Abstract
The presence of unrepaired lesions in DNA represents a challenge for replication. Most, but not all, DNA lesions block the replicative DNA polymerases. The conceptually simplest procedure to bypass lesions during DNA replication is translesion synthesis (TLS), whereby the replicative polymerase is transiently replaced by a specialized DNA polymerase that synthesizes a short patch of DNA across the site of damage. This process is inherently error prone and is the main source of point mutations. The diversity of existing DNA lesions and the biochemical properties of Escherichia coli DNA polymerases will be presented. Our main goal is to deliver an integrated view of TLS pathways involving the multiple switches between replicative and specialized DNA polymerases and their interaction with key accessory factors. Finally, a brief glance at how other bacteria deal with TLS and mutagenesis is presented.
Most DNA lesions block replicative DNA polymerases. In these cases, a specialized DNA polymerase synthesizes DNA across the lesion. However, this process is error prone and is the main source of point mutations.
Within the context of this review, we will limit the notion of DNA lesions to chemically altered bases, although the sugar-phosphodiester backbone is also subject to various types of chemical attack leading, for example, to single-strand breaks. Lesions may be spontaneous (e.g., depurinations), induced endogenously (e.g., by reactive oxygen species), induced by radiations (UV light, X rays) or by chemicals. Treatments that induce DNA lesions cause mutations and cancer and are therefore referred to as mutagens or carcinogens. Carcinogens fall into large chemical families of compounds such as aromatic amides, polycyclic hydrocarbons, and nitrosamines. Carcinogens are not necessarily synthetic; for example, some are natural plant metabolites (e.g., Aflatoxin B1, aristolochic acid, etc.). In addition, some drugs used in cancer chemotherapy such as platinum derivatives form covalent DNA adducts and as such are also carcinogens. Drugs from the thiopurine family, such as azathioprine widely used as immunosuppressants in organ transplant patients, form DNA adducts upon interaction with sunlight and promote skin cancer (Zhang et al. 2007).
HOW DNA DAMAGE INTERFERES WITH REPLICATION: NONCODING VERSUS MISCODING LESIONS
Replicative DNA polymerases are highly specialized enzymes capable of accurately copying DNA templates that contain the four “normal” nucleotides A, G, C, or T. In contrast, replicative DNA polymerases fail to insert a dNTP opposite many damaged bases present in the template. Modified bases that impair the progression of ongoing replication are referred to as noncoding or replication-blocking lesions. Cells have developed tolerance pathways to deal with replication-blocking lesions as discussed throughout this review. On the other hand, there are lesions that do not significantly affect the progression of replicative DNA polymerases. These lesions, referred to as miscoding lesions, are usually small base modifications induced by reactive oxygen species (8-oxo-G) or alkylating agents (O6-meG, O4-meT). Replicative DNA polymerases are able to insert a specific nucleotide opposite these lesions producing a nonconventional base pair compatible with the double helix structure (Fig. 1B). As a consequence, these miscoding lesions induce point mutations with high efficiency (Fig. 1B).
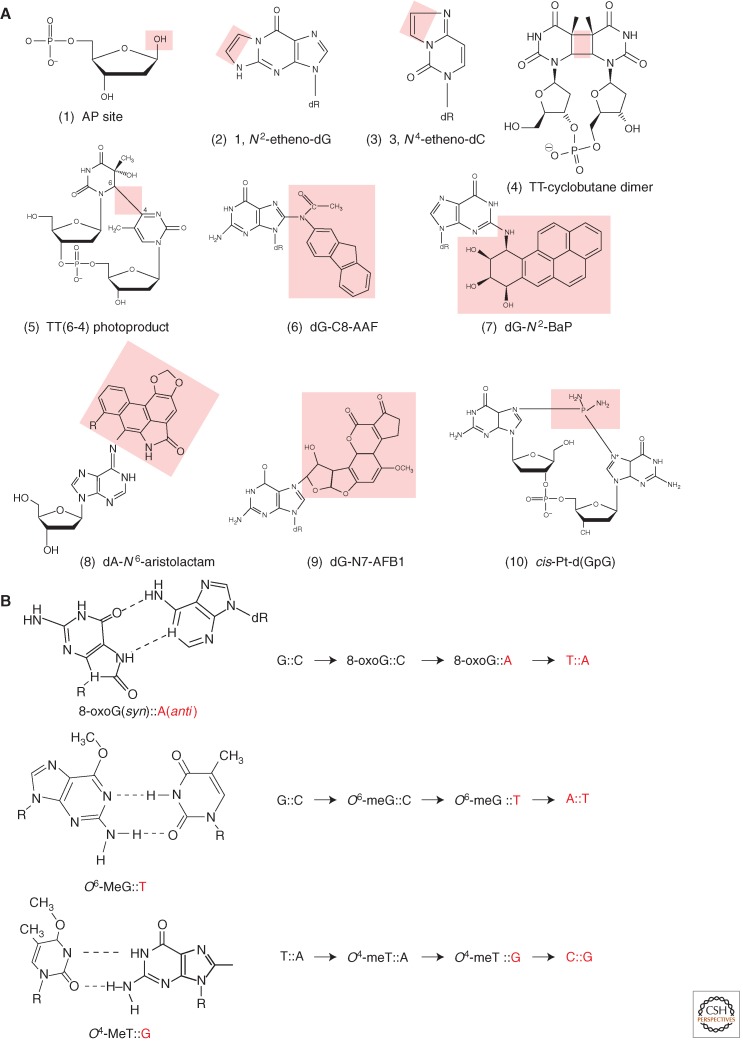
Figure 1.
Diversity of DNA lesions and properties of common miscoding lesions. (A) A glance at the huge diversity of chemical lesions in DNA: Lesions are highlighted by the pink area. (1) Abasic site, a common lesion that can be formed in a variety of ways: spontaneous or alkylation-induced depurination, repair intermediates. (2, 3) G and C etheno-type adducts are formed by various chemicals such as vinyl chloride, lipid peroxidation: A new cycle is formed by double adduction at an exocyclic and an intracyclic nitrogen atom. (4, 5) TT-cyclobutane dimer (CPD) and T(6–4)T photoproduct formed by UV light. (6) dG-C8-AAF is a major adduct formed by an aromatic amine N-2-acetylaminofluorene (AAF), a strong liver carcinogen. (7) B(a)P-N2-dG, the major guanine adduct formed by benzo(a)pyrene, a common polycyclic hydrocarbon found in cigarette smoke and other combustion residues. (8) dA-N6-Aristolactam, an adduct formed by a metabolite of Aristolochia clematitis, a plant that often grows in cultivated fields where its seeds comingle with wheat grain during harvest (Grollman et al. 2007). (9) dG-N7-AFB1 is the major guanine adduct of a potent hepatocarcinogen. Aflatoxin B1, a metabolite produced by a mold that grows on peanuts. (10) cis-Pt-d(GpG) is an intrastrand cross-link produced by the drug cisplatin that is used in human cancer chemotherapy. (B) Examples of direct miscoding lesions. These lesions do not block replicative DNA polymerases. Instead, replicative DNA polymerases efficiently insert a nucleotide opposite these lesions forming a noncanonical base pair that leads to a base substitution at the next replication cycle, as discussed in the text. The nucleotides introduced during replication are shown in red.
The pairing properties of some common miscoding lesions are shown in Figure 1B. During replication, when 8-oxoG is present in the DNA template, replicative DNA polymerases frequently insert A (anti) across 8-oxoG (syn) leading to a GC→TA transversion in the next replication cycle. Similarly, replicative DNA polymerases efficiently misinsert a T residue across an O6-meG lesion in template DNA thus leading to GC→AT transitions. Likewise, O4-meT template lesions readily pair with G during replication leading to TA→CG transitions. It should also be noted that a damaged dNTP, such as 8-oxo-dGTP, can readily be incorporated by replicative DNA polymerases opposite a template A residue, thus leading to AT→CG transversions. In contrast to replication-blocking lesions, miscoding lesions efficiently induce mutations in a process that only involves replicative DNA polymerases. Consequently, dedicated repair systems have evolved to efficiently remove these extremely hazardous lesions (refer to the literature).
LESION-TOLERANCE PATHWAYS: THE CHALLENGE OF DUPLICATING DNA CONTAINING REPLICATION-BLOCKING LESIONS
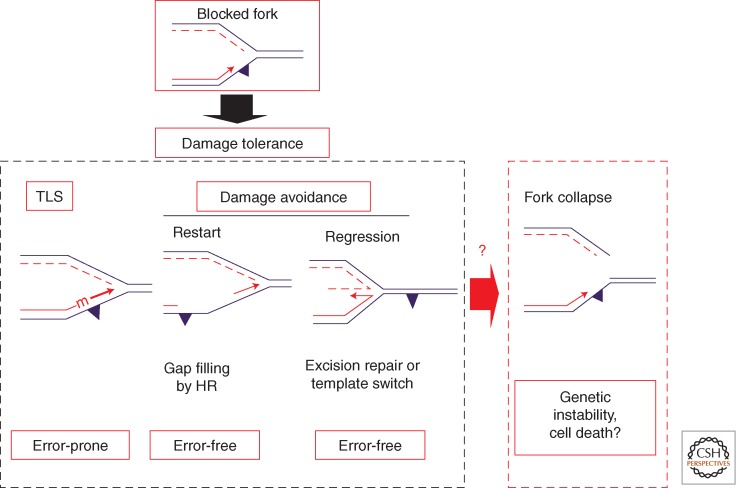
To overcome the challenge of replicating damaged DNA, cells have developed lesion-tolerance mechanisms that enable the replication machinery to bypass sites of damaged DNA. The conceptually simplest procedure of bypassing a lesion that blocks a replication fork is translesion synthesis (TLS), whereby the blocked replicative DNA polymerase is transiently replaced by a specialized DNA polymerase that can extend the nascent strand and synthesize across the site of damage (Fig. 2). This process, although not always mutagenic, is inherently error prone. On the other hand, error-free bypass of DNA lesions is possible by using the information present in the undamaged sister chromatid (Prakash 1989; Frampton et al. 2006; Rudolph et al. 2008; Daigaku et al. 2010; Karras and Jentsch 2010). These processes, collectively referred to as damage avoidance (DA), embrace several pathways related to homologous recombination such as replication fork restart or fork regression (Fig. 2). These pathways are poorly defined both genetically as well as biochemically. Failure to properly achieve these tolerance pathways can lead to replication fork collapse that in turn may result in genetic rearrangements or cell death (Fig. 2).
Figure 2.
Outline of DNA damage-tolerance pathways as triggered by lesions that block the replicative DNA polymerase. There are two DNA damage-tolerance strategies: error-prone translesion synthesis (the topic of the present review) and error-free damage avoidance (DA). DA pathways are still poorly defined and involve either gap filling by homologous recombination or fork regression. If all DNA damage strategies fail, it is likely that the fork collapses leading to gross genetic rearrangements or to cell death.
