Abstract
Mechanical interactions are fundamental to biology. Mechanical forces of chemical origin determine motility and adhesion on the cellular scale, and govern transport and affinity on the molecular scale. Biological sensing in the mechanical domain provides unique opportunities to measure forces, displacements and mass changes from cellular and subcellular processes. Nanomechanical systems are particularly well matched in size with molecular interactions, and provide a basis for biological probes with single-molecule sensitivity. Here we review micro- and nanoscale biosensors, with a particular focus on fast mechanical biosensing in fluid by mass- and force-based methods, and the challenges presented by non-specific interactions. We explain the general issues that will be critical to the success of any type of next-generation mechanical biosensor, such as the need to improve intrinsic device performance, fabrication reproducibility and system integration. We also discuss the need for a greater understanding of analyte–sensor interactions on the nanoscale and of stochastic processes in the sensing environment.
Advances in micro- and nanofabrication technologies are enabling a wide range of new technologies, including the development of mechanical devices with nanosized moving parts. The ability to fabricate such structures using standard wafer-scale semiconductor processing techniques has allowed attention to move from fundamental problems in biological physics and bioengineering towards the development of practical micro- and nanoelectromechanical biosensors that can be produced en masse.
In general, mechanical biosensors capitalize on attributes that scale advantageously as physical size is reduced. First, nanoscale mechanical sensors provide exquisite mass resolution — the minimum detectable added mass is proportional to the total mass of the device. Nanoelectromechanical systems (NEMS) have achieved zeptogramscale mass resolution while operating in vacuum, and nanogram resolution while operating in a fluid environment1.
Second, the mechanical compliance of a device — its ability to be displaced or deformed — greatly increases with uniform reduction of its dimensions. Mechanical compliance converts an applied force into a measurable displacement (and is the mechanical analogue of gain in electronic circuits). This enhanced force responsivity opens new opportunities for measuring the miniscule forces that govern biological interactions. For example, nanomechanical sensors can resolve forces of ∼10 pN, which is sensitive enough to detect the rupturing of individual hydrogen bonds.
Third, small fluidic mechanical devices can exhibit fast response times. This allows biological processes in fluids to be observed on the timescales of milliseconds or shorter over which stochastic molecular interactions begin to evolve.
Mechanical biosensors can generally be delineated into four broad categories based on the chemical interactions between the sensor and the analyte: (1) affinity-based assays where highly selective target identification and capture is achieved by the employing high specificity (that is, affinity) between the target and the ‘functionalization’ at the device surface. Highly specific interactions can exist, for example, between antigens and antibodies; (2) fingerprint assays that rely on a multiplicity of less-selective functionalization layers to identify a target through characteristic binding affinities to an ensemble of sensors; (3) separation-based assays where chemical affinities between immobilized species and flowing analytes permit spatiotemporal separation of analytes; and (4) spectrometric assays where, for example, the mass or optical properties of the target are deduced to enable its identification.
An outstanding challenge in biosensing is to engineer suites of reliable, high-affinity biochemical agents to capture the target biomarkers we are interested in detecting. High affinity binding2 is based on biological molecular recognition, which generally occurs only in liquid phase. After capture, target detection is ideally performed in situ, within the fluid1,3,4. However alternative approaches include removing the detector from the fluid (after the targets are captured), and desiccating it before measurement5. Detection in situ is obviously simpler and immediate, but mechanical sensing in fluid is strongly affected by viscous damping. As described below, this significantly reduces the mass resolution compared with that obtained in gas or vacuum.
Two widely used (non-mechanical) biodetection technologies are lateral flow assays (LFAs) and enzyme-linked immunosorbent assays (ELISAs). LFAs (which are routinely used for urine analysis) provide quick analysis times (∼minutes), ease of use and low cost. However, their concentration sensitivity (that is, the lowest concentration at which target detection is possible) is only ∼0.1 μM, which is not good enough to detect many targets of biological importance. By comparison, ELISA requires a much longer analysis time (∼1 hr), but it offers much better concentration sensitivity (∼1 pM).
Achieving optimal performance for both metrics — an analysis time of less than one minute, and a concentration sensitivity (also known as limit of detection) on the picomolar level or better — is a critical challenge for any new biosensor. Equally important for real applications are practical considerations: can the new technology be mass produced? Can it be integrated with other system components? Can the design of the overall system be kept simple?
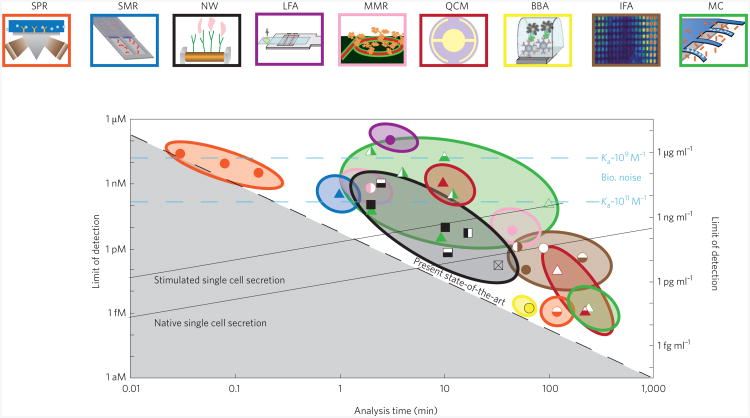
In addition to the four chemistry-based categories outlined above, mechanical biosensors can be subgrouped according to the physical processes that underpin their operation. These are described in the next section. Figure 1 and Table 1 summarize the analysis times and sensitivities of the various existing and emerging biosensing technologies discussed in this Review.
Figure 1. Fluidic detection limits for protein sensing.
The limit of detection in moles (left axis) and grams per millilitre (right axis) versus the analysis time for the different types of biosensor (both mechanical and non-mechanical) shown in the panels at the top of the figure and listed in Table 1. Note that both axes are logarithmic. The black dashed line shows the present state-of-the-art (with longer analysis times leading to lower limits of detection); the ideal biosensor would offer low limits of detection and short analysis time (that is, it would be found in the bottom left region of this graph). For many biomarkers the diagnostic level of significance is within the picomolar to nanomolar range, which can be accessed by conventional immunofluorescence assays (IFAs): the challenge for new biosensors is to achieve this sensitivity while also achieving shorter analysis times than the IFA approach. However, detector performance is frequently limited by non-specific binding effects rather than the intrinsic biosensor performance (see text). Non-specific binding effects lead to a ‘biological noise floor’ below which the analyte of interest cannot be detected. The figure shows the biological noise floors (horizontal blue lines) for target–receptor affinities of 1 nM−1 and 100 nM−1 and a non-specific binding association rate of 104 M−1; this noise floor is less of a problem when the target–receptor affinity is high. Such limitations do not apply to sandwich-type assays (see text). Many microfluidic sensors are now approaching the level of sensitivity that will permit real-time measurements on proteins secreted from individual cells. The figure shows the biosensor performance (solid black sloping lines) needed to detect the secretion of TNF-α from a single human monomyelocytic cell in a 1-nl volume121 for both native single cell (SC) secretion and stimulated SC secretion (in which the rate of secretion is increased by a factor of ∼80); a mass of 34 kDa was used to relate concentration to density. SPR: surface-plasmon resonance; SMR: suspended microchannel resonantor; fNW: nanowire; LFA: lateral flow assay129; MRR: microring resonantor; QCM: quartz crystal microbalance; BBA: biobarcode amplification assay; IFA: immunofluorescent assay; MC: microcantilever. Panels at top of figure reproduced with permission from: SMR, ref. 37, © 2007 NPG; NW, ref. 128, © 2005 NPG; MRR, ref. 130 © 2009 ACS; IFA, ref. 61, © 2004 RSC.
Table 1. Comparison of the analysis time and limit of detection for different types of biosensors.
| Category | Symbol in Fig. 1 Description | Detection conditions Reference | Analysis time Limit of detection | |||
|---|---|---|---|---|---|---|
| Optical detection Label-free real-time detection | ||||||
| MRR: microring resonator |
|
Label-free detection with a microring resonator | 5 protein mixture in BSA-PBS (0.1 mg ml-1 BSA) | 51 | 2 min | 0.6 nM |
| SPR: surface-plasmon resonance |
|
Label-free SPR detection | 0.1 mg ml-1 BSA | 58 | 10 s | 3 nM |
| End point and/or labelled detection | ||||||
| LFA: lateral flow assay |
|
Pregnancy test | Urine | 129 | 3 min | 10 µM |
| IFA: immunofluorescent assay |
|
ELISA | Serum | 60 min | 0.1 pM | |
|
|
Integrated blood barcode chip (IBBC) with DEAL | Whole blood | 62 | 90 min | 1 pM | |
|
|
Microfluidic fluorescent immunoassay | Cell-culture supernatant | 61 | 45 min | 1 pM | |
|
|
Bead-based microfluidic immunoassay with zM sensitivity | 4 protein mixture in PBS with 1% BSA | 63 | 210 min | 0.4 pM | |
| MRR: microring resonator |
|
Labelled detection with a microring resonator | IL-2 in BSA-PBS | 52 | 45 min | 6.5 pM |
|
|
Protein amplification via functionalized nanoparticles | Serum | 72 | 45 min | 0.5 fM | |
| SPR: surface plasmon resonance |
|
Labelled detection with SPR (DNA detection) | TNE | 60 | 2 h | 1.4 fM |
| Mechanical detection Label-free real-time detection | ||||||
| MC: microcantilevers |
|
Static mode (surface-stress sensors, SSS), functionalized reference | HBST buffer | 4 | 10 min | 15 nM |
|
|
SSS, unfunctionalized reference, piezoresistive detection | 0.1 mg ml-1 BSA | 14 | 12 min | 300 pM | |
|
|
SSS, no reference cantilever | 1 mg ml-1 HSA | 7 | 100 min | 100 pM | |
|
|
Dynamic mode detection (mass sensing) | PBS | 30 | 12 min | 0.3 pM | |
| SMR: suspended microchannel resonator |
|
Protein detection in serum | Serum | 3 | 1 min | 300 pM |
| QCM: quartz crystal monitor |
|
Detection of C-reactive protein | 0.1 M sodium phosphate buff | er 46 | 10 min | 1 nM |
| End point and/or labelled detection | ||||||
| MC: microcantilevers |
|
Mass sensing with liquid phase capture and vapour phase detection | Serum | 5 | 4 h | 1.5 fM |
| QCM: quartz crystal monitor |
|
Mass sensing with liquid phase capture and vapour phase detection | Serum | 47 | 180 min | 85 fM |
|
|
DNA detection using a sandwich assay with mass amplification | 0.4 M phosphate buffer | 48 | 220 min | 1 fM | |
| Electrical detection Label-free real-time detection | ||||||
| NW: nanowire |
|
Nanowire FET for DNA detection | Buffer | 55 | 10 min | 10 pM |
|
|
Nanowire FET for detection of PSA, time domain | Buffer | 54 | 17 min | 5 pM | |
|
|
Nanowire FET for detection of PSA, frequency domain | Buffer | 54 | 33 min | 0.15 pM | |
| End point and/or labelled detection | ||||||
| NW: nanowire |
|
Nanoribbon FET | Whole blood | 70 | 11 min | 0.6 pM |
The symbols in the second column correspond to the data points shown in Fig. 1. The analysis time (column six) is the total analysis time including incubation steps.
