Abstract
The skin is the primary physical barrier between the body and the external environment and is also a substrate for the colonization of numerous microbes. Previously, dermatological microbiology research was dominated by culture-based techniques, but significant advances in genomic technologies have enabled the development of less-biased, culture-independent approaches to characterize skin microbial communities. These molecular microbiology approaches illustrate the great diversity of microbiota colonizing the skin and highlight unique features such as site specificity, temporal dynamics, and interpersonal variation. Disruptions in skin commensal microbiota are associated with the progression of many dermatological diseases. A greater understanding of how skin microbes interact with each other and with their host, and how we can therapeutically manipulate those interactions, will provide powerful tools for treating and preventing dermatological disease.
The skin is a complex ecosystem, harboring bacteria, viruses, and fungi that interact extensively with the human body. Advances in genomic technologies have greatly enhanced the characterization of skin microbial communities.
The skin acts not just as a protective physical barrier between the body and the external environment; it is itself an environmental substrate, harboring a rich and diverse community of microorganisms (the microbiome). The human microbiome includes the bacteria, fungi, viruses, archaea, and microeukaryotes that inhabit the various body environments, such as the gut, oral cavity, and skin (Grice and Segre 2012). In recent years, it has become increasingly apparent that the microbiome interacts extensively with the human body and plays roles in immune system development and function, disease etiology and pathology, cancer development, and defense against pathogens.
The skin is a complex ecosystem that maintains topographically distinct microbial populations, as well as distinct environmental niches. Overall, the surface of the skin is cooler than the core body temperature, is slightly acidic, and squames are continuously shed from the skin surface as a result of terminal differentiation (Fuchs and Raghavan 2002). These attributes undoubtedly select for specific microbiota adapted to these unique conditions. The geography of the skin includes sebaceous areas (including face and back), moist areas (including toe/finger web space and arm pit), dry areas (including forearm and buttock), and sites containing varied densities of hair follicles, skin folds, and skin thicknesses. A critical function of the skin microbiota is “colonization resistance,” in which commensal microbiota occupy these distinct niches to block colonization and/or invasion by opportunistic or pathogenic organisms.
Community composition equilibrium across the varied geography of the skin is maintained by nutrient and space competition among microbes, production of antimicrobial peptides (AMPs) by commensal microbes and host cells, and modulation of the host immune response by commensal microbes (Nakatsuji and Gallo 2012). For example, the skin commensal Staphylococcus epidermidis has been reported to modulate the innate immune response by inhibiting skin inflammation through pattern-recognition receptor-mediated cross talk (Lai et al. 2009). Complement, an evolutionarily conserved arm of the innate immune system, was shown to maintain diversity of the skin microbiota in a mouse model, and, conversely, the skin microbiota regulated complement at the gene expression level (Chehoud et al. 2013). The microbiome is also fundamental in adaptive immune system equilibrium at the skin, and skin T-cell function and the local inflammatory milieu appear to be autonomously controlled by the commensal skin microbiota (Naik et al. 2012). These and other findings have contributed to the mounting evidence suggesting that the commensal skin microbiota is intricately involved in both innate and adaptive skin immunity.
For these reasons, a thorough understanding of the commensal skin microbiota is required to gain insight into microbial involvement in skin health and disease. The beneficial role of skin commensals and the pathogenic role of those microbes that cause disease have long been a focus of studies examining the microbial ecology of the skin. Contemporary, culture-independent methods for identifying and characterizing microbial communities have accelerated and added precision to our understanding of host–microbe interactions at the skin surface. In this article, we provide a comprehensive discussion of the human skin microbiome in health and disease states and how this understanding is informing skin disease diagnosis and treatment.
DAWNING OF THE MOLECULAR MICROBIOLOGY ERA
The study of the human cutaneous microbiota has a rich history that spans more than five decades (Marples 1965). Early methods for studying skin-associated bacteria, fungi, and viruses were limited to culturing the microorganism and defining its phylogeny and taxonomy through phenotypic, microscopic, and biochemical relationships. Dependency on generation of pure cultures introduces inherent biases because the procedures select for the most abundant and rapidly growing microorganisms of the community. Culture-based studies of viruses (including bacteriophage) are further limited because they require coculturing with their prokaryotic or eukaryotic hosts, additionally preventing the identification of viruses associated with unknown hosts. Viruses are also not readily visible by basic microscopic methods and thus are very difficult to characterize by direct morphological observation. Although great insight into microbial colonization of cutaneous surfaces in health and disease was gained using culture-dependent methods, there were significant limitations to the conclusions that could be drawn.
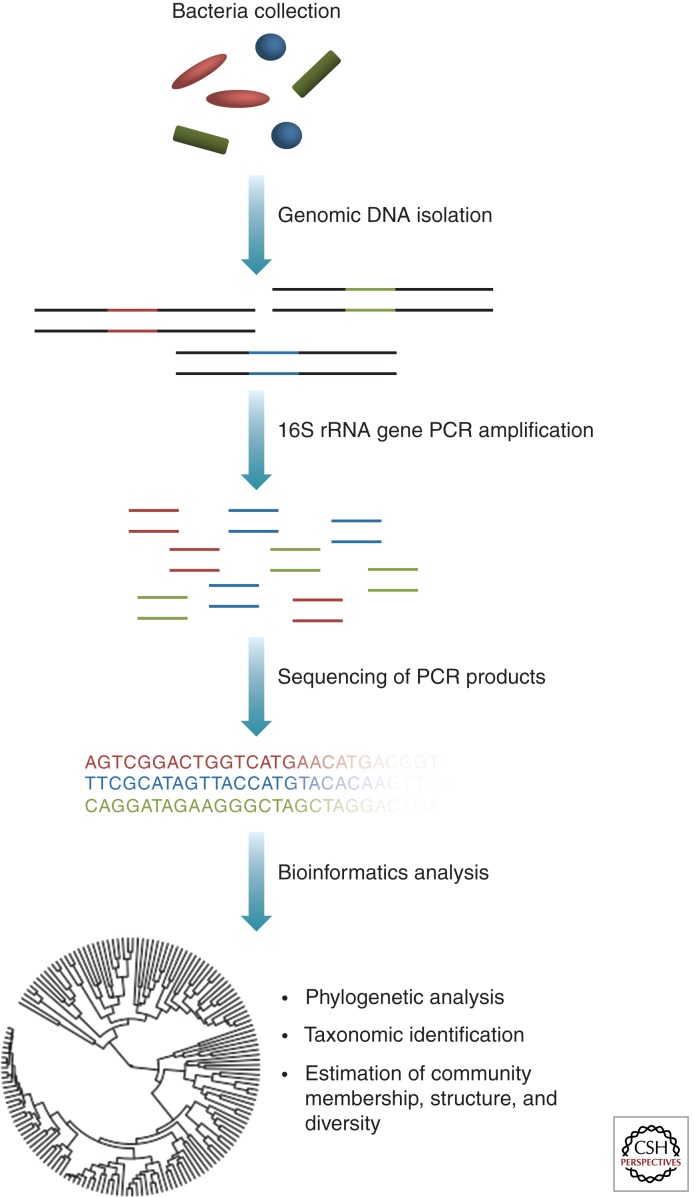
Advances in DNA sequencing technology and culture-independent methods of microbial identification have greatly enabled high-throughput, detailed characterization of microbial communities. These methods are based on surveys of marker genes, generally conserved, universal genes found in all organisms within particular taxonomic levels. Bacterial communities are most commonly classified by the sequence of their small subunit 16S ribosomal RNA (rRNA) gene (Fig. 1) (Lane et al. 1985). These genes contain both conserved regions, which allow for PCR primer binding and phylogenetic analysis, as well as variable regions, whose sequences allow for taxonomic classification. Following amplification and sequencing of 16S rRNA genes, sequence data can be analyzed in a variety of ways, including assignment of taxonomy, phylogenetic analysis, and community analyses (Fig. 1). Fungi are often classified by sequencing of the internal transcribed spacer (ITS) region that lies between the small and large subunit rRNA genes in eukaryotes (Chase and Fay 2009). In addition to offering clearer definitions for determining microbial taxonomy, conserved gene sequence analysis does not require the microorganism to be cultured, which therefore eliminates those biases associated with culturing procedures.
