Abstract
DNA damage is one of many possible perturbations that challenge the mechanisms that preserve genetic stability during the copying of the eukaryotic genome in S phase. This short review provides, in the first part, a general introduction to the topic and an overview of checkpoint responses. In the second part, the mechanisms of error-free tolerance in response to fork-arresting DNA damage will be discussed in some detail.
A cell may encounter 1000 to 100,000 DNA lesions per day. These lesions can arrest replication, activate checkpoint pathways, and set in motion repair or tolerance mechanisms to counter the stalling.
Before eukaryotic cells divide, the successful completion of DNA replication during S phase is essential to preserve genomic integrity from one generation to the next. During this process, the replication apparatus traverses in the form of bidirectionally moving forks to synthesize new daughter strands. Cells use several means to ensure faithful copying of the parental strands—first, by means of regulatory mechanisms a correctly coordinated replication apparatus is established, and second, a high degree of fidelity during DNA synthesis is maintained by replicative polymerases (Kunkel and Bebenek 2000; Reha-Krantz 2010). However, under several stressful circumstances, endogenously or exogenously induced, the replication apparatus can stall (Tourriere and Pasero 2007). Mostly, structural deformations in the form of lesions or special template-specific features arrest the replication process, activate checkpoint pathways and set in motion repair or tolerance mechanisms to counter the stalling (Branzei and Foiani 2009; Zegerman and Diffley 2009). Basic replication mechanism, its regulatory pathways and means to tolerate DNA damage are largely conserved across eukaryotic species (Branzei and Foiani 2010; Yao and O’Donnell 2010). Understanding the mechanisms involved may enable therapeutic intervention to several human conditions arising from an incomplete replication or from the inability to tolerate perturbations (Ciccia et al. 2009; Preston et al. 2010; Abbas et al. 2013). Enhanced replication stress has also been commonly identified in precancerous lesions, and the inactivation of checkpoint responses coping with this presumably oncogene-induced condition is considered necessary to establish the fully malignant phenotype (Bartkova et al. 2005; Negrini et al. 2010).
It is not possible to treat this topic in a comprehensive manner in the allotted space; the reader is referred to excellent recent reviews for more details (Branzei and Foiani 2010; Jones and Petermann 2012). We will attempt to provide an overview of the various strategies that a eukaryotic cell invokes to avoid problems caused by replication stress related to DNA damage and, if problems arise, to tolerate damage without endangering the entire process of genome duplication. In this context, we will only give a brief outline of checkpoint responses that are discussed in more detail in Sirbu and Cortez (2013) and Marechal and Zou (2013). Also, a detailed discussion of translesion synthesis can be reviewed in Sale (2013).
REPLICATING UNDAMAGED DNA
Under normal conditions, replication is a high-fidelity process with an error rate of 10−8 to 10−10 mutations per base pair per cell division (Kunkel and Bebenek 2000). In a general scheme, for the eukaryotic genome to be duplicated, the replication machinery needs to access the DNA from its highly packaged chromatin conformation, move bidirectionally to synthesize new daughter strands, terminate the synthesis, and finally repackage the new strands before mitosis (Bell and Dutta 2002). Replication is subdivided into three major steps: initiation, elongation, and termination.
Regulation of Origin Firing
Before S-phase entry, a prereplication complex (pre-RC) recognizes and assembles at replication origins in late M and early G1 phase of the cell cycle (Dutta and Bell 1997). Pre-RC facilitates unwinding of parental DNA strands and subsequent formation of the replication fork. There are around 500 AT-rich origins of defined sequence in yeast (Dhar et al. 2012) and about 2.5 × 104 origins in metazoan cells (Pope et al. 2013), allowing multiple initiation sites for rapid replication of a complex genome. In metazoans, DNA topology rather than DNA sequence may define an origin (Vashee et al. 2003; Remus et al. 2004; Mechali 2010). Origins are first recognized by the origin recognition complex (ORC) proteins (ORC 1-6). ORC bound at the replication origin serves as a docking site for CDC6 (cell division cycle 6), CDT1 (chromatin licensing and DNA replication factor 1), and then MCM (mini-chromosome maintenance) 2-7 helicase complex, licensing it for the next step of initiation (Tye 1999).
In metazoan cells, around 2–5 adjacent origins are organized in one cluster and are simultaneously initiated for replication (Gillespie and Blow 2010; Aparicio 2013). One or more of these clusters are arranged into DNA domains. Each domain may comprise an individual focus that can be identified microscopically, indicating sites of active replication during S phase termed replication factories, and there is evidence that the sister forks of each replicon stay associated with each other within these factories (Kitamura et al. 2006). In higher eukaryotic cells, even when all origins are licensed similarly, only around 10% are used for replication and the rest remain dormant, albeit fully capable of initiating replication (McIntosh and Blow 2012). During times of replication stress, dormant origins are activated to compensate for fork stalling and to prevent genomic instability (Ge et al. 2007).
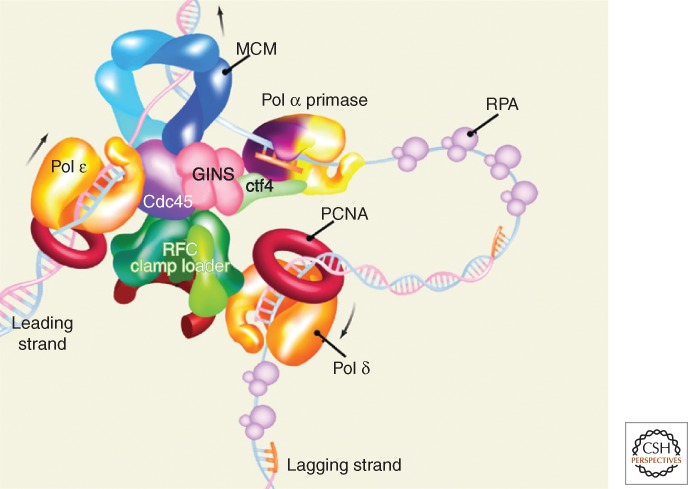
Activation of pre-RC to a preinitiation complex (pre-IC) occurs in S phase by sequential phosphorylation, loading, and activation of initiation complex proteins (Pospiech et al. 2010). In metazoans, two proteins, Treslin/Ticrr (homolog of yeast Sld3) and TopBP1 (Dpb11 homolog), associate in a CDK-phosphorylation-dependent manner with chromatin (Tanaka et al. 2007) to provide a docking site for the next set of proteins. Then, dependent on RPA (replication factor A) and again dependent on CDK phosphorylation, CDC45 binds to Treslin/Ticrr (Zou and Stillman 1998, 2000), and unphosphorylated RecQ4 (Sld2 homolog) binds to TopBP1 to enable subsequent helicase activation and origin unwinding. MCM protein phosphorylation by the CDC7/DBF4 kinase is yet another essential event of origin activation (Zou and Stillman 2000; Masai and Arai 2002). Third, GINS complex loads onto chromatin (Labib and Gambus 2007). Polymerase ε is recruited to pre-RC, enabling formation of CMG (CDC45, MCM2-7, and GINS complex) helicase (Muramatsu et al. 2010). ATP binding activates the CMG helicase activity, which switches dsDNA-bound MCM2-7 helicase to two ssDNA strands. MCM2-7 helicase activity is required throughout replication for fork progression (Labib et al. 2000). At this stage, the DNA polymerase machinery associates with MCM2-7 and begins bidirectional synthesis of complementary strands (Fig. 1).
Figure 1.
A model of the eukaryotic replisome. See text for details. (From Yao and O’Donnell 2010; adapted, with permission, from Elsevier © 2010.)
Although origin use appears to be developmentally regulated, few regulatory molecules are known that determine early or late origin firing, and the replication timing program remains an active area of investigation (Aparicio 2013; Pope et al. 2013). High abundance of CDC45 near early origins as well as its recruitment by Fkh1/2 (forkhead transcription factor) protein, bound to ORCs in G1, facilitates early initiation. On the other hand, late origins are localized and tethered to the nuclear periphery by Rif1/Taz1/Yku70 complex, preventing CDC45 from reaching these origins by chromatin-bound Rpd3.
Elongation and Termination of Replication
As essential players, the replication complex in mammals is composed of RFC (replication factor C complex), PCNA (proliferating cell nuclear antigen), and DNA polymerases—Pol α/primase, Pol ε, or δ—to synthesize the new leading and lagging strand, respectively (Fig. 1) (Stillman 2008; Pope et al. 2013). Generally, Pol α first synthesizes a few nucleotides before synthesis is shifted to the elongating replicative polymerase by the RFC-PCNA complex (Stukenberg et al. 1994), which later also recruits other components such as Fen1 and DNA ligase for completion of replication. Additional proteins such as Claspin (homolog of yeast Mrc1), the complex of Tipin and Timeless (homolog of yeast Tof1), associate with replicons and are required for signaling during the replication stress response (Kumagai and Dunphy 2000; Chini and Chen 2004; Chou and Elledge 2006). Other associated proteins, such as the Rrm3 helicase may come into play to prevent fork stalling by resolving difficult template structures (Torres et al. 2004).
The sister forks of the replication bubble progress in a bidirectional manner until the replication complex encounters a termination signal. The termination signals in higher eukaryotes presumably occur when converging forks meet each other randomly (Santamaria et al. 2000); in yeast, Top-II-dependent resolution of catenated DNA may become the site of termination (Cuvier et al. 2008).
DNA DAMAGE AND REPLICATION
DNA damage has been reviewed in other reviews and so a few general comments will suffice. Lesion formation in DNA can have endogenous (“spontaneous”) or exogenous origins (Friedberg et al. 2006). The lesions that can interfere with replication are typically categorized as bulky or, in more general terms, as a base alteration that does not allow a high-fidelity replicative DNA polymerase to insert a complementary base or, following insertion, to extend from an imperfectly matched base pair. Structural interruptions of the template strand such as single and double-strand breaks or single-stranded gaps represent another class of lesions interfering with replication. These may have been introduced directly, for example, by ionizing radiation or restriction enzymes. They may also represent intermediates of DNA repair taking place before or during replication, or secondary damage as a result of failed damage tolerance and a consequence of replication fork collapse (see below). A special class of DNA lesion that undoubtedly will stall replication by preventing strand separation are interstrand crosslinks between two complementary DNA strands (discussed in Clauson et al. 2013).