DEVELOPMENT OF METHODOLOGIES TO MONITOR LESION-TOLERANCE PATHWAYS
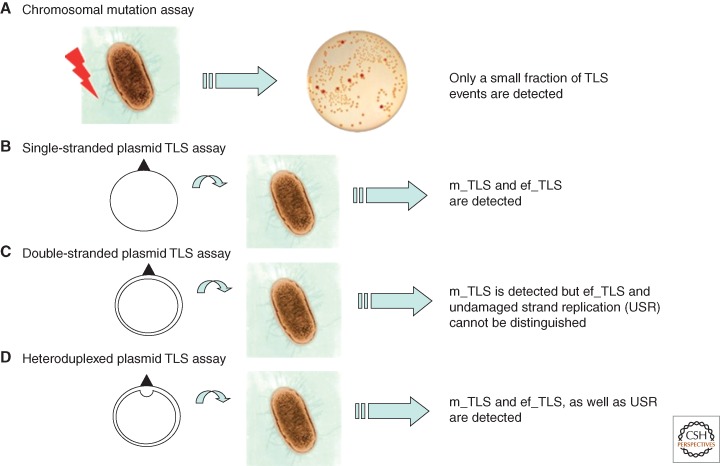
Initially, most studies on the consequences of lesions in DNA in vivo have been limited to the analysis of mutations induced in a given gene. For instance, commonly used mutation assays involve the determination of the frequency of bacteria that become resistant to a given antibiotic following treatment with a mutagen. For instance, resistance to rifampicin or to nalidixic acid is conferred by point mutations in the rpoB or gyrB gene, respectively (Fig. 3A). Such assays only monitor a subfraction of mutagenic TLS events, namely, those that give rise to phenotypically detectable events. Silent and error-free TLS events escape detection. Assays able to monitor all TLS events resulted from the development of single-adducted plasmids in bacteria, yeast, and mammalian cells. Single-adducted, single-stranded plasmids (Fig. 3B) detect all TLS events, the relative frequency of mutagenic and error-free events being subsequently determined by sequence (Banerjee et al. 1988; Napolitano and Fuchs 1997; Napolitano et al. 1997). The major drawback of single-stranded (or gapped plasmid [Gibbs et al. 1995; Paz-Elizur et al. 1996]) assays relates to the fact that TLS does not occur within the context of a bona fide replication fork. Single-adducted double-stranded plasmids (Koehl et al. 1989) represent more adequate tools as they involve actual replication forks (Fig. 3C); however, undamaged strand replication (USR) leads to progeny that cannot be distinguished from error-free TLS unless a genetic marker is introduced. The implementation of double-stranded plasmids with a genetic marker (heteroduplexed plasmids; Fig. 3D) allows the fractions of mutagenic, error-free TLS and USR to be monitored independently (Koffel-Schwartz et al. 1996).
Figure 3.
Methodologies to monitor mutagenesis and TLS. Chromosomal mutation assays (A) monitor, usually within a single target gene, a subset of mutagenic TLS events following the treatment of a cell culture with a mutagen. Several plasmid-based single adduct assays allow mutagenic (m_TLS) and error-free (ef_TLS) TLS events to be monitored quantitatively as well as qualitatively. The specific characteristics of the different assays (B–D) are outlined in the text.
Importantly, plasmid assays are not suitable tools for the determination of DA events. Indeed, plasmid replication patterns show extensive replication fork uncoupling as the USR becomes separated from damaged strand replication in vivo (Pagès and Fuchs 2003; Pagès et al. 2012) and in vitro (Higuchi et al. 2003). Uncoupling of plasmid replication prevents the fork from coming to an arrest, thus precluding DA pathways from taking place (see Fig. 2). As a consequence, to monitor specific DA events requires the introduction of a single replication-blocking lesion into a large replicon such as the E. coli chromosome itself (Pagès et al. 2012).
DISCOVERY AND BIOCHEMICAL PROPERTIES OF E. coli DNA POLYMERASES
Since the discovery of the chemical nature of DNA and its double-helical structure, the existence of enzymes able to duplicate the genetic material was predicted (Watson and Crick 1953). In 1956, Kornberg and colleagues succeeded in purifying an E. coli enzyme, DNA polymerase I (Pol I), satisfying these predictions (Bessman et al. 1956; Lehman et al. 1958). Initially, Pol I was believed to be the replicative polymerase until 1969 when Cairns and colleagues isolated a viable Pol I-deficient strain, thus providing evidence that Pol I could not be the main replicative polymerase (De Lucia and Cairns 1969). Thereafter, Kornberg’s group tried and succeeded in purifying two other DNA polymerases from E. coli in the early 1970s, named DNA polymerase II (Pol II) and DNA polymerase III (Pol III) (Kornberg and Gefter 1970, 1971, 1972). Based on genetics and biochemistry, Pol III was found to be the main replicative polymerase, whereas Pol I was recognized to process Okazaki fragments. On the other hand, the physiological function of Pol II still remains to be fully elucidated. From the discovery of Pol III and for about 30 years it was believed that E. coli only possesses these three polymerases. During that period, the origin of point mutations induced by the action of DNA-damaging agents known as mutagens, were believed to result from the effect of factors that could transiently modify the fidelity of the known DNA polymerases.
Epoch-making discoveries occurred in 1999, when E. coli was shown to encode two additional DNA polymerases, respectively named according to their order of discovery, DNA polymerase IV (Pol IV) (Wagner et al. 1999) and DNA polymerase V (Pol V) (Reuven et al. 1999; Tang et al. 1999). DNA polymerases are categorized into families based on similarities between domain structures. Pol I, II, and III belong to the A, B, and C families, respectively. On the other hand, Pol IV and V showed no similarity to any known family, but had similarities to each other establishing the so-called Y family (Ohmori et al. 2001). Simultaneously, many new DNA polymerases belonging to the Y family were discovered in eukaryotes, including humans, as described in this collection. Typical features of Y-family polymerases are low fidelity, low processivity, and lack of 3′→5′ exonuclease activity (proofreading function); these polymerases are also nonessential for viability. Y-family DNA polymerases were found to be involved in TLS pathways in vivo; they are thus called specialized or TLS polymerase. Thanks to the discovery of these DNA polymerases, research on TLS became more straightforward and the understanding of the molecular mechanisms of TLS has deeply progressed, although there remain many unanswered questions.
First, we will briefly present the properties of the initially characterized “classical” DNA polymerases Pol I, Pol II, and Pol III. Second, the two Y-family DNA polymerases Pol IV and Pol V will be presented in greater detail.
The Initially Characterized “Classical” DNA Polymerases Pol I, Pol II, and Pol III
Pol I
Pol I from E. coli is the first identified DNA polymerase among any kind of species. It is encoded by the polA gene and contains three enzymatic activities: a DNA polymerase, a 3′→5′ exonuclease (proofreading function), and a 5′→3′ exonuclease. Its physiological function is Okazaki fragment maturation and DNA repair synthesis during nucleotide excision repair (NER). Before 1999, many biochemical studies involving Pol I in the context of TLS were conducted. For example, Pol I was found to be able to bypass AP sites more efficiently than Pol III. In particular, the proofreading-deficient form of Pol I was found to efficiently bypass in vitro templates containing AP sites, thymine glycol, and G-AAF adducts making it a potential candidate for bypassing lesions in vivo (Clark and Beardsley 1989; Shibutani and Grollman 1993; Belguise-Valladier et al. 1994, 1996; Paz-Elizur et al. 1997). However, there is no genetic evidence to support such a possibility.
Pol II
The role of Pol II remains enigmatic as cells deficient in polB/dinA lack any clear phenotype under normal growth conditions. Pol II possesses a DNA polymerase and a 3′→5′ exonuclease activity (proofreading function). In contrast to Pol I/III, its expression increases about sevenfold during SOS induction (Qiu and Goodman 1997), suggesting a potential role under stress conditions. There are two interesting polB phenotypes: Although polB cells are not UV sensitive, replication following UV irradiation is essentially blocked during ≈50 min, whereas replication resumes gradually during that period in wild-type cells (Rangarajan et al. 1999, 2002). In addition, polB mutants show a severe reduction in fitness when incubated in stationary phase together with wild-type bacteria (Yeiser et al. 2002). The precise mechanisms underlying both phenomena are unknown.
With respect to TLS activity, Pol II shows efficient bypass across AP sites compared with Pol I/III (Paz-Elizur et al. 1996). It was thus proposed that Pol II is responsible for AP-site bypass although there is no clear genetic evidence. The first example that Pol II acts as a TLS polymerase both in vivo and in vitro comes from the bypass of a single G-AAF adduct located within a particular −2 frameshift hot spot sequence (Napolitano et al. 2000; Becherel and Fuchs 2001). This bypass pathway is almost exclusively dependent on Pol II, and a recent crystal structure of Pol II (Wang and Yang 2009) reveals that this polymerase can accommodate the −2 frameshift intermediate despite Pol II being a high-fidelity polymerase (see below). In this pathway, Pol II absolutely requires interaction with the processivity factor (β-clamp) in vivo but not in vitro (Becherel and Fuchs 2001; Becherel et al. 2002; Fujii and Fuchs 2007). The 3,N4-ethenocytosine lesion (εC) (Fig. 1A) is bypassed by Pol II in vitro as well as in vivo. In vivo, εC bypass requires Pol II to interact with the β-clamp (Al Mamun and Humayun 2006).
Pol III
Pol III is the replicative polymerase duplicating most genomic DNA and one of the largest stable protein complexes that can be purified homogeneously from E. coli extracts. Pol III has several different names (Pol III core, Pol III′, Pol III*, and Pol III-HE) depending on the combination of participating subunits. The largest complex, called Pol III holoenzyme (HE), is composed of 10 different subunits, including the clamp loader (the so-called DnaX complex: either γ complex or τ complex) and the processivity factor (β-clamp). This form, which represents the physiologically relevant functional complex, contains either two Pol III-core subcomplexes (Pol III core) when assembled with the γ complex, or three Pol III cores when the assembly contains the τ complex instead (McHenry 2011b). Pol III-HE possesses DNA polymerase activity and 3′→5′ exonuclease activity (proofreading function). The two core subunits ensure simultaneous leading and lagging strand synthesis at the replication fork in vivo. During replication, when a lesion is present in one of the two template strands, Pol III is obviously the first DNA polymerase to encounter the DNA damage. In the early days, extensive genetic studies revealed a set of key genes essential for induced mutagenesis, namely, RecA, UmuD, UmuC, and Pol III. However, purified Pol III has essentially no capacity to bypass bulky DNA lesions (e.g., UV dimers) in vitro. It was thus believed for many years that the genetically identified proteins (i.e., RecA, UmuD′ derived from UmuD, UmuC) somehow modify the stringency of Pol III to allow it to copy damage-containing templates. This hypothesis turned out to be wrong when the umuDC locus was found to encode a DNA polymerase (see below). It should, however, be stressed that there is a category of lesions, the so-called miscoding lesions, that are efficiently bypassed by Pol III with a high propensity of induced mutations (8-oxo-G, O6-alkyl-G, and O4-alkyl-T) (see Fig. 1B). As lesions such as 8-oxo-G form endogenously, bypass of such lesions largely contributes to what is usually referred to as “spontaneous mutagenesis.” Another class of spontaneous mutations is owing to genuine replication errors that escape proofreading. Proofreading is mediated by the 3′-5′ exonuclease activity associated with replicative DNA polymerases. Proofreading requires melting to the nascent primer-template extremity to allow the primer strand to migrate from polymerase to exonuclease activity sites. Distortions in the primer template caused either by a terminal mismatch or by a misalignment in repetitive sequences delay the next nucleotide addition step and provide a necessary time frame for proofreading to occur. Proofreading thus limits the occurrence of both base substitutions and frameshift mutations within repetitive sequences (Johnson et al. 2003; Hsu et al. 2004).