Figure 1.
The workflow of a bacterial 16S rRNA gene microbiome study. A heterogeneous mixture of genomic DNA is extracted from samples taken from the skin. Primers, containing barcodes that allow for multiplexing, are designed to the desired region of the 16S rRNA gene. 16S rRNA gene PCR products are amplified and sequenced. Low-quality sequences are removed, and various analyses are performed. These analyses can include assignment to taxonomy, analysis of shared phylogeny, and analysis of microbial community membership, structure, and diversity.
Unlike bacteria and fungi, viruses and bacteriophage present a special difficulty because they do not contain a consensus gene that can be used for widespread taxonomic identification. Closely related groups may be phylogenetically analyzed using specific conserved genes, such as the human papillomavirus L1 gene, but this is far less robust than the 16S rRNA and ITS sequencing and identification approaches used to classify bacteria and fungi. Comparative genome analysis is complicated by the high frequency of gene transfer among virus and host genomes and the lack of comprehensive, annotated reference databases and assigned taxonomy. A solution to this problem is the use of whole-genome shotgun metagenomics, which does not rely on amplification and sequencing of marker genes but, rather, allows for sequencing and analysis of the sample’s full genetic potential (National Research Council Committee on Metagenomics 2007). This type of strategy not only bypasses PCR, but also can provide insight into what microbial communities are doing on the skin surface. These types of approaches are still under development in the skin, because high amounts of host DNA and low amounts of microbial DNA present technical limitations for metagenomic approaches.
THE SKIN MICROBIOME IN HEALTH
Bacteria
Before the advent of molecular techniques to characterize skin microbiota, the temporal and topographical diversity of the skin microbiota was still considered vast. Early studies produced variable results in bacterial quantity and taxonomy, hypothesized at the time to be a result of inherent topographical and temporal diversity of skin bacterial communities (Evans et al. 1950). In a comprehensive study, which cultured under both aerobic and anaerobic conditions, skin bacterial colonization differed between anatomical sites, and the highest bacterial load was observed in sebaceous sites (Evans et al. 1950). Furthermore, skin colonization was dominated by a small group of taxa, including Propionibacterium acnes and Staphylococcus epidermidis. Years later, these same features are apparent using sophisticated sequencing-based techniques to characterize skin microbiota.
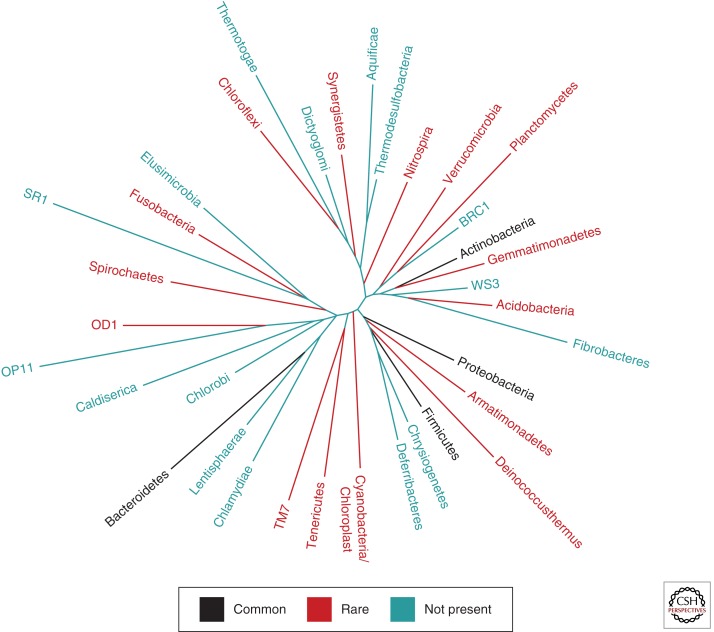
Indeed, site-specific colonization is a key feature of the human skin microbiome. Using a 16S rRNA sequencing approach in healthy adults, sebaceous regions were found to be preferentially colonized by Propionibacterium and Staphylococcus spp.; moist sites predominantly maintained Corynebacterium and Staphylococcus spp.; and dry sites, which, despite general variability and greater diversity, displayed a significant presence of β-Proteobacteria and Flavobacteriales (Grice et al. 2009). In the same study, 19 bacterial phyla were identified, but skin was dominated by four phyla: the Actinobacteria, Firmicutes, Proteobacteria, and Bacteroidetes (Fig. 2). Another key finding was that longitudinal stability was dependent on the skin site, with sebaceous sites being the most stable and dry sites being the most variable over time. Costello et al. (2009) similarly reported that topographical community variability was greater than temporal variability among individuals. Interestingly, greater microbial diversity characterized skin microbiota, as compared with gut or oral microbiota of the same individuals. Key findings of these and other studies show that skin bacterial communities are generally diverse between individuals (Gao et al. 2007; Grice et al. 2008) and may be influenced by ethnicity, lifestyle, and/or geography, as suggested by a study comparing cutaneous microbiota colonizing South American Amerindians and U.S. residents (Blaser et al. 2013). Subject sex, handedness, and time since last hand washing also appear to affect bacterial community composition (Fierer et al. 2008). A subsequent study confirmed the differences in community composition between sexes when investigating the forearm, but found little influence of sex on forehead community composition, thereby supporting early observations of variability among anatomical regions (Staudinger et al. 2011).
Figure 2.
Bacterial diversity of the skin. Phylogenetic tree of the domain Bacteria with each branch representing a phylum. Black branches represent numerically abundant phyla on the skin, red branches represent rare phyla on the skin, and green branches represent phyla that are absent from the skin. (The data are derived from Grice et al. 2009.)
The human skin microbiota is established immediately after birth, and delivery mode seems to influence the neonate’s first skin microbiota. Dominguez-Bello et al. (2010) showed that vaginally delivered neonates were colonized with bacteria similar to those colonizing the mother’s vagina (i.e., Lactobacillus, Prevotella, Sneathia spp.), and neonates delivered by Cesarean section were colonized with those bacteria found on the mother’s skin (i.e., Propionibacterium, Staphylococcus, Corynebacterium spp.). Studies in infants over the first year of life showed that diversity of skin microbiota increases with age, as does site specificity, and is overall characterized by predominance of the phylum Firmicutes (Capone et al. 2011). Upon sexual maturation, the skin becomes colonized by increased amounts of Corynebacterium and Propionibacterium (Oh et al. 2012). Colonization by these lipophilic bacterial taxa is likely a result of hormone-stimulated sebaceous gland activity and increasing sebum production during puberty. Metabolism of the lipids in sebum by these bacterial taxa also decreases the pH of the skin, thus discouraging colonization by other taxa.