Only recently, the incorporation of deoxyribonucleotide triphosphates (rNTPs) during S phase received special attention, and such lesions were found to be the most frequent endogenous lesions in the mammalian genome (Nick McElhinny et al. 2010b; Reijns et al. 2012). Whereas the occurrence of these lesions is minimized by the selectivity of replicative DNA polymerases, RNase H1 and RNase H2 activities are needed to repair incorporated dNMPs and to prevent toxic consequences that may be aggravated by additional replicational stress (Lazzaro et al. 2012; Reijns et al. 2012). Presence of rNMPs in the genome may lead to sensitivity of the DNA backbone, problems during DNA synthesis as a result of altered helix geometry, and short deletions in repeated sequences through a topoisomerase I-dependent mechanism (Nick McElhinny et al. 2010a; Kim et al. 2011; Watt et al. 2011).
Even when cells encounter approximately 103–105 molecular lesions per cell per day, the collection of DNA damage responses that effectively remove such lesions maintains a very low probability of spontaneous mutagenesis.
CHECKPOINT RESPONSES DURING DNA REPLICATION
Checkpoint responses are responsible for the tight feedback regulation of normal progression of the cell cycle and frequently referred to as G1/S, intra-S, and G2/M checkpoints. Controls operate to check for faithful completion of cell cycle phase transitions and respond to genomic perturbations by not only arresting the specific cell-cycle phase but also by improving DNA repair and damage tolerance through direct protein modification or transcriptional regulation (see Marechal and Zou 2013; Sirbu and Cortez 2013 for details; Melo and Toczyski 2002; Sancar et al. 2004; Friedberg et al. 2006; Niida and Nakanishi 2006; Ciccia and Elledge 2010). In the case of DNA-damage-induced checkpoint responses, damage to DNA leading to structural changes is initially detected by sensor molecules (phosphoinositol kinase-like kinases ATM and ATR/ATRIP, the Rad17-RFC-like complex or the 9-1-1/PCNA-like complex) that are largely operative in all phases of the cell cycle. After sensing the damage and with the help of mediator proteins, which may act cell-cycle-stage-specifically (Claspin, BRCA1, 53BP1, MDC1, TOPBP1, MRN complex), the damage signal is passed on to transducer molecules, such as the CHK1 or CHK2 kinases (Bartek et al. 2001; Chen and Sanchez 2004). Mediator proteins provide specificity of signaling because they simultaneously bind both sensors and transducers (Tanaka 2010). Once the damage signal is received by the transducers, they modify direct or indirect mediators of cell-cycle progression (such as p53, CDC25-A, -B, and -C) as their effector molecules by phosphorylation, possibly resulting in proteolytic degradation (Falck et al. 2001; Xiao et al. 2003). If phosphorylated or absent, effector molecules such as CDC25 isoforms cannot promote G1/S or G2/M transitions and CDKs in G1/S and CDC2 in G2 phase remain phosphorylated, thereby establishing a cell-cycle phase-specific arrest.
G1/S Checkpoint
Before DNA replication takes place, the damage that potentially instigates a cell cycle arrest in G1 are double-strand breaks or single-stranded DNA tracts, activating ATM or ATR, respectively. Prereplicatively, single-stranded DNA gaps may be associated with nucleotide excision repair (NER) of bulky lesions, possibly widened by nuclease action (Giannattasio et al. 2010). In vertebrates, the G1/S checkpoint in the presence of DNA damage prevents initiation of replication in G1 by two mechanisms. Activated ATM/ATR phosphorylate effector molecules CHK1/CHK2 and p53 (Lane and Levine 2010), which result in an immediate response and a slower maintenance response that depends on protein synthesis. First, phosphorylated CHK1/CHK2 phosphorylates CDC25-A phosphatase and mediates its degradation by ubiquitination (Falck et al. 2001; Xiao et al. 2003). In the absence of CDC25-A, dephosphorylation of CDK2/Cyclin E complexes is prevented and, for example, CDC45 thus fails to activate pre-IC at otherwise licensed origins, inhibiting initiation of replication. Second, p53 in G1 is phosphorylated at multiple sites by activated ATM/ATR as well as CHK1/CHK2, which allows increased expression of p53 targets like the gene-encoding CDK inhibitor p21WAF-1/Cip1. P21 in turn binds and inhibits CDKs necessary for transition into S phase (El-Deiry et al. 1993; Harper et al. 1993), thus maintaining the G1 arrest.
Intra-S Checkpoint
General Comments
DNA damage introduced in S phase or unrepaired damage from G1, which has escaped the G1/S checkpoint, may trigger intra-S-phase checkpoint(s) (Branzei and Foiani 2009; Zegerman and Diffley 2009; Jones and Petermann 2012; see also Sirbu and Cortez 2013). ATR signals such as single-stranded DNA may arise from uncoupling of leading and lagging strand synthesis, from helicase uncoupling at a stalled replication fork or from gaps opposite base damage (Byun et al. 2005; Lopes et al. 2006; Callegari et al. 2010). Yeast studies suggest independent pathways of activation, one of which is dependent on Pol ε (Navas et al. 1995; Puddu et al. 2011). If not externally introduced, double-strand breaks as ATM-activating signal may arise from fork collapse or topoisomerase malfunctioning.
Checkpoint responses within S phase are multifaceted, using direct protein modification as well as transcriptional regulation to cope with replication stress by controlling replicon initiation, fork progression, and fork stability. They also assist in DNA damage repair and in tolerance mechanisms if fork stalling has occurred. But it is not only DNA damage that can be a source of replication stress. Replication forks may encounter bound proteins, special DNA structures, replication-slow zones, and collisions with RNA polymerase. Replication forks will initially stall with a fully assembled replisome. On extended pausing, however, the replisome dissociates irreversibly and the fork collapses.
Even without induced DNA damage, checkpoint responses are clearly required to successfully cope with areas that are difficult to replicate, preventing chromosome breaks and possibly cycles of chromosome instability (Cha and Kleckner 2002; Admire et al. 2006). A low supply of dNTPs may also cause a slowdown or arrest of replication that may be experimentally triggered by inhibiting ribonucleotide reductase (RNR) with hydroxyurea. Budding yeast defective in the ATR/CHK2 homologs Mec1/Rad53 are unable to recover from such HU-induced arrest (Desany et al. 1998).
Origin Firing
In vertebrates, the mechanisms outlined above for G1/S arrest may affect origin firing throughout S phase because CHK1 also regulates CDK1/Cyclin A complexes (Nakanishi et al. 2010). Another mechanism inactivates the CDC7/DBF4 kinase (Weinreich and Stillman 1999; Costanzo et al. 2003; Heffernan et al. 2007). In yeast, there is evidence for selective inhibition of late origin firing by Rad53 in the presence of DNA damage (Santocanale and Diffley 1998). Interestingly, a single double-strand break in S phase may not trigger the checkpoint system for an extended period of time, and without global checkpoint activation by a higher damage load the effects on origin firing were quite the opposite—firing from a nearby active origin was enhanced and dormant origins were activated (Doksani et al. 2009).
Fork Progression Rate
Here, one has to distinguish between active regulation causing slowdown by checkpoint activation and a passive cessation as a result of damage in the template or any other perturbation; both events may very well occur simultaneously (Minca and Kowalski 2011). Without any applied stresses, vertebrate cells defective in several checkpoint components (ATR, CHK1, Claspin, Timeless) show a slowed fork progression, showing the importance of checkpoint signaling even for normal replication (Petermann et al. 2006; Unsal-Kacmaz et al. 2007). But there is also evidence that the Tipin/Timeless complex initiates such a regulation in response to DNA damage, perhaps through ATM/ATR by regulating MCM2-7 helicase (Cortez et al. 2004; Seiler et al. 2007; Unsal-Kacmaz et al. 2007). In yeast, however, there was no evidence that the Mec1/Rad53 pathway slows down fork progression per se; instead, altered origin use appears to be the predominant reason for a significant extension of overall replication time in methyl methanesulfonate (MMS)-treated cells (Tercero and Diffley 2001).
Fork Stabilization
Using a variety of mechanisms, the checkpoint system promotes fork stability (Jones and Petermann 2012), here defined as preventing accumulation of aberrant fork structures either spontaneously or when challenged by DNA damage or other stresses. These include:
Prevention of dissociation of replication proteins from a stalled fork (Trenz et al. 2006).
Regulation of homologous recombination (HR) activities by phosphorylation of RAD51, BRCA2, FANCD2, and others (Andreassen et al. 2004; Sorensen et al. 2005; Bahassi et al. 2008).
Promotion of sister chromatid cohesion (Kitagawa et al. 2004; Errico et al. 2009; Leman et al. 2010).
Up-regulation of helicases that are beneficial for fork remodeling to promote restart (i.e., BLM and WRN helicases, working together with FANCJ) (Davies et al. 2004; Ammazzalorso et al. 2010; Pichierri et al. 2012).
Down-regulation of nucleases that may damage a stalled fork, such as MUS81-Eme1 in fission yeast or EXO1 in mammals (Kai et al. 2005; El-Shemerly et al. 2008).
Regulation of chromatin modifications and histone supply, the latter by dephosphorylating the histone chaperone ASF1 (Sillje and Nigg 2001; Groth et al. 2003; Clemente-Ruiz and Prado 2009).
Targeting nuclear pore components that tether transcribed genes to release topological strain on replicating DNA (Bermejo et al. 2011).
These are just a few selected insights into an intricate network of checkpoint responses. Space limitations make it impossible to discuss other important aspects such as transcriptional responses, with yeast RNR regulation being the best understood paradigm (Friedberg et al. 2006), or telomere maintenance.
