Significance
Cells attach a methyl group (—CH3) to certain cytosines in DNA to control gene expression. These methylation patterns change over time and can be related to cell differentiation and diseases such as cancer. Existing methylation detection techniques are not ideal for clinical use. We pulled single-stranded DNA molecules through the biological pore MspA and found that ion currents passing through the pore reveal the methylation sites with high confidence. Hydroxymethylation, which differs from methylation by only one oxygen atom, also produces distinct signals. This technique can be developed into a research tool and may ultimately lead to clinical tests.
Keywords: nanopore DNA sequencing, DNA methylation, DNA hydroxymethylation, nanotechnology, next generation sequencing
Abstract
Precise and efficient mapping of epigenetic markers on DNA may become an important clinical tool for prediction and identification of ailments. Methylated CpG sites are involved in gene expression and are biomarkers for diseases such as cancer. Here, we use the engineered biological protein pore Mycobacterium smegmatis porin A (MspA) to detect and map 5-methylcytosine and 5-hydroxymethylcytosine within single strands of DNA. In this unique single-molecule tool, a phi29 DNA polymerase draws ssDNA through the pore in single-nucleotide steps, and the ion current through the pore is recorded. Comparing current levels generated with DNA containing methylated CpG sites to current levels obtained with unmethylated copies of the DNA reveals the precise location of methylated CpG sites. Hydroxymethylation is distinct from methylation and can also be mapped. With a single read, the detection efficiency in a quasirandom DNA strand is 97.5 ± 0.7% for methylation and 97 ± 0.9% for hydroxymethylation.
DNA is often referred to as the “blueprint for life,” but there is more to the code than DNA sequence alone. Epigenetic factors (1) govern the blueprint’s transcription and translation into protein. There is a rapidly growing interest in understanding these epigenetic factors (2).
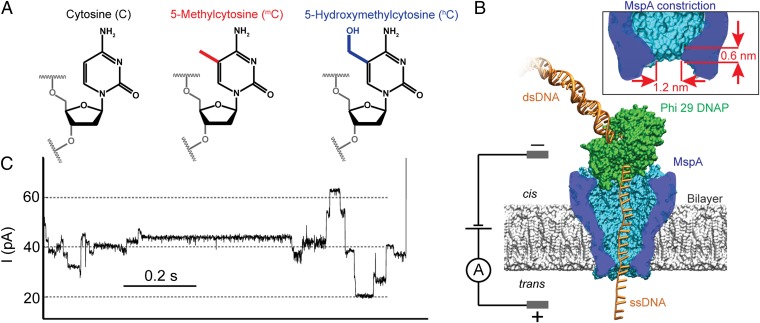
The most common epigenetic DNA modification is the methylation of cytosine leading to 5-methylcytosine (mC) (Fig. 1A). In mammals, most genetically relevant cytosine methylations occur in C-G dinucleotides (CpG). Methylation is associated with gene regulation and therefore has implications for cell development (3), aging, and diseases such as cancer (4–7). Further oxidation of the methyl residue results in 5-hydroxymethylcytosine (hC) (Fig. 1A). Because of its relatively recent discovery in mammalian tissue (8, 9), the function of hC is less well explored. However, there is indication that it also has a role in regulation of chromatin structure and gene expression (10, 11). Unlike DNA sequence, methylation patterns are tissue specific and change over the life of an organism as it develops or is exposed to certain chemicals (11) and environmental conditions (12). In some cases, these changes are heritable through multiple generations (12).
Fig. 1.
Methylated cytosines and schematic setup. (A) Chemical structure of cytosine (C), 5-methylcytosine (mC), and 5-hydroxymethylcytosine (hC). (B) Schematic of a typical MspA–phi29 DNA polymerase (DNAP) experiment. MspA (in blue) is a membrane protein embedded in a phospholipid bilayer. A voltage across the membrane causes an ion current to flow through the pore. We use phi29 DNAP (in green) to feed DNA through the pore in controlled, single-nucleotide steps (see SI Appendix, Fig. S1 for DNA configuration used in phi29 DNAP experiments). (Inset) The short and narrow constriction of MspA concentrates the ion current to resolve the relatively small differences between C, mC, and hC. (C) A typical current trace of DNA being pulled through MspA by phi29 DNAP in synthesis mode (Materials and Methods). As the DNA moves through the pore in single-nucleotide steps, one observes clearly discernible current levels that are associated with DNA sequence. The level duration is stochastic.