The skin was an organ included in the National Institutes of Health Roadmap Human Microbiome Project, in which a cohort of 242 phenotyped healthy adults were subject to sampling of microbiota at various body sites. Their findings confirmed previous smaller-scale studies, by suggesting that the skin microbiota is diverse, but dominated by a small group of genera, in particular Staphylococcus, Propionibacterium, and Corynebacterium (Human Microbiome Project Consortium 2012). Interestingly, the metabolic and functional pathways encoded by the skin microbiota colonizing the retroauricular crease were more constant and diverse than the taxonomic composition, suggesting low metabolic diversity among a taxonomically diverse population. Furthermore, age was associated with differentially encoded metagenomic pathways on the skin, as well as a decrease in the phylum Firmicutes.
Fungi
Although bacteria represent a major focus of past and present microbiome studies, the fungal microbiota is also thought to play a significant role in skin health and disease. Cultivation-based studies identified the major component of the skin fungal community as Malassezia (formerly known as Pityrosporum) genus, consisting primarily of seven of the 14 known species (Gaitanis et al. 2012). These findings have been confirmed by molecular community analysis. Using 18S rRNA gene and ITS region sequencing, Paulino et al. (2006) reported that the skin forearm community is dominated by Malassezia, and further analysis with multiplex real-time PCR (to speciate Malassezia) suggested that the predominant commensal species were Malassezia globosa and Malassezia restricta, with Malassezia furfur (the dominant species identified by culturing methods) contributing relatively little to the overall community abundance (Paulino et al. 2008). Conversely, another recent study of three healthy scalps found that Malassezia spp. only account for a small fraction of the commensal fungi on the scalp (Park et al. 2012). A larger-scale, extensive topographical map of the fungal skin microbiota, based on sequencing of the fungal ITS region, confirmed that Malassezia is dominant in most regions of the skin, but sites on the feet (plantar heel, toenail, and toe web) had the greatest fungal diversity of all body sites (Findley et al. 2013). Databases and other resources for identifying and analyzing fungal sequences, similar to those used for 16S rRNA gene studies, are still under development, and it is expected that our knowledge of the fungal microbiome will expand as these tools become readily available.
Viruses
One of the most extensively studied human skin viruses is the human papillomavirus (HPV). Although it was originally thought that certain strains were found only in skin cancer lesions, PCR quantification of HPV marker genes revealed that healthy skin is also a habitat for a broad spectrum of HPV strains (de Villiers et al. 1997; Astori et al. 1998). Sequence analysis of the conserved L1 open reading frame (ORF) revealed a diverse community of HPV types on healthy skin, while identifying HPVs that were previously unknown (Antonsson et al. 2000). Follow-up studies have confirmed the ubiquity and diversity of HPV types throughout human populations (Antonsson et al. 2003a,b; Forslund 2007).
The other major group of commensal human skin viruses is the human polyomaviruses (HPyVs). Polyomaviruses were first described in mice in 1953 but have since been described in numerous animals, including humans (Moens et al. 2011). Although originally studied in the context of cancer, they, like the papilloma viruses, have been found on healthy human skin (Schowalter et al. 2010). There are many types of HPyVs, with many only recently discovered using molecular techniques; the most common to human skin are HPyV6, HPyV7, and Merkel cell polyomavirus (MCHPyV) (Schowalter et al. 2010; Moens et al. 2011). A recent study using a whole metagenomic analysis of the human skin virome of healthy and cancerous individuals confirmed a cutaneous viral microbiota dominated by HPVs, HPyVs, and circoviruses (Foulongne et al. 2012). These studies are still in their early stages, and as new virus species continue to be discovered and new analysis strategies developed, future studies will likely continue to characterize the viral community diversities and pathogenic/oncogenic potential.
The other viral component of the microbiome is the bacteriophage, about which little is known in the skin. Recent studies have used culture-dependent techniques paired with genomic analyses, as well as analysis of prophage (bacteriophage integrated into the bacterial host genome), to understand and characterize the genomic diversity of subsets of the skin bacteriophage communities, such as the limited diversity of Propionibacterium bacteriophage and the diversity of Staphylococcus bacteriophage (Kwan et al. 2005; Goerke et al. 2009; Marinelli et al. 2012). Whole metagenomic shotgun sequence analysis of skin swabs from five healthy patients and one patient with a previous Merkel cell carcinoma lesion indicate that two families dominate cutaneous bacteriophage communities, the Microviridae and Siphoviridae (Foulongne et al. 2012). Further studies will be required to perform more in-depth and functionally informative analysis of the bacteriophage inhabiting the skin, such as characterization of bacterial antibiotic resistance genes maintained in bacteriophage genomes that may be horizontally transferred among bacteria.
Bacteriophages are also known to be important, yet complicated mediators of bacterial horizontal gene transfer through a process known as transduction. Commensal bacteriophage metagenomes have been shown to maintain antibiotic-resistance genes, as well as mediate their transfer between bacteria, in gut and sputum samples from cystic fibrosis patients (Wang et al. 2010; Fancello et al. 2011; Minot et al. 2011). In the skin, bacteriophage communities have been suggested as mediators of resistance gene transfer between bacteria (Nakaminami et al. 2007; Varga et al. 2012). However, more research is required to understand these complicated interactions between bacteria and skin bacteriophage communities.
SKIN MICROBIOME IN DISEASE
Atopic Dermatitis
Atopic dermatitis (AD) is a chronic, recurring inflammatory skin disease that occurs more frequently in children than in adults, and has been associated with skin colonization by Staphylococcus aureus. Although no clear microbial cause has been established, antibiotics, corticosteroids, and dilute bleach baths have been relatively effective in the treatment of AD (Huang et al. 2009). Furthermore, the enormous increase in incidence over the past three decades with no clear cause raises the interesting possibility that the skin microbiota may modulate gene–environment interactions at the skin surface.
Bacterial virulence factors may in part explain the long-recognized pathogenic association between AD flares and increased colonization by S. aureus. Severe AD development was reported in a mouse model with reduced skin barrier function upon exposure to Staphylococcal protein A (SpA) (Terada et al. 2006). The detection of SpA among 89 children with AD lesions was evaluated as occurring in 91.0% of patients upon presentation, decreasing to 55.6% of patients after antibiotic therapy (Yao et al. 2010). Furthermore, there was a significant positive correlation between the levels of SpA and the clinical severity of the lesions. It has also been reported that AD lesions contain increased levels of lipotechoic acid, a known immune-stimulating molecule derived from Gram-positive bacterial cell walls, whose presence further suggests a role of bacterial components in disease (Travers et al. 2010).