Different types of mechanical biosensor
The central element in many mechanical biosensors is a small cantilever that is sensitive to the biomolecule of interest: such devices can either be surface-stress sensors or dynamic-mode sensors. We will also discuss quartz crystal microbalances and some non-mechanical biosensors.
Surface-stress mechanical biosensors
These devices measure the quasistatic defection of a miniature mechanical device, usually a cantilever, caused by biomolecules binding to functional groups on the surface of the device (Fig. 2a). As the biomolecules bind, surface stress is developed — owing to electrostatic repulsion or attraction, steric interactions, hydration and entropic effects — and this can induce defection of the mechanical element. Reference 6 contains a detailed analysis of the relationship of surface stress to surface free energy. Binding of protein4,, DNA7–9 and mRNA10 have been studied, as have drug interactions11 and conformational changes of proteins12 and DNA13. The amount of defection is usually measured by refecting a laser beam of the cantilever, but electrical (piezoresistive) read out has been employed to measure binding of proteins14 and DNA15. Individual microcantilevers are susceptible to parasitic factors that accompany their exposure to a sample aliquot; spurious defections from proximal changes to index of refraction, temperature and fluidic disturbances can result. These can be partially circumvented by differential measurements4 that enable in situ comparison between the induced strain on cantilevers that have been functionalized and those that have been passivated. Reported sensitivities range from ∼100 pM (ref. 7) to the few nanomolar range4 (Fig. 1, Table 1).
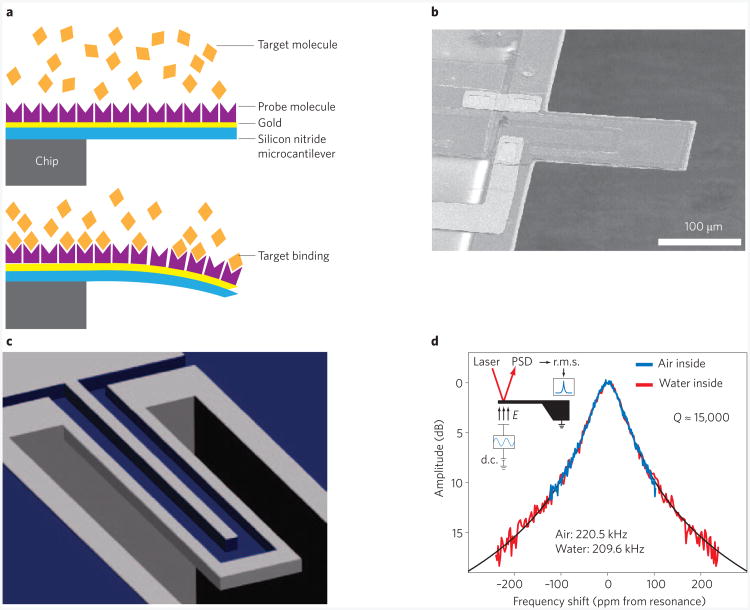
Figure 2. Fluidic micromechanical biosensors.
a, Schematic of static-mode surface-stress sensing MEMS device. Binding of target molecules generates a surface stress, which leads to a quasistatic defection of the cantilever (bottom)7. b, Scanning electron micrograph (SEM) of a dynamic mode MEMS device. Target molecules are detected through their influence on the resonance frequency of the cantilever: when the molecules land on the cantilever, they increase its mass and therefore reduce its resonance frequency25. c, Suspended microchannel resonator (SMR). The fluid containing the target molecules flows through a channel inside the device (the top of the device is not shown in this cutaway schematic) and bind to the inner flow-channel walls, while the resonator oscillates in air or vacuum3. d, Resonance spectrum (oscillation amplitude versus frequency) of a SMR. The quality factor of the device is normally unaffected when the channel is filled with water (red line)37. Figure reproduced with permission from: a, ref. 7, © 2001 NPG; b, ref. 30, © 2004 RSC; c, ref. 3, © 2010 ACS; d, ref. 37, © 2007 NPG.
Stress within self-assembled monolayers can be deduced from surface-stress sensor measurements through Stoney' formula16 for devices with large aspect ratio (that is, length:thickness > 10). For devices with smaller aspect ratio, the more detailed analysis of Sader17 must be used.
Dynamic-mode mechanical biosensors
These devices are not quasi static: rather, they oscillate with a resonance frequency, and this frequency changes when molecules land on the cantilever (Fig. 2b). Below we describe different operating environments and different modes of operation for such sensors
Humid environments
Real-time monitoring of very small-scale bacterial colonies has been achieved by growing them directly on micromechanical mass sensors. This involves maintaining the devices in a humid, gas-phase environment, but obviates the need for their direct immersion in fluid. Monitoring growth of E. coli microcultures in less than one hour18,19 has been demonstrated, which compares favourably with ∼ one day times for conventional methods. Moreover, the detection of antibiotic selective growth has been made in less than two hours19. This approach offers potential for simultaneous multiplexed detection of various bacterial species through device arrays.
Fluid-phase capture and detection in vacuo
Mechanical biosensors can provide exquisite mass resolution in vacuum and in air20,21. An approach that harnesses this level of performance for fluidic biosensing involves operating the device in solution, removing it from solution once the analytes have bound, then desiccating them before mass detection. However, spurious molecules can bind to the device during desiccation, which leads to errors (and continuous monitoring is not possible — see below). Early efforts in this area focused on the detection of relatively massive virus particles22,23, and single-virion resolution was achieved22. More recently, a ‘sandwich assay’ was used to detect prostate-specific antigen (PSA) in serum at femtomolar concentrations5. Sandwich assays employ two affinity-based probes (often two different antibodies) to achieve an effective affinity that is the product of the affinities of the individual agents. A label is often attached to the second probe to enable the readout (for example, fluorescent assays) or to enhance the signal (for example, labelling with relatively massive nanoparticles for mass-based detection5).
Continuous operation
The method described in the previous section is not capable of continuous monitoring and fast detection. However, if a dynamic-mode mechanical biosensor is immersed in the fluid, continuous monitoring with picomolar sensitivity and response times of a few minutes becomes possible. However the concentration sensitivity attained depends on the target mass; subpicomolar detection of T5 virions (molecular weight = 7 × 107 Da) is possible1, whereas micromolar sensitivity is typical for smaller peptides such as ferrichrome (molecular weight = 687.7 Da). Dynamic-mode mass sensing has also been used for mass measurements of individual live cells24,25.
Sensitive frequency-shift-based mass detection requires resonators with high vibrational quality factors, but the quality factor, Q, is compromised in fluid by viscous damping26. However, high- frequency operation increases the effective Reynolds number and can enable operation with higher values of Q27. These higher frequencies can be achieved by reducing the device dimensions, or by operating with high-order vibrational modes28,29. Figure 1 shows the present state-of-the-art performance.
Several groups report discrepancies between adsorbed mass and induced frequency shift, both for liquid-phase30,31 and gas-phase measurements32. This has commonly been attributed to surface stress induced by the adsorbates, yet theoretical estimates of expected magnitudes are far smaller than what is observed. This is especially true for the case of microcantilever sensors, where simple extension or contraction can relieve surface stress33. ‘Strain-dependent’ surface-stress models have been proposed33 but their validity is questioned34. It has been shown34 that clamping effects can significantly affect surface stresses developed in short cantilevers but, again, the expected stress-induced frequency shifts are smaller than experimental observations.
Adsorbed analytes can also potentially induce a frequency shift by changing the composite elasticity of the sensor. It has been proposed that such changes might dominate the frequency shift induced by mass loading, even when the layer of adsorbed species is much thinner than the device35, and both experimental and theoretical evidence have been reported for such a stiffening effect from thin antibody layers on 30-nm-thick microcantilevers36.
Suspended microchannel resonators
An ingenious alternative to immersing dynamic mass sensors in fluid is to constrain the fluid to channels embedded in the mechanical resonator itself37. Such suspended microchannel resonators (SMRs) can be measured in vacuo where values of Q of up to ∼15,000 can be obtained (Fig. 2c,d). Measurements of fluidic dissipation in SMR devices suggest that some Q degradation may occur for fluid-filled nanochannels38,39, but these results are not fully understood theoretically40,41. So far, despite the high values of Q attained, the performance of SMR biosensors is modest. The glycoprotein ALCAM has been detected in undiluted serum at 300 pM concentrations in several minutes3. In this case the detection limit was set by non-specific binding. (The intrinsic performance should be about two orders of magnitude better than this.) SMRs have been applied to measurements of cell mass and density during the cell cycle of yeast42,43 and have been used to measure growth rates from single cells both for bacterial and mammalian cells43. They have also been used for detection of antibiotic resistance44. Elsewhere we have carefully analysed the ultimate practical limits to SMR biosensing45.
Other mechanical biosensors
There is one other widely used mechanical biosensor — the quartz crystal microbalance — and also a variety of non-mechanical biosensors, including whispering-gallery microcavity resonators, optical microring resonators and nanowire biosensors.
Quartz crystal microbalances (QCM)
These are centimetre-scale mechanical resonators that can measure the inertial mass of analytes accreting on their surfaces in vacuum, gas or fluid. A downshift in the resonant frequency occurs with target accretion, which is most reliably tracked electronically, in real time. Fluid-based QCM biodetection spans the nano- to femtomolar range: nanomolar sensitivity is reported for continuous analyte monitoring using an indirect-competitive assay46, whereas ∼100 fM sensitivity has been reported for end-point detection assays involving device removal from fluid, post-capture drying, and subsequent measurement in vacuo. Femtomolar sensitivity is reported by a technique combining this end-point vacuum detection approach with a sandwich assay providing immuno-specific target-mass enhancement47,48. However, as mentioned, the need to remove samples from fluid and desiccate them before measurements in vacuo makes these assays cumbersome and susceptible to measurement artifacts.
Whispering-gallery microcavity (WGM)
Consists of a high-finesse toroidal optical resonator coupled evanescently to an optical fibre. Adsorption of analytes to the surface of the resonator measurably alters its properties. First efforts reported unprecedented ∼100 aM sensitivity with response times ∼1 s (ref. 49), but these results caused significant controversy. The data have not been reproduced, and sub sequent analyses suggest they are incommensurate with expected resonance shifts50 and binding kinetics. Recently, some of the authors of ref. 49 have reported follow-up studies showing reproducible, albeit more conservative, results: detection of the relatively large influenza-A virion at picomolar concentrations within ∼10 s (T. Lu et al., manuscript in preparation).
Optical microring resonators (MRRs)
These devices are similar to WGM devices, but offer the advantage that they can be fabricated by standard methods and, thus, are more readily integrated into multiplexed detection systems. However, this advantage comes at the price of lower optical quality factors and, hence, reduced sensitivity; label-free MRR-based detection is reported in the nanomolar range with response times ∼1 min. MRR biosensors have enabled quantification of unknowns from a mixture of five proteins51, and sandwich-assay detection yielding ∼6.5 pM sensitivity.52
Nanowire biosensors
The conductance of these devices — which are made from semiconductor nanowires and carbon nanotubes — changes when a target molecule binds to the surface of the device. This ‘electrochemical gating’ arises from a change in local surface potential induced by target binding or changes in solution pH. Even for nominally similar systems, the concentration sensitivities reported for nanowire biosensors span a large range of values, from the femtomolar53 to the few-picomolar scale54,55. An initial sensitivity of 5 pM can be improved to 0.15 pM through the use of frequency-domain detection54, and optimization by subthreshold biasing can improve this further, to 1.5 fM (ref. 56). Some reported results are not consistent with recent estimates of binding kinetics57 — given the minuscule surface areas available on the surface of a nanowire for binding, estimates suggest that at fM concentrations there should be only one capture event every few days!