Because of methylation’s proven link to gene expression, precise methylation mapping may yield more pertinent information to research and ultimately to clinical diagnosis (6) than sequencing the standard four bases alone. Clinical uses will require fast, inexpensive, and reliable detection methods to map methylation. Because methylation patterns vary between cells (13), it is preferable to use small, native, unamplified DNA samples, making this task suitable for single-molecule techniques.
Currently available techniques for mapping of DNA methylation include the following: bisulfite sequencing, methylation-specific enzyme restriction, affinity enrichment, and various single-molecule techniques. In bisulfite sequencing, all unmethylated cytosines are converted to deoxyuridine. Converted samples are amplified, sequenced, and compared with unmodified sequence information. Converted Cs become Ts in the amplified DNA, whereas mCs remain unchanged. Conditions required to bring this conversion close to 100% completion cause DNA damage by fragmentation (14). Conventional bisulfite sequencing cannot differentiate between mC and hC (15). Oxidative bisulfite sequencing (16, 17) can distinguish between mC and hC; however, this assay has significant sample losses with only 0.5% of the original DNA fragments remaining intact (16). In methylation-specific enzyme restriction, proteins recognize and cut DNA strands at mCs, and subsequent sequencing and alignment of the strands to the known genomic sequence reveal the locations of the mCs (18, 19). This technique works for broadly spaced mCs but lacks sensitivity for densely packed mCs (14). Affinity enrichment assays are bulk assays and are unable to resolve mCs with nucleotide precision (20).
Single-molecule real-time sequencing (SMRT) (21) exploits polymerase incorporation kinetics to detect methylation while also sequencing via fluorescently tagged dNTPs. When encountering a mC, a polymerase pauses longer on average to incorporate deoxyguanosine triphosphate than when it encounters an unmethylated C (22). The durations of the pauses are stochastically distributed, and the change in kinetics caused by mC is subtle (22, 23). Thus, detection requires averaging over dozens of reads, complicating methylation detection. Recently, nanochannels have been used as nano-Coulter counters to measure the correlation between DNA methylation and certain histone modifications for single chromatin molecules (24). This method lacks single-nucleotide resolution.
Nanopore sequencing (25–28) is an emerging single-molecule technique that has shown promise for simultaneous DNA sequencing and methylation detection (28–31). In nanopore sequencing, a thin membrane containing a single nanometer-sized pore divides a salt solution into two wells, cis and trans. A voltage across the membrane causes an ion current through the pore and also drives DNA into the pore. As the DNA passes through the pore, the nucleotides at the narrowest section of the pore modulate the ion current. In principle, one can determine the identity and sequence of the nucleotides from the current recording (32). Solid-state nanopores have been used to detect the bulk presence of mC and hC in double-stranded DNA (dsDNA) (30). Recently, solid-state nanopores were also used to detect dsDNA complexed with methyl-binding proteins and thereby indirectly measured the approximate location of individual methylation sites (31). Experiments with ssDNA held statically in biological pores have distinguished C, mC, or hC directly (28, 29).
Previously, we used phi29 DNA polymerase (DNAP) to draw ssDNA through a mutated Mycobacterium smegmatis porin A (MspA) protein pore (33) in single-nucleotide steps (Fig. 1B). This yielded resolved current levels (Fig. 1C) that could be associated with the DNA sequence (32). Here, we show that this system directly detects and maps mC and hC along single molecules of ssDNA with single-nucleotide resolution.