The “Specialized” DNA Polymerases Pol IV and Pol V
Since 1970 and for about 30 years, it was strongly believed that all E. coli DNA polymerases had been discovered, as there was neither biochemical, nor bioinformatic evidence to support the existence of additional DNA polymerases. Genetics of induced mutagenesis highlighted the absolute requirement of both recA and the umuDC operon to support induced mutagenesis in vivo. As none of the purified DNA polymerases could efficiently copy in vitro templates containing UV-induced photoproducts or AP sites, it was assumed that UmuDC and RecA help Pol III replicate through lesions. The dogma that the umuDC locus encodes factors that modify Pol III became overwhelmingly strong and prevented many scientists involved in this field to consider that UmuDC could actually be a DNA polymerase per se. This situation changed when it was shown that the dinB gene product shares strong local sequence homologies with UmuC-like proteins, including REV1 protein from Saccharomyces cerevisiae (Larimer et al. 1989; Ohmori et al. 1995; Kulaeva et al. 1996). Woodgate and his colleagues (Kulaeva et al. 1996) also noted that “the high level of sequence conservation between UmuC-like proteins from bacteria, archaea and eukaryotes suggests that these proteins may have an enzymatic activity, the nature of which remains to be determined.” This prediction was first supported by the discovery that the REV1 protein was endowed with a highly specific deoxycitidyl transferase activity in vitro (Nelson et al. 1996).
Pol IV
Pol IV, encoded by the SOS-controlled dinB/dinP gene, possesses DNA polymerase activity but lacks 3′→5′ proofreading exonuclease activity. During SOS induction, its expression level increases about 10-fold, from about 250 to 2500 molecules/cell (Kim et al. 2001). In contrast to umuDC, before the formal demonstration that dinB encodes a DNA polymerase, it was not regarded as a factor directly involved in TLS. Instead it was known to be involved in a process known as untargeted phage λ mutagenesis (Brotcorne-Lannoye and Maenhaut-Michel 1986). Later on, it was found that robust overexpression of DinB leads to an increase in −1 frameshift mutations within short repeats of G residues, a property known as the dinB-mutator phenotype (Kim et al. 1997). When DinB was recognized as a DNA polymerase (Wagner et al. 1999), its potential function as a TLS polymerase was documented in vivo and in vitro. From biochemical experiments, Pol IV is reported to bypass various lesions (e.g., 8-oxo-dG, O6-medG, AP site, AAF, AF, CPD, 6-4 PP, BaP, NFZ, and 4-NQO) either efficiently or inefficiently (Tang et al. 2000; Suzuki et al. 2001; Shen et al. 2002; Maor-Shoshani et al. 2003). However, genetic evidence for the bypass of only a few N2-guanine adducts (e.g., BaP, NFZ, or 4-NQO) by Pol IV has been obtained so far (Lenne-Samuel et al. 2000; Napolitano et al. 2000; Kim et al. 2001; Shen et al. 2002; Yin et al. 2004; Jarosz et al. 2006; Seo et al. 2006). It should be stressed that these lesions have in common their location in the minor groove of DNA. Polymerization activity of Pol IV per se is distributive (one nucleotide insertion per binding event), its processivity highly increases (∼30–400 nucleotides) upon interaction with the β-clamp (Wagner et al. 2000). Under physiological levels of expression, Pol IV was shown not to contribute to chromosomal mutation rates (Kuban et al. 2004; Wolff et al. 2004). In contrast, upon Pol IV overexpression (>10,000 molecules/cell), cells show minor growth defects (Kuban et al. 2005); under these conditions, a decrease in replication fidelity has been observed (Kim et al. 1997; Wagner and Nohmi 2000; Kuban et al. 2005). Kuban and colleagues suggest that under high levels of expression, Pol IV may specifically extend replication errors made by Pol III in the lagging strand (Kuban et al. 2005). Massive overproduction of Pol IV (>100,000 molecules/cell) was shown to arrest replication forks and to be lethal (Uchida et al. 2008). In vitro, Pol IV can mediate a dynamic DNA polymerase exchange replacing Pol III on the sliding clamp (Indiani et al. 2005; Furukohri et al. 2008). As for Pol II, it was shown that the interaction of Pol IV with the β-clamp is essential to support its TLS activity in vivo (Becherel et al. 2002). Until recently, it was thought that Pol III only contacts a single monomer of the dimeric β-clamp as shown in vitro (Sutton et al. 2010). Together these observations appeared to comfort the so-called tool-belt model predicting simultaneous binding to the clamp of the replicative polymerase and a specialized polymerase (Pagès and Fuchs 2002; Indiani et al. 2005). However, two recent papers showed that under normal conditions Pol III occupies both β-clamp-binding pockets, one being occupied by the α subunit and the other by the ε proofreading subunit (Jergic et al. 2013; Toste Rêgo et al. 2013). It is suggested that the ε-β interaction selected during evolution is weak, and thus suited for transient disruption allowing, for instance, the recruitment of a specialized DNA polymerase such as Pol IV or Pol V. In addition to its role in lesion bypass, Pol IV appears to be involved in several other physiological functions. First, under stress conditions, the expression of Pol IV is up-regulated and plays an essential role in adaptive mutagenesis (McKenzie et al. 2001; Slechta et al. 2003). Second, Pol IV (as well as Pol V), confers a competitive fitness advantage during the stationary phase of the bacterial life cycle (Yeiser et al. 2002). Third, although the molecular mechanism remains unknown, Pol IV contributes to the recovery of arrested transcription events caused by DNA damage through its interaction with NusA (Cohen et al. 2010). Fourth, Pol IV is an essential factor to prevent hydroxyurea-induced cell death in a umuC122 (a mutant allele of the catalytic subunit of Pol V) background in vivo (Godoy et al. 2006). Interestingly, this phenomenon not only requires UmuD′ (a subunit of Pol V) but also UmuD (the precursor of UmuD′). Thus, besides their role in TLS, Y-family polymerases play distinct role(s) under stress conditions. Fifth, it is reported that Pol IV (and also Pol V) possess an intrinsic AP lyase activity, although there is no genetic evidence related to their participation in the base excision repair (BER) pathway (Shen et al. 2005).
Pol V
Genetically, Pol V is clearly the main TLS polymerase in E. coli as umuDC strains show a dramatic decrease of UV-induced mutation frequency (Kato and Shinoura 1977; Steinborn 1978). Surprisingly though, umuDC strains show only moderate UV sensitivity. Even triple TLS polymerase strains umuDC, dinB, and polB are not very UV sensitive (Courcelle et al. 2005), strongly suggesting that TLS plays a minor role in terms of survival to genotoxic agents (see below). In contrast to Pol II and Pol IV, Pol V could not be detected biochemically in non-SOS-induced E. coli strains. In addition, there is no functional evidence for the presence of Pol V in non-SOS-induced cells. Indeed, the level of bypass of a single TT(6-4) lesion is similarly low in a ΔumuDC (0.26%) and in a wild-type (0.35%) strain (Becherel and Fuchs 1999) suggesting that there is no functional Pol V molecule in non-SOS-induced E. coli cells. Expression of Pol V is SOS controlled and occurs ∼50 min after UV irradiation. As for Pol II and Pol IV, the interaction of Pol V with the β-clamp is essential for Pol V-mediated TLS activities (Becherel et al. 2002). Pol V, the UmuD′2C heterotrimer encoded by the SOS-controlled umuDC operon, contains UmuC and a UmuD′ homodimer, an accessory subunit derived from UmuD (Reuven et al. 1999; Tang et al. 1999). Livneh and colleagues showed that a soluble form of UmuC, amino-terminal fusion to the maltose-binding domain, possesses weak DNA polymerase activity (Reuven et al. 1999). Unfortunately, the MBP-UmuC fusion turned out to lack important properties of native Pol V as it fails to be stimulated by the β-clamp (Reuven et al. 1999). Pol V lacks 3′→5′ exonuclease activity (proofreading function). Strikingly, under optimal conditions in the presence of both RecA and the β-clamp, its velocity is only ≈0.3 nucleotide(s) (Fujii and Fuchs 2004). It is thus the slowest E. coli polymerase (e.g., Pol III-HE, >650 nucleotides(s); Pol IV with the β-clamp, ≈2 nucleotides(s) [Wagner et al. 2000]). On a lesion-free template, its average processivity, in the presence of the β-clamp, is ≈25 nucleotides. Although this value slightly decreases in the presence of DNA damage, Pol V appears to be able to replicate across many different lesions with efficiencies similar to replication of undamaged DNA (Fujii and Fuchs 2004).
Historically, UV-induced damage and AP sites are regarded as the most representative DNA damage. Consequently, there was a lot of interest in the study of umuDC mutants because these mutants were isolated as UV nonmutable strains (Kato and Shinoura 1977; Steinborn 1978). In contrast to polB and dinB, the two other SOS-inducible E. coli DNA polymerases, there is essentially no expression of Pol V in the absence of SOS induction (see above). Such tight regulation is achieved by the following successive control steps: at the level of transcription, the promoter being strongly repressed by LexA; at the posttranslational level, as UmuD needs to be processed into UmuD′ (Burckhardt et al. 1988; Nohmi et al. 1988; Shinagawa et al. 1988); and at the level of protein degradation by limiting their half-life (Goodman 2002; Jarosz et al. 2007). When UmuD′2C was recognized as a DNA polymerase, its biochemical properties triggered a lot of interest.
The laboratories of Goodman and Woodgate, and our own team, succeeded in purifying native Pol V. The two studies led to rather distinct models for the mode of action of Pol V during TLS; we will thus carefully review the main conclusions that emerged from both laboratories as reported in the literature.
Pol V Works from the Goodman and Woodgate Laboratories
Over the years, these investigators proposed a series of models, which they referred to as “evolution of translesion synthesis models” describing various models as to how Pol V may function (Schlacher et al. 2006). Most of their efforts aimed at finding experimental conditions where Pol V shows robust polymerase activity despite the fact that Pol V is expected to have only weak activity in vivo. Here we summarize the trajectory of the successive Pol V working models as published over the years:
-
1.
In 1999, it was proposed that, for the bypass of an AP site, Pol V requires single-stranded binding protein (SSB), the β-clamp, and the presence of an RecA-nucleoprotein filament in the presence of ATP (Tang et al. 1999). Although the requirements of RecA and the β-clamp are in good agreement with genetic data, there is no genetic evidence for the requirement of SSB.
-
2.
In 2000, the same group showed that the β-clamp is dispensable for Pol V-mediated TLS provided ATP-γS, a poorly hydrolyzable ATP analog, is used instead of ATP (Tang et al. 2000). This ATP analog is known to “freeze” the otherwise dynamic structure of the RecA filament. It is possible that the more rigid RecA-ATP-γS complex can somehow “compensate” for the absence of β-clamp by providing to Pol V the additional stability that is normally conferred by the β-clamp. Genetically, however, the interaction of Pol V with the β-clamp is an absolute requirement for TLS (Becherel et al. 2002).