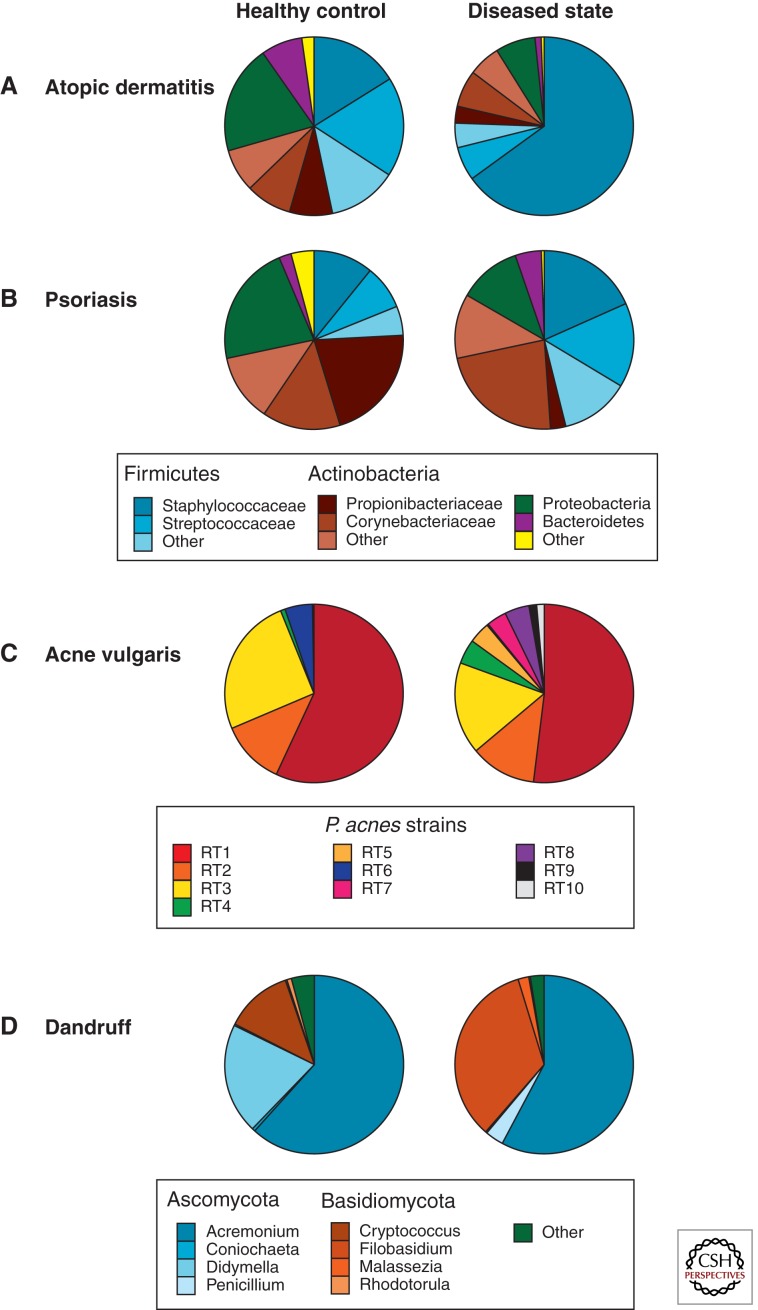
Although S. aureus likely in part contributes to disease pathogenesis, a role for the greater microbial community has recently been investigated. In a 16S-rRNA-based study that analyzed skin microbiota during the course of AD flares and improvement, a correlation between increased disease severity and decreased bacterial diversity was observed, along with altered microbial community structure in AD patients as compared with healthy controls (Fig. 3A) (Kong et al. 2012). Bacterial community diversity was also shown to increase after standard AD treatment. Additionally, fungal communities have been shown to change in composition as disease severity progresses (Kaga et al. 2011; Zhang et al. 2011). Infants who develop AD maintained early fecal microbiota with less diversity than the early fecal microbiotas of patients who did not develop the disease (Wang et al. 2008). Other studies have also shown that altered bacterial compositions of the infant gut microbiota precede the development of AD (Penders et al. 2006, 2007).
Figure 3.
Changes in skin microbiota are associated with disease. (A) Relative abundance of bacteria (16S rRNA) in 12 children with AD flares as compared with 11 healthy controls (Kong et al. 2012). (B) Relative abundance of bacteria (16S rRNA) in six patients with psoriasis, in the lesional area as compared with unaffected skin as a control (Gao et al. 2008). (C) Relative abundance of P. acnes strains in 49 acne patients and 52 healthy individuals (Fitz-Gibbon et al. 2013). (D) Relative abundance of fungi (26S rRNA) in three healthy scalps and four dandruff-afflicted scalps (Park et al. 2012).
Mutations in the gene-encoding filaggrin, an epidermal structural and hydration protein, have been associated with atopic dermatitis and other ichthyotic disorders (Sandilands et al. 2007). Analysis of skin microbiota of a mouse model with a hypomorphic mutation in St14, encoding the serine protease matriptase that regulates filaggrin processing, showed a selective shift in bacterial populations, with increased Corynebacterium and Streptococcus and decreased Pseudomonas species (Scharschmidt et al. 2009). These findings provide a link between filaggrin deficiency, a common genetic feature of AD, and changes in the skin microbiota.
Psoriasis
Multiple clinical observations support a role for dysbiosis of the skin microbiota in the pathogenesis of psoriasis including the clinical efficacy of topical corticosteroids in the treatment of psoriasis (Gottlieb 2005) and the observation that a variant of plaque psoriasis, guttate psoriasis, is triggered by Streptococcus infection. Xenograft models of psoriasis, in which unaffected skin from psoriasis patients was grafted on immunodeficient mice, showed that stimulation with bacterial antigen could trigger the skin to become lesional (Boehncke et al. 1996). Early culture-based studies examining microorganisms associated with psoriasis identified Malassezia, group A and B β-hemolytic streptococci, S. aureus, and Enterococcus faecalis (Aly et al. 1976; Rosenberg et al. 1994; Nickoloff et al. 2000). Culture-independent analysis of fungal microbiota found no conclusive evidence to link Malassezia with psoriasis (Paulino et al. 2006, 2008). Analyses of the bacterial microbiota by 16S rRNA gene-based approaches in cross-sectional studies suggest underrepresentation of Propionibacterium and increased representation of the phylum Firmicutes in psoriatic plaques as compared with healthy controls or uninvolved limb skin (Fig. 3B) (Gao et al. 2008; Fahlen et al. 2012). Longitudinal studies of the skin microbiota in psoriasis plaques may provide insight into the role of microbes in triggering, propagation, and maintenance of plaques.
Acne Vulgaris
Acne vulgaris is a common skin disorder characterized by abnormalities of sebum production by the pilosebaceous unit, bacterial proliferation, and inflammation. The etiology and pathogenesis of acne remain unclear, but there has been significant evidence supporting microbial roles in the disease. The primary microbe associated with development of acne is Propionibacterium acnes, also a prominent member of the commensal skin microbiota. Topical and systemic antibacterial drugs have long been used to treat acne, with the efficacy commonly attributed to decreased P. acnes colonization and/or activity (Tan 2003). Strain-level analysis of the 16S rRNA gene showed that, although the relative abundances of P. acnes did not significantly differ between healthy and acne patients, the relative abundances of different strains did differ between skin states (Fig. 3C) (Fitz-Gibbon et al. 2013). Additionally, genomic comparison of 71 different P. acnes strains shows that the acne-associated genomes maintained different chromosomal genomic region loci and a linear plasmid, thereby suggesting that there may be specific genes at these loci that contribute to acne pathology (Fitz-Gibbon et al. 2013). These findings suggest that strain-level analysis of the skin microbiota may be instrumental in explaining disease pathogenesis.
Dandruff
Dandruff is a mild inflammatory condition that is characterized by scaling of skin on the scalp. Malassezia fungi were proposed as the primary cause of dandruff in 1874, and this idea is still prevalent today. In fact, dandruff therapeutic shampoos are made with strong antifungal compounds in an attempt to target fungal causes of the disease (Bulmer and Bulmer 1999). Although Malassezia is the dominant fungal genus cultured from the skin and has been shown to increase in abundance on dandruff-afflicted skin (McGinley et al. 1975), recent work has suggested that the dandruff microbial communities are more complex. A molecular analysis of the 26S rRNA gene of the fungal communities associated with healthy and dandruff-afflicted scalps showed that, similar to what was shown in previous studies, the relative abundance of Malassezia was increased in the dandruff-afflicted scalp skin (Fig. 3D) (Park et al. 2012). The study also reported that dandruff-afflicted skin harbored increased relative abundances of Penicillium and Filoblasidium floriforme that correlated with increased severity of dandruff. Furthermore, because Malassezia is found in the commensal fungal microbiota, it is not a likely cause of disease on its own, and thus there may be other interactive mechanisms involved in etiology.