Results
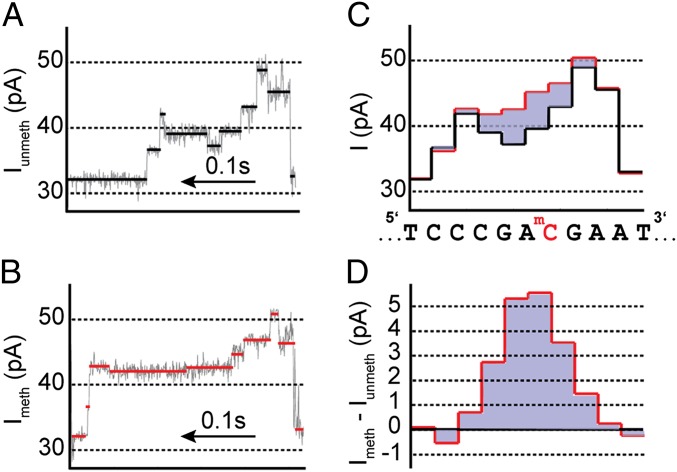
We measured current traces for methylated, hydroxymethylated, and unmethylated DNA passing through the pore. The effect of methylation on the ion current can be seen by comparing the current level sequence from reads of methylated DNA with the current level sequence from reads of unmethylated DNA of the same sequence. The single methyl group on a mC increases the current relative to unmethylated C. This increase persists for several current levels as the DNA passes through the pore. Fig. 2 A and B show raw current traces for unmethylated and methylated DNA, respectively. See SI Appendix, Fig. S2, for additional raw current traces. The extracted average current levels are shown in Fig. 2C for unmethylated (methylated) DNA in black (red). As in Manrao et al. (32), these current level sequences are aligned to their DNA sequence (shown below Fig. 2C). The difference between the methylated and unmethylated current level sequences (Fig. 2D) isolates the effect.
Fig. 2.
Methylation detection. (A and B) Segments of raw current traces. Ion current changes as DNA passes through the pore in single-nucleotide steps. Average current values for each current level are shown in black or red. The traces shown in A and B are for DNA with identical nucleotide sequence. The current trace shown in A contains a single unmethylated CpG site, whereas the trace in B contains a single mCpG. (C) Extracted average current values from each level in A in black and B in red. The stochastic duration of current levels has been removed so that the DNA base sequence can be aligned to the observed current levels. The DNA sequence is shown below with the modified C indicated in red. (D) Current difference plot. The current levels obtained with methylated DNA were subtracted from the current levels obtained with unmethylated DNA. The effect of a single mCpG causes an ion current increase that persists over approximately four steps of the DNA through the pore. The magnitude and shape of the current difference is determined by the nucleotides adjacent to the methylated C (Figs. 3 and 4).
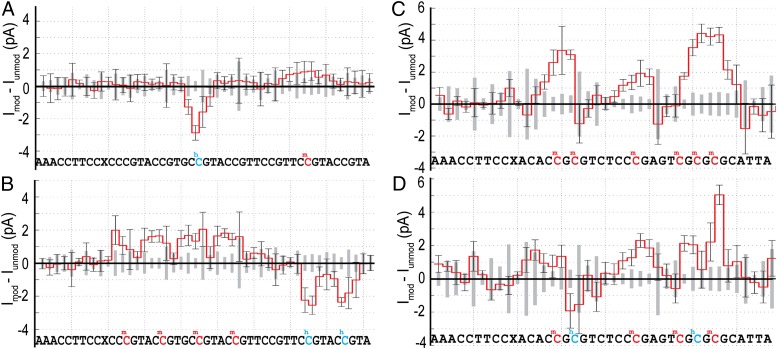
We investigated eight different DNA sequences each with multiple CpGs. We compared several reads of unmethylated DNA to reads of methylated and hydroxymethylated DNA. Fig. 3 shows the average current level differences for 20 or more single-molecule comparisons for four different DNA constructs. Across all such comparisons, we observed that mC consistently increases current relative to C, whereas hC generally decreases current relative to C.
Fig. 3.
Differences in the ion current level sequences taken with DNA containing methylation (hydroxymethylation) and DNA without methylation. (A and B) Current differences [∆I = Imeth – Iunmeth; in red, where Imeth (Iunmeth) is the average current for at least 20 reads of methylated (unmethylated) DNA] obtained with two DNA strands each containing three methylated CpG sites, indicated by red letters in the associated sequence. X is an abasic site. The methylated positions are marked by a significant current increase that persists over approximately four steps of the DNA through the pore. The amplitude and shape of the current difference depend on the nucleotides adjacent to the mC. In regions containing no methylation, current differences are insignificant. (C and D) Current difference obtained with two DNA strands each containing three hCpG sites [∆I = Ihydroxy − Iunmeth; in blue, where Imeth (Iunmeth) is the average current for at least 23 reads of hydroxymethylated (unmethylated) DNA]. In most cases, hC results in a small reduction in current, although the magnitude of the current difference is less than observed for mC. In a few cases, hC results in a current increase. Error bars are the observed SD for single-molecule reads of methylated DNA and indicate the variation in single-molecule reads. The gray boxes along the x axis are the SDs for reads of unmethylated DNA. See SI Appendix, Table S1, for exact numbers of events.