-
3.
In 2001, the same laboratory showed that efficient Pol V-mediated TLS requires the β-clamp, SSB, RecA, and ATP-γS. To explain the role of SSB, a novel model, the “cowcatcher” model was proposed (Pham et al. 2001). In this model, SSB directly interacts with Pol V and actively dissociates the RecA molecules from the RecA filament formed on the front of the Pol V-SSB complex. In the same paper, it was shown that DNA synthesis by Pol V on normal template DNA is highly stimulated by the presence of SSB or SSB with β-clamp even in the absence of RecA. This paper clearly showed that Pol V itself is active as a DNA polymerase on nondamaged template and that the role of RecA is likely to endow Pol V with additional properties to function in TLS.
-
4.
In 2002, using a short hairpin template oligonucleotide that contains only a 3-nucleotide single-stranded overhang downstream from an AP site, Pol V-mediated TLS was shown to require RecA-ATP-γS but no SSB and no β-clamp. RecA1730, a recA allele known to be defective in SOS-induced mutagenesis in vivo, was shown to support the insertion step opposite the AP site but not the extension steps, suggesting that the process of Pol V-mediated TLS may be dividable into two steps: insertion and extension. From these biochemical observations, a novel model was proposed in which Pol V functions as a TLS polymerase when two RecA monomers individually interact with Pol V. It also suggested that the role of the RecA filament is to supply RecA monomers to Pol V (Pham et al. 2002). This hypothesis is in good agreement with genetic data proposing that the 3′ tip of the RecA filament is directly required for Pol V-mediated TLS (the so-called third role of RecA in mutagenesis) (Blanco et al. 1982; Dutreix et al. 1989; Sweasy et al. 1990).
-
5.
In 2005, the same group reported that Pol V interacts with RecA independently of ATP, and that both UmuD and UmuD′ interact with RecA depending on ATP-γS and DNA. From that difference in interaction with RecA, a so-called “minimal mutasome” model, similar to the 2002 model, was proposed (Schlacher et al. 2005). In this model, UmuC and UmuD′ individually interact with a single RecA monomer each. The resulting complex between Pol V and two RecA monomers is necessary and sufficient to support Pol V-mediated TLS. As a consequence, the requirements of both SSB and the β-clamp disappeared from the new model.
-
6.
In 2006, the Pol V TLS model further evolved when it was shown that Pol V behaves as a TLS polymerase provided it is preincubated with the free 3′ end of a RecA filament formed on single-stranded DNA. Note that this RecA filament, referred to as the “trans-RecA filament,” does not serve as the template for synthesis but merely acts as a Pol V-activating factor. Neither SSB nor the β-clamp is required. ATP-γS appears to be a nearly essential cofactor in the formation of the “trans-RecA filament.” A novel model, referred to as the “transactivation model,” was thus proposed (Schlacher et al. 2006). However, the physiological source of “trans-RecA filaments” with free 3′ ends to activate Pol V in vivo fully remains to be determined. In contrast to the transactivation model, when Pol V interacts with the RecA filament assembled downstream from the replication-blocking lesion, it is referred to as the “cis-activation model.” Using molecular modeling, Chandani and Loechler have recently suggested a model of Pol V activation by the addition of a RecA monomer that is more likely to originate from a RecA filament in cis rather than trans (Chandani and Loechler 2013).
-
7.
In 2009, Goodman and Woodgate reported the isolation of a Pol V-RecA complex with a 1:1 stoichiometry upon incubation of Pol V with a “trans-RecA filament” under ATP-γS condition. This complex, referred to as the “minimal mutasome” behaves as a TLS polymerase (Jiang et al. 2009). It also suggests that the RecA filament itself is not an essential factor.
-
8.
Interestingly in 2012, the same group published a paper showing that Pol V lesion bypass can be mediated by either cis- or trans-RecA filaments (Karata et al. 2012). Using a single-stranded circular substrate, Pol V was shown to bypass a single TT-CPD lesion in the presence of both the β-clamp and a cis-RecA filament in good agreement with previously published data (Fujii et al. 2004; Fujii and Fuchs 2009). On the other hand, efficient lesion bypass was also possible when Pol V was activated with trans-RecA provided ATP-γS was added to stabilize the trans-RecA filament.
Pol V Work from the Fuchs/Fujii Team
Beyond the mere purification of Pol V and the determination of its intrinsic biochemical properties, our main goals were (1) to reconstitute the whole pathway of Pol V-mediated TLS including the polymerase switches from Pol III to Pol V and back to Pol III, and (2) to mimic as much as possible the physiological conditions taking into account the requirements for essential accessory factors as uncovered over the years by numerous genetic studies.
Our team reported the purification of native Pol V in 2004 (Fujii et al. 2004) and aimed at obtaining an integrated picture for Pol V-mediated TLS in the presence of Pol III (Fujii and Fuchs 2004). Using a primed, large single-stranded circular plasmid template, Pol V was shown to possess TLS activities across several replication-blocking lesions in the presence of RecA filament. The presence of the loaded form of the β-clamp dramatically stimulated the TLS activity. SSB is not required under ATP conditions, although it is an essential factor under ATP-γS conditions most likely because SSB assists dissociation of RecA molecules from the RecA/ATP-γS filament. In good agreement with known genetic requirements, it was shown that indeed Pol V becomes a TLS polymerase provided both the β-clamp (Becherel et al. 2002) and a correctly formed RecA filament are present (Blanco et al. 1982; Dutreix et al. 1989; Sweasy et al. 1990).
Main Factors in Pol V-Mediated TLS: RecA* and the β-Clamp
Based on genetic evidence, the overall scenario of TLS can be viewed as follows. When the replicative DNA polymerase (i.e., Pol III) encounters a replication-blocking lesion, it either stops one nucleotide before the lesion or it may eventually idle at the lesion site, performing futile cycles of insertion/excision (see below). When Pol III stops at the lesion site and dissociates from the template, the replicative DNA helicase (DnaB) continues to open the parental duplex for some distance generating a stretch of single-stranded DNA downstream from the lesion. The stretch of single-stranded DNA may first be covered by SSB. The SSB-DNA filament is then converted into a RecA-nucleoprotein filament (called RecA*) by means of the action of the recombination mediator proteins (RecFOR; see below). RecA* possesses the following functions: (1) up-regulation of a number of SOS-induced gene products through enhancing the autocleavage reaction of LexA; (2) activation of Pol V through the autocleavage of UmuD into UmuD′ (Burckhardt et al. 1988; Nohmi et al. 1988; Shinagawa et al. 1988); (3) activation of Pol V as a TLS polymerase through direct contact with the 3′ tip of the RecA filament (the third role of RecA); (4) activation of the damage avoidance (DA) pathways possibly via its homologous recombination function; and (5) inhibition of cellular division via the SOS-induced gene products (e.g., SfiA) thus giving time for cells to recover from stress (George et al. 1975). If Pol V engages in TLS, it accesses the 3′-OH end of the nascent strand freed by the dissociating Pol III. Following a short patch of synthesis by Pol V (TLS patch), Pol III regains access to the nascent strand and resumes elongation.
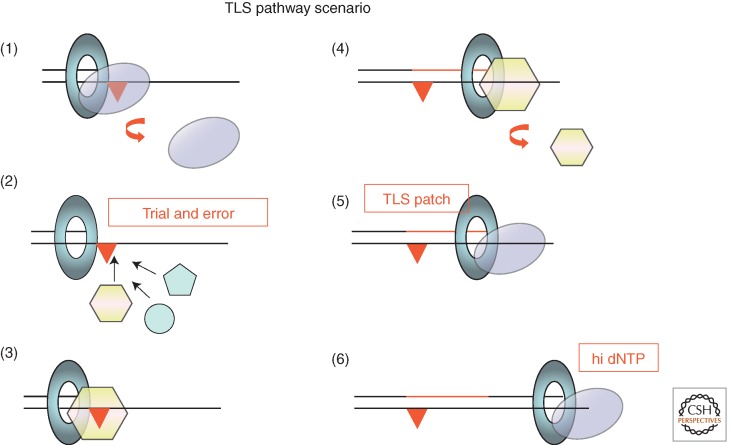
Based on the present scenario, we attempted to reconstitute Pol V-mediated TLS in the presence of Pol III (Fujii and Fuchs 2004). In this experiment, Pol III was found not to function as a passive player; rather, it actively takes part in the process (Fig. 4). A striking feature of Pol III transpires in its capacity to recognize the internal distortion induced by the lesion in nascent primer-template DNA. If the TLS patch made by Pol V extends beyond the lesion site by ≤4 nucleotides, Pol III degrades the nascent strand by means of its proofreading activity despite the presence of three correct base pairs at the terminus. In contrast, when the TLS patch is ≥5 nucleotides long, Pol III extends the primer and thus successfully completes the TLS pathway. Similarly, Walker and colleagues observed that efficient bypass of an N2-furfuryl-guanine adduct by Pol IV requires both insertion across the lesion and subsequent extension by at least four bases to prevent degradation by proofreading (Jarosz et al. 2009; Foti and Walker 2011). It should thus be stressed that the most important factor that determines success or failure of a TLS pathway resides in the length of the TLS patch given that polymerase is able to synthesize under single-hit conditions. It turns out that the β-clamp endows TLS polymerases with sufficient stability to synthesize a TLS patch that is long enough to resist degradation by the proofreading function associated with Pol III. Additionally, proofreading is attenuated by dNTP pool size increase during the DNA damage response as discussed below. Under single-hit conditions, in the presence of RecA and the β-clamp, the bypass of a G-AAF adduct by Pol V leads to an average TLS patch size of ≈18 nucleotides. About 75% of the bypass products are beyond the critical size for efficient elongation by Pol III. In the absence of β-clamp, Pol V appears to be entirely distributive in vitro, thus substantiating the genetic requirement of the β-clamp for TLS in vivo. It is likely that the β-clamp that will be used by Pol V originates from the β-clamp initially bound by Pol III-HE before its dissociation. Therefore, under SOS-induced conditions, all components (the β-clamp and RecA*) required for Pol V-mediated TLS appear to be prepositioned before Pol V accesses to the primer terminus.
Figure 4.
An integrated view of TLS pathways: (1) The replicative polymerase dissociates from the primer template upon encounter with a noncoding template base. (2) The vacant primer template becomes the substrate for binding by specialized DNA polymerases; to the best of our present knowledge, there is no active selection process for the binding of a specific polymerase; binding is stochastic and obeys classical mass-action law. (3, 4) The successful specialized polymerase is one that is able to synthesize, in a single binding event, a patch long enough to resist proofreading. The interaction of the TLS polymerase with the loaded β-clamp is essential for that purpose. For all three SOS polymerases (Pol II, Pol IV, and Pol V), mutations that inactivate the β-clamp-binding motif abrogate their TLS activity in vivo. (5, 6) Upon dissociation of the TLS polymerase, the “TLS patch” is extended upon reloading of the replicative polymerase. The balance between exonucleolytic degradation and polymerization by the replicative DNA polymerase is modulated by the dNTP pool size. Increased dNTP pools that arise as a consequence of genotoxic stress favor elongation over proofreading.