Merkel Cell Carcinoma
Merkel cell carcinoma (MCC) is a rare but aggressive malignant, neuroendocrine tumor that has been increasing in incidence in past decades (Hodgson 2005). In 2008, Feng et al. (2008) showed that there was a novel virus (Merkel cell polyomavirus [MCPyV]) associated with MCC tissue, but not healthy tissue. Numerous additional studies, across diverse populations, also found strong associations between the presence of MCPyV and MCC (Foulongne et al. 2008, 2010; Kassem et al. 2008; Becker et al. 2009; Duncavage et al. 2009; Katano et al. 2009; Touze et al. 2009; Mangana et al. 2010; Jung et al. 2011; Paolini et al. 2011; Haitz et al. 2012). Investigation into the virus’s role in health and disease have shown that MCPyVs are members of the commensal skin viral communities, are often asymptomatically carried, and can be shed from the skin as assembled virus particles (Foulongne et al. 2008, 2010; Kassem et al. 2008; Becker et al. 2009; Duncavage et al. 2009; Katano et al. 2009; Touze et al. 2009; Mangana et al. 2010; Jung et al. 2011; Paolini et al. 2011; Haitz et al. 2012). Because this virus is a commensal microbe on healthy skin that does not develop MCC, there are likely other factors that interact with MCPyV to cause disease, including host immune function.
DIAGNOSTIC AND THERAPEUTIC POTENTIAL OF THE MICROBIOME IN SKIN DISEASE
It is clear that the microbiome plays a broad, intricate, and complicated role in both human skin health and disease. In light of the many translational opportunities to use these findings in the clinic, a great amount of research has been devoted to clinical applications of microbiome research (Table 1). Probiotics, live microorganisms or microorganism components that confer health benefits, have long been administered therapeutically and prophylactically to the gastrointestinal tract, even before their mechanism was known. Effective and safe probiotics for use on the skin is an area of active investigation with great promise (Muizzuddin et al. 2012; Lew and Liong 2013; Shu et al. 2013). For those skin diseases that may be influenced by the gut microbiota, there is evidence that probiotic intervention may provide benefit. The efficacy of probiotics in treating AD remains somewhat controversial, but evidence suggests that administration of some Lactobacillus rhamnosus strains to mothers before and after birth reduces the occurrence and frequency of infantile AD (Kalliomaki et al. 2001; Wickens et al. 2008, 2012; Dotterud et al. 2010).
Table 1.
Therapeutic approaches based on the microbiome
| Therapeutic | Disease target example | Summary |
|---|---|---|
| Probiotics | Atopic dermatitis (Batchelor et al. 2010) | Administration of microorganisms, or their components, to confer health benefits |
| Prebiotics | Atopic dermatitis (Foolad et al. 2008) | Administration of a substance to promote growth and/or action of therapeutically beneficial indigenous microbes |
| Bacteria-mediated gene delivery | Vibrio cholera infection (Duan and March 2010) | Inhibition of pathogen virulence by administrating bacteria engineered to express genes absent from the commensal community |
| Bacteriophage-mediated antibiotic susceptibility | Antibiotic-resistant bacteria (Edgar et al. 2012) | Reduction in antibiotic-resistant bacteria populations by using bacteriophage to deliver gene constructs that promote evolution toward antibiotic susceptibility |
| Bacteriophage antimicrobial peptides | P. acnes infection (Marinelli et al. 2012) | Use of bacteriophage peptides, such as endolysin, which promotes bacteria lysis during the lytic bacteriophage life cycle, against bacterial infections |
| Direct bacteriophage therapy | Escherichia coli biofilm infections (Lu and Collins 2007) | Administration of bacteriophages, which can be engineered to express enzymes for biofilm destruction, to combat biofilm-forming bacteria infections |
Another microbiome-related approach to treating disease may be the use of prebiotics, which consist of substrates that promote the growth and/or metabolic activity of beneficial indigenous microbiota. Current prebiotics are primarily associated with ingestion and consequent manipulation of the gut microbiome. Different types of gut prebiotics such as galacto- and long-chain fructo-oligosaccharides show promise in treating infants with AD (Moro et al. 2006; Arslanoglu et al. 2008). But imaginable prebiotic approaches such as treating the skin with substrates to alter the environmental conditions and thus promote or discourage the growth of certain microbiota may offer promise for the treatment of skin disorders whose pathogenesis is clearly linked to a microbial cause.
Genetic engineering of microorganisms as vectors for delivery of therapeutic genes is another area of active investigation. The potential utility of such approaches was shown by a study in which E. coli was engineered to express a quorum-sensing peptide that is naturally expressed by Vibrio cholera and inhibits V. cholera virulence (Duan and March 2010). Administration of the genetically engineered microbe to the gut of infant mice before challenge with V. cholera significantly increased survival while decreasing cholera toxin binding to the intestines. Bacteriophage can also be engineered and administered for therapeutic benefit. For example, bacteriophage have been used to deliver gene constructs to reverse antibiotic resistance in bacteria populations (Edgar et al. 2012). This approach provides the first steps toward applying evolutionary pressure against antibiotic resistance, while reversing the pressure toward antibiotic resistance from decades of antibiotic use. Of particular concern to the skin is multidrug resistance in skin-associated opportunistic pathogens, such as S. epidermidis, S. aureus, and Pseudomonas aeruginosa (Branski et al. 2009; Otto 2009). Bacteriophages have also been engineered to degrade bacterial biofilms (Lu and Collins 2007), a significant therapeutic challenge because antibiotics are often not able to physically access the bacteria comprising the biofilm, and are therefore not effective in treating them.
An in-depth understanding of the human skin microbiota may also have important implications in informing synthetic biology therapeutics. For example, comparative genomic analysis of P. acnes bacteriophage genomes led to the discovery of a highly conserved gene-encoding endolysin, an enzyme with broad lytic potential for P. acnes hosts (Marinelli et al. 2012). The utility of endolysin as an antibacterial has been shown in other phage–host systems, and bacterial resistance to the recombinant protein was not observed even after repeated exposure (Fischetti 2008).
CONCLUDING REMARKS
The skin acts as both a protective physical barrier between the body and the external environment, as well as an environmental substrate that harbors rich and diverse communities of microorganisms that contribute to skin health and disease. The recent advent of molecular and metagenomic techniques for microbial community analysis has addressed many culture-based limitations. As a result, a greater appreciation of the microbial diversity across different skin sites as well as the diversity between people, over time, has emerged. It is becoming increasingly apparent that certain microbes promote healthy skin equilibrium, and contemporary molecular approaches have also provided greater information about the role of microbial community disturbances in disease pathogenesis.
The commensal fungal and viral communities in either health or disease remain largely uncharacterized, and future investigations are likely to focus on these knowledge gaps. Most molecular studies up to this point have focused on taxonomic characterization of microbial skin communities. Although this approach is valuable because taxonomy provides a functional proxy for patterns of the genomes present, new techniques will allow for more in-depth investigations, beyond taxonomic identification. In light of increasingly feasible whole metagenomic shotgun sequencing approaches, investigations will be able to focus directly on the genetic functional potential and assess the community compositions of relevant genes. We also expect that, with the ever-advancing technologies and bioinformatics associated with mRNA sequencing (the metatranscriptome) and protein community analysis (the metaproteome), significant effort will be directed toward the functional aspects of microbiomes associated with skin health and disease.