Nanomolar sensitivity has also been reported for label-free protein biosensing with surface-plasmon resonance (SPR) biosensors58 and photonic-bandgap (PBG) biosensors59. This level of performance can be improved to the femtomolar range by using sandwich-assay end-point detection (in which label probes incorporating gold nano-particles that enhance SPR are employed60). Optical fluorescence detection methods are routinely used to achieve picomolar sensitivity, but they typically require incubation times on the order of hours61–63.
Sensitivity versus other performance metrics
The detection of rare biomarkers in blood plasma is an archetypal goal for advanced biosensors. For many biomedical targets of interest, existing sensors are capable of reaching the diagnostic level of significance: this is ∼4 ng ml−1 (120 pM) for PSA. A number of other cancer biomarkers have similar thresholds, that is, they are within the sensitivity range of other technologies (Fig. 1). However, there is a clear need for biosensors that can simultaneously detect a number of different biomarkers (that is, fingerprint assays). The case of prostate cancer illustrates some of these challenges: recent studies haves shown that 70% of males with PSA levels at or below the current diagnostic level of significance do not develop this form of cancer64, so there is a need for better diagnostics.
For biosensor applications, it is necessary to focus on both the intrinsic device performance and on the performance of the overall sensor system. Important considerations include: ease of biofunction-alization and potential for multiplexing; complexity of fabrication and integration; device robustness and shelf life; the trade-off between sensitivity and frequency of false positives65; and the readiness and adaptability for production en masse.
Non-specific binding and the biological noise floor
It is not often appreciated that biosensing is more complex than simply finding ‘a needle in a haystack’ because of the problem of non-specific binding. Other species are present at much higher concentrations than the target biomolecule (perhaps at concentrations a billion times greater than the target), and these species can also bind to the sensor, which results in most of the sensor interactions being non-specific. Even if the residence times associated with these non-specific binding events are much shorter than those for specific binding events, non-specific interactions impose a fundamental biological noise ‘floor’ to achievable limits of detection.
We illustrate this problem with a simple hypothetical example. Non-specific interactions can take place at functionalized, passivated and untreated regions of a device; and all can have a role in limiting detection sensitivity. Representative rates of protein association for non-specific binding66,67, typically fall within the range of 104 – 105 M−1, whereas generic target/receptor interactions, such as TNF and TNFR1, have binding affinities of ∼1011 M−1. Albumin, the most prevalent protein in blood plasma, is present at concentration cprev ∼600 μM. We assume the number of specific and non-specific binding sites of the sensor — represented as bS and bNS, respectively — are comparable, and define the limit of detection as yielding a 3:1 signal-to-background ratio. For the example of TNF in plasma, these considerations result in a background biological noise floor, that is equivalent to ∼1.8 nM target concentration. Many other targets of interest have much weaker binding affinities and will correspond to higher biological noise levels.
Measurements of ALCAM in serum with suspended microchannel resonators demonstrate that non-specific binding can be central in determining ultimate detection limits3. In these measurements, the practical detection limit (defined as the standard deviation of the response to negative controls) was roughly 200 times worse than expected from the mass resolution of the device. Although non-specific binding is not the only factor that determines this detection limit (the measurements are performed over a period of approximately 20 min, so sensor drift might also have a role), these measurements demonstrate that state-of-the-art technologies have already reached a level where the detection limit is determined not by the intrinsic device sensitivity but by other factors. Understanding and controlling non-specific binding is likely to be key to further gains in sensitivity.
Despite the importance of these considerations, little systematic experimental investigation has been undertaken to quantify biological noise arising from non-specific interactions in practical situations. Nair and Alam have modelled physisorption onto unpassivated regions of devices, assuming the rate constants between non-specific and specific binding differ by a factor of 109. Even though this ratio is somewhat arbitrary, their conclusions underscore the importance of dense biofunctionalization surface coverage to achieving high selec-tivity68. Their model indicates that target discrimination remains possible with high coverage of specific receptors (∼2 × 1012 cm−2), even when other species that we are not interested in are 109 times more abundant in solution. They suggest discrimination can be enhanced further by back-filling ‘voids’ in functionalization with, for example, PEG or other biopassivation species. Finally, it has also been shown that differential sufficiently to circumvent false positives arising from biological noise10.
Practical signal enhancement
The limitations imposed by non-specific binding can be overcome, at least in part, at the cost of more complex procedures such as the use of sandwich assays52,69 to increase target capture specificity. Variations on the traditional sandwich assays can also be used, such as the two-step process used to detect PSA at ∼60 pM (2ng mlminus;1) in whole blood within ∼20 min using a nanoribbon sensor70.
Another approach is to amplify the target analyte so that its concentration rises above the biological noise floor. The widely used polymerase chain reaction (PCR) exponentially amplifies initial tar get species and has enabled measurements of DNA from individual cells in volumes > 100 μl (ref. 71). At present, protein assays do not achieve such profound species amplification, but enhancement methods have been developed that provide some level of signal amplification. The ELISA assay, perhaps the archetypal example, employs an enzyme bound to a detection antibody. Each enzyme molecule acts as a signal amplifier, typically producing thousands of signal molecules per second. Although the ELISA process provides only a linear (rather than exponential) increase in the signal with time, it can still achieve subpicomolar detection sensitivities (Fig. 1).
Labelling provides another form of signal amplification. A label can serve two purposes: to enhance detection specificity through sandwich-assay mechanisms, and to directly amplify the detected signal. For example, SPR sensors achieve nanomolar-concentration sensitivity in their basic, label-free form (Fig. 1, Table 1). However, substantial enhancement of the induced plasmonic signal, reportedly to enable femtomolar sensitivity, is possible through immunospecific attachment of gold nanoparticles to the target in a final labelling step (although this approach also involved a two-hour incubation period)60. Labelling enhancement is possible with optical (MRR) biosensors; 0.6-nM label-free detection within several minutes is typical51, and labelled detection with 6.5-pM sensitivity, which enables detection of smaller proteins such as cytokines, has been reported (albeit with a 45-min incubation period)52.
Labelling can also enhance the signals detected by fluidic mechanical biosensors. Gold nanoparticle labels have been used to seed additional gold precipitation, sufficiently enhancing their QCM mass signal to enable femtomolar detection of DNA48.
‘Biobarcode’ (BBA) sensors combine both amplification and nanoparticle labelling and have achieved record sensitivities of ∼500 aM (Fig. 1)72.
In general, it is complex to scale labelling and non-PCR amplification methods to highly multiplexed assays. Also, labelling and sandwich assays are inherently one-shot detection techniques — they are not readily adaptable to continuous, real-time monitoring. Furthermore, the most selective sandwich-type assays are predicated on the availability of two high-affinity capture agents, for example, antibodies. In this context, it is noteworthy that obtaining robust and effective capture agents is often a limiting factor in immunoassay development2.
Diffusion, convection, reaction kinetics and response time
Capture kinetics have a critical and underappreciated role in determining the overall sensor system performance. For most applications, a very fast flow rate is optimal for microfluidic devices — although this leads to a reduced percentage of captured target molecules, it increases the actual number of captured molecules per unit time. Although this might seem wasteful, the small volumes of microfluidic devices and their tiny maximal flow rates result in the use of very small sample aliquots — often in the range of microlitres, or less.
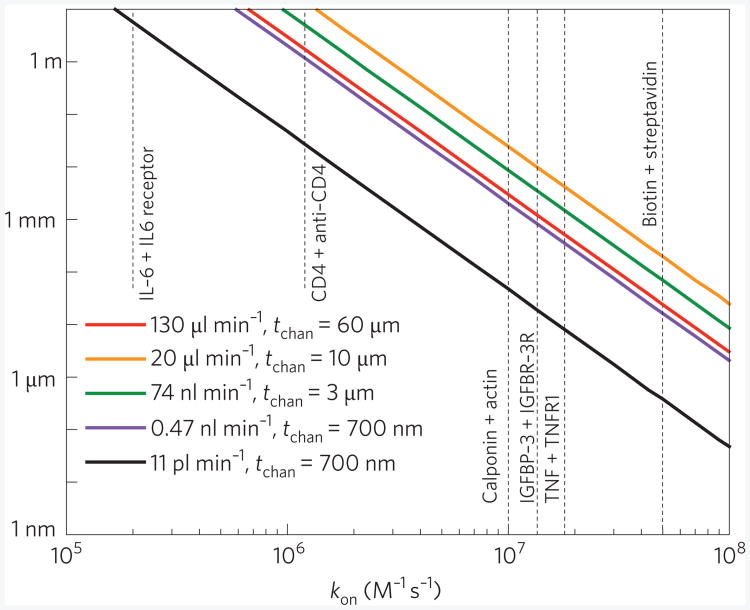
To illustrate these considerations we summarize the kinetics of analyte capture in a microfluidic channel. (See Box 1 and ref. 57). We define a critical length, , where D is diffusion rate, QV is flow rate, bm is the number of receptor binding sites, hchan is channel height, kon is the rate of association and wchan is channel width, over which analytes, owing to binding, become depleted near the functionalized surface to 50% of their initial (bulk) concentration. For sensors shorter than L*, such depletion and, hence, mass transport itself, can safely be ignored. Conversely, for sensors significantly longer than L*, depletion plays an increasingly important role. Figure 3 shows the strong dependence L* has on kon for a variety of microfluidic device geometries. For typical biological binding affinities — for example, kon ∼107 M−1 s−1, characteristic of TNF binding to TNF-R173 — L* ranges from micrometres to tens of millimetres depending on the flow geometry.
Box 1. Kinetics of microfluidic systems.
In microfluidic systems, it is essential to determine whether ana-lytes are depleted near the active surfaces of sensors. Following Squires, Messinger and Manalis57 we employ the Damkohler number, Da = (c0 - <cs>)/<cs>, to characterize the importance of depletion at the surface, where c0 and <cs> are the bulk and aver age concentration at the device surface. For Da << 1, the kinetics of capture are entirely governed by reaction kinetics; for Da >> 1, the kinetics become mass-transport limited. The Damkohler number can also be expressed as Da = konbmAsensor/(J/c0), where kon is the rate of association, bm the surface concentration of receptors on the sensor, Asensor the surface area of the sensor, and J the flux of target molecules reaching the device through mass transport. This flux is given by57:
where D is the diffusion rate, ws is the sensor width, PeH∼ QV/Dwchan is the Peclet number with respect to the thick ness of the microfluidic channel, hchan, PeS∼ 6(lsensor/hchan)2PeH is the Peclet number with respect to the width of the channel, wchan,QV is the volumetric flow rate, and lsensor is the sensor length in the direction of fluid flow. See ref. 57 for a more detailed discussion.
Figure 3. Depletion in microfluidic structures.
