessed August 7, 2013
- 26.Das K. Antivirals targeting influenza A virus. J Med Chem. 2012;55(14):6263–6277. doi: 10.1021/jm300455c. [DOI] [PubMed] [Google Scholar]
- 27.Furuta Y, Takahashi K, Shiraki K, et al. T-705 (favipiravir) and related compounds: Novel broad-spectrum inhibitors of RNA viral infections. Antiviral Res. 2009;82(3):95–102. doi: 10.1016/j.antiviral.2009.02.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samson M, Pizzorno A, Abed Y, Boivin G. Influenza virus resistance to neuraminidase inhibitors. Antiviral Res. 2013;98(2):174–185. doi: 10.1016/j.antiviral.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 29.Varghese JN, Laver WG, Colman PM. Structure of the influenza virus glycoprotein antigen neuraminidase at 2.9 A resolution. Nature. 1983;303(5912):35–40. doi: 10.1038/303035a0. [DOI] [PubMed] [Google Scholar]
- 30.Colman PM, Varghese JN, Laver WG. Structure of the catalytic and antigenic sites in influenza virus neuraminidase. Nature. 1983;303(5912):41–44. doi: 10.1038/303041a0. [DOI] [PubMed] [Google Scholar]
- 31.Ison MG. Clinical use of approved influenza antivirals: therapy and prophylaxis. Influenza Other Respi Viruses. 2013;7(Suppl 1):7–13. doi: 10.1111/irv.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burmeister WP, Ruigrok RW, Cusack S. The 2.2 A resolution crystal structure of influenza B neuraminidase and its complex with sialic acid. EMBO J. 1992;11(1):49–56. doi: 10.1002/j.1460-2075.1992.tb05026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng E, Ye D, Li J, et al. Recent advances in neuraminidase inhibitor development as anti-influenza drugs. ChemMedChem. 2012;7(9):1527–1536. doi: 10.1002/cmdc.201200155. [DOI] [PubMed] [Google Scholar]
- 34.Gong JZ, Liu Y, Xu WF. Pharmacophore model of influenza neuraminidase inhibitors – a systematic review. Pharmazie. 2009;64(10):627–632. [PubMed] [Google Scholar]
- 35.Russell RJ, Haire LF, Stevens DJ, et al. The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature. 2006;443(7107):45–49. doi: 10.1038/nature05114. [DOI] [PubMed] [Google Scholar]
- 36.Xu J, Davis CT, Christman MC, et al. Evolutionary history and phylodynamics of influenza A and B neuraminidase (NA) genes inferred from large-scale sequence analyses. PloS ONE. 2012;7(7):e38665. doi: 10.1371/journal.pone.0038665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.von Itzstein M, Wu WY, Kok GB, et al. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature. 1993;363(6428):418–423. doi: 10.1038/363418a0. [DOI] [PubMed] [Google Scholar]
- 38.GlaxoSmithKline Relenza: Prescribing information Available from: http://us.gsk.com/products/assets/us_relenza.pdfAccessed August 7, 2013
- 39.Kim CU, Lew W, Williams MA, et al. Influenza neuraminidase inhibitors possessing a novel hydrophobic interaction in the enzyme active site: design, synthesis, and structural analysis of carbocyclic sialic acid analogues with potent anti-influenza activity. J Am Chem Soc. 1997;119(4):681–690. doi: 10.1021/ja963036t. [DOI] [PubMed] [Google Scholar]
- 40.Beck CR, Sokal R, Arunachalam N, et al. UK Antiviral Effectiveness Review Group Neuraminidase inhibitors for influenza: a review and public health perspective in the aftermath of the 2009 pandemic. Influenza Other Respi Viruses. 2013;7(Suppl 1):14–24. doi: 10.1111/irv.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boltz DA, Aldridge JR, Jr, Webster RG, Govorkova EA. Drugs in development for influenza. Drugs. 2010;70(11):1349–1362. doi: 10.2165/11537960-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chairat K, Tarning J, White NJ, et al. Pharmacokinetic properties of anti-influenza neuraminidase inhibitors. J Clin Pharmacol. 2013;53(2):119–139. doi: 10.1177/0091270012440280. [DOI] [PubMed] [Google Scholar]