Secondary Factors Involved in TLS: RecFOR and dNTP Pool Size
The so-called recombination mediator proteins RecFOR are instrumental in converting SSB-bound single-stranded DNA to RecA-bound single-stranded DNA (Beernink and Morrical 1999). Genetically, it was shown that recFOR gene products are quasi-essential factors to support UV-induced mutagenesis (Schaaper et al. 1982; Wood and Stein 1986). Biochemically, Pol V-mediated TLS is completely abolished in the presence of SSB and RecA in amounts able to fully cover the available single-stranded template DNA (Fujii et al. 2004). The addition of optimal amounts of RecFOR fully restores Pol V-mediated TLS (Fujii et al. 2006). Thus, in good agreement with the genetic requirements of recFOR for Pol V-dependent mutagenesis, RecFOR-mediated formation of a RecA filament in cis fully supports Pol V-mediated TLS in vitro. The role of the RecA filament in Pol V-mediated TLS is thought to supply RecA monomers from the 3′ tip of the filament to Pol V (the so-called third role of RecA). However, we suggest that the RecA filament itself contributes to Pol V-mediated TLS by insuring smooth elongation across natural DNA sequences that otherwise trigger strong pause sites. This “fourth role” of RecA allows successful TLS events to occur in all sequence contexts (Fujii and Fuchs 2009).
More recently, the role of dNTP pool size in TLS was highlighted. It has been known for many years that, following genotoxic stress such as UV irradiation, cells increase the expression level of their ribonucleotide reductase gene (nrdAB) resulting in a three- to fourfold increase in dNTP pool size. The mechanism of transcriptional activation of nrdAB by UV irradiation is not controlled by the SOS response and remains to be established (Courcelle et al. 2001). We have shown that the increase in dNTP pool size strongly synergizes with the expression of the TLS polymerases to promote TLS (Gon et al. 2011). The increase of dNTP pool size is likely to act at the level of Pol III by attenuation of its proofreading function. We suggest that the increase in dNTP pool size shifts the internal exo/pol equilibrium within Pol III toward more synthesis (i.e., less proofreading). Indeed, it is shown that inactivation by a point mutation of Pol III’s proofreading function mimics the dNTP pool increase by strongly stimulating TLS across many different lesions without alleviating the requirements for the TLS polymerase per se. It should also be noted that either a slight increase in dNTP pool size (via modest overexpression of NrdAB), or an imbalance in dNTP pools as in ndk or dcd mutant strains increases spontaneous mutation rates (Miller et al. 2002; Wheeler et al. 2005; Nordman and Wright 2008; Gon et al. 2011; Schaaper and Mathews 2012). Conversely, a slight decrease in dNTP pool size as in ndh strains decreases spontaneous mutation rates (Laureti et al. 2013).
Pol III Actively Shapes the Nature of TLS Events Produced In Vivo
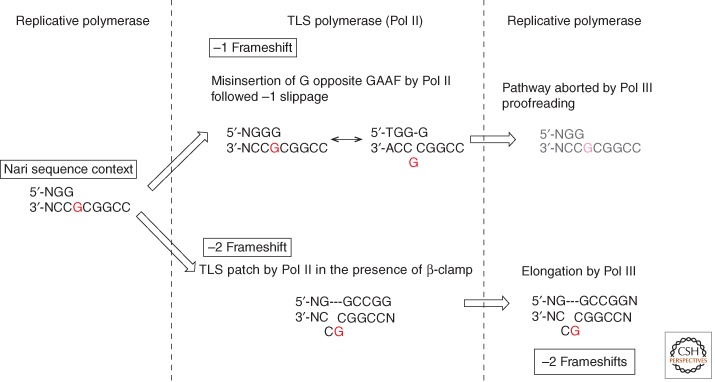
Let us focus on a Pol II-mediated frameshift mutation hot spot induced within (GpC)2 sequences by many chemical carcinogens that bind to the C8 position of guanine. In vivo, Pol II is an essential factor for the production of the −2 frameshift mutation hot spot triggered by a single G-AAF adduct located within a specific sequence context (i.e., GGCGAAFCC, the NarI site) (Fig. 5) (Fuchs et al. 1981; Koffel-Schwartz et al. 1984). It is also known that the interaction of Pol II with the β-clamp is essential for the −2 frameshift pathway in vivo (Becherel et al. 2002).
Figure 5.
DNA polymerase III actively shapes the outcome of TLS reaction in vivo. This notion is illustrated by the Pol II-mediated TLS pathway at the NarI site in the presence of a GAAF adduct (shown in red). In vitro, Pol II generates both −1 and −2 frameshift TLS products, whereas in vivo only −2 frameshift events are made. Minus 1 frameshift events result from frequent misinsertion by Pol II of G opposite GAAF, slippage, and subsequent elongation. It turns out that these −1 frameshift events are aborted by Pol III proofreading. The addition of Pol III to the Pol II-mediated bypass reaction in vitro allows the in vivo situation to be mimicked.
In vitro, Pol II possesses the capacity to produce −2 frameshifts within the NarI containing the G-AAF adduct sequence, but unexpectedly, Pol II also produces a significant amount of −1 frameshifts that are not detected in vivo (Fuchs et al. 1981; Koffel-Schwartz et al. 1984). Another surprising observation is that the presence of the β-clamp strongly stimulates the production of −1 frameshift mutations but not −2 frameshifts (Fujii and Fuchs 2007). Consequently, in vitro in the presence of the β-clamp, Pol II produces more −1 than −2 frameshifts, in striking contrast with in vivo results. Interestingly, these inconsistencies between in vitro/in vivo results are fixed by the addition of Pol III to the in vitro reaction mixture. Indeed, Pol III was found to suppress Pol II-mediated −1 frameshifts but not −2 frameshifts (Fujii and Fuchs 2007). Our data show that the −1 frameshifts result from misincorporation by Pol II of G across GAAF followed by slippage. It turns out that this intermediate is suppressed by the proofreading activity of Pol III (Fig. 5). In contrast, Pol III enhances the production of −2 frameshift intermediates in view of its capacity to insert C opposite GAAF. Moreover, under single-hit conditions, the processivity of Pol II to extend the slipped −2 frameshift intermediate is increased from 2, in the absence of β-clamp, to 3 nucleotides in its presence. The additional nucleotide, conferred by the presence of the β-clamp, is shown to be critical to support further extension of the slipped intermediate by Pol III rather than its degradation. The fact that Pol II, a “classical” DNA polymerase, is directly involved in lesion bypass despite its proficient proofreading activity appears to be puzzling. However, recent structural studies showed that DNA Pol II possesses small cavities outside of the active site that can accommodate looped out template nucleotides of up to two base pairs, supporting the −2 frameshift pathway (Wang and Yang 2009). In conclusion, we were able to derive experimental conditions allowing the in vitro data to properly reflect the in vivo results. These conditions involve the presence of both the β-clamp and Pol III in the reaction mixture. In most cases, meaningful reconstitution of a given TLS pathway goes far beyond the simple lesion bypass reaction mediated by a TLS polymerase; the present example highlights the notion that in vivo TLS is an integrated pathway that involves a cross talk between replicative, specialized DNA polymerase and accessory factors. It highlights the fact that Pol III has an active role in shaping the pattern TLS events.
TLS Pathways: A Trial and Error Process?
The question as to whether there are specific lesion-to-polymerase matches during TLS entails at least two distinct facets: specificity at the recruitment step and proficiency at the bypass step.
Specialized DNA polymerases are likely to bind blocked primer-template termini in a way that is not “instructed” by the nature of the lesion. Indeed, there is no genetic evidence for a model invoking a cognate lesion-to-polymerase relationship. Such a model is unlikely given the huge diversity of DNA lesions and the relatively small number of TLS polymerases. Genetically, it is clear that the efficiencies of TLS events are positively correlated with the amounts of available TLS polymerases (Becherel and Fuchs 2001). In this respect, the NarI mutation hot spot offers a remarkable model in which a single G-AAF lesion is bypassed by two different DNA polymerases, Pol V and Pol II, leading to two distinct molecular events, error-free and −2 frameshift events, respectively. Both in vivo and in vitro, the respective amounts of error-free and −2 frameshift events reflect the corresponding amounts of Pol II and Pol V, respectively (Becherel and Fuchs 2001; Fujii and Fuchs 2007). TLS polymerases are necessarily recruited to the β-clamp to become proficient in TLS (Becherel et al. 2002). In conclusion, polymerase recruitment to blocked primer-template termini follows classical mass-action parameters, such as the affinity of the polymerase to the β-clamp and its concentration.
On the other hand, there are limited data showing that polymerases show specificity with respect to the lesions they are able or unable to bypass. Selectivity is not based on precise chemical determinants of the lesion itself but appears to depend on the location of the adduct with respect to the geometry of the double helix. Indeed, lesions that are located in the major groove appear to be preferentially bypassed by Pol V, whereas minor groove lesions are substrate for Pol IV. For example, Pol V efficiently bypasses UV-induced lesions (Fig. 1A: (4) and (5)) (Tang et al. 2000; Fujii et al. 2004), although it cannot bypass the N2-BP-G (Fig. 1A: (7)) adduct located in the minor groove. Conversely, Pol IV efficiently bypasses N2-BP-G or N-2-furfuryl adducts (Napolitano et al. 2000; Lenne-Samuel et al. 2002; Shen et al. 2002; Yin et al. 2004; Jarosz et al. 2006; Seo et al. 2006) but not UV-induced photoproducts. Insights into the architecture of Y-family DNA polymerases in relation to TLS have recently been revealed by structural and molecular modeling studies (see review by Chandani et al. 2010).
In conclusion, trial and error, a heuristic problem-solving method, may represent a good approximation for the way TLS polymerases get involved in TLS pathways. TLS will be successful provided the polymerase that gets recruited is able to synthesize during the time it remains bound to the blocked replication termini, a long enough TLS patch to resist proofreading by the replicative polymerase (Fig. 4). On the other hand, TLS will fail if the recruited polymerase is unable to bypass the lesion or if the TLS patch size is too small. A failed TLS pathway will start all over with another TLS polymerase-binding event.
OTHER BACTERIA, OTHER SOLUTIONS TO INDUCED MUTAGENESIS
Bacteria that Possess an imuABC Mutagenesis Cassette
The mechanisms of induced mutagenesis and TLS as discussed above reflect the situation in E. coli. In recent years, it has become clear that other bacteria do not follow the paradigm provided by the widely studied E. coli (for a short review, see McHenry 2011a). In fact, bacteria such as Pseudomonas aeruginosa, Caulobacter crescentus, and Mycobacterium tuberculosis express two dnaE genes (dnaE genes encode the α subunit of Pol III), DnaE1 and DnaE2, DnaE2 being used instead of Pol V for induced mutagenesis (Boshoff et al. 2003; Galhardo et al. 2005; Sanders et al. 2006). Because dnaE2 is the distal gene in an operon preceded by a small gene (imuA) that has a weak similarity to E. coli sulA and recA, and a gene similar to a TLS DNA polymerase (imuB), it was recently suggested to rename dnaE2 as imuC. All three genes in this operon are required for induced mutagenesis and are epistatic to each other. Surprisingly, M. tuberculosis, imuB, despite being homologous to Y-family DNA polymerases, does not contain the triad of acidic residues that are conserved in the active site of DNA polymerases (Warner et al. 2010). In contrast, mutants in catalytic acidic residues in imuC abolish induced mutagenesis suggesting that ImuC rather than ImuB acts as the bypass polymerase. As ImuB binds to both the β-clamp and ImuC, one might suggest that the role of ImuB in mutagenesis is to recruit ImuC to the blocked replication fork. Available data suggest that a similar model applies for the imuABC mutagenesis cassette present in P. aeruginosa and C. crescentus.