The current difference caused by a mC or a hC is strongly affected by the sequence context in which it is embedded. In particular, the nucleotides immediately adjacent to a mC or hC have the greatest influence on the size and shape of the current difference. We varied the nucleotide on the 5′ side and chose to keep the nucleotide on the 3′ side of the C fixed as a G because of the biological relevance of CpG sites. In exploratory experiments, we found that the nucleotide two positions to the 5′ side of the modified cytosine had a bigger influence than the nucleotide two positions toward the 3′ side, which had a lesser effect (SI Appendix, Fig. S11). Therefore, we measured all 16 two-nucleotide combinations of the form XYCpG, where X and Y represent A, C, G, or T. We performed experiments with 115 MspA pores, using 22 different DNA constructs each containing several CpG regions. These CpGs were either unmethylated (CpG), methylated (mCpG), or hydroxymethylated (hCpG). In total, we analyzed 814 translocation events that contained full reads of the given DNA strand. These events contained a total of 2,857 passages of various CpGs through the pore (strand-specific statistics can be found in SI Appendix, Table S1).
Results for all 16 XYmCpGs and XYhCpGs are summarized in Fig. 4. The maximum difference is up to 7 pA depending on sequence context. On average, the maximum difference caused by mC is ∼2.5 pA (Fig. 4A, Bottom Right). Previously we found that four nucleotides within MspA’s constriction affect each current level (Figs. 1B, Inset, and 5), with the two nucleotides centered in the pore’s constriction affecting the current the most (28, 32). Here, we also find the replacement of C for mC or hC affects approximately four consecutive current levels. The current difference is maximal when the mC is positioned immediately to cis of the constriction and the shape of the difference peak exhibits skewness.
Fig. 4.
DNA sequence context changes the current difference pattern when a modified cytosine replaces a cytosine at a CpG site. Shown are the current difference patterns caused by the sequence XYmCpG in A or XYhCpG in B, where X and Y are any of the four nucleotides A, C, G, and T. The rightmost column and bottom row of each figure display the current differences averaged over the nucleotides X or Y, respectively, and the bottom right box displays the average current difference for all studied sequence contexts. Both the amplitude and the shape of current difference change with sequence context. (A) The maximum difference reaches 7 pA for AAmCpG and is only 1–2 pA when XY contains a thymine. The average maximum difference is ∼2 pA. The number of levels showing a significant current difference varies from 3 to 5. The difference is maximal when the mC is immediately above the constriction and the distribution is skewed. (B) Current deviations due to hC are more complex. Generally, when the hC is centered within MspA’s constriction, the difference is −2 to −1 pA. However, some contexts involve positive differences. The differences associated with sequences containing XThCpG, XAhCpG, AYhCpG, and CYhCpG are small, with only ∼1σ differences. As seen for mC, difference patterns caused by hC involve between 3 and 5 levels and are also skewed. The average difference patterns due to mC and hC are similar; both difference patterns map out a single tight recognition site within MspA’s constriction (Fig. 5).
Fig. 5.
Spatial methylation sensitivity of MspA. Schematic cross-section of MspA with mC held just above (A) and just below (B) MspA’s constriction. Orange shading indicates the region of higher electric field within MspA. (A) When mC is cis of the constriction, it is in a high field region and it modulates the ion current. Other nucleotides that are also within the high field region determine the magnitude of the mC-specific signal. (B) When mC is trans of the constriction, it is outside the high field region and no longer affects the current.
Current differences due to hC are more complex than differences due to mC. Typically, when hC is centered within MspA’s constriction, the difference is −2 to −1 pA. In some cases (GGhCpG, AAhCpG, AThCpG, TGhCpG, and CAhCpG), the current difference includes some levels with positive difference. The differences associated with some sequence contexts are small, with only ∼1σ differences per level. Averaging over all sequence contexts (Fig. 4B, Bottom Right), the difference reaches a negative peak when the hC is near the cis side of the constriction. The locations of maximal difference caused by mC and hC differ by ∼1 nt. Average difference patterns for both mC and hC map out a single, sharp recognition site within MspA’s constriction (Fig. 1B, Inset). All current profiles caused by hC are very distinct from current profiles caused by mC within the same sequence context. (Data from which plots in Fig. 4 are derived are available in SI Appendix, Figs. S3–S9.)