The length needed for 50% depletion L* versus the rate of association kon in an open-loop fluidic configuration for five different combinations of flow rate and microchannel geometry: details of four of these combinations are shown in Table 2; for the fifth combination (black line) t = 700 nm, w = 4 μm and l = 2.05 cm. The dotted vertical lines show the values of kon for the six target–receptor pairs listed in Table 2. Significant depletion can be achieved for lengths of hundreds of nanometres for very small channels (in which the flow rate is reduced) for the highest values of kon (such as for biotin–streptavidin binding), but tens of micrometres or more are needed to achieve Significant depletion for larger channels (with much greater flow rates), even for the highest values of kon. For much lower values of kon (such as IL-6 binding to its receptor) it is not possible to achieve Significant depletion within practical length scales for microfluidic sensors, implying that the kinetics are always reaction limited. Depletion length scales shown here are for short timescales, that is, far from equilibrium. Near equilibrium the kinetics are always dominated by reaction kinetics (see Table 2). Depletion is strongly dependent on the flux of molecules to the surface, which depends on both the flow rate and the channel geometry; here depletion has the greatest role for the combination shown by the black line.
These expressions also allow us to estimate the time required to reach steady-state, τss. For the geometries and targets of Fig. 3, τSS can range from seconds (for interactions with the lowest affinities) to hours.
Concentration sensitivity versus absolute sensitivity
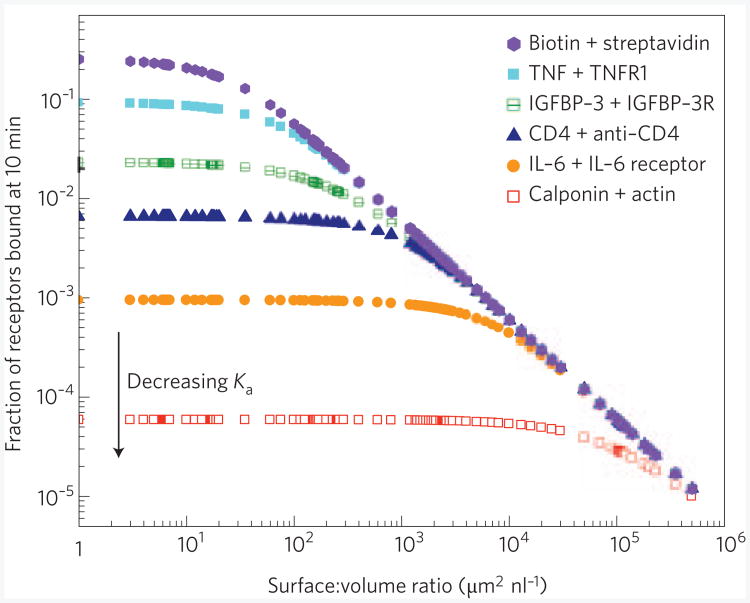
For very small sample volumes, one may also need to consider depletion in the bulk solution. Table 2 summarizes the smallest volume at which the bulk concentration remains within 90% of the initial value at steady state. As microfluidic sample volumes are generally ∼μl, bulk depletion is often negligible. However, recent work on microfluidic single cell analysis exemplifies an important situation where depletion becomes relevant63: a sensitivity of ∼2 zeptomoles (10−20 moles or ∼1,000 copies) has been achieved by confining individual cells in a 5-nl chamber in which a bead-based immunofluorescence assay (IFA) was implemented. These are very small volumes compared with typical ∼μl-scale microfluidic assays. For reaction-limited systems (see Box 1), the time-dependent capture profile can be described by the expression, Here b(t) is the number of target molecules bound to the surface at time, t, V is the total (limited) volume of sample, and NA is Avogadro' number. For detection at very low concentrations, we ignore terms of order . Figure 4 shows the fraction of receptors bound after 10 min in such an experiment for a range of target molecules and capture areas.
Table 2. Timescales for reaching biochemical steady state.
| Analyte(s) | kon (M−1 s−1) | Ka (M−1) | QV=130μlmin−1 tchan=60μm wchan=100μm |
QV=20μlmin−1 tchan = 10μm wchan =70μm |
QV = 74 nl min−1 tchan = 3μm wchan = 8μm |
QV = 47nl min−1 tchan =700nm wchan =4μm |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||||
| Equil. time (min) | Volume (10% bound) | Volume (10% bound) | Volume (10% bound) | Volum (10% bound) | |||||||
| Lsensor | 10 μm | 200 μm | 10 μm | 200 μm | 10 μm | 200 μm | 10 μm | 200 μm | |||
| Analyte | |||||||||||
| Calponin + actin | 1 × 107 | 6 × 107 | 0.1 | 5 pl | 98 pl | 3 pl | 68 pl | 0.4 pl | 7.8 pl | 0.2 pl | 3.9 pl |
| IL-6 + IL-6R | 2 × 105 | 2.4 × 108 | 20 | 0.2 nl | 4 nl | 0.1 nl | 2.8 nl | 16 pl | 0.3 nl | 8 pl | 0.2 nl |
| IGFBP-3 + IGFBP-3R | 1.4 × 107 | 2.6 × 109 | 3 | 2 nl | 40 nl | 1 nl | 28 nl | 0.2 nl | 3.2 nl | 80 pl | 1.6 nl |
| CD4 + anti-CD4R | 1.2 × 106 | 4.1 × 109 | 55 | 3 nl | 66 nl | 2 nl | 47 nl | 0.3 nl | 5.3 nl | 0.1 nl | 2.7 nl |
| TNF + TNFR1 | 1.8 × 107 | 5.3 × 1010 | 32 | 29 nl | 0.6 μl | 20 nl | 0.4 μl | 2 nl | 46 nl | 1 nl | 23 nl |
| Biotin + streptavidin | 5 × 107 | 2.9 × 1013 | 33 | 83 nl | 2 μl | 58 nl | 1.2 μl | 7 nl | 0.1 μl | 3 nl | 66 nl |
An important goal for many microfluidic-embedded sensors is achieving ‘fast detection’. Here we provide estimates of the equilibration time, τeq, for six analytes (in order of increasing affinity Ka) for detection at a concentration of 10 pM for two sensor lengths (10 μm and 200 μm) and four different microfluidic geometries (in order of decreasing flow rate and channel cross-section). For all but the highest affinity analyte (biotin + streptavidin), all the devices are reaction limited at these flow rates and τeq ∼ Ka/kon(1 + c0Ka); in other words τeq does not depend on the sensor length or microfluidic geometry. For biotin + streptavidin, τeq increases from 33 min (length = 10 μm; flow = 130 μl min−1; channel cross-section = 60 μm × 100 μm) to 55 min (length = 200 μm; flow = 0.47 nl min−1; channel cross-section = 700 nm × 4 μm; τeq not shown in Table) as the system changes from being reaction limited to transport limited. We also provide examples of the sample volumes below which more than 10% of the analyte molecules are bound to the device in equilibrium and bulk depletion must be considered (see main text and Fig. 4). Above these volumes only surface depletion need be considered (Fig. 3), and analyte can be recirculated without degradation of performance. For all cases, we assume that the sensor width is half the channel width and that its thickness is neglible. The flow rates QV were chosen under the assumption that the channel has the dimensions listed above over a length of 500 μm, and that it is in series with a channel with lchan = 2 cm; tchan = 60 μm and wchan = 100 μm (included to represent the region of the microfluidic channel in which sample processing would occur). The channel is presumed to be pressurized to 5 psi.
Figure 4. Effect of surface-area: volume ratio on bulk target depletion.
The fraction of receptors bound at 10 min versus the surface-area: volume ratio for the six target–receptor pairs listed in Table 2 under reaction-limited conditions (Box 1): the affinity Ka of the pairs decreases from top to bottom. The fraction of bound receptors can be increased by reducing the surface-area: volume ratio. However, below a threshold (determined by Ka), there is no further gain.
These considerations illustrate that nanoscale, and even microscale, sensors cannot capture sufficient targets from solution to become depletion-limited for most applications (Fig. 3). In cases where the analysis volume is extremely minute (for example, for the single-cell analyses mentioned previously), depletion can play a role for surface-to-volume ratios on the order of 100 μm2 nl−1 or less (Fig. 4). Thus, for a sample volume ∼nl, Significant gains in surface density of target molecules (and hence limits of detection) can be realized by scaling the active sensor surface to an area of roughly 100 μm2. Except for the highest affinity targets, further gains cannot be realized with smaller capture cross-sections (Fig. 4). The volumes at which depletion begins to play a role are summarized in Table 2 for several sensor geometries.
Force and energy sensing
Mechanical devices can perform other types of sensing, especially molecular-force and energy-based sensing. The ability to access other modes of operation highlights the potential of new sensors to open different avenues of fundamental biological research, and to enable applications beyond the simple ‘on/off’ indication of target analyte capture.
Chemically functionalized atomic force microscopes have been employed to measure the force of intramolecular interactions74–78, arrays of polydimethylsiloxane (PDMS) posts have been used to measure forces exerted by cells79, and optical tweezers have been used to measure the elasticity of cells and have measured Significant (three fold) differences in deformability between cancerous and normal cells80,81. Measurement of forces, elasticity and displacement is ideally suited to the mechanical domain and, in particular, the unprecedented sensitivity of nanoelectromechanical systems (NEMS) devices. Many of these applications are just beginning to be explored — a recent example is the use of surface-stress sensors to measure conformational changes of proteins12 and DNA13. In the energy domain, micro-fluidic calorimeters with potential for resolving the metabolic output of individual cells are on the horizon82. Here we highlight several of these promising new areas of research.
Fluid-based force sensing
The atomic force microscope (AFM) is best know for probing various systems with atomic resolution in vacuum, but it can also image samples at atmospheric pressure and immersed in fluid. Measurement of the elastic properties of live cells has also been demonstrated83. As with mass sensitivity, improvements in force resolution are achieved by reducing the dimensions of the sensor (Fig. 5). Current microcantilevers have the sensitivity to resolve forces at the level of individual hydrogen bonds and to investigate biological molecules based on their force– extension profile as the molecule is stretched84,77,78 or ruptured74. Bond lifetime and dynamic force spectroscopy experiments have enabled measurements of bond formation and dissociation at the single-molecule level, yielding new insights to molecular behaviour, binding states and reaction pathways. In particular, unbinding force measurements have been used to study receptor–ligand dissociation rates, koff75,85. However, care must be taken in interpreting these rates, as the initial ‘bound state’ and hence the measured rupture force, is strongly dependent on its history86. With careful study, Significant information on the energy landscape for receptor–ligand bonds can be obtained, yielding good agreement between simulations87 and experiments76.
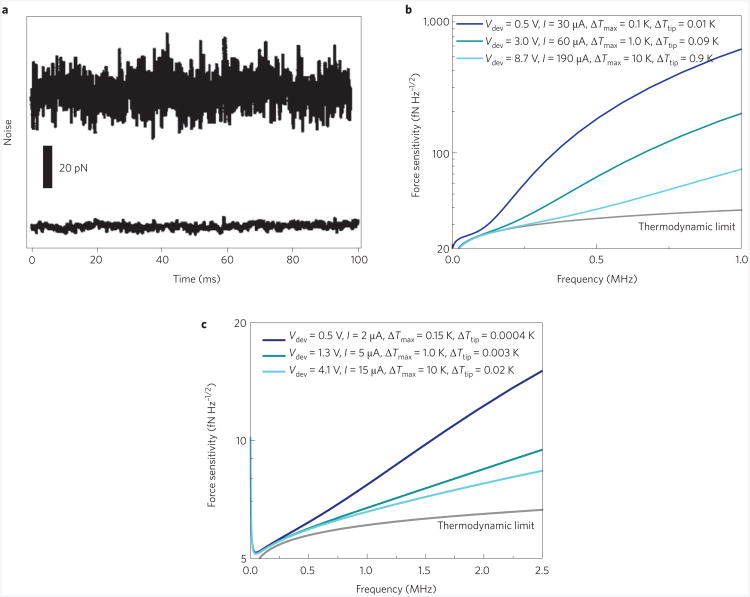
Figure 5. Fluidic nanomechanical biosensors.