Bacteria that Use a Proofreading-Deficient polI Gene to Promote Genetic Instability
Helicobacter pylori, a human pathogen infecting about half of the world population, is characterized by its considerable genome plasticity that appears to be the basis for its high adaptation capacity. Consistent with its small genome, H. pylori possesses only two DNA polymerases, Pol I and the replicative Pol III, lacking homologs of TLS DNA polymerases. It was shown that although Pol I plays its crucial role in replication and repair, it also contributes to genomic instability (García-Ortíz et al. 2011). Indeed, strains defective in the DNA polymerase activity of the protein display reduced mutation frequencies. Conversely, overexpression of Pol I leads to a mutator phenotype. Although the overall structure of the 3′-5′ exonuclease domain appears to be preserved, at least three conserved acidic residues involved in metal binding and essential for exonuclease catalytic activity are missing (Derbyshire et al. 1988). Consistently, the purified protein lacks proofreading activity, allowing it to efficiently elongate mismatched primers and perform TLS. This work supports the idea that the proofreading activity present in replicative DNA polymerases constitutes the main barrier for lesion bypass by these polymerases. Bacteria with small genomes that lack bona fide TLS polymerases may use a proofreading-deficient version of a replicative DNA polymerase, DNA Pol I for instance, to generate genomic plasticity (Liu et al. 2006).
CONCLUSION: SIGNIFICANCE OF TLS AND MUTAGENESIS FOR PROKARYOTES
During the last 10 years, the process of TLS has been solidly established at the biochemical level, mostly based on the E. coli paradigm. In contrast, the strategies implemented by other bacteria to deal with lesions and induce mutations largely remains to be explored. A major challenge for the future will be to unravel the regulation of TLS in vivo in the context of all other lesion-tolerance pathways. Despite its prime importance as a generator of genetic diversity, TLS across replication-blocking lesions represents a minor lesion-tolerance pathway, representing ≈1%–2% and up to 10%–20% under non-SOS- and SOS-induced conditions, respectively (Pagès et al. 2012; K Naiman et al., unpubl.). DA pathways process all remaining lesions. The modest contribution of TLS to lesion tolerance is also evidenced by the relatively moderate UV sensitivity of strains that are deficient in all three SOS-inducible DNA polymerases.
More work is required to understand fine-tuning of TLS in response to dNTP pool size changes that are elicited in response to genotoxic stress.
The process of TLS is induced as part of the response to DNA-damaging agents and represents the main source of point mutations. Its physiological role can thus be viewed as a beneficial source of genetic diversity and thus as an engine for evolution under stress conditions. In recent years, it was also shown that subinhibitory concentrations of antibiotics trigger the SOS response; the ensuing induction of mutations is thus likely to contribute to the emergence of antibiotic resistance and to virulence (see Kreuzer 2013).
ACKNOWLEDGMENTS
The present work is funded partly by the LIGUE Contre le Cancer (Labellisation 2011) and by ANR grant ForkRepair (ANR 11 BSV8 017 01). The authors thank the laboratory members for critical reading.
Footnotes
Editors: Errol C. Friedberg, Stephen J. Elledge, Alan R. Lehmann, Tomas Lindahl, and Marco Muzi-Falconi
Additional Perspectives on DNA Repair, Mutagenesis, and Other Responses to DNA Damage available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Al Mamun AA, Humayun MZ 2006. Escherichia coli DNA polymerase II can efficiently bypass 3,N(4)-ethenocytosine lesions in vitro and in vivo. Mutat Res 593: 164–176 [DOI] [PubMed] [Google Scholar]
- Banerjee SK, Christensen RB, Lawrence CW, LeClerc JE 1988. Frequency and spectrum of mutations produced by a single cis-syn thymine-thymine cyclobutane dimer in a single-stranded vector. Proc Natl Acad Sci 85: 8141–8145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becherel OJ, Fuchs RP 1999. SOS mutagenesis results from up-regulation of translesion synthesis. J Mol Biol 294: 299–306 [DOI] [PubMed] [Google Scholar]
- Becherel OJ, Fuchs RP 2001. Mechanism of DNA polymerase II-mediated frameshift mutagenesis. Proc Natl Acad Sci 98: 8566–8571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becherel OJ, Fuchs RPP, Wagner J 2002. Pivotal role of the β-clamp in translesion DNA synthesis and mutagenesis in E. coli cells. DNA Repair (Amst) 1: 703–708 [DOI] [PubMed] [Google Scholar]
- Beernink HT, Morrical SW 1999. RMPs: Recombination/replication mediator proteins. Trends Biochem Sci 24: 385–389 [DOI] [PubMed] [Google Scholar]
- Belguise-Valladier P, Maki H, Sekiguchi M, Fuchs RP 1994. Effect of single DNA lesions on in vitro replication with DNA polymerase III holoenzyme. Comparison with other polymerases. J Mol Biol 236: 151–164 [DOI] [PubMed] [Google Scholar]
- Bessman MJ, Kornberg A, Lehman IR, Simms ES 1956. Enzymic synthesis of deoxyribonucleic acid. Biochim Biophys Acta 21: 197–198 [DOI] [PubMed] [Google Scholar]
- Blanco M, Herrera G, Collado P, Rebollo JE, Botella LM 1982. Influence of RecA protein on induced mutagenesis. Biochimie 64: 633–636 [DOI] [PubMed] [Google Scholar]
- Boshoff HIM, Reed MB, Barry CE, Mizrahi V 2003. DnaE2 polymerase contributes to in vivo survival and the emergence of drug resistance in Mycobacterium tuberculosis. Cell 113: 183–193 [DOI] [PubMed] [Google Scholar]
- Brotcorne-Lannoye A, Maenhaut-Michel G 1986. Role of RecA protein in untargeted UV mutagenesis of bacteriophage λ: Evidence for the requirement for the dinB gene. Proc Natl Acad Sci 83: 3904–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckhardt SE, Woodgate R, Scheuermann RH, Echols H 1988. UmuD mutagenesis protein of Escherichia coli: Overproduction, purification, and cleavage by RecA. Proc Natl Acad Sci 85: 1811–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandani S, Loechler EL 2013. Structural model of the Y-Family DNA polymerase V/RecA mutasome. J Mol Graph Model 39: 133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandani S, Jacobs C, Loechler EL 2010. Architecture of y-family DNA polymerases relevant to translesion DNA synthesis as revealed in structural and molecular modeling studies. J Nucleic Acids 2010: 784081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark JM, Beardsley GP 1989. Template length, sequence context, and 3″–5″ exonuclease activity modulate replicative bypass of thymine glycol lesions in vitro. Biochemistry 28: 775–779 [DOI] [PubMed] [Google Scholar]
- Cohen SE, Lewis CA, Mooney RA, Kohanski MA, Collins JJ, Landick R, Walker GC 2010. Roles for the transcription elongation factor NusA in both DNA repair and damage tolerance pathways in Escherichia coli. Proc Natl Acad Sci 107: 15517–15522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC 2001. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics 158: 41–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle CT, Belle JJ, Courcelle J 2005. Nucleotide excision repair or polymerase V-mediated lesion bypass can act to restore UV-arrested replication forks in Escherichia coli. J Bacteriol 187: 6953–6961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigaku Y, Davies AA, Ulrich HD 2010. Ubiquitin-dependent DNA damage bypass is separable from genome replication. Nature 465: 951–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lucia P, Cairns J 1969. Isolation of an E. coli strain with a mutation affecting DNA polymerase. Nature 224: 1164–1166 [DOI] [PubMed] [Google Scholar]
- Derbyshire V, Freemont PS, Sanderson MR, Beese L, Friedman JM, Joyce CM, Steitz TA 1988. Genetic and crystallographic studies of the 3′,5′-exonucleolytic site of DNA polymerase I. Science 240: 199–201 [DOI] [PubMed] [Google Scholar]
- Dutreix M, Moreau PL, Bailone A, Galibert F, Battista JR, Walker GC, Devoret R 1989. New recA mutations that dissociate the various RecA protein activities in Escherichia coli provide evidence for an additional role for RecA protein in UV mutagenesis. J Bacteriol 171: 2415–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foti JJ, Walker GC 2011. Efficient extension of slipped DNA intermediates by DinB is required to escape primer template realignment by DnaQ. J Bacteriol 193: 2637–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frampton J, Irmisch A, Green CM, Neiss A, Trickey M, Ulrich HD, Furuya K, Watts FZ, Carr AM, Lehmann AR 2006. Postreplication repair and PCNA modification in Schizosaccharomyces pombe. Mol Biol Cell 17: 2976–2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs RP, Schwartz N, Daune MP 1981. Hot spots of frameshift mutations induced by the ultimate carcinogen N-acetoxy-N-2-acetylaminofluorene. Nature 294: 657–659 [DOI] [PubMed] [Google Scholar]
- Fujii S, Fuchs RP 2004. Defining the position of the switches between replicative and bypass DNA polymerases. EMBO J 23: 4342–4352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Fuchs RP 2007. Interplay among replicative and specialized DNA polymerases determines failure or success of translesion synthesis pathways. J Mol Biol 372: 883–893 [DOI] [PubMed] [Google Scholar]
- Fujii S, Fuchs RP 2009. Biochemical basis for the essential genetic requirements of RecA and the β-clamp in Pol V activation. Proc Natl Acad Sci 106: 14825–14830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Gasser V, Fuchs RP 2004. The biochemical requirements of DNA polymerase V-mediated translesion synthesis revisited. J Mol Biol 341: 405–417 [DOI] [PubMed] [Google Scholar]
- Fujii S, Isogawa A, Fuchs RP 2006. RecFOR proteins are essential for Pol V-mediated translesion synthesis and mutagenesis. EMBO J 25: 5754–5763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukohri A, Goodman MF, Maki H 2008. A dynamic polymerase exchange with Escherichia coli DNA polymerase IV replacing DNA polymerase III on the sliding clamp. J Biol Chem 283: 11260–11269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galhardo RS, Rocha RP, Marques MV, Menck CFM 2005. An SOS-regulated operon involved in damage-inducible mutagenesis in Caulobacter crescentus. Nucleic Acids Res 33: 2603–2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Ortíz M-V, Marsin S, Arana ME, Gasparutto D, Guérois R, Kunkel TA, Radicella JP 2011. Unexpected role for Helicobacter pylori DNA polymerase I as a source of genetic variability. PLoS Genet 7: e1002152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George J, Castellazzi M, Buttin G 1975. Prophage induction and cell division in E. coli. III: Mutations sfiA and sfiB restore division in tif and lon strains and permit the expression of mutator properties of tif. Mol Gen Genet 140: 309–332 [PubMed] [Google Scholar]