The schematic in Fig. 5 shows how sequence context dependence arises. MspA’s cross-section is shown in blue, and orange shading indicates the region of high electric field. Nucleotides within the region of high electric field affect the ion current. As mC or hC pass through the pore, their location relative to the pore constriction determines how much they affect the current. All nucleotides within the high-field region of the constriction will influence the current, and therefore alter the influence of a mC and hC modification. Apart from the two nucleotides positioned immediately to the 5′ of the CpG, the nucleotide to the 3′ of the CpG is also relevant, albeit to a smaller extent. For example, when a T follows the CpG, as in TGmCpGT, the 3′ side of the current difference peak caused by mC is reduced (Fig. 4). Ultimately, a wider sequence context will need to be measured for high-precision MspA-based methylation detection. However, the data in Fig. 4 demonstrate that the four nucleotides X, Y, C, and G dominate the magnitude of the current difference caused by mCpG and hCpG (SI Appendix, Fig. S11).
We also investigated the effect of several mCpGs near one another. Fig. 6B shows a construct with several modified Cs spaced only five nucleotides apart. The current difference peaks associated with the four mCs and two hCs are still easily distinguishable. When two or three mCpGs are immediately adjacent to one another, as in Fig. 6C, the difference peak is wider and higher than the signal for just one mCpG within the same context. Placing a hCpG immediately adjacent to a mCpG (Fig. 6D) reduces the signal of the nearby mCpGs. The signal is approximately a superposition of the individual mC and hC signals.
Fig. 6.
Multiple adjacent mCs and hCs. Current differences (Imodified − Iunmodified) for four DNA strands containing different methylation patterns. Although CpGs rarely occur in such high density, it is possible to discern multiple adjacent mCpGs and hCpGs. (A) Data from a strand containing one mC (indicated in red) and one hC (indicated in blue) demonstrates that one can simultaneously detect mC and hC. (B) A strand with identical sequence to that shown in A but containing four mCs (red) as well as two hCs (blue). Individual mCs and hCs can be resolved. (C) Adjacent mCpG sites result in wide and large current difference profiles. The current difference profiles for individual mCs seemingly superimpose. The current increase in the middle of the trace is due to only one mCpG and compares well with a mCpG embedded in the same sequence context shown in Fig. 4B above. (D) Current differences for a strand with identical sequence to that in C but with two mCs (red) replaced by two hCs (blue). Here, the effects of mC and hC counteract one another. As in C, the result is approximately a superposition of the signals shown in Fig. 4 (SI Appendix, Figs. S9 and S10 show data from which these plots are derived).
Using the current differences shown in Fig. 4, we implemented a simple Bayesian probability methylation detection algorithm. We compared three consecutive current differences from single-molecule measurements to the current difference patterns in Fig. 4 (Materials and Methods and SI Appendix). We used this algorithm to call C, mC, and hC at known CpG sites. We found a mCpG true-positive detection rate of 97.5 ± 0.7% and a hCpG true-positive detection rate of 97.0 ± 0.9%. The true-negative detection rate for unmethylated CpGs was 98.4 ± 0.6%. Many XYmCpGs, such as AAmCpG, were always properly called. Methylated sites with smaller current differences, such as CTmCpG and TCmCpG, were detected with lower accuracy: ∼86% and ∼88%, respectively (see SI Appendix, Table S1, for individual context-dependent detection rates). As one would expect based on the comparatively smaller current differences shown, hC true-positive rates were lower than for mC. In all sequence contexts, mC was distinct from hC; mCpGs were miscalled as hCpGs in 3 out of 478 occurrences, whereas hCpGs were never miscalled as mCpGs in 609 reads.
Current differences are also effective in locating mCpG sites relative to one another without using our prior knowledge of the DNA sequence. Current level sequences of DNA containing CpG sites were aligned via Needleman–Wunsch alignment (32) to current level recordings of unmethylated DNA. We searched for methylation sites within the current difference between methylated event traces and an unmethylated consensus using a peak detection algorithm. Detecting a mC within two nucleotides of its known position was considered a true-positive detection. We found a true-positive detection rate of 92.7% for mC and a true-negative rate of 99.1% for all unmethylated regions. In this method, true negatives included non-CpG regions in addition to CpGs tested above, resulting in a higher true-negative detection rate than in the method described in the preceding paragraph. Rates from these two methods are not directly comparable. In another sequence-independent technique, we used a Bayesian classification measure to find mCs, yielding similar detection efficiencies (SI Appendix). mC detection without reference to DNA sequence is useful for hypermethylation or hypomethylation detection and is comparable to other nanopore methylation detection techniques (30, 31).