Finally, a looming challenge is applying this knowledge to develop therapeutic and diagnostic tools for the clinic. Enhanced understanding of the skin microbiome will continue to inform research toward probiotic and prebiotic development, prevention of antibiotic resistance gene transfer, bacteriophage-mediated treatments, and gene delivery using bacterial vectors. New therapeutic developments will allow for a type of “microbiome engineering” in which the community composition will be stimulated and/or manipulated to include beneficial components. Additionally, in light of increasing antibiotic resistance across medically relevant bacterial populations, there will likely be an increased interest in alternative approaches to treating infections, as well as slowing the spread of resistance.
ACKNOWLEDGMENTS
We thank members of the Grice laboratory for their underlying contributions. G.D.H. is supported by the Department of Defense through the National Defense Science and Engineering Graduate Fellowship Program. E.A.G. gratefully acknowledges the support of the National Institutes of Health (AR060873) and the University of Pennsylvania Skin Disease Research Center (supported by AR057217).
Footnotes
Editors: Anthony E. Oro and Fiona M. Watt
Additional Perspectives on The Skin and Its Diseases available at www.perspectivesinmedicine.org
REFERENCES
- Aly R, Maibach HE, Mandel A 1976. Bacterial flora in psoriasis. Br J Dermatol 95: 603–606 [DOI] [PubMed] [Google Scholar]
- Antonsson A, Forslund O, Ekberg H, Sterner G, Hansson BG 2000. The ubiquity and impressive genomic diversity of human skin papillomaviruses suggest a commensalic nature of these viruses. J Virol 74: 11636–11641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonsson A, Erfurt C, Hazard K, Holmgren V, Simon M, Kataoka A, Hossain S, Hakangard C, Hansson BG 2003a. Prevalence and type spectrum of human papillomaviruses in healthy skin samples collected in three continents. J Gen Virol 84: 1881–1886 [DOI] [PubMed] [Google Scholar]
- Antonsson A, Karanfilovska S, Lindqvist PG, Hansson BG 2003b. General acquisition of human papillomavirus infections of skin occurs in early infancy. J Clin Microbiol 41: 2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arslanoglu S, Moro GE, Schmitt J, Tandoi L, Rizzardi S, Boehm G 2008. Early dietary intervention with a mixture of prebiotic oligosaccharides reduces the incidence of allergic manifestations and infections during the first two years of life. J Nutr 138: 1091–1095 [DOI] [PubMed] [Google Scholar]
- Astori G, Lavergne D, Benton C, Hockmayr B, Egawa K, Garbe C, de Villiers EM 1998. Human papillomaviruses are commonly found in normal skin of immunocompetent hosts. J Invest Dermatol 110: 752–755 [DOI] [PubMed] [Google Scholar]
- Batchelor JM, Grindlay DJ, Williams HC 2010. What's new in atopic dermatitis? An analysis of systematic reviews published in 2008 and 2009. Clin Exp Dermatol 35: 823–827 [DOI] [PubMed] [Google Scholar]
- Becker JC, Houben R, Ugurel S, Trefzer U, Pfohler C, Schrama D 2009. MC polyomavirus is frequently present in Merkel cell carcinoma of European patients. J Invest Dermatol 129: 248–250 [DOI] [PubMed] [Google Scholar]
- Blaser MJ, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Estrada I, Gao Z, Clemente JC, Costello EK, Knight R 2013. Distinct cutaneous bacterial assemblages in a sampling of South American Amerindians and US residents. ISME J 7: 85–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehncke WH, Dressel D, Zollner TM, Kaufmann R 1996. Pulling the trigger on psoriasis. Nature 379: 777. [DOI] [PubMed] [Google Scholar]
- Branski LK, Al-Mousawi A, Rivero H, Jeschke MG, Sanford AP, Herndon DN 2009. Emerging infections in burns. Surg Infect (Larchmt) 10: 389–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulmer AC, Bulmer GS 1999. The antifungal action of dandruff shampoos. Mycopathologia 147: 63–65 [DOI] [PubMed] [Google Scholar]
- Capone KA, Dowd SE, Stamatas GN, Nikolovski J 2011. Diversity of the human skin microbiome early in life. J Invest Dermatol 131: 2026–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase MW, Fay MF 2009. Ecology. Barcoding of plants and fungi. Science 325: 682–683 [DOI] [PubMed] [Google Scholar]
- Chehoud C, Rafail S, Tyldsley AS, Seykora JT, Lambris JD, Grice EA 2013. Complement modulates the cutaneous microbiome and inflammatory milieu. Proc Natl Acad Sci 110: 15061–15066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R 2009. Bacterial community variation in human body habitats across space and time. Science 326: 1694–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Villiers EM, Lavergne D, McLaren K, Benton EC 1997. Prevailing papillomavirus types in non-melanoma carcinomas of the skin in renal allograft recipients. Int J Cancer 73: 356–361 [DOI] [PubMed] [Google Scholar]
- Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R 2010. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci 107: 11971–11975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotterud CK, Storro O, Johnsen R, Oien T 2010. Probiotics in pregnant women to prevent allergic disease: A randomized, double-blind trial. Br J Dermatol 163: 616–623 [DOI] [PubMed] [Google Scholar]
- Duan F, March JC 2010. Engineered bacterial communication prevents Vibrio cholerae virulence in an infant mouse model. Proc Natl Acad Sci 107: 11260–11264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncavage EJ, Zehnbauer BA, Pfeifer JD 2009. Prevalence of Merkel cell polyomavirus in Merkel cell carcinoma. Mod Pathol 22: 516–521 [DOI] [PubMed] [Google Scholar]
- Edgar R, Friedman N, Molshanski-Mor S, Qimron U 2012. Reversing bacterial resistance to antibiotics by phage-mediated delivery of dominant sensitive genes. Appl Environ Microbiol 78: 744–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans CA, Smith WM, Johnston EA, Giblett ER 1950. Bacterial flora of the normal human skin. J Invest Dermatol 15: 305–324 [DOI] [PubMed] [Google Scholar]
- Fahlen A, Engstrand L, Baker BS, Powles A, Fry L 2012. Comparison of bacterial microbiota in skin biopsies from normal and psoriatic skin. Arch Dermatol Res 304: 15–22 [DOI] [PubMed] [Google Scholar]
- Fancello L, Desnues C, Raoult D, Rolain JM 2011. Bacteriophages and diffusion of genes encoding antimicrobial resistance in cystic fibrosis sputum microbiota. J Antimicrob Chemother 66: 2448–2454 [DOI] [PubMed] [Google Scholar]
- Feng H, Shuda M, Chang Y, Moore PS 2008. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 319: 1096–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Hamady M, Lauber CL, Knight R 2008. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci 105: 17994–17999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, Schoenfeld D, Nomicos E, Park M, Becker J, et al. 2013. Topographic diversity of fungal and bacterial communities in human skin. Nature 498: 367–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischetti VA 2008. Bacteriophage lysins as effective antibacterials. Curr Opin Microbiol 11: 393–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitz-Gibbon S, Tomida S, Chiu BH, Nguyen L, Du C, Liu M, Elashoff D, Erfe MC, Loncaric A, Kim J, et al. 2013. Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J Invest Dermatol 133: 2152–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foolad N, Brezinski EA, Chase EP, Armstrong AW 2013. Effect of nutrient supplementation on atopic dermatitis in children: A systematic review of probiotics, prebiotics, formula, and fatty acids. JAMA Dermatol 149: 350–355 [DOI] [PubMed] [Google Scholar]