DNA DAMAGE TOLERANCE PATHWAYS AT ARRESTED REPLICATION FORKS
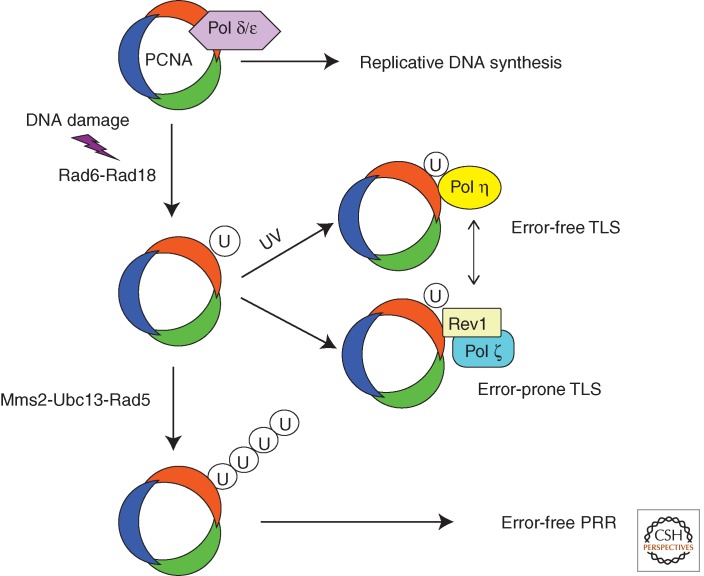
After mostly considering higher eukaryotes, we will revisit the yeast model to gain a mechanistic understanding of the various tolerance pathways that allow completion of replication in the presence of fork-arresting DNA base damage. A similar degree of mechanistic resolution has not been achieved in any other system. In the yeast Saccharomyces cerevisiae, it has now been firmly established that essentially three mechanisms of DNA damage bypass exist whose activation is initially triggered by the ubiquitin-conjugating enzyme (E2) Rad6 that acts in a complex with Rad18, its E3 partner (Fig. 2) (Jentsch et al. 1987; Friedberg et al. 2006). Rad18 may localize this activity to stalled replication forks through its single-stranded DNA-binding activity (Bailly et al. 1997). One essential target appears to be the sliding clamp PCNA, which is monoubiquitinated at Lys164, whose interaction with Rev1 sets the stage to initiate error-prone translesion synthesis (TLS) by Pol ζ (the complex of Rev3-Rev7) (Fig. 2) (Hoege et al. 2002; Stelter and Ulrich 2003; Acharya et al. 2006; Guo et al. 2006a,b). Pol ζ acts frequently at the extension step after another polymerase has inserted the first nucleotide opposite a lesion (Prakash et al. 2005). PCNA also attracts Pol η, an enzyme catalyzing an error-free bypass of the most frequent UV lesions. However, the significance of PCNA monoubiquitination and the ubiquitin-binding domain of Pol η has been subject to controversies (Fig. 2) (Garg and Burgers 2005; Acharya et al. 2007). Independent of its catalytic activity, Pol η may also have a role in recruiting Rad18 (Durando et al. 2013). A detailed discussion of TLS can be found in Sale (2013).
Figure 2.
Significance of PCNA and its ubiquitination for DNA damage tolerance pathways in budding yeast. Depicted is the repurposing of PCNA from its role as a processivity factor in normal replication by Rad6-Rad18 dependent monoubiquitination or Mms2-Ubc13-Rad5 polyubiquitination for error-free or error-prone translesion synthesis (TLS) and error-free postreplication repair (PRR). (From Zhang et al. 2011; adapted, with permission, from Elsevier © 2011.)
The other option, frequently termed error-free postreplication repair (PRR), requires polyubiquitination of PCNA via Lys63 linkage performed by the ubiquitin ligase activity of Rad5 protein together with its E2 enzyme component Ubc13–Mms21 (Fig. 2) (Broomfield et al. 1998). Rad5 itself forms contacts with the E3 enzyme Rad18, which starts the initial PCNA monoubiquitination (Ulrich and Jentsch 2000). This scheme of pathway choice is not only applicable to exogenously induced DNA damage but the Pol ζ and Rad5 pathway (not Pol η, however) also act as backup pathways to tolerate rNMPs in the genome if repair by RNase is nonfunctional (Lazzaro et al. 2012).
There is very clearly cross talk on multiple levels with the checkpoint system (e.g., CHK1 and Claspin up-regulate PCNA ubiquitination in response to UV or HU) (Yang et al. 2008). Independent of its checkpoint function, the PCNA-related 9-1-1 complex activates the Rad5 pathway in yeast (Karras et al. 2013) and suppresses TLS under conditions of chronic DNA damage (Murakami-Sekimata et al. 2010).
The Rad5 pathway is clearly overall error-free in budding yeast and contributes to genetic stability. Under continuous low-dose UV exposure, this pathway is a major determinant of UV resistance (Hishida et al. 2009). Many years ago, the possibility of a transient template switch by the replication machinery for overcoming an impediment in the template was suggested—a copy-choice mechanism directed to the sister chromatid. The predicted mechanism requires a certain degree of replication uncoupling because a newly synthesized strand at the sister chromatid needs to be present to serve as a template. In a yeast plasmid system, it was indeed shown that the Rad5 mechanism involves recombination between partly replicated sister strands but neither TLS polymerase ζ nor mismatch repair (Zhang and Lawrence 2005).
All available information suggests that the overall outline of this model is indeed correct. In the following, we will provide an update on Rad5 activities in yeast and mammalian cells and will then describe recent mechanistic models for the pathway.
RAD5 AND ITS ACTIVITIES
Besides its activity as an E3-type ubiquitin ligase, Rad5 is also a member of the SWI/SNF family of proteins and shows DNA-dependent ATPase activity. A Rad5 deletion mutant is much more UV-sensitive than a deletion of its E2-partners Ubc13 or Mms2, so both activities are clearly required for efficient damage tolerance. Thus, its roles turned out to be multifaceted and also extend to certain aspects of TLS (Gangavarapu et al. 2006; Pages et al. 2008). Although promoting genetic stability through overall error-free damage tolerance, budding yeast Rad5 is required for efficient mutagenic TLS in certain mutation systems, possibly by playing a structural role mediated through interaction with Rev1 (see Sale 2013). Interestingly and in contrast to Saccharomyces cerevisiae, it is the polyubiquitination of PCNA by Schizosaccharomyces pombe Rad5 homolog Rad8 that is clearly required for various modes of TLS, possibly facilitating the formation of multipolymerase complexes (Coulon et al. 2010). Thus, it is to be noted that gene products and their primary activities within DNA damage tolerance pathways may well be evolutionarily conserved but their significance for pathway choice may vary greatly from organism to organism.
In mammalian cells, two homologs of Rad5 were identified, termed SHPRH (for SNF2 histone-linker PHD-finger RING-finger helicase) and HLTF (for helicase-like transcription factor), both with the ability to polyubiquitinate PCNA (Unk et al. 2006, 2008, 2010; Motegi et al. 2008). Complex formation with the homologs of yeast Rad18 and Mms2-Ubc13 was confirmed. Loss of either Rad5 homolog increases the frequency of chromosomal abnormalities as well as point mutations in response to DNA damage (Motegi et al. 2006; Lin et al. 2011), which characterizes this pathway also as overall error-free and contributing to genetic stability in mammalian cells. Frequent silencing of HLTF and changes of SHPRH in various cancers points in the same direction (Unk et al. 2010). The roles of these homologs are clearly not only nonredundant but even antagonistic, depending on the nature of the DNA damage (Lin et al. 2011). In response to MMS, HLTF is degraded and Pol κ is recruited by SHPRH for error-free TLS; in response to UV, HLTF suppresses SHPRH and, surprisingly, promotes monoubiquitination of PCNA, which in turn may facilitate error-free TLS by Pol η.
Helicase activity of purified yeast Rad5 had never been identified in traditional DNA unwinding assays. However, if certain model substrates were tested that mimic replication forks, remodeling activities were identified for yeast Rad5 and mammalian HLTF that proceeded without exposing single-stranded DNA (Blastyak et al. 2007, 2010; Unk et al. 2010). Additionally, on four-way substrates mimicking Holliday junctions, branch migration was detected.
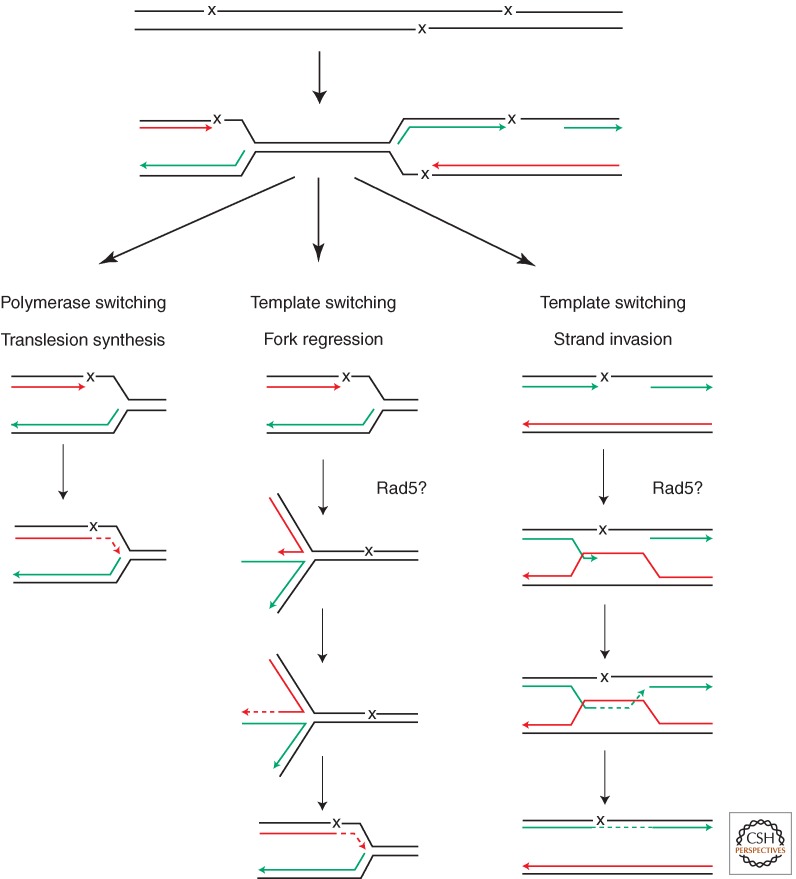
If one considers a structure where template switching can successfully operate to overcome a replication block (e.g., a leading-strand arrested fork with some uncoupling of lagging strand synthesis), one would model a homologous fork structure with a single-stranded gap. Here, Rad5 activity can indeed catalyze fork regression into a so-called chicken foot structure (Fig. 3). Chicken foot structures will allow the use of the newly synthesized daughter strand as a template for the arrested leading strand. Is this then a viable model for the template switch pathway?
Figure 3.
A model of various tolerance mechanisms for DNA damage in S phase to support ongoing replication, with special consideration of Rad5 activities. Shown are translesion synthesis and two possible modes of Rad5-mediated template switching, by fork regression or strand invasion. (From Unk et al. 2010; adapted, with permission, from Elsevier © 2010.)
TOWARD A MODEL FOR THE TEMPLATE SWITCH PATHWAY
At this point, a few words on the origin of DNA strand discontinuities as a consequence of replicating DNA with bulky DNA damage are in order. It is a classic idea from studies in E. coli that DNA containing UV damage is first synthesized in smaller pieces because gaps are left opposite photoproducts and later closed by recombination with the daughter strand (Rupp and Howard-Flanders 1968; Rupp et al. 1971). In mammals, though, there was little evidence for sister strand exchanges (Lehmann 1972). Given the bidirectional mode of replication from multiple origins, earlier ideas on overall replication patterns are still worth considering (Spivak and Hanawalt 1992). Because silent origins can be activated, it is feasible that DNA replication can be largely completed, with only single-stranded gaps opposite photoproducts left. Only an origin-free region in which replicons approaching from either side are arrested because of damage in the leading strand template will result in unreplicated areas. The latter may even be avoidable if uncoupling of leading and lagging strand synthesis occurs.