- 43.Koyama K, Takahashi M, Nakai N, et al. Pharmacokinetics and disposition of CS-8958, a long-acting prodrug of the novel neuraminidase inhibitor laninamivir in rats. Xenobiotica. 2010;40(3):207–216. doi: 10.3109/00498250903447691. [DOI] [PubMed] [Google Scholar]
- 44.Thorlund K, Awad T, Boivin G, Thabane L. Systematic review of influenza resistance to the neuraminidase inhibitors. BMC Infect Dis. 2011;11:134. doi: 10.1186/1471-2334-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferraris O, Lina B. Mutations of neuraminidase implicated in neuraminidase inhibitors resistance. J ClinVirol. 2008;41(1):13–19. doi: 10.1016/j.jcv.2007.10.020. [DOI] [PubMed] [Google Scholar]
- 46.Earhart KC, Elsayed NM, Saad MD, et al. Oseltamivir resistance mutation N294S in human influenza A(H5N1) virus in Egypt. Journal of Infection and Public Health. 2009;2(2):74–80. doi: 10.1016/j.jiph.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 47.Baranovich T, Webster RG, Govorkova EA. Fitness of neuraminidase inhibitor-resistant influenza A viruses. Curr Opin Virol. 2011 Dec;1(6):574–581. doi: 10.1016/j.coviro.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rameix-Welti MA, Munier S, Le Gal S, et al. Neuraminidase of 2007–2008 influenza A(H1N1) viruses shows increased affinity for sialic acids due to the D344N substitution. Antivir Ther. 2011;16(4):597–603. doi: 10.3851/IMP1804. [DOI] [PubMed] [Google Scholar]
- 49.Bloom JD, Gong LI, Baltimore D. Permissive secondary mutations enable the evolution of influenza oseltamivir resistance. Science. 2010;328(5983):1272–1275. doi: 10.1126/science.1187816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Varghese JN, Smith PW, Sollis SL, et al. Drug design against a shifting target: a structural basis for resistance to inhibitors in a variant of influenza virus neuraminidase. Structure. 1998;6(6):735–746. doi: 10.1016/s0969-2126(98)00075-6. [DOI] [PubMed] [Google Scholar]
- 51.Pizzorno A, Bouhy X, Abed Y, Boivin G. Generation and characterization of recombinant pandemic influenza A(H1N1) viruses resistant to neuraminidase inhibitors. J Infect Dis. 2011;203(1):25–31. doi: 10.1093/infdis/jiq010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hurt AC, Lee RT, Leang SK, et al. Increased detection in Australia and Singapore of a novel influenza A(H1N1)2009 variant with reduced oseltamivir and zanamivir sensitivity due to a S247N neuraminidase mutation. Euro Surveill. 2011;16(23) article Id=19884. [PubMed] [Google Scholar]
- 53.Nguyen HT, Fry AM, Loveless PA, et al. Recovery of a multidrug-resistant strain of pandemic influenza A 2009 (H1N1) virus carrying a dual H275Y/I223R mutation from a child after prolonged treatment with oseltamivir. Clin Infect Dis. 2010;51(8):983–984. doi: 10.1086/656439. [DOI] [PubMed] [Google Scholar]
- 54.van der Vries E, Stelma FF, Boucher CA. Emergence of a multidrug-resistant pandemic influenza A (H1N1) virus. N Eng J Med. 2010;363:1381–1382. doi: 10.1056/NEJMc1003749. [DOI] [PubMed] [Google Scholar]
- 55.Simon P, Holder BP, Bouhy X, et al. The I222V neuraminidase mutation has a compensatory role in replication of an oseltamivir-resistant influenza virus A/H3N2 E119V mutant. J Clin Microbiol. 2011;49(2):715–717. doi: 10.1128/JCM.01732-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ghedin E, Laplante J, DePasse J, et al. Deep sequencing reveals mixed infection with 2009 pandemic influenza A (H1N1) virus strains and the emergence of oseltamivir resistance. J Infect Dis. 2011;203(2):168–174. doi: 10.1093/infdis/jiq040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weinstock DM, Gubareva LV, Zuccotti G. Prolonged shedding of multidrug-resistant influenza A virus in an immunocompromised patient. N Eng J Med. 2003;348(9):867–868. doi: 10.1056/NEJM200302273480923. [DOI] [PubMed] [Google Scholar]