Demonstration of reduction in force noise through the overall reduction of cantilever dimensions. a, Noise versus time for a large cantilever (length = 200 μm; spring constant k = 0.060 N m−1; top trace) and a small cantilever (length = 10 μm; k = 0.060 N m−1; bottom trace)131. b, Theoretical predictions for total force sensitivity (including thermomechanical Brownian noise, Johnson noise and typical readout amplifier noise) on a logarithmic scale versus frequency for a silicon piezoresistive cantilever immersed in water and operating at room temperature for three different sets of conditions; the thermodynamic limit (that is, just Brownian noise) is also shown for reference. The sensitivity depends on the maximum tolerable temperature rise both at the tip ΔTtip and the position of maximum heating ΔTmax. As the bias voltage Vdev and bias current I increase, both ΔTtip and ΔTmax also increase, and the sensitivity improves, approaching the thermodynamic limit. The cantilever device dimensions are: t = 130 nm, w = 2.5 μm, l = 15 μm.c, Analogous plot to b for a smaller cantilever132 showing qualitatively similar behaviour but substantially higher sensitivity (note that the scale on the y-axis is different): t = 30 nm, w = 100 nm, l = 3 μm. At frequencies below 1 MHz, the system approaches the thermodynamic limit, and the sensitivity remains within about 20% of the fluidic noise floor at the relatively low bias voltage of 0.5 V. Below 0.25 MHz, the total sensitivity is (for reasonable bias voltages). Figure reproduced with permission from: a, ref. 131, © 1999 AIP; b,c, ref. 132, © 2007 Springer
Of particular interest in this domain have been studies of cell adhesion and the interaction between mechanical stimuli and chemical circuitry in the cell88. Single-molecule atomic force microscopy techniques, in which the bonds are stretched but not ruptured, have allowed studies of the dynamic rearrangement of the active site of an enzyme during catalysis77, and have also been used to investigate protein78,84 and RNA89 folding. In single-cell force spectroscopy, a cell is attached to an AFM cantilever and brought into contact with a substrate at a predetermined contact force, kept stationary for a fixed time, and then pulled away from the substrate. Individual bond-breaking events can be resolved, enabling the investigation of adhesion forces — down to the level of individual receptor interactions. This has been used to investigate a wide variety of phenomena, from the properties of cell adhesion itself90, to force interaction in cancer91 and immune response92. Most recently, functionalized surfaces have been used to investigate receptor crosstalk93.
Fluid-based energy sensing devices
The inherently small heat capacities of suspended nanoscale devices make them ideal candidates for ultrasensitive calorimetry. Indeed, vacuum-based nanoscale devices have achieved a resolution of 0.5 aJ K−1 at 2 K (ref. 94). Scaling these chip calorimeters up to room temperature operation, and embed ding them in integrated microfluidics, offers the prospect of high-throughput measurements requiring very low sample consumption. In particular, a power sensitivity on the order of nanowatts, on sample volumes of a few nanolitres, has been achieved82. Next-generation improvements on the horizon suggest that sensitivities on the scale of picowatts are feasible; this will enable metabolic measurements at the level of individual cells.
Practical aspects of fluidic mechanical biosensors
A major challenge for all NEMS devices has been development of efficient actuation and transduction methods. Here we provide a brief overview of common techniques and describe recent advances (see ref. 95 for a comprehensive discussion).
Optical detection, a cornerstone of microelectromechanical devices such as AFM probes, becomes increasingly challenging to implement as the device dimensions scale below an optical wave length. Nevertheless, devices with widths as small as 50 nm have been measured optically through the use of optical interferometry. Measurements have been performed both on individual devices96,97 and on grating-based systems98. Recently, near-field, non-interferometric optical transduction has been identified as a promising alter native for arrays of nanocantilevers99. The latter holds Significant potential for co-integration with on-chip light sources, because non-interferometric techniques do not require a coherent light source. Evanescent coupling to the substrate of a propagating light field has also been used to drive NEMS100.
Electrostatic detection and actuation, used ubiquitously in integrated microelectromechanical systems (MEMS), generally lose efficiency for nanoscale devices. Capacitance scales as area/separation but practical limits on drive–gate gaps ultimately limit reduction of their dimensions. Given the higher frequencies of NEMS compared with MEMS, a large fraction of the electro statically based drive and detection signals are lost through parasitic capacitances. However, with an appropriate LC network for impedance transformation it is possible, on resonance, to couple electrostatically through a gate electrode to the device with reason able efficiency. This technique has been used to measure a NEMS array with closely spaced resonance frequencies (above 10 MHz) using a single RF circuit (Fig. 6a,b)101.
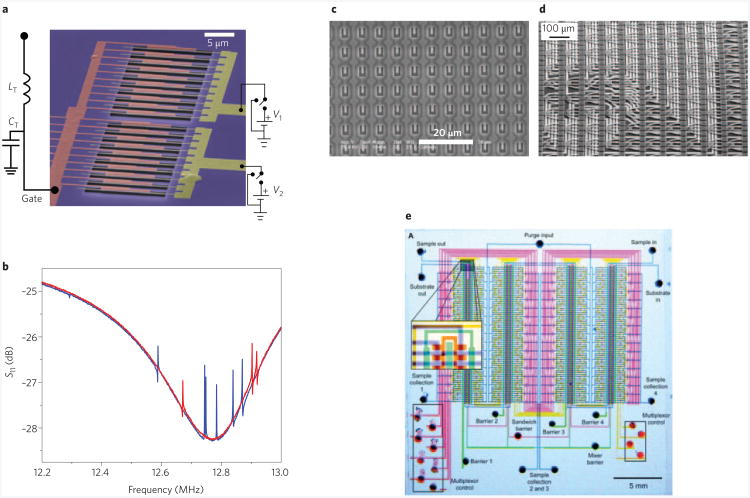
Figure 6. NEMS arrays and system integration.
a, False-colour SEM image of an array of 20 silicon nitride nanomechanical resonators (in two separately biased banks) with capacitive readout and actuation101; the resonant frequencies of the resonators are ∼12 MHz. b, Resonance spectrum (oscillation amplitude (S11) versus frequency) of the array in a. It is possible to read out the array with a single radiofrequency readout circuit101. Seven resonators in bank 1 (blue trace) and three resonators in bank 2 (red trace) were detected in this frequency range. c, Array of silicon cantilevers: each cantilever is 2.8-μm long and 0.7-μm wide, with the ‘legs’ being 200-nm wide. A piezoresistive approach was used for readout. Image courtesy of P. Andreucci (Minatec, Leti, CEA). d, SEM of a section of a 4,096 silicon cantilever array, transferred onto a wiring wafer. The transfer is done on the 100-mm wafer scale, with approximately 50 such arrays per wafer114. These cantilevers were designed for memory storage applications, with resistors at the base to induce (the write step) and measure (read step) the defection. e, Multiplexed microfluidics. PDMS microvalves enable independent compartmentalization, purging and pairwise mixing for each of the 256 chambers on the chip115. Figure reproduced with permission from: a,b, ref. 101, © 2007 ACS; d, ref. 114, © 2004 IEEE; e, ref. 115, © 2002 AAAS.
Termoelastic actuation has been demonstrated — through photo thermal heating in air102 and liquid103, and through integrated electrothermal heating in both air104 and liquid105. The thermally induced elastic strain, a measure of stored energy density, is generally constant as the device dimensions are uniformly scaled down. This makes thermoelastic actuation promising for increasingly smaller NEMS devices.
Piezoelectric actuation has also been used extensively for MEMS devices in both air106 and liquid30. Advances in the quality of piezoelectric ultra-thin film (< 20 nm) materials have recently enabled their application to NEMS107. An important benefit of pie zoelectric actuation is its exceptionally small power consumption, owing to the minimal current flow through the device — especially compared with that of thermoelastic actuation. As the piezo electric effect itself generates voltages when the NEMS vibrates, it can be used for detection as well as actuation. As with capacitive detection, however, direct signal transduction faces the challenge of relatively small-magnitude, high-frequency signals originating from high-source impedances, in the presence of substantial parasitic capacitances.
Piezoresistive detection technique is now widely used for room temperature NEMS applications. Devices with doped semicon ductor piezoresistive sensors have a long history in MEMS108–110. However, the use of these materials in nanoscale devices is challenging because the doped layer must remain thin compared with the total device thickness. Requisite structures at nanoscale dimensions also require exceptionally careful processing to avoid damage to the ultra-thin doped surface layer required. The displacement transducers that result suffer from high Johnson noise given their relatively high impedance, and from very high 1/f noise owing to their low carrier concentrations and small volumes111. Recent work shows these difficulties become exacerbated with semiconducting transducers as their size is scaled downwards, but can be overcome through the use of metallic piezoresistors112.
Future nanosystems for complex biosensing
Te ultimate mechanical biosensing systems will combine precise microfluidic sample handling, automated and complex pre paratory protocols, and highly sensitive nanomechanical sensing elements in multiplexed device arrays that can be readily mass-producible by microelectronic fabrication technologies. Although most results published so far describe measurements from one or, at most, a few biosensors, it has already been shown that thou sands of suspended cantilevers can be fabricated to ft on a chip measuring a few millimetres by a few millimetres (Fig. 6c). The outstanding challenges, therefore, are the difficulty of differentially functionalizing closely packed sensors (which is a front-end issue) and the complexity of multiplexing the electrical readout of a dense array of devices (which is a back-end issue).
Effort is at present focused on leveraging the existing infra structure for the very large-scale integration of silicon micro electronics (that is, complementary metal oxide semiconductor (CMOS) devices) to facilitate the very large-scale integration of NEMS. Routes being taken include development of a unified, monolithic NEMS–CMOS process113 and a multilayer, multichip, three-dimensional stacking or hybridization process for NEMS and CMOS114. The IBM Millipede project demonstrates that wafer-level transfer of MEMS devices to the surface of other wafers is both achievable and robust. This project has achieved device densities ∼100 cantilevers mm−2 and interconnect densities ∼300 mm-2. (Fig. 6d; ref. 114). Highly multiplexed microfluidics have also been demonstrated (Fig. 6e)115 and are leading to a growing range of applications116,117. The outstanding task is the integration of complex microfluidics and dense arrays of nanoscale biosensors.
In terms of performance for diagnostic applications, we have already discussed examples where mechanical biosensors have reached the stage where non-specific binding and other factors such as sensor drift — rather than the inherent device mass sensitivity — set the limit of detection. For applications involving the detection of rare biomarkers in serum, enhancing the limits of detection requires confronting the problem that the concentrations of the most abundant proteins in samples are many orders of magnitude higher than those of the least abundant targets. Pre-concentration and/or immunoaffinity depletion can help to some extent, but the ultimate efficacy of such approaches is compromised by the tendency for competing molecules to be concentrated along with the target of interest and/or for the target to be depleted along with the competing molecules.