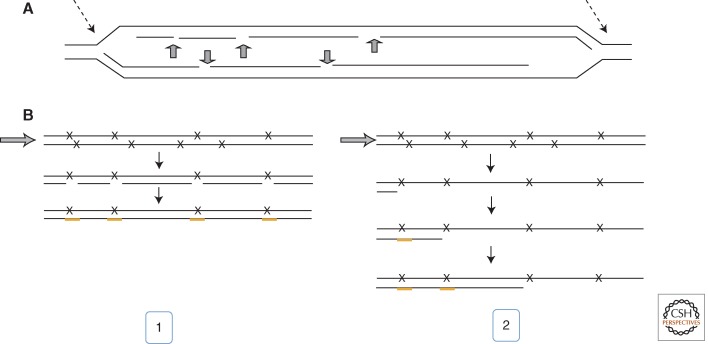
A more recent study on replication in the presence of UV damage in yeast, using electron microscopy and 2D gel electrophoresis, added important insights (Lopes et al. 2006). Evidence for uncoupling of leading and lagging strand synthesis was found, creating extended single-stranded DNA tracts on one side of the replication fork, on opposing sides within bubble structure (Fig. 4). Accumulation of single-stranded gaps during progression of replication was noted on both strands. It is thus very clear that single-stranded gaps were left behind, most likely to be filled after replication was completed (Fig. 4). The activity of TLS and homologous recombination (HR) seemed to reduce their accumulation but these processes did not affect overall fork progression itself; thus, efficient repriming must occur (Lopes et al. 2006; Callegari et al. 2010). Chicken foot intermediates, however, were classified as pathological structures that accumulated only in checkpoint-deficient cells—fork stabilization appears to prevent fork regression. Additionally, Exo1 processing counteracted extended fork regression by creating single-stranded DNA intermediates (Cotta-Ramusino et al. 2005). So, whereas Rad5-mediated fork regression is certainly an appealing mechanism to preserve ongoing leading strand synthesis, its significance awaits confirmation. Fork regression events may conceivably escape detection if quickly resolved.
Figure 4.
Concepts for origin and repair of single-stranded DNA gaps during replication of damaged DNA. (A) Pattern of single-stranded gaps during DNA replication of UV-irradiated budding yeast, as found in an EM study (Lopes et al. 2006). Gaps are detectable in both daughter strands of the same replicon. Note the position of single gaps at the diverging fork boundaries (narrow arrows), indicating arrested leading strand synthesis and ongoing uncoupled lagging strand synthesis. (B) Two possible modes of filling gaps opposite photoproducts (x) are outlined: entirely postreplicatively after replication fork (arrow) has passed and created an otherwise full-length daughter strand (thin line) (1) or concomitantly with fork progression, which may depend on this gap-filling process (2). (From Lehmann and Fuchs 2006; adapted, with permission, from Elsevier © 2006.)
The bulk of DNA replication appears to be advanced by efficient repriming of DNA synthesis downstream of the lesion, most likely at otherwise silent origins, to prevent large regions of single-stranded or unreplicated DNA. The other sources of small gaps are, of course, lesions in the lagging strand template. Accumulating gaps may be filled by TLS or the HR machinery (or a subset of HR proteins) whose connection with the Rad5 pathway was initially unknown. We will be addressing these interrelationships in the next section.
ROLE OF RECOMBINATION IN ERROR-FREE POSTREPLICATION REPAIR
It is useful to continue confining this discussion to the tolerance of unrepaired bulky or fork-arresting DNA damage. Here, we are not concerned with the details of homologous recombination provoked by double-strand breaks (as discussed in Clauson et al. 2013).
SUMOylation of various DNA repair and recombination targets has emerged as a major response to DNA damage that is required for successful replication of a damage-containing genome (Cremona et al. 2012). The response is activated independently of the checkpoint system (represented in yeast by the ATR homolog Mec1) but is instead positively regulated by the MRX (Mre11-Rad50-Xrs2) complex. Besides being ubiquitinated, PCNA is also subject to Siz1-mediated SUMOylation at the same lysine (164) that is ubiquitinated and a minor site (Lys127). SUMO-modified PCNA recruits the Srs2 helicase, which in turn disrupts Rad51 filaments to prevent “unwanted” (but ill-defined) HR events that appear to involve sister chromatids (Pfander et al. 2005). This mechanism can, however, substitute to some extent for the Rad5 mechanism because the UV resistance of rad6, rad18, or rad5 mutants is increased if Srs2 (or Siz1) is inactive (Friedberg et al. 2006).
During treatment of yeast with MMS and other agents, X-shaped sister chromatid junctions (representing pseudo-double Holliday junctions) have been identified by 2D gel electrophoresis (Branzei et al. 2008; Minca and Kowalski 2010). Their resolution depends on the WRN/BLM-related, RecQ-type helicase complex Sgs1-Top3 (Liberi et al. 2005), of which Sgs1 is also SUMOylated (Branzei et al. 2006). Studied in an sgs1 defective background, these molecules were formed through a pathway that involves Rad5-Rad18 and their PCNA polyubiquitination activity as well as Rad51-Rad52 if PCNA SUMOylation is functional; without it, the appearance of these junctions is solely dependent on the HR pathway (Branzei et al. 2008). This supports the assumption of a cooperatively acting Rad5/HR pathway of gap filling by template switch (a model is shown in Fig. 5) and an “unwanted” pathway that involves HR only and may not be strictly DNA damage dependent.
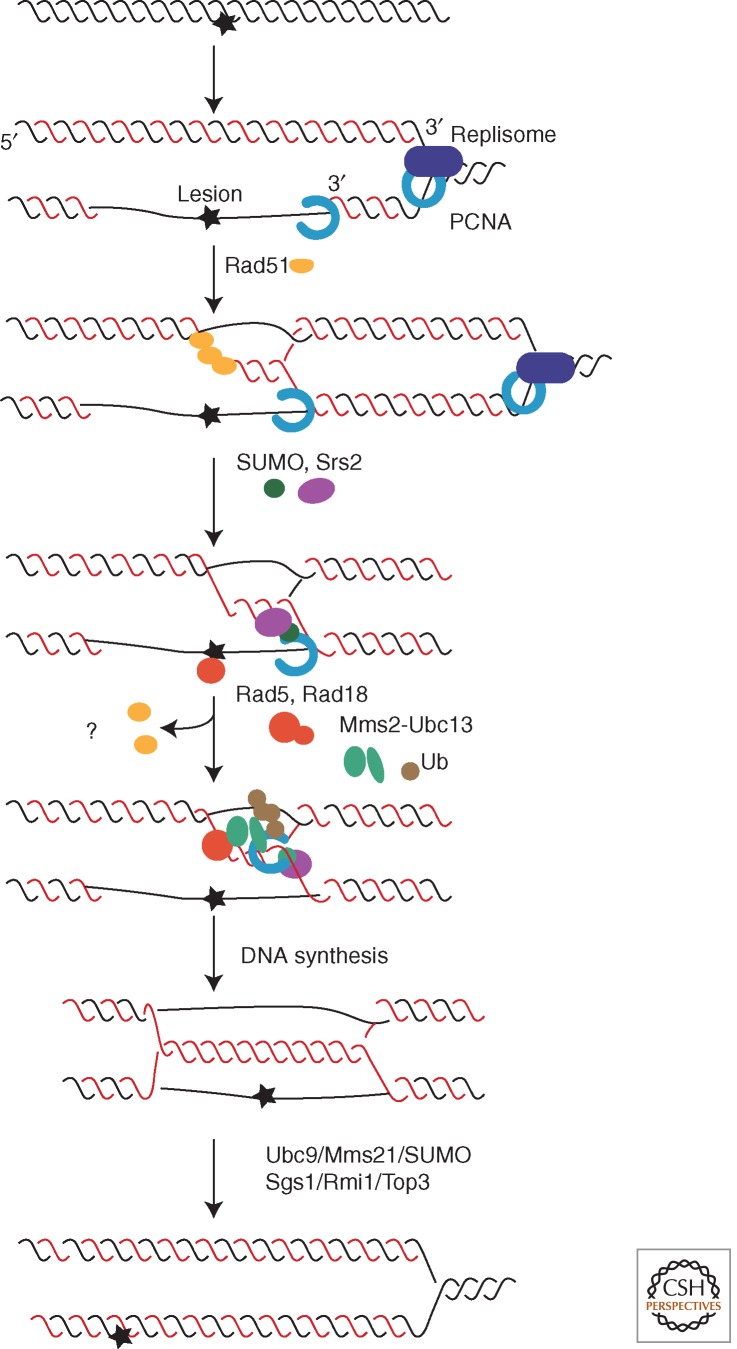
Figure 5.
A detailed model of the Rad5-mediate template switch pathway, assuming gap filling dependent on a subset of HR functions after the replication fork has passed. First, Siz1-dependent, SUMOylated PCNA stimulates Srs2 helicase to disrupt Rad51 filaments that have initiated D-loop formation. The role of Rad51 and other HR proteins still needs to be defined; they may rejoin and support downstream events in a remodeled complex. Depending on Rad18, PCNA is polyubiquitinated by Rad5-Mms2-Ubc13 (see Fig. 2), which, in an unknown way, promotes gap filling by DNA synthesis. (SUMOylation and polyubiquitination do not have to occur sequentially but may persist simultaneously on different subunits of the PCNA trimer.) The resulting hemicatenane structure is resolved by Sgs1-Top3-Rmi1 helicase, itself dependent on SUMOylation by Ubc9-Mms21. (Based on data from Branzei et al. 2008.)
Further studies have defined the subset of HR proteins involved more closely and also identified Pol δ, but none of the TLS polymerases were identified as an important component (Vanoli et al. 2010). Mention should also be made of a complex of proteins termed Shu that is required for efficient HR, and genetic arguments suggest a role in facilitating template switching and junction resolution (Ball et al. 2009).
Uncertainties remain. For UV damage, Rad5 and HR pathways interact nonepistatically, arguing against an exclusive participation in a joint pathway. It has been proposed that Rad5 may act on the leading strand through fork regression, whereas HR proteins may fill gaps on the lagging strand through a noncanonical mechanism (synthesis-dependent strand annealing) (Gangavarapu et al. 2006).