- 58.Meijer A, Lackenby A, Hungnes O, et al. Oseltamivir-resistant influenza virus A (H1N1), Europe, 2007–2008 season. Emerg Infect Dis. 2009;15(4):552–560. doi: 10.3201/eid1504.081280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dharan NJ, Gubareva LV, Meyer JJ, et al. Oseltamivir-Resistance Working Group Oseltamir Infections with oseltamivir-resistant influenza A (H1N1) virus in the United States. JAMA. 2009;301(10):1034–1041. doi: 10.1001/jama.2009.294. [DOI] [PubMed] [Google Scholar]
- 60.Lackenby A, Hungnes O, Dudman SG, et al. Emergence of resistance to oseltamivir among influenza A(H1N1) viruses in Europe. Euro Surveill. 2008;13(5) doi: 10.2807/ese.13.05.08026-en. article Id=8026. [DOI] [PubMed] [Google Scholar]
- 61.Baz M, Abed Y, Simon P, et al. Effect of the neuraminidase mutation H274Y conferring resistance to oseltamivir on the replicative capacity and virulence of old and recent human influenza A(H1N1) viruses. J Infect Dis. 2010;201(5):740–745. doi: 10.1086/650464. [DOI] [PubMed] [Google Scholar]
- 62.Govorkova EA. Consequences of resistance: in vitro fitness, in vivo infectivity, and transmissibility of oseltamivir-resistant influenza A viruses. Influenza Other Respi Viruses. 2013;7(Suppl 1):50–57. doi: 10.1111/irv.12044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Duan S, Boltz DA, Seiler P, et al. Competitive transmissibility and fitness of oseltamivirsensitive and resistant pandemic influenza H1N1 viruses in ferrets. Influenza Other Respi Viruses. 2011;5(Suppl 1):79–82. [PubMed] [Google Scholar]
- 64.Hu Y, Lu S, Song Z, et al. Association between adverse clinical outcome in human disease caused by novel influenza A H7N9 virus and sustained viral shedding and emergence of antiviral resistance. Lancet. 2013;381(9885):2273–2279. doi: 10.1016/S0140-6736(13)61125-3. [DOI] [PubMed] [Google Scholar]
- 65.Okomo-Adhiambo M, Sheu TG, Gubareva LV. Assays for monitoring susceptibility of influenza viruses to neuraminidase inhibitors. Influenza Other Respi Viruses. 2013;7(Suppl 1):44–49. doi: 10.1111/irv.12051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gubareva LV, Webster RG, Hayden FG. Detection of influenza virus resistance to neuraminidase inhibitors by an enzyme inhibition assay. Antiviral Res. 2002;53(1):47–61. doi: 10.1016/s0166-3542(01)00192-9. [DOI] [PubMed] [Google Scholar]
- 67.Biosystems A.NA-Star Influenza Neuraminidase Inhibitor Resistance Detection Kit Available from: http://www3.appliedbiosystems.com/cms/groups/applied_markets_marketing/documents/generaldocuments/cms_042653.pdfAccessed August 7, 2013
- 68.McKimm-Breschkin J, Trivedi T, Hampson A, et al. Neuraminidase sequence analysis and susceptibilities of influenza virus clinical isolates to zanamivir and oseltamivir. Antimicrob Agents Chemother. 2003;47(7):2264–2272. doi: 10.1128/AAC.47.7.2264-2272.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Duan S, Boltz DA, Li J, et al. Novel genotyping and quantitative analysis of neuraminidase inhibitor resistance-associated mutations in influenza a viruses by single-nucleotide polymorphism analysis. Antimicrob Agents Chemother. 2011;55(10):4718–4727. doi: 10.1128/AAC.00316-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Suzuki Y, Saito R, Sato I, et al. Identification of oseltamivir resistance among pandemic and seasonal influenza A (H1N1) viruses by an His275Tyr genotyping assay using the cycling probe method. J Clin Microbio. 2011;49(1):125–130. doi: 10.1128/JCM.01401-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Deyde VM, Okomo-Adhiambo M, Sheu TG, et al. Pyrosequencing as a tool to detect molecular markers of resistance to neuraminidase inhibitors in seasonal influenza A viruses. Antiviral Res. 2009;81(1):16–24. doi: 10.1016/j.antiviral.