Another problem is that small, low-abundance target proteins (such as cytokines) can be sequestered by proteins that are abundant in serum (such as albumin). Indeed it has been shown that sequestered biomarkers may exist at concentrations that are 10–500 times greater than that of their free counterparts118. Standard procedures for the depletion of albumin can lead to Significant depletion of cytokine119. Effective solutions to these issues will probably transcend the simplest of label-free approaches — and involve slower, complex and multi-step protocols such as high-affinity sandwich assays. For laboratory applications such as rapid, high-throughput drug screening, however, it may be possible to work with reasonably pure solutions where the range of concentrations is smaller. For such cases, there will always be Significant benefit to improving the device sensitivity.
We have seen how microfluidic technology provides researchers with the capability to place individual cells in chambers with picolitre to nanolitre scale volumes117,120. This circumvents the massive dilution of samples that is inherent to conventional approaches and can therefore maintain proteins obtained from individual cells — be it by secretion or cell lysis — at concentrations that are readily detected with the most sensitive technologies, represented in Fig. 1. Although secretion rates from individual cells are highly variable, and depend on the specific molecules secreted, detection on the picomolar scale serves as an important initial benchmark. We illustrate this with the example of native (unstimulated) human monomyelectic cells, which secrete an average rate of ∼7,000 TNF-α molecules per minute per cell121. For an individual cell sequestered in a volume of 1 nl, this would correspond to a concentration increase of 40 fM min−1, and this rate can be increased by a factor of ∼80 if the cells are stimulated. The levels of detector performance needed to measure these processes are included in Fig. 1 as an example of an application that requires sensitivity beyond that needed for many diagnostic assays.
Single-cell analyses also have the potential to improve our under standing of cellular heterogeneity by exploring in detail variations in the responses of genetically identical cells to identical stimuli63,117. No existing technology can perform simultaneous, real-time, quantitative assays on large populations (arrays) of individual cells, but Fig. 1 makes it evident that micro- and nanoscale sensors may soon make this feasible.
Critical to achieving such goals is development of new methods for functionalization, especially approaches enabling proximal multiplexing. For example, Huber et al. have demonstrated simultaneous protein and DNA detection in a single microcantilever surface-stress sensor array122. Detection of numerous DNA123 and protein124 targets has also been demonstrated. However, existing approaches typically employ methods (such as functionalization in separate microcapillar-ies123 or ink-jet spotting,4,125) that cannot be reduced in size to nanos-cale dimensions or scaled upwards to make large, multiplexed arrays with, say, thousands of elements. Photolabile crosslinkers and photo lithographic light-directed synthesis, as in gene chips and release pro-tocols57, show promise for the functionalization of arrays of devices, but the diffraction limit makes it difficult to scale this approach down to the nanoscale. More elaborate techniques, such as scanning-probe-based coating deposition126 or localized electrochemical growth127, may prove helpful.
Many challenges remain — from the development of better capture agents (see ref. 2 for a review) to the integration of arrays of advanced nanosensors with conventional microelectronic fabrication techniques — but the ultimate goal of developing tools that are capable of high-throughput studies of biological systems at the level of single cells and individual molecules will continue to drive the field forwards.
Acknowledgments
The authors thank the Defense Advanced Research Projects Agency (HR00110610043 and N66001-08-1-2043) and the Fondation pour la Recherche et l'Enseignement Superieur for support. M.L.R. acknowledges a Director' Pioneer Award from the National Institutes of Health (1DP1OD006924). We also thank P. Puget for many discussions.
Footnotes
Additional information: The authors declare no competing financial interests
References
- 1.Braun T, et al. Quantitative time-resolved measurement of membrane protein- ligand interactions using microcantilever array sensors. Nature Nanotech. 2009;4:179–185. doi: 10.1038/nnano.2008.398. [DOI] [PubMed] [Google Scholar]
- 2.Phelan ML, Nock S. Generation of bioreagents for protein chips. Proteomics. 2003;3:2123–2134. doi: 10.1002/pmic.200300596. [DOI] [PubMed] [Google Scholar]
- 3.von Muhlen MG, Brault ND, Knudsen SM, Jiang S, Manalis SR. Label-free biomarker sensing in undiluted serum with suspended microchannel resonators. Anal Chem. 2010;82:1905–1910. doi: 10.1021/ac9027356. This work is notable for the high sensitivity (300 pM) and fast response time (∼1 min) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Backmann N, et al. A label-free immunosensor array using single-chain antibody fragments. Proc Natl Acad Sci USA. 2005;102:14587–14952. doi: 10.1073/pnas.0504917102. The authors use a microcantilever with static-mode defection to achieve a sensitivity of≈1 nM, and include a detailed discussion of device functionalization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waggoner PS, Varshney M, Craighead HG. Detection of prostate specific antigen with nanomechanical resonators. Lab Chip. 2009;9:3095–3099. doi: 10.1039/b907309b. The authors report femtomolar detection from serum through fluid phase capture and detection in vacuo. [DOI] [PubMed] [Google Scholar]
- 6.Ibach H. The role of surface stress in reconstruction, epitaxial growth and stabilization of mesoscopic structures. Surf Sci. 1997;29:193–263. [Google Scholar]
- 7.Wu G, et al. Bioassay of prostrate-specific antigen (PSA) using microcantilevers. Nature Biotechnol. 2001;19:856–860. doi: 10.1038/nbt0901-856. [DOI] [PubMed] [Google Scholar]
- 8.Fritz J, et al. Translating biomolecular recognition into nanomechanics. Science. 2000;288:316–318. doi: 10.1126/science.288.5464.316. [DOI] [PubMed] [Google Scholar]
- 9.Mertens J, et al. Label-free detection of DNA hybridization based on hydration-induced tension in nucleicacid films. Nature Nanotech. 2008;3:302–307. doi: 10.1038/nnano.2008.91. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J, et al. Rapid and label-free nanomechanical detection of biomarker transcripts in human RNA. Nature Nanotech. 2006;1:214–220. doi: 10.1038/nnano.2006.134. [DOI] [PubMed] [Google Scholar]
- 11.Ndieyira JW, et al. Nanomechanical detection of antibiotic-mucopeptide binding in a model for superbug drug resistance. Nature Nanotech. 2008;3:691–696. doi: 10.1038/nnano.2008.275. [DOI] [PubMed] [Google Scholar]
- 12.Braun T, et al. Conformational change of bacteriorhodopsin quantitatively monitored by microcantilever sensors. Biophys J. 2006;90:2970–2977. doi: 10.1529/biophysj.105.072934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shu W, et al. DNA molecular motor driven micromechanical cantilever arrays. J Am Chem Soc. 2005;127:17054–17060. doi: 10.1021/ja0554514. [DOI] [PubMed] [Google Scholar]
- 14.Wee KW, et al. Novel electrical detection of label-free disease marker proteins using peizoresistive self-sensing micro-cantilevers. Biosens Bioelectron. 2005;20:1932–1938. doi: 10.1016/j.bios.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 15.Rasmussen PA, Taysen J, Hansen O, Eriksen SC, Boisen A. Optimised cantilever biosensor with piezoresistive read-out. Ultramicroscopy. 2003;97:371–376. doi: 10.1016/S0304-3991(03)00063-9. [DOI] [PubMed] [Google Scholar]
- 16.Stoney GG. The tension of metallicfilms deposited by electrolysis. Proc R Soc Lond A. 1909;82:172–175. [Google Scholar]
- 17.Sader JE. Surface stress induced defections of cantilever plates with applications to the atomic force microscopy: Rectangular plates. J Appl Phys. 2001;89:2911–2921. [Google Scholar]
- 18.Gfeller KY, Nugaeva N, Hegner M. Micromechanical oscillators as rapid biosensor for the detection of active growth of Escherichia coli. Biosens Bioelectron. 2005;21:528–533. doi: 10.1016/j.bios.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 19.Gfeller KY, Nugaeva N, Hegner M. Rapid biosensor for detection of antibiotic-selective growth of Escherichia coli. Appl Environ Microbiol. 2005;71:2626–2631. doi: 10.1128/AEM.71.5.2626-2631.2005. The growth and detection of E. coli was performed using dynamic mode microcantilevers: the sensitivity was ∼100 cells and the detection times were less than one hour. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang YT, Callegari C, Feng XL, Ekinci KL, Roukes ML. Zeptogram-scale nanomechanical mass sensing. Nano Lett. 2006;6:583–586. doi: 10.1021/nl052134m. [DOI] [PubMed] [Google Scholar]
- 21.Jensen K, Kim K, Zettl A. An atomic-resolution nanomechanical mass sensor. Nature Nanotech. 2008;3:533–537. doi: 10.1038/nnano.2008.200. [DOI] [PubMed] [Google Scholar]
- 22.Gupta A, Akin D, Bashir R. Single virus particle mass detection using microresonators with nanoscale thickness. Appl Phys Lett. 2004;84:1976–1978. [Google Scholar]
- 23.Ilic B, Yang Y, Craighead HG. Virus detection using nanoelectromechanical devices. Appl Phys Lett. 2004;85:2604–2606. [Google Scholar]
- 24.Park K, et al. Measurement of adherent cell mass and growth. Proc Natl Acad Sci USA. 2010;107:20691–20696. doi: 10.1073/pnas.1011365107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park K, et al. Concentration, growth and mass measurements of mammalian cells on silicon cantilevers. Lab Chip. 2008;8:1034–1041. doi: 10.1039/b803601b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sader JE. Frequency response of cantilever beams immersed in viscous fluids with applications to the atomic force microscope. J Appl Phys. 1998;84:64–76. [Google Scholar]
- 27.Van Eysden CA, Sader JE. Frequency response of cantilever beams immersed in viscous fluids with applications to the atomic force microscope: Arbitrary mode order. J Appl Phys. 2007;101:044908. This paper includes a detailed theoretical analysis of the viscous damping of fluid-immersed cantilevers. [Google Scholar]
- 28.Braun T, et al. Micromechanical mass sensors for biomolecular detection in a physiological environment. Phys Rev E. 2005;72:031907. doi: 10.1103/PhysRevE.72.031907. The authors use dynamic mode mass detection with microcantilevers to achieve a resolution of 7 ng. [DOI] [PubMed] [Google Scholar]