- Forslund O 2007. Genetic diversity of cutaneous human papillomaviruses. J Gen Virol 88: 2662–2669 [DOI] [PubMed] [Google Scholar]
- Foulongne V, Kluger N, Dereure O, Brieu N, Guillot B, Segondy M 2008. Merkel cell polyomavirus and Merkel cell carcinoma, France. Emerg Infect Dis 14: 1491–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulongne V, Dereure O, Kluger N, Moles JP, Guillot B, Segondy M 2010. Merkel cell polyomavirus DNA detection in lesional and nonlesional skin from patients with Merkel cell carcinoma or other skin diseases. Br J Dermatol 162: 59–63 [DOI] [PubMed] [Google Scholar]
- Foulongne V, Sauvage V, Hebert C, Dereure O, Cheval J, Gouilh MA, Pariente K, Segondy M, Burguiere A, Manuguerra JC, et al. 2012. Human skin microbiota: High diversity of DNA viruses identified on the human skin by high throughput sequencing. PLoS ONE 7: e38499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E, Raghavan S 2002. Getting under the skin of epidermal morphogenesis. Nat Rev Genet 3: 199–209 [DOI] [PubMed] [Google Scholar]
- Gaitanis G, Magiatis P, Hantschke M, Bassukas ID, Velegraki A 2012. The Malassezia genus in skin and systemic diseases. Clin Microbiol Rev 25: 106–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Tseng CH, Pei Z, Blaser MJ 2007. Molecular analysis of human forearm superficial skin bacterial biota. Proc Natl Acad Sci 104: 2927–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Tseng CH, Strober BE, Pei Z, Blaser MJ 2008. Substantial alterations of the cutaneous bacterial biota in psoriatic lesions. PLoS ONE 3: e2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goerke C, Pantucek R, Holtfreter S, Schulte B, Zink M, Grumann D, Broker BM, Doskar J, Wolz C 2009. Diversity of prophages in dominant Staphylococcus aureus clonal lineages. J Bacteriol 191: 3462–3468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb AB 2005. Therapeutic options in the treatment of psoriasis and atopic dermatitis. J Am Acad Dermatol 53: S3–S16 [DOI] [PubMed] [Google Scholar]
- Grice EA, Segre JA 2012. The human microbiome: Our second genome. Annu Rev Genomics Hum Genet 13: 151–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Kong HH, Renaud G, Young AC, Bouffard GG, Blakesley RW, Wolfsberg TG, Turner ML, Segre JA 2008. A diversity profile of the human skin microbiota. Genome Res 18: 1043–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Kong HK, Conlan S, Deming CB, Davis J, Young AC, Program NCS, Bouffard GG, Blakesley RW, Murray PR, et al. 2009. Topographical and temporal diversity of the human skin microbiome. Science 324: 1190–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haitz KA, Rady PL, Nguyen HP, He Q, Prieto VG, Tyring SK, Ciurea AM 2012. Merkel cell polyomavirus DNA detection in a patient with Merkel cell carcinoma and multiple other skin cancers. Int J Dermatol 51: 442–444 [DOI] [PubMed] [Google Scholar]
- Hodgson NC 2005. Merkel cell carcinoma: Changing incidence trends. J Surg Oncol 89: 1–4 [DOI] [PubMed] [Google Scholar]
- Huang JT, Abrams M, Tlougan B, Rademaker A, Paller AS 2009. Treatment of Staphylococcus aureus colonization in atopic dermatitis decreases disease severity. Pediatrics 123: e808–e814 [DOI] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium 2012. Structure, function and diversity of the healthy human microbiome. Nature 486: 207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HS, Choi YL, Choi JS, Roh JH, Pyon JK, Woo KJ, Lee EH, Jang KT, Han J, Park CS, et al. 2011. Detection of Merkel cell polyomavirus in Merkel cell carcinomas and small cell carcinomas by PCR and immunohistochemistry. Histol Histopathol 26: 1231–1241 [DOI] [PubMed] [Google Scholar]
- Kaga M, Sugita T, Nishikawa A, Wada Y, Hiruma M, Ikeda S 2011. Molecular analysis of the cutaneous Malassezia microbiota from the skin of patients with atopic dermatitis of different severities. Mycoses 54: e24–e28 [DOI] [PubMed] [Google Scholar]
- Kalliomaki M, Salminen S, Arvilommi H, Kero P, Koskinen P, Isolauri E 2001. Probiotics in primary prevention of atopic disease: A randomised placebo-controlled trial. Lancet 357: 1076–1079 [DOI] [PubMed] [Google Scholar]
- Kassem A, Schopflin A, Diaz C, Weyers W, Stickeler E, Werner M, Zur Hausen A 2008. Frequent detection of Merkel cell polyomavirus in human Merkel cell carcinomas and identification of a unique deletion in the VP1 gene. Cancer Res 68: 5009–5013 [DOI] [PubMed] [Google Scholar]
- Katano H, Ito H, Suzuki Y, Nakamura T, Sato Y, Tsuji T, Matsuo K, Nakagawa H, Sata T 2009. Detection of Merkel cell polyomavirus in Merkel cell carcinoma and Kaposi’s sarcoma. J Med Virol 81: 1951–1958 [DOI] [PubMed] [Google Scholar]
- Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, Nomicos E, Polley EC, Komarow HD, Murray PR, et al. 2012. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res 22: 850–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan T, Liu J, DuBow M, Gros P, Pelletier J 2005. The complete genomes and proteomes of 27 Staphylococcus aureus bacteriophages. Proc Natl Acad Sci 102: 5174–5179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y, Di Nardo A, Nakatsuji T, Leichtle A, Yang Y, Cogen AL, Wu ZR, Hooper LV, Schmidt RR, von Aulock S, et al. 2009. Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nat Med 15: 1377–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR 1985. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci 82: 6955–6959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew LC, Liong MT 2013. Bioactives from probiotics for dermal health: Functions and benefits. J Appl Microbiol 114: 1241–1253 [DOI] [PubMed] [Google Scholar]
- Lu TK, Collins JJ 2007. Dispersing biofilms with engineered enzymatic bacteriophage. Proc Natl Acad Sci 104: 11197–11202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangana J, Dziunycz P, Kerl K, Dummer R, Cozzio A 2010. Prevalence of Merkel cell polyomavirus among Swiss Merkel cell carcinoma patients. Dermatology 221: 184–188 [DOI] [PubMed] [Google Scholar]
- Marinelli LJ, Fitz-Gibbon S, Hayes C, Bowman C, Inkeles M, Loncaric A, Russell DA, Jacobs-Sera D, Cokus S, Pellegrini M, et al. 2012. Propionibacterium acnes bacteriophages display limited genetic diversity and broad killing activity against bacterial skin isolates. mBio 3: e00279–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marples MJ 1965. The ecology of the human skin. Thomas, Springfield, IL [Google Scholar]
- McGinley KJ, Leyden JJ, Marples RR, Kligman AM 1975. Quantitative microbiology of the scalp in non-dandruff, dandruff, and seborrheic dermatitis. J Invest Dermatol 64: 401–405 [DOI] [PubMed] [Google Scholar]
- Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, Wu GD, Lewis JD, Bushman FD 2011. The human gut virome: Inter-individual variation and dynamic response to diet. Genome Res 21: 1616–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens U, Ludvigsen M, Van Ghelue M 2011. Human polyomaviruses in skin diseases. Patholog Res Int 2011: 123491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro G, Arslanoglu S, Stahl B, Jelinek J, Wahn U, Boehm G 2006. A mixture of prebiotic oligosaccharides reduces the incidence of atopic dermatitis during the first six months of age. Arch Dis Child 91: 814–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muizzuddin N, Maher W, Sullivan M, Schnittger S, Mammone T 2012. Physiological effect of a probiotic on skin. J Cosmet Sci 63: 385–395 [PubMed] [Google Scholar]
- Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, Deming C, Quinones M, Koo L, Conlan S, et al. 2012. Compartmentalized control of skin immunity by resident commensals. Science 337: 1115–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaminami H, Noguchi N, Nishijima S, Kurokawa I, So H, Sasatsu M 2007. Transduction of the plasmid encoding antiseptic resistance gene qacB in Staphylococcus aureus. Biol Pharm Bull 30: 1412–1415 [DOI] [PubMed] [Google Scholar]
- Nakatsuji T, Gallo RL 2012. Antimicrobial peptides: Old molecules with new ideas. J Invest Dermatol 132: 887–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council Committee on Metagenomics 2007. The new science of metagenomics: Revealing the secrets of our microbial planet. The National Academies Press, Washington, DC. [PubMed] [Google Scholar]
- Nickoloff BJ, Schroder JM, von den Driesch P, Raychaudhuri SP, Farber EM, Boehncke WH, Morhenn VB, Rosenberg EW, Schon MP, Holick MF 2000. Is psoriasis a T-cell disease? Exp Dermatol 9: 359–375 [DOI] [PubMed] [Google Scholar]
- Oh J, Conlan S, Polley EC, Segre JA, Kong HH 2012. Shifts in human skin and nares microbiota of healthy children and adults. Genome Med 4: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M 2009. Staphylococcus epidermidis—The “accidental” pathogen. Nat Rev Microbiol 7: 555–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolini F, Donati P, Amantea A, Bucher S, Migliano E, Venuti A 2011. Merkel cell polyomavirus in Merkel cell carcinoma of Italian patients. Virol J 8: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HK, Ha MH, Park SG, Kim MN, Kim BJ, Kim W 2012. Characterization of the fungal microbiota (mycobiome) in healthy and dandruff-afflicted human scalps. PLoS ONE 7: e32847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulino LC, Tseng CH, Strober BE, Blaser MJ 2006. Molecular analysis of fungal microbiota in samples from healthy human skin and psoriatic lesions. J Clin Microbiol 44: 2933–2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulino LC, Tseng CH, Blaser MJ 2008. Analysis of Malassezia microbiota in healthy superficial human skin and in psoriatic lesions by multiplex real-time PCR. FEMS Yeast Res 8: 460–471 [DOI] [PubMed] [Google Scholar]
- Penders J, Stobberingh EE, Thijs C, Adams H, Vink C, van Ree R, van den Brandt PA 2006. Molecular fingerprinting of the intestinal microbiota of infants in whom atopic eczema was or was not developing. Clin Exp Allergy 36: 1602–1608 [DOI] [PubMed] [Google Scholar]
- Penders J, Thijs C, van den Brandt PA, Kummeling I, Snijders B, Stelma F, Adams H, van Ree R, Stobberingh EE 2007. Gut microbiota composition and development of atopic manifestations in infancy: The KOALA Birth Cohort Study. Gut 56: 661–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg EW, Noah PW, Skinner RB Jr 1994. Microorganisms and psoriasis. J Natl Med Assoc 86: 305–310 [PMC free article] [PubMed] [Google Scholar]
- Sandilands A, Terron-Kwiatkowski A, Hull PR, O’Regan GM, Clayton TH, Watson RM, Carrick T, Evans AT, Liao H, Zhao Y, et al. 2007. Comprehensive analysis of the gene encoding filaggrin uncovers prevalent and rare mutations in ichthyosis vulgaris and atopic eczema. Nat Genet 39: 650–654 [DOI] [PubMed] [Google Scholar]
- Scharschmidt TC, List K, Grice EA, Szabo R, Renaud G, Lee CC, Wolfsberg TG, Bugge TH, Segre JA 2009. Matriptase-deficient mice exhibit ichthyotic skin with a selective shift in skin microbiota. J Invest Dermatol 129: 2435–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schowalter RM, Pastrana DV, Pumphrey KA, Moyer AL, Buck CB 2010. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe 7: 509–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu M, Wang Y, Yu J, Kuo S, Coda A, Jiang Y, Gallo RL, Huang CM 2013. Fermentation of Propionibacterium acnes, a commensal bacterium in the human skin microbiome, as skin probiotics against methicillin-resistant Staphylococcus aureus. PLoS ONE 8: e55380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudinger T, Pipal A, Redl B 2011. Molecular analysis of the prevalent microbiota of human male and female forehead skin compared to forearm skin and the influence of make-up. J Appl Microbiol 110: 1381–1389 [DOI] [PubMed] [Google Scholar]
- Tan HH 2003. Antibacterial therapy for acne: A guide to selection and use of systemic agents. Am J Clin Dermatol 4: 307–314 [DOI] [PubMed] [Google Scholar]
- Terada M, Tsutsui H, Imai Y, Yasuda K, Mizutani H, Yamanishi K, Kubo M, Matsui K, Sano H, Nakanishi K 2006. Contribution of IL-18 to atopic-dermatitis-like skin inflammation induced by Staphylococcus aureus product in mice. Proc Natl Acad Sci 103: 8816–8821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touze A, Gaitan J, Maruani A, Le Bidre E, Doussinaud A, Clavel C, Durlach A, Aubin F, Guyetant S, Lorette G, et al. 2009. Merkel cell polyomavirus strains in patients with Merkel cell carcinoma. Emerg Infect Dis 15: 960–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers JB, Kozman A, Mousdicas N, Saha C, Landis M, Al-Hassani M, Yao W, Yao Y, Hyatt AM, Sheehan MP, et al. 2010. Infected atopic dermatitis lesions contain pharmacologic amounts of lipoteichoic acid. J Allergy Clin Immunol 125: 146–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga M, Kuntova L, Pantucek R, Maslanova I, Ruzickova V, Doskar J 2012. Efficient transfer of antibiotic resistance plasmids by transduction within methicillin-resistant Staphylococcus aureus USA300 clone. FEMS Microbiol Lett 332: 146–152 [DOI] [PubMed] [Google Scholar]
- Wang M, Karlsson C, Olsson C, Adlerberth I, Wold AE, Strachan DP, Martricardi PM, Aberg N, Perkin MR, Tripodi S, et al. 2008. Reduced diversity in the early fecal microbiota of infants with atopic eczema. J Allergy Clin Immunol 121: 129–134 [DOI] [PubMed] [Google Scholar]
- Wang X, Kim Y, Ma Q, Hong SH, Pokusaeva K, Sturino JM, Wood TK 2010. Cryptic prophages help bacteria cope with adverse environments. Nat Commun 1: 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens K, Black PN, Stanley TV, Mitchell E, Fitzharris P, Tannock GW, Purdie G, Crane J 2008. A differential effect of 2 probiotics in the prevention of eczema and atopy: A double-blind, randomized, placebo-controlled trial. J Allergy Clin Immunol 122: 788–794 [DOI] [PubMed] [Google Scholar]
- Wickens K, Black P, Stanley TV, Mitchell E, Barthow C, Fitzharris P, Purdie G, Crane J 2012. A protective effect of Lactobacillus rhamnosus HN001 against eczema in the first 2 years of life persists to age 4 years. Clin Exp Allergy 42: 1071–1079 [DOI] [PubMed] [Google Scholar]
- Yao Y, Kozman A, Al-Hassani M, Saha CK, Yi Q, Yao W, Mousdicas N, Kaplan MH, Travers JB 2010. Identification of staphylococcal protein A in infected atopic dermatitis lesions. J Invest Dermatol 130: 2502–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang E, Tanaka T, Tajima M, Tsuboi R, Nishikawa A, Sugita T 2011. Characterization of the skin fungal microbiota in patients with atopic dermatitis and in healthy subjects. Microbiol Immunol 55: 625–632 [DOI] [PubMed] [Google Scholar]