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 72.Flaherty P, Natsoulis G, Muralidharan O, et al. Ultrasensitive detection of rare mutations using next-generation targeted resequencing. Nucleic Acids Res. 2012;40(1):e2. doi: 10.1093/nar/gkr861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Perelson AS, Rong L, Hayden FG. Combination antiviral therapy for influenza: predictions from modeling of human infections. J Infect Dis. 2012;205(11):1642–1645. doi: 10.1093/infdis/jis265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nguyen JT, Hoopes JD, Le MH, et al. Triple combination of amantadine, ribavirin, and oseltamivir is highly active and synergistic against drug resistant influenza virus strains in vitro. PLoS ONE. 2010;5(2):e9332. doi: 10.1371/journal.pone.0009332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nguyen JT, Smee DF, Barnard DL, et al. Efficacy of combined therapy with amantadine, oseltamivir, and ribavirin in vivo against susceptible and amantadine-resistant influenza A viruses. PLoS ONE. 2012;7(1):e31006. doi: 10.1371/journal.pone.0031006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Smee DF, Hurst BL, Wong MH, et al. Effects of the combination of favipiravir (T-705) and oseltamivir on influenza A virus infections in mice. Antimicrob Agents Chemother. 2010;54(1):126–133. doi: 10.1128/AAC.00933-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tarbet EB, Maekawa M, Furuta Y, et al. Combinations of favipiravir and peramivir for the treatment of pandemic influenza A/California/04/2009 (H1N1) virus infections in mice. Antiviral Res. 2012;94(1):103–110. doi: 10.1016/j.antiviral.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Duval X, van der Werf S, Blanchon T, et al. Bivir Study Group Efficacy of oseltamivir-zanamivir combination compared to each monotherapy for seasonal influenza: a randomized placebo-controlled trial. PLoS Med. 2010;7(11):e1000362. doi: 10.1371/journal.pmed.1000362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Escuret V, Cornu C, Boutitie F, et al. Oseltamivir-zanamivir bitherapy compared to oseltamivir monotherapy in the treatment of pandemic 2009 influenza A(H1N1) virus infections. Antiviral Res. 2012;96(2):130–137. doi: 10.1016/j.antiviral.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 80.Hoopes JD, Driebe EM, Kelley E, et al. Triple combination antiviral drug (TCAD) composed of amantadine, oseltamivir, and ribavirin impedes the selection of drug-resistant influenza A virus. PLoS ONE. 2011;6(12):e29778. doi: 10.1371/journal.pone.0029778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim WY, Young Suh G, Huh JW, et al. Korean Society of Critical Care Medicine H1N1 Collaborative Triple-combination antiviral drug for pandemic H1N1 influenza virus infection in critically ill patients on mechanical ventilation. Antimicrob Agents Chemother. 2011;55(12):5703–5709. doi: 10.1128/AAC.05529-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fred Hutchinson Cancer Research Center TCAD vs. Monotherapy for Influenza A in Immunocompromised Patients Available from: http://clinicaltrials.gov/show/NCT00867139. NLM identifier: NCT00867139Accessed August 23, 2013
- 83.Adamas Pharmaceuticals, Inc Oral Triple Combination Antiviral Drug Therapy for Treatment of Influenza A in Immunocompromised Subjects (PO206) Available from: http://clinicaltrials.gov/show/NCT00979251. NLM identifier: NCT00979251Accessed August 23, 2013
- 84.Fred Hutchinson Cancer Research Center TCAD vs. Monotherapy for Influenza A in Immunocompromised Patients Available from: http://clinicaltrials.gov/show/NCT00867139. NLM Identifier: NCT00867139Accessed August 23, 2013