Discussion
We have shown that MspA-based nanopore sequencing can locate DNA methylation sites with near unity efficiency. In this work, we compared single reads of DNA molecules containing mCpGs and hCpGs to measured current references acquired with unmethylated DNA of the same sequence. We found that mC and hC have distinct current signatures. We expect our detection probabilities to be reasonable estimates for mC and hC detection in genomic DNA because the constructs studied simulate a heterogeneous sequence. Although mC is distinct from both C and hC, confident detection of mC and hC in some contexts may require repeated reads. The nanopore strand sequencing method used in this work produces a second read of the same DNA molecule (32) (Materials and Methods). Using this second read will improve calling accuracies. In contrast to other mC and hC detection techniques that rely on mC-specific chemical reactions and/or enzymatic kinetics, our system detects the methylation directly. Unlike single-molecule detection via SMRT (22, 34), the methylation signal in MspA-based nanopore sequencing is carried in the primary signal: the ion current. Polymerase kinetics (22) may be used as an additional indicator of modified bases in our method.
As was seen previously (32, 35), the phi29 DNAP exhibited toggling/backstepping behavior that is thought to be related to the polymerase’s proofreading function. This behavior and the stochastic level durations complicate level extraction. Optimized DNA translation control will be necessary for the industrial application of our method. New or modified motor enzymes will be the subject of future studies.
Because MspA demonstrated well-resolved signals for nucleotides that are differentiated by only a single methyl group, it is expected that other modified bases such as 8-oxo-guanine (36), thymine dimers, 5-carboxylcytosine, and 5-formylcytosine (37, 38) will have equally well-resolved current signatures. MspA has already proved to be extremely sensitive to abasic residues, one of the most common DNA lesions (32).
This methylation detection method does not require de novo sequencing with the nanopore to detect methylation. Given a previously measured reference current sequence for unmethylated DNA and known context-dependent methylation patterns as in Fig. 4, one can then take a single read of a methylated DNA molecule and detect methylation with confidence for most sequence contexts. To map methylation in genomic DNA, we propose comparison of native DNA with DNA generated by PCR amplification. Because PCR does not copy methylation, nanopore reads of amplified copies would serve as the unmethylated reference. Genomic DNA would then be extracted, given adapters to enable polymerase control, and then be presented to the pore. Individual reads of methylated DNA could then be aligned to the current level reference using a Smith–Waterman alignment algorithm (32). Once aligned, current level comparisons could be made and methylations detected. The unmethylated current reference would only need to be made once and could be reused as a reference detection in other genomic samples. Because of the low copy number of DNA obtainable from mammalian samples, efficient transport of the DNA to the nanopore remains a technical challenge before clinical application of the technique. Industrial implementation will include miniaturization and parallelization of the experimental setup as well as optimization of operating conditions or engineering of even more sensitive pores.
All of the intrinsic advantages of nanopore sequencing, such as long read lengths, speed, and minimal sample preparation, are transferable to MspA-based methylation mapping. The conceptual and practical simplicity, as well as the high sensitivity and robust data interpretation, favor conversion of this concept into an industrial platform. It is anticipated that fast and confident methylation detection will accelerate research and ultimately improve health care.
Materials and Methods
Our experimental setup was as previously described (32). Briefly, we used phi29 DNAP as a molecular motor to control the motion of DNA through a single MspA pore established in an unsupported phospholipid bilayer. Our buffer was 300 mM KCl, 10 mM Hepes buffered at pH 8.00 ± 0.05. Currents were recorded on an Axopatch 200B amplifier with custom Labview software (National Instruments) at a voltage bias of 180 mV.
Before each experiment, the DNA template, primer, and blocking oligomer were mixed together in a 1:1:1.2 ratio to a final concentration of 50 µM. DNA was then annealed by heating to 95 °C for 5 min, cooling to 60 °C for 2 min, and then cooling to 4 °C. Experimental concentrations were ∼500 nM for DNA, ∼500 nM for phi29 DNAP, ∼500 µM for dNTPs, ∼10 mM for MgCl2, and ∼1 mM for DTT.