TIMING OF DNA DAMAGE TOLERANCE MECHANISMS
We already mentioned that budding yeast cells seem to have the option to fix single-stranded gaps long after bulk DNA replication is over, which may not be significantly perturbed by the absence of tolerance pathways (Lopes et al. 2006). In yeast, the timing of error-free PRR and TLS has been addressed in two sophisticated studies, using cell-cycle-specific targeting of expression of key proteins (Daigaku et al. 2010; Karras and Jentsch 2010). For example, whereas the peak of PCNA ubiquitination normally occurs in S phase, it is clearly also possible to delay this process until G2/M if Rad18 expression is targeted to that stage. Overall, these studies agree that there is no disadvantage for a yeast cell to delay all UV-damage-tolerance processes until G2/M, but is this what happens under physiological conditions? For error-prone TLS of UV photoproducts, this may indeed be the case because yeast Rev1 expression is strongly increased during G2/M (Waters and Walker 2006). In vertebrates, however, two activities have been delineated for Rev1-dependent TLS—a role during S phase in advancing replication and a postreplicative gap-filling function (Edmunds et al. 2008; Jansen et al. 2009). At least for very low doses of MMS, the Rad5 pathway in yeast enables normal fork progression without any need for checkpoint activation, preventing accumulation of single-strand gaps that would need to be repaired postreplicatively by TLS (Huang et al. 2013).
It is to be expected that the structure of damage needs to be taken into consideration. A study focusing on adozelesin causing bulky minor-groove damage shows a clear requirement for budding yeast Rad5 in resolving stalled forks in a recombination-dependent manner so that replication can progress normally and be completed (Minca and Kowalski 2010).
Most of these DNA damage tolerance studies have been performed in NER-deficient cells. However, one should remember that a largely prereplicative fixation of UV mutations has been inferred from genetic data for NER-proficient budding yeast (Friedberg et al. 2006). A recent detailed genetic characterization, in conjunction with the described nuclease-dependent extension of single-stranded gaps in G1, may finally provide a mechanistic basis (Giannattasio et al. 2010; Kozmin and Jinks-Robertson 2013).
In conclusion, many open questions remain. It should be noted that a precise mechanistic role for polyubiquitination of PCNA has yet to be delineated. Does this modification create a new interaction surface or is its duty the repelling of replication fork components that interfere with the tolerance mechanism? If error-free PRR and TLS are temporally separated, does polyubiquitination of PCNA actually precede the accumulation of monoubiquitinated PCNA? Or, alternatively, are different PCNA molecules assigned to keep one modification or the other, depending on unknown criteria? Do poly- and monoubiquitinated PCNA monomers coexist within the same trimer? Detailed kinetic measurements in budding yeast were consistent with sequential ubiquitination steps and, even at low UV doses, the fast emergence of polyubiquitinated PCNA together with a lower but persistent fraction of monoubiquitinated PCNA (Amara et al. 2013). Active deubiquitination also clearly occurred but this process is insufficiently understood—Ubp10 participates but it is clearly not the only PCNA deubiquitinating enzyme.
REPLICATION FORK RESTART
In the preceding sections, we have focused on the bypass mechanisms of bulky DNA damage. Even without any necessity for damage bypass, recombination activities play a complex role in the response to replication perturbation. Ultimately, terminally collapsed replicons will be converted into double-strand breaks that trigger HR (Petermann et al. 2010). Whereas this phenomenon would invoke the canonical HR pathway, another role is played in the restart of arrested forks, which in vertebrates does not require RAD51 foci formation (Petermann et al. 2010). RAD51, BRCA2, MRE11, XRCC3, FANCA, and FANCD2 have all been implicated in replication fork restart or stabilization—if defective, double-strand breaks are accumulated when challenged with replication inhibitors and even during normal replication (Sonoda et al. 1998; Costanzo et al. 2001; Henry-Mowatt et al. 2003; Lomonosov et al. 2003; Sobeck et al. 2006). The involvement of MRE11 seems to hint at the need for (limited) single-strand degradation at DNA ends. MRE11 interacts with BLM helicase (Robison et al. 2004), and a variety of helicases (WRN, BLM, FANCM, and SMARCAL1), appear to be supportive of fork restart (Davies et al. 2007; Sidorova et al. 2008; Luke-Glaser et al. 2010). The SMARCAL1 helicase is targeted to stalled replication forks through its RPA-binding motif and functions as an annealing enzyme, possibly reannealing excessively unwound DNA (Bansbach et al. 2009; Ciccia et al. 2009). Figure 6 incorporates these findings into a model based on fork regression, resection, and restart after Rad51-mediated D-loop formation but there may very well be more than one mechanism at work.
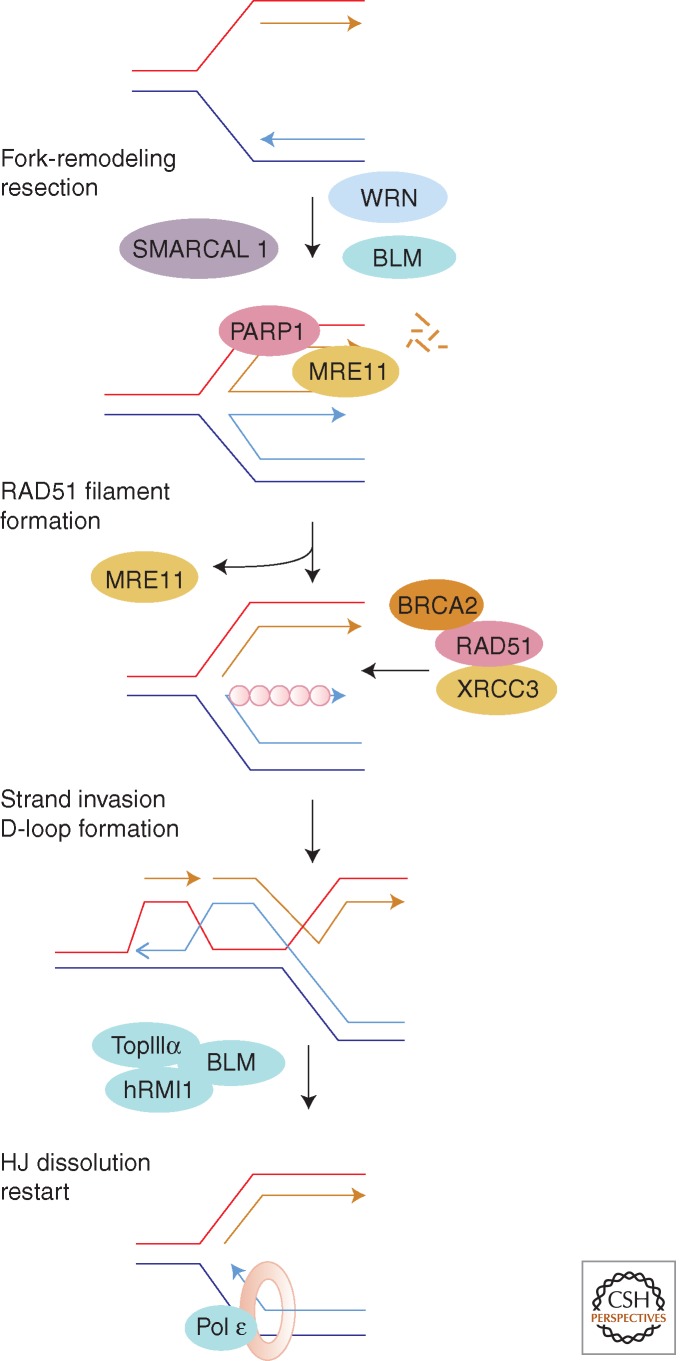
Figure 6.
A possible scheme for replication fork restart in vertebrates. Some fork regression catalyzed by SMARCAL is assumed, followed by limited single-strand resection by MRE11 (of the MRN complex). D-loop formation by HR proteins and Holliday junction resolution (without crossover) by the BLM helicase will enable fork restart. (Based on data from Jones and Petermann 2012.)
PARP INHIBITORS IN CANCER THERAPY
In this last section, we would like to provide an example of how knowledge of certain DNA repair defects in S phase found in cancer cells can be exploited to provide chemotherapy with a high degree of selectivity. PARP-1 (poly-ADP-ribose polymerase) was already shown in the replication restart model (Fig. 5) where it may interact with MRE11 (Bryant et al. 2009). PARP-1 has the ability to bind single-strand breaks or gaps in which its poly-ADP-ribosylation activity may generate important signals that are indispensable for repair, especially single-strand break repair. Inhibitors of PARP-1 were found to be synthetically lethal with HR defects, typically found in BRCA1- and BRCA2-deficient cancer cells (i.e., these cells are much more sensitive toward the inhibitor than normal cells) (Farmer et al. 2005; Martin et al. 2008). Initially, it was suggested that a PARP inhibitor prevents the repair of spontaneous or base-excision repair-related single-strand breaks that are converted into double-strand breaks by an approaching replication fork. Such damage will then require repair by HR that is compromised in these cancer cells, hence the cells are more sensitive.
Interestingly, the critical synergizing event does not appear to be the absence of single-strand break repair in S phase, but instead the trapping on DNA of PARP-1 by the applied inhibitor (Murai et al. 2012). Besides BRCA1 or BRCA2, a broad range of HR proteins as well the FANC system are required for repair or tolerance of these lesions, and thus PARP inhibitors may very well be successful in a wider range of cancers.
We close in reiterating that replication stress and responses to such stress have been implicated in early events of tumorigenesis (Bartkova et al. 2005; Negrini et al. 2010) and, hopefully, there are significant rewards associated with an understanding of the admittedly highly complex pathways that deal with challenges to eukaryotic DNA replication.