- 85.National Institute of Allergy and Infectious Diseases Comparing the Efficacy, Safety, and Tolerability of Combination Antivirals (Amantadine, Ribavirin, Oseltamivir) Versus Oseltamivir for the Treatment of Influenza in Adults at Risk for Complications (IRC003) Available from: http://clinicaltrials.gov/ct2/show/NCT01227967. NLM Identifier: NCT01227967Accessed August 23, 2013
- 86.Seo S, Englund JA, Nguyen JT, et al. Combination therapy with amantadine, oseltamivir and ribavirin for influenza A infection: safety and pharmacokinetics. Antivir Ther. 2013;18(3):377–386. doi: 10.3851/IMP2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yamashita M, Tomozawa T, Kakuta M, et al. CS-8958, a prodrug of the new neuraminidase inhibitor R-125489, shows long-acting anti-influenza virus activity. Antimicrob Agents Chemother. 2009;53(1):186–192. doi: 10.1128/AAC.00333-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Krueger AC, Xu Y, Kati WM, et al. Synthesis of potent pyrrolidine influenza neuraminidase inhibitors. Bioorg Med Chem Lett. 2008;18(5):1692–1695. doi: 10.1016/j.bmcl.2008.01.048. [DOI] [PubMed] [Google Scholar]
- 89.Kim JH, Resende R, Wennekes T, et al. Mechanism-based covalent neuraminidase inhibitors with broad-spectrum influenza antiviral activity. Science. 2013;340(6128):71–75. doi: 10.1126/science.1232552. [DOI] [PubMed] [Google Scholar]
- 90.Chamni S, De-Eknamkul W. Recent progress and challenges in the discovery of new neuraminidase inhibitors. Expert Opin Ther Pat. 2013;23(4):409–423. doi: 10.1517/13543776.2013.765861. [DOI] [PubMed] [Google Scholar]
- 91.Moss RB, Hansen C, Sanders RL, et al. A phase II study of DAS181, a novel host directed antiviral for the treatment of influenza infection. J Infect Dis. 2012;206(12):1844–1851. doi: 10.1093/infdis/jis622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kiso M, Takahashi K, Sakai-Tagawa Y, et al. T-705 (favipiravir) activity against lethal H5N1 influenza A viruses. Proc Natl Acad Sci U S A. 2010;107(2):882–887. doi: 10.1073/pnas.0909603107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sleeman K, Mishin VP, Deyde VM, et al. In vitro antiviral activity of favipiravir (T-705) against drug-resistant influenza and 2009 A(H1N1) viruses. Antimicrob Agents Chemother. 2010;54(6):2517–2524. doi: 10.1128/AAC.01739-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jackson RJ, Cooper KL, Tappenden P, et al. Oseltamivir, zanamivir and amantadine in the prevention of influenza: a systematic review. J Infect. 2011;62(1):14–25. doi: 10.1016/j.jinf.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 95.Centers for Disease Control and Prevention Seasonal Influtenza (Flu) Use of Antivirals Available from: http://www.cdc.gov/flu/professionals/antivirals/antiviral-use-influenza.htm#postexposureAccessed August 7, 2013
- 96.Harper SA, Bradley JS, Englund JA, et al. Expert Panel of the INfectious Diseases Society of America Seasonal influenza in adults and children – diagnosis, treatment, chemoprophylaxis, and institutional outbreak management: clinical practice guidelines of the Infectious Diseases Society of America. Clin Infect Dis. 2009;48(8):1003–1032. doi: 10.1086/604670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Reisinger K, Shu D, Cupelli L, et al. Safety and tolerability of a 6-week course of oseltamivir prophylaxis for seasonal influenza in children. Influenza Other Respi Viruses. 2013;7(1):11–13. doi: 10.1111/j.1750-2659.2012.00367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Baz M, Abed Y, Papenburg J, et al. Emergence of oseltamivir-resistant pandemic H1N1 virus during prophylaxis. N Eng J Med. 2009;361(23):2296–2297. doi: 10.1056/NEJMc0910060. [DOI] [PubMed] [Google Scholar]
- 99.Moscona A. Neuraminidase inhibitors for influenza. N Engl J Med. 2005;353(13):1363–1373. doi: 10.1056/NEJMra050740. [DOI] [PubMed] [Google Scholar]