- Gibbs PE, Borden A, Lawrence CW 1995. The T-T pyrimidine (6-4) pyrimidinone UV photoproduct is much less mutagenic in yeast than in Escherichia coli. Nucleic Acids Res 23: 1919–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy VG, Jarosz DF, Walker FL, Simmons LA, Walker GC 2006. Y-family DNA polymerases respond to DNA damage-independent inhibition of replication fork progression. EMBO J 25: 868–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gon S, Napolitano R, Rocha W, Coulon S, Fuchs RP 2011. Increase in dNTP pool size during the DNA damage response plays a key role in spontaneous and induced-mutagenesis in Escherichia coli. Proc Natl Acad Sci 108: 19311–19316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman MF 2002. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu Rev Biochem 71: 17–50 [DOI] [PubMed] [Google Scholar]
- Grollman AP, Shibutani S, Moriya M, Miller F, Wu L, Moll U, Suzuki N, Fernandes A, Rosenquist T, Medverec Z, et al. 2007. Aristolochic acid and the etiology of endemic (Balkan) nephropathy. Proc Natl Acad Sci 104: 12129–12134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi K, Katayama T, Iwai S, Hidaka M, Horiuchi T, Maki H 2003. Fate of DNA replication fork encountering a single DNA lesion during oriC plasmid DNA replication in vitro. Genes Cells 8: 437–449 [DOI] [PubMed] [Google Scholar]
- Hsu GW, Kiefer JR, Burnouf D, Becherel OJ, Fuchs RPP, Beese LS 2004. Observing translesion synthesis of an aromatic amine DNA adduct by a high-fidelity DNA polymerase. J Biol Chem 279: 50280–50285 [DOI] [PubMed] [Google Scholar]
- Indiani C, McInerney P, Georgescu R, Goodman MF, O’Donnell M 2005. A sliding-clamp toolbelt binds high- and low-fidelity DNA polymerases simultaneously. Mol Cell 19: 805–815 [DOI] [PubMed] [Google Scholar]
- Jarosz DF, Godoy VG, Delaney JC, Essigmann JM, Walker GC 2006. A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature 439: 225–228 [DOI] [PubMed] [Google Scholar]
- Jarosz DF, Beuning PJ, Cohen SE, Walker GC 2007. Y-family DNA polymerases in Escherichia coli. Trends Microbiol 15: 70–77 [DOI] [PubMed] [Google Scholar]
- Jarosz DF, Cohen SE, Delaney JC, Essigmann JM, Walker GC 2009. A DinB variant reveals diverse physiological consequences of incomplete TLS extension by a Y-family DNA polymerase. Proc Natl Acad Sci 106: 21137–21142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jergic S, Horan NP, Elshenawy MM, Mason CE, Urathamakul T, Ozawa K, Robinson A, Goudsmits JMH, Wang Y, Pan X, et al. 2013. A direct proofreader-clamp interaction stabilizes the Pol III replicase in the polymerization mode. EMBO J 32: 1322–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q, Karata K, Woodgate R, Cox MM, Goodman MF 2009. The active form of DNA polymerase V is UmuD′2C-RecA-ATP. Nature 460: 359–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SJ, Taylor JS, Beese LS 2003. Processive DNA synthesis observed in a polymerase crystal suggests a mechanism for the prevention of frameshift mutations. Proc Natl Acad Sci 100: 3895–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karata K, Vaisman A, Goodman MF, Woodgate R 2012. Simple and efficient purification of Escherichia coli DNA polymerase V: Cofactor requirements for optimal activity and processivity in vitro. DNA Repair (Amst) 11: 431–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karras GI, Jentsch S 2010. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell 141: 255–267 [DOI] [PubMed] [Google Scholar]
- Kato T, Shinoura Y 1977. Isolation and characterization of mutants of Escherichia coli deficient in induction of mutations by ultraviolet light. Mol Gen Genet 156: 121–131 [DOI] [PubMed] [Google Scholar]
- Kim SR, Maenhaut-Michel G, Yamada M, Yamamoto Y, Matsui K, Sofuni T, Nohmi T, Ohmori H 1997. Multiple pathways for SOS-induced mutagenesis in Escherichia coli: An overexpression of dinB/dinP results in strongly enhancing mutagenesis in the absence of any exogenous treatment to damage DNA. Proc Natl Acad Sci 94: 13792–13797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SR, Matsui K, Yamada M, Gruz P, Nohmi T 2001. Roles of chromosomal and episomal dinB genes encoding DNA pol IV in targeted and untargeted mutagenesis in Escherichia coli. Mol Genet Genomics 266: 207–215 [DOI] [PubMed] [Google Scholar]
- Koehl P, Burnouf D, Fuchs RP 1989. Construction of plasmids containing a unique acetylaminofluorene adduct located within a mutation hot spot. A new probe for frameshift mutagenesis. J Mol Biol 207: 355–364 [DOI] [PubMed] [Google Scholar]
- Koffel-Schwartz N, Verdier JM, Bichara M, Freund AM, Daune MP, Fuchs RP 1984. Carcinogen-induced mutation spectrum in wild-type, uvrA and umuC strains of Escherichia coli. Strain specificity and mutation-prone sequences. J Mol Biol 177: 33–51 [DOI] [PubMed] [Google Scholar]
- Koffel-Schwartz N, Coin F, Veaute X, Fuchs RP 1996. Cellular strategies for accommodating replication-hindering adducts in DNA: Control by the SOS response in Escherichia coli. Proc Natl Acad Sci 93: 7805–7810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg T, Gefter ML 1970. DNA synthesis in cell-free extracts of a DNA polymerase-defective mutant. Biochem Biophys Res Commun 40: 1348–1355 [DOI] [PubMed] [Google Scholar]
- Kornberg T, Gefter ML 1971. Purification and DNA synthesis in cell-free extracts: Properties of DNA polymerase II. Proc Natl Acad Sci 68: 761–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg T, Gefter ML 1972. Deoxyribonucleic acid synthesis in cell-free extracts. IV. Purification and catalytic properties of deoxyribonucleic acid polymerase III. J Biol Chem 247: 5369–5375 [PubMed] [Google Scholar]
- *.Kreuzer KN 2013. DNA damage responses in prokaryotes: Regulating gene expression, modulating growth patterns, and manipulating replication forks. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a012674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuban W, Jonczyk P, Gawel D, Malanowska K, Schaaper RM, Fijalkowska IJ 2004. Role of Escherichia coli DNA polymerase IV in in vivo replication fidelity. J Bacteriol 186: 4802–4807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuban W, Banach-Orlowska M, Bialoskorska M, Lipowska A, Schaaper RM, Jonczyk P, Fijalkowska IJ 2005. Mutator phenotype resulting from DNA polymerase IV overproduction in Escherichia coli: Preferential mutagenesis on the lagging strand. J Bacteriol 187: 6862–6866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulaeva OI, Koonin EV, McDonald JP, Randall SK, Rabinovich N, Connaughton JF, Levine AS, Woodgate R 1996. Identification of a DinB/UmuC homolog in the archeon Sulfolobus solfataricus. Mutat Res 357: 245–253 [DOI] [PubMed] [Google Scholar]
- Larimer FW, Perry JR, Hardigree AA 1989. The REV1 gene of Saccharomyces cerevisiae: Isolation, sequence, and functional analysis. J Bacteriol 171: 230–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laureti L, Selva M, Dairou J, Matic I 2013. Reduction of dNTP levels enhances DNA replication fidelity in vivo. DNA Repair (Amst) 12: 300–305 [DOI] [PubMed] [Google Scholar]
- Lehman IR, Bessman MJ, Simms ES, Kornberg A 1958. Enzymatic synthesis of deoxyribonucleic acid. I. Preparation of substrates and partial purification of an enzyme from Escherichia coli. J Biol Chem 233: 163–170 [PubMed] [Google Scholar]
- Lenne-Samuel N, Janel-Bintz R, Kolbanovskiy A, Geacintov NE, Fuchs RP 2000. The processing of a Benzo(a)pyrene adduct into a frameshift or a base substitution mutation requires a different set of genes in Escherichia coli. Mol Microbiol 38: 299–307 [DOI] [PubMed] [Google Scholar]
- Lenne-Samuel N, Wagner J, Etienne H, Fuchs RPP 2002. The processivity factor β controls DNA polymerase IV traffic during spontaneous mutagenesis and translesion synthesis in vivo. EMBO Rep 3: 45–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Hou J, Liu J 2006. Chlamydial DNA polymerase I can bypass lesions in vitro. Biochem Biophys Res Commun 345: 1083–1091 [DOI] [PubMed] [Google Scholar]
- Maor-Shoshani A, Hayashi K, Ohmori H, Livneh Z 2003. Analysis of translesion replication across an abasic site by DNA polymerase IV of Escherichia coli. DNA Repair (Amst) 2: 1227–1238 [DOI] [PubMed] [Google Scholar]
- McHenry CS 2011a. Breaking the rules: Bacteria that use several DNA polymerase IIIs. EMBO Rep 12: 408–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHenry CS 2011b. DNA replicases from a bacterial perspective. Annu Rev Biochem 80: 403–436 [DOI] [PubMed] [Google Scholar]
- McKenzie GJ, Lee PL, Lombardo MJ, Hastings PJ, Rosenberg SM 2001. SOS mutator DNA polymerase IV functions in adaptive mutation and not adaptive amplification. Mol Cell 7: 571–579 [DOI] [PubMed] [Google Scholar]
- Miller JH, Funchain P, Clendenin W, Huang T, Nguyen A, Wolff E, Yeung A, Chiang J-H, Garibyan L, Slupska MM, et al. 2002. Escherichia coli strains (ndk) lacking nucleoside diphosphate kinase are powerful mutators for base substitutions and frameshifts in mismatch-repair-deficient strains. Genetics 162: 5–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolitano RL, Fuchs RPP 1997. New strategy for the construction of single stranded plasmids with single mutagenic lesions. Chem Res Toxicol 10: 667–671 [DOI] [PubMed] [Google Scholar]
- Napolitano RL, Lambert IB, Fuchs RP 1997. SOS factors involved in translesion synthesis. Proc Natl Acad Sci 94: 5733–5738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolitano R, Janel-Bintz R, Wagner J, Fuchs RP 2000. All three SOS-inducible DNA polymerases (Pol II, Pol IV and Pol V) are involved in induced mutagenesis. EMBO J 19: 6259–6265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JR, Lawrence CW, Hinkle DC 1996. Deoxycytidyl transferase activity of yeast REV1 protein. Nature 382: 729–731 [DOI] [PubMed] [Google Scholar]