During strand sequencing (32, 35), the DNA is passed through the pore twice, once in the 5′-to-3′ direction (unzipping mode) and once in the 3′-to-5′ direction (synthesis mode). In this report, we used data from the synthesis mode of phi29 DNAP motion. [See Cherf et al. (35) and Manrao et al. (32) for further details.] All strands included the sequence 5′-PAAAAAAACCTTCCX-3′ at the 5′ end of the strand (where P is a phosphorylated 5′ end and X is an abasic residue). This sequence creates a reproducible current motif that signals the end of the read. We use this region to calibrate currents to control for small changes in buffer conductivity due to evaporation or temperature variation. The sequence of interest followed this calibration sequence. We designed the DNA to contain a variety of nucleotides adjacent to the CpGs. Each strand had at least three CpGs embedded in a random sequence, sufficiently spaced so that their current signatures did not overlap. In each strand, three of these CpGs were uniformly either unmethylated, methylated, or hydroxymethylated. We examined eight different DNA sequences (PAN Laboratories, Stanford University, Stanford, CA) containing various methylation patterns (see SI Appendix, Table S1, for sequences used). Some experiments were performed with a mixture of methylated, hydroxymethylated, and unmethylated DNA. Without calibration, these strands could still be sorted by methylation-specific currents (SI Appendix).
Data analysis is described in greater detail in SI Appendix. Events were determined using a thresholding method on current data. A feedforward neural network removed events that did not correspond with phi29 polymerase activity. Once appropriate events were determined, raw current levels were discerned using a custom-written graphical user interface. Current level transition boundaries were selected, and the median current levels were extracted in the time order that they occurred for each event. The phi29 DNAP occasionally exhibited backstepping, causing repeated levels that were removed. Consensus current level sequences were found for each sequence type, and event levels associated with that sequence were automatically aligned using a Needleman–Wunsch algorithm. For experiments with DNA mixtures, a quality score from the Needleman–Wunsch algorithm was used to distinguish DNA with different types of methylation. Once aligned, levels from methylated and unmethylated DNA were examined with a Bayesian probability measure to classify mCpG, hCpG, and CpG sites. The algorithm used current level differences for three consecutive levels, centered on the level associated with XYCpG.
Supplementary Material
Acknowledgments
Thanks to Graham Naylor and Kenji Doering for their help in data acquisition. Thanks to Vikram Dhawan for coding our event analysis graphical user interface. Thanks also to Brian Ross and Andrew Adey for insight and helpful discussion. This work was supported by the National Institutes of Health, National Human Genome Research Institute $1,000 Genome Program Grants R01HG005115 and R01HG006321. M.P. was also supported by Marie and Emmett Carmichael Fund for Graduate Students in Biosciences, University of Alabama at Birmingham.
Footnotes
Conflict of interest statement: A.H.L., I.M.D., and J.H.G have filed a provisional patent on the detection strategy described herein. I.M.D. and J.H.G. have an interest with Illumina Inc. through licensed technology.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1310240110/-/DCSupplemental.
References
- 1.Bird A. Perceptions of epigenetics. Nature. 2007;447(7143):396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 2.Marx V. Epigenetics: Reading the second genomic code. Nature. 2012;491(7422):143–147. doi: 10.1038/491143a. [DOI] [PubMed] [Google Scholar]
- 3.Iqbal K, Jin SG, Pfeifer GP, Szabó PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci USA. 2011;108(9):3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Das PM, Singal R. DNA methylation and cancer. J Clin Oncol. 2004;22(22):4632–4642. doi: 10.1200/JCO.2004.07.151. [DOI] [PubMed] [Google Scholar]
- 5.Gal-Yam EN, Saito Y, Egger G, Jones PA. Cancer epigenetics: Modifications, screening, and therapy. Annu Rev Med. 2008;59:267–280. doi: 10.1146/annurev.med.59.061606.095816. [DOI] [PubMed] [Google Scholar]
- 6.Heyn HE, Esteller M. DNA methylation profiling in the clinic: Applications and challenges. Nat Rev Genet. 2012;13(10):679–692. doi: 10.1038/nrg3270. [DOI] [PubMed] [Google Scholar]
- 7.Aran D, Sabato S, Hellman A. DNA methylation of distal regulatory sites characterizes dysregulation of cancer genes. Genome Biol. 2013;14(3):R21. doi: 10.1186/gb-2013-14-3-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324(5929):929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mellén M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151(7):1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Thomson JP, et al. (2013) Dynamic changes in 5-hydroxymethylation signatures underpin early and late events in drug exposed liver. Nucleic Acids Res 41(11):5639–5654. [DOI] [PMC free article] [PubMed]