Footnotes
Editors: Errol C. Friedberg, Stephen J. Elledge, Alan R. Lehmann, Tomas Lindahl, and Marco Muzi-Falconi
Additional Perspectives on DNA Repair, Mutagenesis, and Other Responses to DNA Damage available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Abbas T, Keaton MA, Dutta A 2013. Genomic instability in cancer. Cold Spring Harb Perspect Biol 5: a012914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya N, Johnson RE, Prakash S, Prakash L 2006. Complex formation with Rev1 enhances the proficiency of Saccharomyces cerevisiae DNA polymerase ζ for mismatch extension and for extension opposite from DNA lesions. Mol Cell Biol 26: 9555–9563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya N, Brahma A, Haracska L, Prakash L, Prakash S 2007. Mutations in the ubiquitin binding UBZ motif of DNA polymerase η do not impair its function in translesion synthesis during replication. Mol Cell Biol 27: 7266–7272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Admire A, Shanks L, Danzl N, Wang M, Weier U, Stevens W, Hunt E, Weinert T 2006. Cycles of chromosome instability are associated with a fragile site and are increased by defects in DNA replication and checkpoint controls in yeast. Genes Dev 20: 159–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amara F, Colombo R, Cazzaniga P, Pescini D, Csikasz-Nagy A, Muzi Falconi M, Besozzi D, Plevani P 2013. In vivo and in silico analysis of PCNA ubiquitylation in the activation of the post replication repair pathway in S. cerevisiae. BMC Syst Biol 7: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammazzalorso F, Pirzio LM, Bignami M, Franchitto A, Pichierri P 2010. ATR and ATM differently regulate WRN to prevent DSBs at stalled replication forks and promote replication fork recovery. EMBO J 29: 3156–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassen PR, D’Andrea AD, Taniguchi T 2004. ATR couples FANCD2 monoubiquitination to the DNA-damage response. Genes Dev 18: 1958–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio OM 2013. Location, location, location: It’s all in the timing for replication origins. Genes Dev 27: 117–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahassi EM, Ovesen JL, Riesenberg AL, Bernstein WZ, Hasty PE, Stambrook PJ 2008. The checkpoint kinases Chk1 and Chk2 regulate the functional associations between hBRCA2 and Rad51 in response to DNA damage. Oncogene 27: 3977–3985 [DOI] [PubMed] [Google Scholar]
- Bailly V, Lauder S, Prakash S, Prakash L 1997. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J Biol Chem 272: 23360–23365 [DOI] [PubMed] [Google Scholar]
- Ball LG, Zhang K, Cobb JA, Boone C, Xiao W 2009. The yeast Shu complex couples error-free post-replication repair to homologous recombination. Mol Microbiol 73: 89–102 [DOI] [PubMed] [Google Scholar]
- Bansbach CE, Betous R, Lovejoy CA, Glick GG, Cortez D 2009. The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes Dev 23: 2405–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartek J, Falck J, Lukas J 2001. Chk2 kinase—A busy messenger. Nat Rev Mol Cell Biol 2: 877–886 [DOI] [PubMed] [Google Scholar]
- Bartkova J, Horejsi Z, Koed K, Krämer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et al. 2005. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434: 864–870 [DOI] [PubMed] [Google Scholar]
- Bell SP, Dutta A 2002. DNA replication in eukaryotic cells. Annu Rev Biochem 71: 333–374 [DOI] [PubMed] [Google Scholar]
- Bermejo R, Capra T, Jossen R, Colosio A, Frattini C, Carotenuto W, Cocito A, Doksani Y, Klein H, Gomez-Gonzalez B, et al. 2011. The replication checkpoint protects fork stability by releasing transcribed genes from nuclear pores. Cell 146: 233–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blastyak A, Pinter L, Unk I, Prakash L, Prakash S, Haracska L 2007. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol Cell 28: 167–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blastyak A, Hajdu I, Unk I, Haracska L 2010. Role of double-stranded DNA translocase activity of human HLTF in replication of damaged DNA. Mol Cell Biol 30: 684–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Foiani M 2009. The checkpoint response to replication stress. DNA Repair (Amst) 8: 1038–1046 [DOI] [PubMed] [Google Scholar]
- Branzei D, Foiani M 2010. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol 11: 208–219 [DOI] [PubMed] [Google Scholar]
- Branzei D, Sollier J, Liberi G, Zhao X, Maeda D, Seki M, Enomoto T, Ohta K, Foiani M 2006. Ubc9- and Mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell 127: 509–522 [DOI] [PubMed] [Google Scholar]
- Branzei D, Vanoli F, Foiani M 2008. SUMOylation regulates Rad18-mediated template switch. Nature 456: 915–920 [DOI] [PubMed] [Google Scholar]
- Broomfield S, Chow BL, Xiao W 1998. MMS2, encoding a ubiquitin-conjugating-enzyme-like protein, is a member of the yeast error-free postreplication repair pathway. Proc Natl Acad Sci 95: 5678–5683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant HE, Petermann E, Schultz N, Jemth AS, Loseva O, Issaeva N, Johansson F, Fernandez S, McGlynn P, Helleday T 2009. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J 28: 2601–2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun TS, Pacek M, Yee M-C, Walter JC, Cimprich KA 2005. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev 19: 1040–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callegari AJ, Clark E, Pneuman A, Kelly TJ 2010. Postreplication gaps at UV lesions are signals for checkpoint activation. Proc Natl Acad Sci 107: 8219–8224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha RS, Kleckner N 2002. ATR Homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science 297: 602–606 [DOI] [PubMed] [Google Scholar]
- Chen Y, Sanchez Y 2004. Chk1 in the DNA damage response: Conserved roles from yeasts to mammals. DNA Repair (Amst) 3: 1025–1032 [DOI] [PubMed] [Google Scholar]
- Chini C, Chen J 2004. Claspin, a regulator of Chk1 in replication stress pathway. DNA Repair (Amst) 3: 1033–1037 [DOI] [PubMed] [Google Scholar]
- Chou DM, Elledge SJ 2006. Tipin and Timeless form a mutually protective complex required for genotoxic stress resistance and checkpoint function. Proc Natl Acad Sci 103: 18143–18147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ 2010. The DNA damage response: Making it safe to play with knives. Mol Cell 40: 179–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Bredemeyer AL, Sowa ME, Terret ME, Jallepalli PV, Harper JW, Elledge SJ 2009. The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart. Genes Dev 23: 2415–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Clauson C, Schärer OD, Niedernhofer L 2013. Advances in understanding the complex mechanisms of DNA interstrand cross-link repair. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a012732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente-Ruiz M, Prado F 2009. Chromatin assembly controls replication fork stability. EMBO Rep 10: 790–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Glick G, Elledge SJ 2004. Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc Natl Acad Sci 101: 10078–10083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo V, Robertson K, Bibikova M, Kim E, Grieco D, Gottesman M, Carroll D, Gautier J 2001. Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol Cell 8: 137–147 [DOI] [PubMed] [Google Scholar]
- Costanzo V, Shechter D, Lupardus PJ, Cimprich KA, Gottesman M, Gautier J 2003. An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol Cell 11: 203–213 [DOI] [PubMed] [Google Scholar]
- Cotta-Ramusino C, Fachinetti D, Lucca C, Doksani Y, Lopes M, Sogo J, Foiani M 2005. Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol Cell 17: 153–159 [DOI] [PubMed] [Google Scholar]
- Coulon S, Ramasubramanyan S, Alies C, Philippin G, Lehmann A, Fuchs RP 2010. Rad8Rad5/Mms2-Ubc13 ubiquitin ligase complex controls translesion synthesis in fission yeast. EMBO J 29: 2048–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremona CA, Sarangi P, Yang Y, Hang LE, Rahman S, Zhao X 2012. Extensive DNA damage-induced sumoylation contributes to replication and repair and acts in addition to the Mec1 checkpoint. Mol Cell 45: 422–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuvier O, Stanojcic S, Lemaitre JM, Mechali M 2008. A topoisomerase II-dependent mechanism for resetting replicons at the S-M-phase transition. Genes Dev 22: 860–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigaku Y, Davies AA, Ulrich HD 2010. Ubiquitin-dependent DNA damage bypass is separable from genome replication. Nature 465: 951–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies SL, North PS, Dart A, Lakin ND, Hickson ID 2004. Phosphorylation of the Bloom’s syndrome helicase and its role in recovery from S-phase arrest. Mol Cell Biol 24: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies SL, North PS, Hickson ID 2007. Role for BLM in replication-fork restart and suppression of origin firing after replicative stress. Nat Struct Mol Biol 14: 677–679 [DOI] [PubMed] [Google Scholar]
- Desany BA, Alcasabas AA, Bachant JB, Elledge SJ 1998. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev 12: 2956–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar MK, Sehgal S, Kaul S 2012. Structure, replication efficiency and fragility of yeast ARS elements. Res Microbiol 163: 243–253 [DOI] [PubMed] [Google Scholar]
- Doksani Y, Bermejo R, Fiorani S, Haber JE, Foiani M 2009. Replicon dynamics, dormant origin firing, and terminal fork integrity after double-strand break formation. Cell 137: 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durando M, Tateishi S, Vaziri C 2013. A non-catalytic role of DNA polymerase η in recruiting Rad18 and promoting PCNA monoubiquitination at stalled replication forks. Nucleic Acids Res 41: 3079–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta A, Bell SP 1997. Initiation of DNA replication in eukaryotic cells. Annu Rev Cell Dev Biol 13: 293–332 [DOI] [PubMed] [Google Scholar]
- Edmunds CE, Simpson LJ, Sale JE 2008. PCNA ubiquitination and REV1 define temporally distinct mechanisms for controlling translesion synthesis in the avian cell line DT40. Mol Cell 30: 519–529 [DOI] [PubMed] [Google Scholar]
- El-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75: 817–825 [DOI] [PubMed] [Google Scholar]
- El-Shemerly M, Hess D, Pyakurel AK, Moselhy S, Ferrari S 2008. ATR-dependent pathways control hEXO1 stability in response to stalled forks. Nucleic Acids Res 36: 511–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errico A, Cosentino C, Rivera T, Losada A, Schwob E, Hunt T, Costanzo V 2009. Tipin/Tim1/And1 protein complex promotes Pol α chromatin binding and sister chromatid cohesion. EMBO J 28: 3681–3692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falck J, Mailand N, Syljuåsen RG, Bartek J, Lukas J 2001. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature 410: 842–847 [DOI] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. 2005. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434: 917–921 [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T 2006. DNA repair and mutagenesis, 2nd ed. American Society of Microbiology, Washington, DC [Google Scholar]