- 29.Ghatkesar MK, et al. Higher modes of vibration increase mass sensitivity in nanomechanical microcantilevers. Nanotechnology. 2007;18:445502. [Google Scholar]
- 30.Hwang KS, et al. In-situ quantitative analysis of a prostate-specific antigen (PSA) using a nanomechanical PZT cantilever. Lab Chip. 2004;4:547–552. doi: 10.1039/b410905h. [DOI] [PubMed] [Google Scholar]
- 31.Lee JH, Kim TS, Yoon KH. Effect of mass and stress on resonant frequency shift of functionalized Pb(Zr0.52Ti0.48)O3 thin film microcantilever for the detection of C-reactive protein. Appl Phys Lett. 2004;84:3187–3189. [Google Scholar]
- 32.McFarland AW, Poggi MA, Doyle MJ, Bottomley LA, Colton JS. Infuence of surface stress on the resonance behavior of microcantilevers. Appl Phys Lett. 2005;87:053505. [Google Scholar]
- 33.Lu P, Lee HP, Lu C, O'hea SJ. Surface stress effects on the resonance properties of cantilever sensors. Phys Rev B. 2005;72:085405. [Google Scholar]
- 34.Lachut MJ, Sader JE. Effect of surface stress on the stifness of cantilever plates. Phys Rev Lett. 2007;99:206102. doi: 10.1103/PhysRevLett.99.206102. [DOI] [PubMed] [Google Scholar]
- 35.Tamayo J, Ramos D, Mertens J, Calleja M. Effect of the adsorbate stiffness on the resonance response of microcantilever sensors. Appl Phys Lett. 2006;89:224104. [Google Scholar]
- 36.Gupta AK, et al. Anomalous resonance in a nanomechanical biosensor. Proc Natl Acad Sci USA. 2006;103:13362–13367. doi: 10.1073/pnas.0602022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burg TP, et al. Weighing of biomolecules, single cells and single nanoparticles in fluid. Nature. 2007;446:1066–1069. doi: 10.1038/nature05741. [DOI] [PubMed] [Google Scholar]
- 38.Barton RA, et al. Fabrication of a nanomechanical mass sensor containing a nanofluidic channel. Nano Lett. 2010;10:2058–2063. doi: 10.1021/nl100193g. [DOI] [PubMed] [Google Scholar]
- 39.Zuniga C, Rinaldi M, Piazza G. High frequency piezoelectric resonant nanochannels for bio-sensing applications in liquid environment. IEEE Sensors 2010. 2010 Nov;:52–55. [Google Scholar]
- 40.Burg TP, Sader JE, Manalis SR. Nonmonotonic energy dissipation in microfluidic resonators. Phys Rev Lett. 2009;102:228103. doi: 10.1103/PhysRevLett.102.228103. [DOI] [PubMed] [Google Scholar]
- 41.Sader JE, Burg TP, Manalis SR. Energy dissipation in microfluidic beam resonators. J Fluid Mech. 2010;650:215–250. [Google Scholar]
- 42.Bryan AK, Goranov A, Amon A, Manalis SR. Measurement of mass, density, and volume during the cell cycle of yeast. Proc Natl Acad Sci USA. 2010;107:999–1004. doi: 10.1073/pnas.0901851107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Godin M, et al. Using buoyant mass to measure the growth of single cells. Nature Methods. 2010;7:387–390. doi: 10.1038/nmeth.1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Knudsen SM, von Muhlen MG, Schauer DB, Manalis SR. Determination of bacterial antibiotic resistance based on osmotic shock response. Anal Chem. 2009;81:7087–7090. doi: 10.1021/ac900968r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arlett JL, Roukes ML. Ultimate sensitivity limits for mass sensing with hollow micro/nanochannel resonators. J Appl Phys. 2010;108:084701. [Google Scholar]
- 46.Kim N, Kim DK, Cho YJ. Development of indirect-competitive quartz crystal microbalance immunosensor for C-reactive protein. Sensor Actuat B-Chem. 2009;143:444–448. [Google Scholar]
- 47.Kurosawa S, et al. Evaluation of a high-affinity QCM immunosensor using antibody fragmentation and 2-methacryloyloxyethyl phosphorylcholine (MPC) polymer. Biosens Bioelectron. 2004;20:1134–1139. doi: 10.1016/j.bios.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 48.Weizmann Y, Patolsky F, Willner I. Amplified detection of DNA and analysis of single-base mismatches by the catalyzed deposition of gold on Au-nanoparticles. Analyst. 2001;126:1502–1504. doi: 10.1039/b106613g. [DOI] [PubMed] [Google Scholar]
- 49.Armani AM, Kulkarni RP, Fraser SE, Flagan RC, Vahala KJ. Label-free, single-molecule detection with optical microcavities. Science. 2007;314:783–787. doi: 10.1126/science.1145002. [DOI] [PubMed] [Google Scholar]
- 50.Arnold S, Shopova SI, Holler S. Whispering gallery mode bio-sensor for label-free detection of single molecules: thermo-optic vs. reactive mechanism. Opt Express. 2010;18:281–287. doi: 10.1364/OE.18.000281. [DOI] [PubMed] [Google Scholar]
- 51.Washburn AL, Luchansky MS, Bowman AL, Bailey RC. Quantitative, label-free detection of five protein biomarkers using muliplexed arrays of silicon photonic microring resonators. Anal Chem. 2010;82:69–72. doi: 10.1021/ac902451b. The authors report quantitative, parallel detection from five protein mixtures. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luchansky MS, Bailey RC. Silicon photonic microring resonators for quantitative cytokine detection and T-cell secretion anslysis. Anal Chem. 2010;82:1975–1981. doi: 10.1021/ac902725q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stern E, et al. Label-free immunodetection with CMOS-compatible semiconducting nanowires. Nature. 2007;445:519–522. doi: 10.1038/nature05498. [DOI] [PubMed] [Google Scholar]
- 54.Zheng G, Gao XPA, Lieber CM. Frequency domain detection of biomolecules using silicon nanowire biosensors. Nano Lett. 2010;10:3179–3183. doi: 10.1021/nl1020975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bunimovich YL, et al. Quantitative real-time measurement of DNA hybridization with alkylated nonoxidized silicon nanowires in electrolyte solution. J Am Chem Soc. 2006;128:16323–16331. doi: 10.1021/ja065923u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gao XPA, Zheng G, Lieber CM. Subthreshold regime has the optimal sensitivity for nanowire FET biosensors. Nano Lett. 2010;10:547–552. doi: 10.1021/nl9034219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Squires TM, Messinger RJ, Manalis SR. Making it stick: Convection reaction and difusion in surface-based biosensors. Nature Biotechnol. 2008;26:417–426. doi: 10.1038/nbt1388. This review article contains a detailed kinetics analysis that is relevant to all biosensors. [DOI] [PubMed] [Google Scholar]
- 58.Rich R, Myszka DG. Advances in surface plasmon resonance biosensor analysis. Curr Opin Biotechnol. 2000;11:54–61. doi: 10.1016/s0958-1669(99)00054-3. [DOI] [PubMed] [Google Scholar]
- 59.Skivesen N, et al. Photonic-crystal waveguide biosensor. Opt Express. 2007;15:3169–3176. doi: 10.1364/oe.15.003169. [DOI] [PubMed] [Google Scholar]
- 60.Yao X, et al. Sub-attomole oligonucleotide and p53 cDNA determinations via a high-resolution surface plasmon resonance combined with oligonucleotide-capped gold nanoparticle signal amplification. Anal Biochem. 2006;354:220–228. doi: 10.1016/j.ab.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 61.Cesaro-Tadic S, et al. High-sensitivity miniaturized immunoassays for tumor necrosis factor α using microfluidic systems. Lab Chip. 2004;4:563–569. doi: 10.1039/b408964b. [DOI] [PubMed] [Google Scholar]
- 62.Fan R, et al. Integrated barcode chips for rapid, multiplexed analysis of proteins in microliter quantities of blood. Nature Biotechnol. 2008;26:1373–1378. doi: 10.1038/nbt.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Diercks AH, et al. A microfluidic device for multiplexed protein detection in nano-liter volumes. Anal Biochem. 2009;386:30–35. doi: 10.1016/j.ab.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harris R, Lohr KN. Screening for Prostate Cancer: An Update of the Evidence for the U. S. Preventitive Services Task Force. Ann Intern Med. 2002;137:915–916. doi: 10.7326/0003-4819-137-11-200212030-00014. See also: http://en.wikipedia.org/wiki/Prostate_specifc_antigen. [DOI] [PubMed] [Google Scholar]
- 65.Carrano J. Chemical and Biological Sensor Standards Study. DARPA; 2005. Available at http://go.nature.com/JNBVGN. [Google Scholar]
- 66.Steward CC, Steward SJ. In: Cytometry. 3rd. Darzynkiewicz Z, editor. Vol. 63. 2001. p. 223. [Google Scholar]
- 67.Konopka K, Neilands JB. Effect of serum albumin on siderophore-mediated utilization of transferring iron. Biochem. 1984;10:2122–2127. doi: 10.1021/bi00305a003. [DOI] [PubMed] [Google Scholar]
- 68.Nair PR, Alam MA. Teory of “selectivity” of label-free nanobiosensors: A geometro-physical perspective. J Appl Phys. 2010;107:064701. doi: 10.1063/1.3310531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fan R, et al. Integrated barcode chips for rapid, multiplexed analysis of proteins in microliter quantities of blood. Nature Biotechnol. 2008;26:1373–1378. doi: 10.1038/nbt.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stern E, et al. Label-free biomarker detection from whole blood. Nature Nanotech. 2010;5:138–142. doi: 10.1038/nnano.2009.353. A two-stage detection approach leads to Significantly enhanced specificity, enabling subpicomolar detection in whole blood with nanoribbon sensors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hirrlinger J, Moeller H, Kirchof F, Dringen R. Expression of multidrug resistance proteins (Mrps) in astrocytes of the mouse brain: A single cell RT-PCR study. Neurochem Res. 2005;30:1237–1244. doi: 10.1007/s11064-005-8795-y. [DOI] [PubMed] [Google Scholar]
- 72.Goluch ED, et al. A biobarcode assay for on-chip attomolar-sensitivity protein detection. Lab Chip. 2006;6:1293–1299. doi: 10.1039/b606294f. [DOI] [PubMed] [Google Scholar]
- 73.Grell M, Wajant H, Zimmermann G, Scheurich P. The type 1 receptor (CD120a) is the high-affinity receptor for solube tumor necrosis factor. Proc Natl Acad Sci USA. 1998;95:570–575. doi: 10.1073/pnas.95.2.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Florin EL, Moy VT, Gaub HE. Intermolecular forces and energies between ligands and receptors. Science. 1994;264:415–417. doi: 10.1126/science.7939660. [DOI] [PubMed] [Google Scholar]
- 75.Fritz J, Katopodis AG, Kolbinger F, Anselmetti D. Force-mediated kinetics of single P-selectin/ligand complexes observed by atomic force microscopy. Proc Natl Acad Sci USA. 1998;95:12283–12288. doi: 10.1073/pnas.95.21.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Merkel R, Nassoy P, Leung A, Ritchie K, Evans E. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature. 1999;397:50–53. doi: 10.1038/16219. [DOI] [PubMed] [Google Scholar]
- 77.Wiita AP, et al. Probing the chemistry of thioredoxin catalysis with force. Nature. 2007;450:124–127. doi: 10.1038/nature06231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fernandez JM, Hongbin L. Force spectroscopy monitors the folding trajectory of a single protein. Science. 2004;303:1674–1678. doi: 10.1126/science.1092497. [DOI] [PubMed] [Google Scholar]