- 12.Skinner MK, Manikkam M, Guerrero-Bosagna C. Epigenetic transgenerational actions of environmental factors in disease etiology. Trends Endocrinol Metab. 2010;21(4):214–222. doi: 10.1016/j.tem.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silva AJ, Ward K, White R. Mosaic methylation in clonal tissue. Dev Biol. 1993;156(2):391–398. doi: 10.1006/dbio.1993.1086. [DOI] [PubMed] [Google Scholar]
- 14.Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nat Rev Genet. 2010;11(3):191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 15.Jin SG, Kadam S, Pfeifer GP. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 2010;38(11):e125. doi: 10.1093/nar/gkq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Booth MJ, et al. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336(6083):934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- 17.Yu M, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149(6):1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khulan B, et al. Comparative isoschizomer profiling of cytosine methylation: The HELP assay. Genome Res. 2006;16(8):1046–1055. doi: 10.1101/gr.5273806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Irizarry RA, et al. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome Res. 2008;18(5):780–790. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pastor WA, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473(7347):394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323(5910):133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 22.Flusberg BA, et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods. 2010;7(6):461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clark TA, et al. Enhanced 5-methylcytosine detection in single-molecule, real-time sequencing via Tet1 oxidation. BMC Biol. 2013;11:4. doi: 10.1186/1741-7007-11-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy PJ, et al. Single-molecule analysis of combinatorial epigenomic states in normal and tumor cells. Proc Natl Acad Sci USA. 2013;110(19):7772–7777. doi: 10.1073/pnas.1218495110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci USA. 1996;93(24):13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Branton D, et al. The potential and challenges of nanopore sequencing. Nat Biotechnol. 2008;26(10):1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wanunu M. Nanopores: A journey towards DNA sequencing. Phys Life Rev. 2012;9(2):125–158. doi: 10.1016/j.plrev.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manrao EA, Derrington IM, Pavlenok M, Niederweis M, Gundlach JH. Nucleotide discrimination with DNA immobilized in the MspA nanopore. PLoS One. 2011;6(10):e25723. doi: 10.1371/journal.pone.0025723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wallace EVB, et al. Identification of epigenetic DNA modifications with a protein nanopore. Chem Commun (Camb) 2010;46(43):8195–8197. doi: 10.1039/c0cc02864a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wanunu M, et al. Discrimination of methylcytosine from hydroxymethylcytosine in DNA molecules. J Am Chem Soc. 2011;133(3):486–492. doi: 10.1021/ja107836t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shim J, et al. Detection and quantification of methylation in DNA using solid-state nanopores. Sci Rep. 2013;3:1389. doi: 10.1038/srep01389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manrao EA, et al. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat Biotechnol. 2012;30(4):349–353. doi: 10.1038/nbt.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Butler TZ, Pavlenok M, Derrington IM, Niederweis M, Gundlach JH. Single-molecule DNA detection with an engineered MspA protein nanopore. Proc Natl Acad Sci USA. 2008;105(52):20647–20652. doi: 10.1073/pnas.0807514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lluch-Senar M, et al. Comprehensive methylome characterization of Mycoplasma genitalium and Mycoplasma pneumoniae at single-base resolution. PLoS Genet. 2013;9(1):e1003191. doi: 10.1371/journal.pgen.1003191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cherf GM, et al. Automated forward and reverse ratcheting of DNA in a nanopore at 5-Å precision. Nat Biotechnol. 2012;30(4):344–348. doi: 10.1038/nbt.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schibel AE, et al. Nanopore detection of 8-oxo-7,8-dihydro-2′-deoxyguanosine in immobilized single-stranded DNA via adduct formation to the DNA damage site. J Am Chem Soc. 2010;132(51):17992–17995. doi: 10.1021/ja109501x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ito S, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333(6047):1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nabel CS, Manning SA, Kohli RM. The curious chemical biology of cytosine: Deamination, methylation, and oxidation as modulators of genomic potential. ACS Chem Biol. 2012;7(1):20–30. doi: 10.1021/cb2002895. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.