- Gangavarapu V, Haracska L, Unk I, Johnson RE, Prakash S, Prakash L 2006. Mms2-Ubc13-dependent and -independent roles of Rad5 ubiquitin ligase in postreplication repair and translesion DNA synthesis in Saccharomyces cerevisiae. Mol Cell Biol 26: 7783–7790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg P, Burgers PM 2005. Ubiquitinated proliferating cell nuclear antigen activates translesion DNA polymerases η and REV1. Proc Natl Acad Sci 102: 18361–18366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge XQ, Jackson DA, Blow JJ 2007. Dormant origins licensed by excess Mcm2–7 are required for human cells to survive replicative stress. Genes Dev 21: 3331–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannattasio M, Follonier C, Tourriere H, Puddu F, Lazzaro F, Pasero P, Lopes M, Plevani P, Muzi-Falconi M 2010. Exo1 competes with repair synthesis, converts NER intermediates to long ssDNA gaps, and promotes checkpoint activation. Mol Cell 40: 50–62 [DOI] [PubMed] [Google Scholar]
- Gillespie PJ, Blow JJ 2010. Clusters, factories and domains: The complex structure of S-phase comes into focus. Cell Cycle 9: 3218–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A, Lukas J, Nigg EA, Sillje HH, Wernstedt C, Bartek J, Hansen K 2003. Human Tousled like kinases are targeted by an ATM- and Chk1-dependent DNA damage checkpoint. EMBO J 22: 1676–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Sonoda E, Tang T-S, Parker JL, Bielen AB, Takeda S, Ulrich HD, Friedberg EC 2006a. REV1 protein interacts with PCNA: Significance of the REV1 BRCT domain in vitro and in vivo. Mol Cell 23: 265–271 [DOI] [PubMed] [Google Scholar]
- Guo C, Tang T-S, Bienko M, Parker JL, Bielen AB, Sonoda E, Takeda S, Ulrich HD, Dikic I, Friedberg EC 2006b. Ubiquitin-binding motifs in REV1 protein are required for its role in the tolerance of DNA damage. Mol Cell Biol 26: 8892–8900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ 1993. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75: 805–816 [DOI] [PubMed] [Google Scholar]
- Heffernan TP, Unsal-Kacmaz K, Heinloth AN, Simpson DA, Paules RS, Sancar A, Cordeiro-Stone M, Kaufmann WK 2007. Cdc7-Dbf4 and the human S checkpoint response to UVC. J Biol Chem 282: 9458–9468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry-Mowatt J, Jackson D, Masson JY, Johnson PA, Clements PM, Benson FE, Thompson LH, Takeda S, West SC, Caldecott KW 2003. XRCC3 and Rad51 modulate replication fork progression on damaged vertebrate chromosomes. Mol Cell 11: 1109–1117 [DOI] [PubMed] [Google Scholar]
- Hishida T, Kubota Y, Carr AM, Iwasaki H 2009. RAD6-RAD18-RAD5-pathway-dependent tolerance to chronic low-dose ultraviolet light. Nature 457: 612–615 [DOI] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan G-L, Pyrowolakis G, Jentsch S 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141 [DOI] [PubMed] [Google Scholar]
- Huang D, Piening BD, Paulovich AG 2013. The preference for error-free or error-prone postreplication repair in Saccharomyces cerevisiae exposed to low-dose methyl methanesulfonate is cell cycle dependent. Mol Cell Biol 33: 1515–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen JG, Tsaalbi-Shtylik A, Hendriks G, Gali H, Hendel A, Johansson F, Erixon K, Livneh Z, Mullenders LH, Haracska L, et al. 2009. Separate domains of Rev1 mediate two modes of DNA damage bypass in mammalian cells. Mol Cell Biol 29: 3113–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch S, McGrath JP, Varshavsky A 1987. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature 329: 131–134 [DOI] [PubMed] [Google Scholar]
- Jones RM, Petermann E 2012. Replication fork dynamics and the DNA damage response. Biochem J 443: 13–26 [DOI] [PubMed] [Google Scholar]
- Kai M, Boddy MN, Russell P, Wang TS 2005. Replication checkpoint kinase Cds1 regulates Mus81 to preserve genome integrity during replication stress. Genes Dev 19: 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karras GI, Jentsch S 2010. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell 141: 255–267 [DOI] [PubMed] [Google Scholar]
- Karras GI, Fumasoni M, Sienski G, Vanoli F, Branzei D, Jentsch S 2013. Noncanonical role of the 9-1-1 clamp in the error-free DNA damage tolerance pathway. Mol Cell 49: 536–546 [DOI] [PubMed] [Google Scholar]
- Kim N, Huang SN, Williams JS, Li YC, Clark AB, Cho JE, Kunkel TA, Pommier Y, Jinks-Robertson S 2011. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science 332: 1561–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa R, Bakkenist CJ, McKinnon PJ, Kastan MB 2004. Phosphorylation of SMC1 is a critical downstream event in the ATM-NBS1-BRCA1 pathway. Genes Dev 18: 1423–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura E, Blow JJ, Tanaka TU 2006. Live-cell imaging reveals replication of individual replicons in eukaryotic replication factories. Cell 125: 1297–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozmin SG, Jinks-Robertson S 2013. The mechanism of nucleotide excision repair-mediated UV-induced mutagenesis in nonproliferating cells. Genetics 193: 803–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Dunphy WG 2000. Claspin, a novel protein required for the activation of Chk1 during a DNA replication checkpoint response in Xenopus egg extracts. Mol Cell 6: 839–849 [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Bebenek K 2000. DNA replication fidelity. Annu Rev Biochem 69: 497–529 [DOI] [PubMed] [Google Scholar]
- Labib K, Gambus A 2007. A key role for the GINS complex at DNA replication forks. Trends Cell Biol 17: 271–278 [DOI] [PubMed] [Google Scholar]
- Labib K, Tercero JA, Diffley JF 2000. Uninterrupted MCM2–7 function required for DNA replication fork progression. Science 288: 1643–1647 [DOI] [PubMed] [Google Scholar]
- Lane D, Levine A 2010. p53 Research: The past thirty years and the next thirty years. Cold Spring Harb Perspect Biol 2: a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro F, Novarina D, Amara F, Watt DL, Stone JE, Costanzo V, Burgers PM, Kunkel TA, Plevani P, Muzi-Falconi M 2012. RNase H and postreplication repair protect cells from ribonucleotides incorporated in DNA. Mol Cell 45: 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann AR 1972. Postreplication repair of DNA in ultraviolet-irradiated mammalian cells. J Mol Biol 66: 319–337 [DOI] [PubMed] [Google Scholar]
- Lehmann AR, Fuchs RP 2006. Gaps and forks in DNA replication: Rediscovering old models. DNA Repair (Amst) 5: 1495–1498 [DOI] [PubMed] [Google Scholar]
- Leman AR, Noguchi C, Lee CY, Noguchi E 2010. Human Timeless and Tipin stabilize replication forks and facilitate sister-chromatid cohesion. J Cell Sci 123: 660–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberi G, Maffioletti G, Lucca C, Chiolo I, Baryshnikova A, Cotta-Ramusino C, Lopes M, Pellicioli A, Haber JE, Foiani M 2005. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev 19: 339–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JR, Zeman MK, Chen JY, Yee MC, Cimprich KA 2011. SHPRH and HLTF act in a damage-specific manner to coordinate different forms of postreplication repair and prevent mutagenesis. Mol Cell 42: 237–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomonosov M, Anand S, Sangrithi M, Davies R, Venkitaraman AR 2003. Stabilization of stalled DNA replication forks by the BRCA2 breast cancer susceptibility protein. Genes Dev 17: 3017–3022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M, Foiani M, Sogo JM 2006. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell 21: 15–27 [DOI] [PubMed] [Google Scholar]
- Luke-Glaser S, Luke B, Grossi S, Constantinou A 2010. FANCM regulates DNA chain elongation and is stabilized by S-phase checkpoint signalling. EMBO J 29: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Maréchal A, Zou L 2013. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a012716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SA, Lord CJ, Ashworth A 2008. DNA repair deficiency as a therapeutic target in cancer. Curr Opin Genet Dev 18: 80–86 [DOI] [PubMed] [Google Scholar]
- Masai H, Arai K 2002. Cdc7 kinase complex: A key regulator in the initiation of DNA replication. J Cell Physiol 190: 287–296 [DOI] [PubMed] [Google Scholar]
- McIntosh D, Blow JJ 2012. Dormant origins, the licensing checkpoint, and the response to replicative stresses. Cold Spring Harb Perspect Biol 4: a012955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechali M 2010. Eukaryotic DNA replication origins: Many choices for appropriate answers. Nat Rev Mol Cell Biol 11: 728–738 [DOI] [PubMed] [Google Scholar]
- Melo JA, Toczyski DP 2002. A unified view of the DNA-damage checkpoint. Curr Opin Cell Biol 14: 237–245 [DOI] [PubMed] [Google Scholar]
- Minca EC, Kowalski D 2010. Multiple Rad5 activities mediate sister chromatid recombination to bypass DNA damage at stalled replication forks. Mol Cell 38: 649–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minca EC, Kowalski D 2011. Replication fork stalling by bulky DNA damage: Localization at active origins and checkpoint modulation. Nucleic Acids Res 39: 2610–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motegi A, Kuntz K, Majeed A, Smith S, Myung K 2006. Regulation of gross chromosomal rearrangements by ubiquitin and SUMO ligases in Saccharomyces cerevisiae. Mol Cell Biol 26: 1424–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motegi A, Liaw HJ, Lee KY, Roest HP, Maas A, Wu X, Moinova H, Markowitz SD, Ding H, Hoeijmakers JH, et al. 2008. Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks. Proc Natl Acad Sci 105: 12411–12416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JH, Ji J, Takeda S, Pommier Y 2012. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res 72: 5588–5599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami-Sekimata A, Huang D, Piening BD, Bangur C, Paulovich AG 2010. The Saccharomyces cerevisiae RAD9, RAD17 and RAD24 genes are required for suppression of mutagenic post-replicative repair during chronic DNA damage. DNA Repair (Amst) 9: 824–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu S, Hirai K, Tak YS, Kamimura Y, Araki H 2010. CDK-dependent complex formation between replication proteins Dpb11, Sld2, Pol ε, and GINS in budding yeast. Genes Dev 24: 602–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi M, Katsuno Y, Niida H, Murakami H, Shimada M 2010. Chk1-cyclin A/Cdk1 axis regulates origin firing programs in mammals. Chromosome Res 18: 103–113 [DOI] [PubMed] [Google Scholar]
- Navas TA, Zhou Z, Elledge SJ 1995. DNA polymerase ε links the DNA replication machinery to the S phase checkpoint. Cell 80: 29–39 [DOI] [PubMed] [Google Scholar]
- Negrini S, Gorgoulis VG, Halazonetis TD 2010. Genomic instability—An evolving hallmark of cancer. Nat Rev Mol Cell Biol 11: 220–228 [DOI] [PubMed] [Google Scholar]