- 79.Tan JL, et al. Cells lying on a bed of microneedles: An approach to isolate mechanical force. Proc Natl Acad Sci USA. 2003;100:1484–1489. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guck J, et al. Optical deformability as an inherent cell marker for testing malignant transformation and metastatic competence. Biophys J. 2005;88:3689–3698. doi: 10.1529/biophysj.104.045476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Suresh S. Biomechanics and biophysics of cancer cells. Acta Mater. 2007;55:3989–4014. doi: 10.1016/j.actbio.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee W, Fon W, Axelrod BW, Roukes ML. High-sensitivity microfluidic calorimeters for biological and chemical applications. Proc Natl Acad Sci USA. 2009;106:15225–15230. doi: 10.1073/pnas.0901447106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cross SE, Jin YS, Rao J, Gimzewski JK. Nanomechanical analysis of cells from cancer patients. Nature Nanotech. 2007;2:780–783. doi: 10.1038/nnano.2007.388. [DOI] [PubMed] [Google Scholar]
- 84.Brujić J, Hermans RI, Walther KA, Fernandez JM. Single-molecule force spectroscopy reveals signatures of glassy dynamics in the energy landscape of ubiquitin. Nature Phys. 2006;2:282–286. [Google Scholar]
- 85.Evans E, Leung A, Heinrich V, Zhu Ch. Mechanical switching and coupling between two dissociation pathways in a P-selecti0n adhesion bond. Proc Natl Acad Sci USA. 2004;101:11281–11286. doi: 10.1073/pnas.0401870101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Robert P, Benoliel AM, Pierres A, Bongrand P. What is the biological relevance of the specific bond properties revealed by single-molecule studies? J Mol Recognit. 2007;20:432–447. doi: 10.1002/jmr.827. [DOI] [PubMed] [Google Scholar]
- 87.Zhou J, et al. Unbinding of the streptavidin-biotin complex by atomic force microscopy: a hybrid simulation study. J Chem Phys. 2006;125:104905. doi: 10.1063/1.2337629. [DOI] [PubMed] [Google Scholar]
- 88.Evans EA, Calderwood DA. Forces and bond dynamics in cell adhesion. Science. 2007;316:1148–1153. doi: 10.1126/science.1137592. [DOI] [PubMed] [Google Scholar]
- 89.Liphardt J, Onoa B, Smith SB, Tinoco I, Jr, Bustamante C. Reversible unfolding of single RNA molecules by mechanical force. Science. 2001;292:733–737. doi: 10.1126/science.1058498. [DOI] [PubMed] [Google Scholar]
- 90.Krieg M, et al. Tensile forces govern germ-layer organization in zebrafsh. Nature Cell Biol. 2008;10:429–436. doi: 10.1038/ncb1705. [DOI] [PubMed] [Google Scholar]
- 91.Fierro FA, et al. BCR/ABL expression of myeloid progenitors increases beta1-integrin mediated adhesion to stromal cells. J Mol Biol. 2008;377:1082–1093. doi: 10.1016/j.jmb.2008.01.085. [DOI] [PubMed] [Google Scholar]
- 92.Wjcikiewicz E, et al. LFA-1 binding destabilizes the JAM-A homophilic interaction during leukocyte transmigration. Biophys J. 2009;96:285–293. doi: 10.1529/biophysj.108.135491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Friedrichs J, Helenius J, Müller DJ. Stimulated single-cell force spectroscopy to quantify cell adhesion receptor crosstalk. Proteomics. 2010;10:1455–1462. doi: 10.1002/pmic.200900724. [DOI] [PubMed] [Google Scholar]
- 94.Fon WC, Schwab KC, Worlock JM, Roukes ML. Nanoscale, phonon-coupled calorimetry with sub-attojoule/Kelvin resolution. Nano Lett. 2005;5:1968–1971. doi: 10.1021/nl051345o. [DOI] [PubMed] [Google Scholar]
- 95.Ekinci KL. Electromechanical transducers at the nanoscale: actuation and sensing of motion in nanoelectromechanical systems (NEMS) Small. 2005;1:786–797. doi: 10.1002/smll.200500077. [DOI] [PubMed] [Google Scholar]
- 96.Carr DW, Evoy S, Sekaric L, Craighead HG, Parpia JM. Measurement of mechanical resonance and losses in nanometer scale silicon wires. Appl Phys Lett. 1999;75:920–922. [Google Scholar]
- 97.Kouh T, Karabacak D, Kim DH, Ekinci KL. Difraction efects in optical interferometric displacement detection in nanoelectromechanical systems. Appl Phys Lett. 2005;86:013106. [Google Scholar]
- 98.Keeler BEN, Carr DW, Sullivan JP, Friedmann TA, Wendt JR. Experimental demonstration of a laterally deformable optical nanoelectromechanical system grating transducer. Opt Lett. 2004;29:1182–1184. doi: 10.1364/ol.29.001182. [DOI] [PubMed] [Google Scholar]
- 99.Li M, Pernice WHP, Tang HX. Broadband all-photonic transduction of nanocantilevers. Nature Nanotech. 2009;4:377–382. doi: 10.1038/nnano.2009.92. [DOI] [PubMed] [Google Scholar]
- 100.Li M, et al. Harnessing optical forces in integrated photonic circuits. Nature. 2008;467:480–484. doi: 10.1038/nature07545. [DOI] [PubMed] [Google Scholar]
- 101.Truitt PA, Hertzberg JB, Huang CC, Ekinci KL, Schwab KC. Efficient and sensitive capacitive readout of nanomechanical resonator arrays. Nano Lett. 2007;7:120–126. doi: 10.1021/nl062278g. [DOI] [PubMed] [Google Scholar]
- 102.Sampathkumar A, Murray TW, Ekinci KL. Photothermal operation of high frequency nanoelectromechanical systems. Appl Phys Lett. 2006;88:223104. [Google Scholar]
- 103.Verbridge SS, Bellan LM, Parpia JM, Craighead HG. Optically driven resonance of nanoscale flexural oscillators in liquid. Nano Lett. 2006;6:2109–2114. doi: 10.1021/nl061397t. [DOI] [PubMed] [Google Scholar]
- 104.Bargatin I, Kokinsky I, Roukes ML. Efficient electrothermal actuation of multiple modes of high-frequency nanoelectromechanical resonators. Appl Phys Lett. 2007;90:093116. [Google Scholar]
- 105.Seo JH, Brand O. High Q-factor in-plane-mode resonant microsensor platform for gaseous/liquid environment. J Microelectromech Syst. 2008;17:483–493. [Google Scholar]
- 106.Piazza G, Stephanou PJ, Pisano AP. Piezoelectric aluminum nitride vibrating contour-mode MEMS resonators. J Microelectromech Syst. 2006;15:1406–1418. [Google Scholar]
- 107.Karabalin RB, et al. Piezoelectric nanoelectromechanical resonators based on aluminum nitride thin films. Appl Phys Lett. 2009;95:103111. [Google Scholar]
- 108.Bargatin I, Myers EB, Arlett J, Gudlewski B, Roukes ML. Sensitive detection of nanomechanical motion using piezoresistive signal downmixing. Appl Phys Lett. 2005;86:133109. [Google Scholar]
- 109.Harley JA, Kenny TW. High-sensitivity piezoeresistive cantilevers under 1000Å thick. Appl Phys Lett. 1999;75:289–291. [Google Scholar]
- 110.Arlett JL, Maloney JR, Gudlewski B, Muluneh M, Roukes ML. Self-sensing micro- and nanocantilevers with attonewton-scale force resolution. Nano Lett. 2006;6:1000–1006. [Google Scholar]
- 111.Harley JA, Kenny TW. 1/f noise considerations for the design and process optimization of piezoresistive cantilevers. J Microelectromech Syst. 2000;9:226–235. [Google Scholar]
- 112.Li M, Tang HX, Roukes ML. Ultra-sensitive NEMS-based cantilevers for sensing, scanned probe and very high-frequency applications. Nature Nanotech. 2007;2:114–120. doi: 10.1038/nnano.2006.208. [DOI] [PubMed] [Google Scholar]
- 113.Nguyen CTC, Howe RT. An integrated CMOS micromechanical resonator high-Q oscillator. IEEE J Solid-St Circ. 1999;34:440–455. [Google Scholar]
- 114.Despont M, Drechsler U, Yu R, Pogge HB, Vettiger P. Wafer-scale microdevice transfer/interconnect: its application in an AFM-based data-storage system. J Microelectromech Syst. 2004;13:895–901. [Google Scholar]
- 115.Torsen T, Maerkl SJ, Quake SR. Microfluidic large-scale integration. Science. 2002;298:580–584. doi: 10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- 116.Zhong JF, et al. A microfluidic processor for gene expression profiling of single human embryonic stem cells. Lab Chip. 2008;8:68–74. doi: 10.1039/b712116d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gómez-Sjöberg R, Leyrat AA, Pirone DM, Chen CS, Quake SR. Versatile, fully automated, microfluidic cell culture system. Anal Chem. 2007;79:8557–8563. doi: 10.1021/ac071311w. [DOI] [PubMed] [Google Scholar]
- 118.Mehta AI, et al. Biomarker amplification by serum carrier protein binding. Dis Markers. 2003;19:1–10. doi: 10.1155/2003/104879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Granger J, Siddiqui J, Copeland S, Remick D. Albumin depletion of human plasma also removes low abundance proteins including the cytokines. Proteomics. 2005;5:4713–4718. doi: 10.1002/pmic.200401331. [DOI] [PubMed] [Google Scholar]
- 120.Köster S, et al. Drop-based microfluidic devices for encapsulation of single cells. Lab Chip. 2008;8:1110–1115. doi: 10.1039/b802941e. [DOI] [PubMed] [Google Scholar]
- 121.Pradines-Figueres A, Raetz CRH. Processing and secretion of tumor necrosis factor α in endotoxin-treated Mono Mac 6 cells are dependent on phorbol myristate acetate. J Biol Chem. 1992;267:23261–23268. [PubMed] [Google Scholar]
- 122.Huber F, et al. Analyzing gene expression using combined nanomechanical cantilever sensors. J Phys Conf Ser. 2007;61:450–453. [Google Scholar]
- 123.McKendry R, et al. Multiple label-free biodetection and quantitative DNA-binding assays on a nanomechanical cantilever array. Proc Natl Acad Sci USA. 2002;99:9783–9788. doi: 10.1073/pnas.152330199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Huber F, Hegner M, Gerber C, Güntherodt HJ, Lang HP. Label free analysis of transcription factors using microcantilever arrays. Biosens Bioelectron. 2006;21:1599–1605. doi: 10.1016/j.bios.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 125.Bietsch A, Hegner M, Lang HP, Gerber C. Inkjet deposition of alkanethiolate monolayers and DNA Oligonucleotides on gold: evaluation of spot uniformity by wet etching. Langmuir. 2004;20:5119–5122. doi: 10.1021/la049621m. [DOI] [PubMed] [Google Scholar]
- 126.Zhang H, Lee KB, Li Z, Mirkin CA. Biofunctionalized nanoarrays of inorganic structures prepared by dip-pen nanolithography. Nanotechnology. 2003;14:1113–1117. [Google Scholar]
- 127.Harper JC, Polsky R, Dirk SM, Wheeler DR, Brozik SM. Electroaddressable selective functionalization of electrode arrays: Catalytic NADH detection using ardyl diazonium modified gold electrodes Electroanalysis. 2007;19:1268–1274. [Google Scholar]
- 128.Zheng G, Patolsky F, Cui Y, Wang WU, Lieber CM. Multiplexed electrical detection of cancer markers with nanowire sensor arrays. Nature Biotechnol. 2005;23:1294–1301. doi: 10.1038/nbt1138. [DOI] [PubMed] [Google Scholar]
- 129.Lipsitz R. Diagnostics at home: Pregnancy tests. Sci Amer. 2000 Nov;283:110–111. [Google Scholar]
- 130.Washburn AL, Gunn LC, Bailey RC. Label-free quantitation of a cancer biomarker in complex media using silicon photonic microring resonators. Anal Chem. 2009;81:9499–9506. doi: 10.1021/ac902006p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Viani MB, et al. Small cantilevers for force spectroscopy of single molecules. J Appl Phys. 1999;86:2258–2262. [Google Scholar]
- 132.Arlett JL, et al. BioNEMS: Nanomechanical systems for single-molecule biophysics. Lect Notes Phys. 2007;711:241–270. [Google Scholar]