- Nohmi T, Battista JR, Dodson LA, Walker GC 1988. RecA-mediated cleavage activates UmuD for mutagenesis: Mechanistic relationship between transcriptional derepression and posttranslational activation. Proc Natl Acad Sci 85: 1816–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordman J, Wright A 2008. The relationship between dNTP pool levels and mutagenesis in an Escherichia coli NDP kinase mutant. Proc Natl Acad Sci 105: 10197–10202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori H, Hatada E, Qiao Y, Tsuji M, Fukuda R 1995. dinP, a new gene in Escherichia coli, whose product shows similarities to UmuC and its homologues. Mutat Res 347: 1–7 [DOI] [PubMed] [Google Scholar]
- Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, et al. 2001. The Y-family of DNA polymerases. Mol Cell 8: 7–8 [DOI] [PubMed] [Google Scholar]
- Pagès V, Fuchs RPP 2002. How DNA lesions are turned into mutations within cells? Oncogene 21: 8957–8966 [DOI] [PubMed] [Google Scholar]
- Pagès V, Fuchs RP 2003. Uncoupling of leading- and lagging-strand DNA replication during lesion bypass in vivo. Science 300: 1300–1303 [DOI] [PubMed] [Google Scholar]
- Pagès V, Mazon G, Naiman K, Philippin G, Fuchs RP 2012. Monitoring bypass of single replication-blocking lesions by damage avoidance in the Escherichia coli chromosome. Nucleic Acids Res 40: 9036–9043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Elizur T, Takeshita M, Goodman M, O’Donnell M, Livneh Z 1996. Mechanism of translesion DNA synthesis by DNA polymerase II. Comparison to DNA polymerases I and III core. J Biol Chem 271: 24662–24669 [DOI] [PubMed] [Google Scholar]
- Paz-Elizur T, Takeshita M, Livneh Z 1997. Mechanism of bypass synthesis through an abasic site analog by DNA polymerase I. Biochemistry 36: 1766–1773 [DOI] [PubMed] [Google Scholar]
- Pham P, Bertram JG, O’Donnell M, Woodgate R, Goodman MF 2001. A model for SOS-lesion-targeted mutations in Escherichia coli. Nature 409: 366–370 [DOI] [PubMed] [Google Scholar]
- Pham P, Seitz EM, Saveliev S, Shen X, Woodgate R, Cox MM, Goodman MF 2002. Two distinct modes of RecA action are required for DNA polymerase V-catalyzed translesion synthesis. Proc Natl Acad Sci 99: 11061–11066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash L 1989. The structure and function of RAD6 and RAD18 DNA repair genes of Saccharomyces cerevisiae. Genome 31: 597–600 [DOI] [PubMed] [Google Scholar]
- Qiu Z, Goodman MF 1997. The Escherichia coli polB locus is identical to dinA, the structural gene for DNA polymerase II. Characterization of Pol II purified from a polB mutant. J Biol Chem 272: 8611–8617 [DOI] [PubMed] [Google Scholar]
- Rangarajan S, Woodgate R, Goodman MF 1999. A phenotype for enigmatic DNA polymerase II: A pivotal role for pol II in replication restart in UV-irradiated Escherichia coli. Proc Natl Acad Sci 96: 9224–9229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan S, Woodgate R, Goodman MF 2002. Replication restart in UV-irradiated Escherichia coli involving pols II, III, V, PriA, RecA, and RecFOR proteins. Mol Microbiol 43: 617–628 [DOI] [PubMed] [Google Scholar]
- Reuven NB, Arad G, Maor-Shoshani A, Livneh Z 1999. The mutagenesis protein UmuC is a DNA polymerase activated by UmuD′, RecA, and SSB and is specialized for translesion replication. J Biol Chem 274: 31763–31766 [DOI] [PubMed] [Google Scholar]
- Rudolph CJ, Upton AL, Lloyd RG 2008. Maintaining replication fork integrity in UV-irradiated Escherichia coli cells. DNA Repair (Amst) 7: 1589–1602 [DOI] [PubMed] [Google Scholar]
- Sanders LH, Rockel A, Lu H, Wozniak DJ, Sutton MD 2006. Role of Pseudomonas aeruginosa dinB-encoded DNA polymerase IV in mutagenesis. J Bacteriol 188: 8573–8585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaper RM, Mathews CK 2012. Mutational consequences of dNTP pool imbalances in E. coli. DNA Repair (Amst) 12: 73–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaper RM, Glickman BW, Loeb LA 1982. Mutagenesis resulting from depurination is an SOS process. Mutat Res 106: 1–9 [DOI] [PubMed] [Google Scholar]
- Schlacher K, Leslie K, Wyman C, Woodgate R, Cox MM, Goodman MF 2005. DNA polymerase V and RecA protein, a minimal mutasome. Mol Cell 17: 561–572 [DOI] [PubMed] [Google Scholar]
- Schlacher K, Cox MM, Woodgate R, Goodman MF 2006. RecA acts in trans to allow replication of damaged DNA by DNA polymerase V. Nature 442: 883–887 [DOI] [PubMed] [Google Scholar]
- Seo KY, Nagalingam A, Miri S, Yin J, Chandani S, Kolbanovskiy A, Shastry A, Loechler EL 2006. Mirror image stereoisomers of the major benzo[a]pyrene N2-dG adduct are bypassed by different lesion-bypass DNA polymerases in E. coli. DNA Repair (Amst) 5: 515–522 [DOI] [PubMed] [Google Scholar]
- Shen X, Sayer JM, Kroth H, Ponten I, O’Donnell M, Woodgate R, Jerina DM, Goodman MF 2002. Efficiency and accuracy of SOS-induced DNA polymerases replicating benzo[a]pyrene-7,8-diol 9,10-epoxide A and G adducts. J Biol Chem 277: 5265–5274 [DOI] [PubMed] [Google Scholar]
- Shen X, Woodgate R, Goodman MF 2005. Lyase activities intrinsic to Escherichia coli polymerases IV and V. DNA Repair (Amst) 4: 1368–1373 [DOI] [PubMed] [Google Scholar]
- Shibutani S, Grollman AP 1993. On the mechanism of frameshift (deletion) mutagenesis in vitro. J Biol Chem 268: 11703–11710 [PubMed] [Google Scholar]
- Shinagawa H, Iwasaki H, Kato T, Nakata A 1988. RecA protein-dependent cleavage of UmuD protein and SOS mutagenesis. Proc Natl Acad Sci 85: 1806–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slechta ES, Bunny KL, Kugelberg E, Kofoid E, Andersson DI, Roth JR 2003. Adaptive mutation: General mutagenesis is not a programmed response to stress but results from rare coamplification of dinB with lac. Proc Natl Acad Sci 100: 12847–12852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinborn G 1978. Uvm mutants of Escherichia coli K12 deficient in UV mutagenesis. I: Isolation of uvm mutants and their phenotypical characterization in DNA repair and mutagenesis. Mol Gen Genet 165: 87–93 [DOI] [PubMed] [Google Scholar]
- Sutton MD, Duzen JM, Scouten Ponticelli SK 2010. A single hydrophobic cleft in the Escherichia coli processivity clamp is sufficient to support cell viability and DNA damage-induced mutagenesis in vivo. BMC Mol Biol 11: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Ohashi E, Hayashi K, Ohmori H, Grollman AP, Shibutani S 2001. Translesional synthesis past acetylaminofluorene-derived DNA adducts catalyzed by human DNA polymerase κ and Escherichia coli DNA polymerase IV. Biochemistry 40: 15176–15183 [DOI] [PubMed] [Google Scholar]
- Sweasy JB, Witkin EM, Sinha N, Roegner-Maniscalco V 1990. RecA protein of Escherichia coli has a third essential role in SOS mutator activity. J Bacteriol 172: 3030–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Shen X, Frank EG, O’Donnell M, Woodgate R, Goodman MF 1999. UmuD′2C is an error-prone DNA polymerase, Escherichia coli pol V. Proc Natl Acad Sci 96: 8919–8924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Pham P, Shen X, Taylor JS, O’Donnell M, Woodgate R, Goodman MF 2000. Roles of E. coli DNA polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature 404: 1014–1018 [DOI] [PubMed] [Google Scholar]
- Toste Rêgo A, Holding AN, Kent H, Lamers MH 2013. Architecture of the Pol III-clamp-exonuclease complex reveals key roles of the exonuclease subunit in processive DNA synthesis and repair. EMBO J 32: 1334–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida K, Furukohri A, Shinozaki Y, Mori T, Ogawara D, Kanaya S, Nohmi T, Maki H, Akiyama M 2008. Overproduction of Escherichia coli DNA polymerase DinB (Pol IV) inhibits replication fork progression and is lethal. Mol Microbiol 70: 608–622 [DOI] [PubMed] [Google Scholar]
- Wagner J, Nohmi T 2000. Escherichia coli DNA polymerase IV mutator activity: Genetic requirements and mutational specificity. J Bacteriol 182: 4587–4595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner J, Gruz P, Kim SR, Yamada M, Matsui K, Fuchs RP, Nohmi T 1999. The dinB gene encodes a novel E. coli DNA polymerase, DNA pol IV, involved in mutagenesis. Mol Cell 4: 281–286 [DOI] [PubMed] [Google Scholar]
- Wagner J, Fujii S, Gruz P, Nohmi T, Fuchs RP 2000. The β clamp targets DNA polymerase IV to DNA and strongly increases its processivity. EMBO Rep 1: 484–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Yang W 2009. Structural insight into translesion synthesis by DNA Pol II. Cell 139: 1279–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner DF, Ndwandwe DE, Abrahams GL, Kana BD, Machowski EE, Venclovas C, Mizrahi V 2010. Essential roles for imuA′- and imuB-encoded accessory factors in DnaE2-dependent mutagenesis in Mycobacterium tuberculosis. Proc Natl Acad Sci 107: 13093–13098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JD, Crick FH 1953. Genetical implications of the structure of deoxyribonucleic acid. Nature 171: 964–967 [DOI] [PubMed] [Google Scholar]
- Wheeler LJ, Rajagopal I, Mathews CK 2005. Stimulation of mutagenesis by proportional deoxyribonucleoside triphosphate accumulation in Escherichia coli. DNA Repair (Amst) 4: 1450–1456 [DOI] [PubMed] [Google Scholar]
- Wolff E, Kim M, Hu K, Yang H, Miller JH 2004. Polymerases leave fingerprints: Analysis of the mutational spectrum in Escherichia coli rpoB to assess the role of polymerase IV in spontaneous mutation. J Bacteriol 186: 2900–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood RD, Stein J 1986. Role of the RecF gene product in UV mutagenesis of λ phage. Mol Gen Genet 204: 82–84 [DOI] [PubMed] [Google Scholar]
- Yeiser B, Pepper ED, Goodman MF, Finkel SE 2002. SOS-induced DNA polymerases enhance long-term survival and evolutionary fitness. Proc Natl Acad Sci 99: 8737–8741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Seo KY, Loechler EL 2004. A role for DNA polymerase V in G→T mutations from the major benzo[a]pyrene N2-dG adduct when studied in a 5′-TGT sequence in E. coli. DNA Repair (Amst) 3: 323–334 [DOI] [PubMed] [Google Scholar]
- Zhang X, Jeffs G, Ren X, O’Donovan P, Montaner B, Perrett CM, Karran P, Xu Y-Z 2007. Novel DNA lesions generated by the interaction between therapeutic thiopurines and UVA light. DNA Repair (Amst) 6: 344–354 [DOI] [PubMed] [Google Scholar]