- Nick McElhinny SA, Kumar D, Clark AB, Watt DL, Watts BE, Lundstrom EB, Johansson E, Chabes A, Kunkel TA 2010a. Genome instability due to ribonucleotide incorporation into DNA. Nat Chem Biol 6: 774–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Watts BE, Kumar D, Watt DL, Lundstrom EB, Burgers PM, Johansson E, Chabes A, Kunkel TA 2010b. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc Natl Acad Sci 107: 4949–4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niida H, Nakanishi M 2006. DNA damage checkpoints in mammals. Mutagenesis 21: 3–9 [DOI] [PubMed] [Google Scholar]
- Pages V, Bresson A, Acharya N, Prakash S, Fuchs RP, Prakash L 2008. Requirement of Rad5 for DNA polymerase ζ-dependent translesion synthesis in Saccharomyces cerevisiae. Genetics 180: 73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann E, Maya-Mendoza A, Zachos G, Gillespie DA, Jackson DA, Caldecott KW 2006. Chk1 requirement for high global rates of replication fork progression during normal vertebrate S phase. Mol Cell Biol 26: 3319–3326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T 2010. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell 37: 492–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfander B, Moldovan G-L, Sacher M, Hoege C, Jentsch S 2005. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436: 428–433 [DOI] [PubMed] [Google Scholar]
- Pichierri P, Nicolai S, Cignolo L, Bignami M, Franchitto A 2012. The RAD9-RAD1-HUS1 (9.1.1) complex interacts with WRN and is crucial to regulate its response to replication fork stalling. Oncogene 31: 2809–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope BD, Aparicio OM, Gilbert DM 2013. SnapShot: Replication timing. Cell 152: 1390–1390.e1 [DOI] [PubMed] [Google Scholar]
- Pospiech H, Grosse F, Pisani FM 2010. The initiation step of eukaryotic DNA replication. Subcell Biochem 50: 79–104 [DOI] [PubMed] [Google Scholar]
- Prakash S, Johnson RE, Prakash L 2005. Eukaryotic translesion synthesis DNA polymerases: Specificity of structure and function. Annu Rev Biochem 74: 317–353 [DOI] [PubMed] [Google Scholar]
- Preston BD, Albertson TM, Herr AJ 2010. DNA replication fidelity and cancer. Semin Cancer Biol 20: 281–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puddu F, Piergiovanni G, Plevani P, Muzi-Falconi M 2011. Sensing of replication stress and Mec1 activation act through two independent pathways involving the 9-1-1 complex and DNA polymerase ε. PLoS Genet 7: e1002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reha-Krantz LJ 2010. DNA polymerase proofreading: Multiple roles maintain genome stability. Biochim Biophys Acta 1804: 1049–1063 [DOI] [PubMed] [Google Scholar]
- Reijns MA, Rabe B, Rigby RE, Mill P, Astell KR, Lettice LA, Boyle S, Leitch A, Keighren M, Kilanowski F, et al. 2012. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell 149: 1008–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus D, Beall EL, Botchan MR 2004. DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC-DNA binding. EMBO J 23: 897–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison JG, Elliott J, Dixon K, Oakley GG 2004. Replication protein A and the Mre11.Rad50.Nbs1 complex co-localize and interact at sites of stalled replication forks. J Biol Chem 279: 34802–34810 [DOI] [PubMed] [Google Scholar]
- Rupp WD, Howard-Flanders P 1968. Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J Mol Biol 31: 291–304 [DOI] [PubMed] [Google Scholar]
- Rupp WD, Wilde CE, Reno DL, Howard-Flanders P 1971. Exchanges between DNA strands in ultraviolet irradiated Escherichia coli. J Mol Biol 61: 25–44 [DOI] [PubMed] [Google Scholar]
- *.Sale JE 2013. Translesion DNA synthesis and mutagenesis in eukaryotes. Cold Spring Harb Perspect Biol 5: a012708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S 2004. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem 73: 39–85 [DOI] [PubMed] [Google Scholar]
- Santamaria D, Viguera E, Martinez-Robles ML, Hyrien O, Hernandez P, Krimer DB, Schvartzman JB 2000. Bi-directional replication and random termination. Nucleic Acids Res 28: 2099–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santocanale C, Diffley JFX 1998. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395: 615–618 [DOI] [PubMed] [Google Scholar]
- Seiler JA, Conti C, Syed A, Aladjem MI, Pommier Y 2007. The intra-S-phase checkpoint affects both DNA replication initiation and elongation: Single-cell and -DNA fiber analyses. Mol Cell Biol 27: 5806–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorova JM, Li N, Folch A, Monnat RJ Jr 2008. The RecQ helicase WRN is required for normal replication fork progression after DNA damage or replication fork arrest. Cell Cycle 7: 796–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillje HH, Nigg EA 2001. Identification of human Asf1 chromatin assembly factors as substrates of Tousled-like kinases. Curr Biol 11: 1068–1073 [DOI] [PubMed] [Google Scholar]
- *.Sirbu BM, Cortez D 2013. DNA damage response: Three levels of DNA repair regulation. Cold Spring Harb Perspect Biol 5: a012724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobeck A, Stone S, Costanzo V, de Graaf B, Reuter T, de Winter J, Wallisch M, Akkari Y, Olson S, Wang W, et al. 2006. Fanconi anemia proteins are required to prevent accumulation of replication-associated DNA double-strand breaks. Mol Cell Biol 26: 425–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E, Sasaki MS, Buerstedde J-M, Bezzubova O, Shinohara A, Ogawa H, Takata M, Yamaguchi-Iwai Y, Takeda S 1998. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J 17: 598–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, Bartek J, Helleday T 2005. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat Cell Biol 7: 195–201 [DOI] [PubMed] [Google Scholar]
- Spivak G, Hanawalt PC 1992. Translesion DNA synthesis in the dihydrofolate reductase domain of UV-irradiated CHO cells. Biochemistry 31: 6794–6800 [DOI] [PubMed] [Google Scholar]
- Stelter P, Ulrich HD 2003. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425: 188–191 [DOI] [PubMed] [Google Scholar]
- Stillman B 2008. DNA polymerases at the replication fork in eukaryotes. Mol Cell 30: 259–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenberg PT, Turner J, O’Donnell M 1994. An explanation for lagging strand replication: Polymerase hopping among DNA sliding clamps. Cell 78: 877–887 [DOI] [PubMed] [Google Scholar]
- Tanaka K 2010. Multiple functions of the S-phase checkpoint mediator. Biosci Biotechnol Biochem 74: 2367–2373 [DOI] [PubMed] [Google Scholar]
- Tanaka S, Tak YS, Araki H 2007. The role of CDK in the initiation step of DNA replication in eukaryotes. Cell Div 2: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero JA, Diffley JFX 2001. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412: 553–557 [DOI] [PubMed] [Google Scholar]
- Torres JZ, Schnakenberg SL, Zakian VA 2004. Saccharomyces cerevisiae Rrm3p DNA helicase promotes genome integrity by preventing replication fork stalling: Viability of rrm3 cells requires the intra-S-phase checkpoint and fork restart activities. Mol Cell Biol 24: 3198–3212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourriere H, Pasero P 2007. Maintenance of fork integrity at damaged DNA and natural pause sites. DNA Repair (Amst) 6: 900–913 [DOI] [PubMed] [Google Scholar]
- Trenz K, Smith E, Smith S, Costanzo V 2006. ATM and ATR promote Mre11 dependent restart of collapsed replication forks and prevent accumulation of DNA breaks. EMBO J 25: 1764–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye BK 1999. MCM proteins in DNA replication. Annu Rev Biochem 68: 649–686 [DOI] [PubMed] [Google Scholar]
- Ulrich HD, Jentsch S 2000. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J 19: 3388–3397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unk I, Hajdu I, Fatyol K, Szakal B, Blastyak A, Bermudez V, Hurwitz J, Prakash L, Prakash S, Haracska L 2006. Human SHPRH is a ubiquitin ligase for Mms2-Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen. Proc Natl Acad Sci 103: 18107–18112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unk I, Hajdu I, Fatyol K, Hurwitz J, Yoon JH, Prakash L, Prakash S, Haracska L 2008. Human HLTF functions as a ubiquitin ligase for proliferating cell nuclear antigen polyubiquitination. Proc Natl Acad Sci 105: 3768–3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unk I, Hajdu I, Blastyak A, Haracska L 2010. Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance. DNA Repair (Amst) 9: 257–267 [DOI] [PubMed] [Google Scholar]
- Unsal-Kacmaz K, Chastain PD, Qu PP, Minoo P, Cordeiro-Stone M, Sancar A, Kaufmann WK 2007. The human Tim/Tipin complex coordinates an intra-S checkpoint response to UV that slows replication fork displacement. Mol Cell Biol 27: 3131–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanoli F, Fumasoni M, Szakal B, Maloisel L, Branzei D 2010. Replication and recombination factors contributing to recombination-dependent bypass of DNA lesions by template switch. PLoS Genet 6: e1001205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashee S, Cvetic C, Lu W, Simancek P, Kelly TJ, Walter JC 2003. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev 17: 1894–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LS, Walker GC 2006. The critical mutagenic translesion DNA polymerase Rev1 is highly expressed during G2/M phase rather than S phase. Proc Natl Acad Sci 103: 8971–8976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt DL, Johansson E, Burgers PM, Kunkel TA 2011. Replication of ribonucleotide-containing DNA templates by yeast replicative polymerases. DNA Repair (Amst) 10: 897–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich M, Stillman B 1999. Cdc7p-Dbf4p kinase binds to chromatin during S-phase and is regulated by both the APC and the RAD53 checkpoint pathway. EMBO J 18: 5334–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Chen Z, Gunasekera AH, Sowin TJ, Rosenberg SH, Fesik S, Zhang H 2003. Chk1 mediates S and G2 arrests through Cdc25A degradation in response to DNA-damaging agents. J Biol Chem 278: 21767–21773 [DOI] [PubMed] [Google Scholar]
- Yang XH, Shiotani B, Classon M, Zou L 2008. Chk1 and Claspin potentiate PCNA ubiquitination. Genes Dev 22: 1147–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao NY, O’Donnell M 2010. SnapShot: The replisome. Cell 141: 1088, 1088.e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF 2009. DNA replication as a target of the DNA damage checkpoint. DNA Repair (Amst) 8: 1077–1088 [DOI] [PubMed] [Google Scholar]
- Zhang H, Lawrence CW 2005. The error-free component of the RAD6/RAD18 DNA damage tolerance pathway of budding yeast employs sister-strand recombination. Proc Natl Acad Sci 102: 15954–15959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Qin Z, Zhang X, Xiao W 2011. Roles of sequential ubiquitination of PCNA in DNA-damage tolerance. FEBS Lett 585: 2786–2794 [DOI] [PubMed] [Google Scholar]
- Zou L, Stillman B 1998. Formation of a preinitiation complex by S-phase cyclin CDK-dependent loading of Cdc45p onto chromatin. Science 280: 593–596 [DOI] [PubMed] [Google Scholar]
- Zou L, Stillman B 2000. Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p-Dbf4p kinase. Mol Cell Biol 20: 3086–3096 [DOI] [PMC free article] [PubMed] [Google Scholar]