Significance
The gene responsible for the X-linked form of Opitz syndrome (OS), Midline-1 (MID1), encodes an E3 ubiquitin ligase and was reported to guide the degradation of the catalytic subunit of protein phosphatase 2A (PP2Ac). But whether and how it is involved in neural development is unclear. We demonstrate here that Mid1-dependent PP2Ac turnover is involved in axon development. Knocking down or knocking out Mid1 not only promotes axon growth and branching in vitro, but also accelerates axon elongation and disrupts the pattern of callosal projection in mouse cortex. These defects can be reversed by down-regulating the accumulated PP2Ac in Mid1-depleted cells. Dysfunction of this Mid1–PP2Ac pathway may underlie neural symptoms of OS patients.
Abstract
Opitz syndrome (OS) is a genetic neurological disorder. The gene responsible for the X-linked form of OS, Midline-1 (MID1), encodes an E3 ubiquitin ligase that regulates the degradation of the catalytic subunit of protein phosphatase 2A (PP2Ac). However, how Mid1 functions during neural development is largely unknown. In this study, we provide data from in vitro and in vivo experiments suggesting that silencing Mid1 in developing neurons promotes axon growth and branch formation, resulting in a disruption of callosal axon projections in the contralateral cortex. In addition, a similar phenotype of axonal development was observed in the Mid1 knockout mouse. This defect was largely due to the accumulation of PP2Ac in Mid1-depleted cells as further down-regulation of PP2Ac rescued the axonal phenotype. Together, these data demonstrate that Mid1-dependent PP2Ac turnover is important for normal axonal development and that dysregulation of this process may contribute to the underlying cause of OS.
Opitz G/BBB syndrome (OS) is a genetically heterogeneous disorder characterized by distinctive facial and genital features, as well as a spectrum of variably penetrant phenotypes, including structural heart defects, structural brain anomalies, intellectual disability, and developmental delay (1). The gene responsible for the X-linked form of OS has been identified and named Midline-1 (MID1) (2). Mid1 encodes a 667-amino acid protein of the RBCC/TRIM family and has been shown to exhibit E3 ligase activity (3). The Mid1 protein is highly conserved between rodents and human (4), and its transcript is expressed ubiquitously in embryonic tissues, with the highest levels observed in the progenitor cells of the central nervous system and where cell proliferation is active (4, 5). Full-length Mid1 protein is associated with microtubules (6, 7), and the most frequently reported cases of mutations in Mid1 occur at its C terminus (8), which is known to disrupt its association with microtubules, leading to the clustering of truncated Mid1 (6). Additional researches have implicated Mid1 in the development of Caenorhabditis elegans (9, 10), Xenopus (11), and chicken (12). However, whether and how Mid1 plays a role in the development of the mammalian central nervous system, and especially in neuronal development, remains largely unknown.
Previous studies revealed that Mid1 has multiple binding partners (13–15), including the α4 subunit of the major cellular phosphatase protein phosphatase 2A (PP2A). It guides the degradation of the catalytic subunit in the PP2A complex (PP2Ac) (13). Recently, the Mid1–PP2Ac complex has been shown to be involved in asthma (16) and in regulating mRNA translation (17). Here, we report that Mid1-dependent turnover of PP2Ac is required for proper axonal development, specifically for the control of axonal growth speed and branch formation. We reported that Mid1 is highly expressed in the cerebral cortex during development and enriched in the axon segment of developing neurons. Silencing Mid1 in cultured neurons increased axonal growth and branch formation whereas down-regulation of Mid1 in the developing cortex accelerated callosal axon growth and altered the projection pattern of callosal axons. In addition, a similar defect of axon development was observed in Mid1 knockout (KO) mice. Consistent with an observed increase in PP2Ac levels following loss of Mid1, knockdown of PP2Ac in Mid1-depleted cells rescued the axonal phenotypes both in vitro and in vivo. Together, we report the importance of Mid1-regulated PP2Ac turnover in axon development and provide a possible mechanism underlying the pathological outcomes observed in OS.
Results
Mid1 Is Expressed in the Developing Brain and Enriched in the Axonal Segment of Developing Neurons.
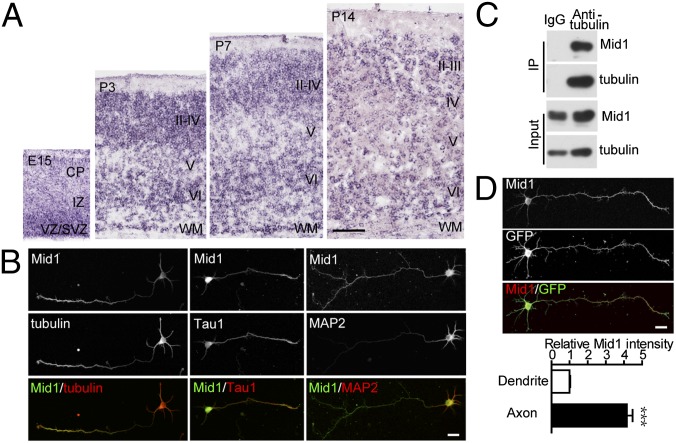
Real-time PCR showed that Mid1 mRNA was strongly expressed, and at similar levels from embryonic day (E) 14 to adult in the mouse cerebral cortex (Fig. S1A). Similarly, the level of Mid1 protein remained relatively high throughout development and peaked between E15 and postnatal day (P) 3, and then slightly decreased at later postnatal stages (Fig. S1B). In situ hybridization demonstrated that Mid1 mRNA was abundant in the ventricular/subventricular zone (VZ/SVZ) at E15 and extended to the cortical plate (CP) at E18 (Fig. 1A and Fig. S1C). After birth, Mid1 mRNA was primarily located in the cerebral cortex, olfactory bulb, hippocampus, and cerebellum (Fig. S1C), and it was mainly expressed in cortical layers II/III and VI at P7 and P14 (Fig. 1A).
Fig. 1.
Mid1 is expressed in the developing cortex and enriched in axons. (A) Expression of Mid1 mRNA in the developing mouse cortex. CP, cortical plate; I-VI, cortical layer I-VI; IZ, intermediate zone; SVZ, subventricular zone; VZ, ventricular zone; WM, white matter. (B) Mid1 is associated with microtubules and mainly located in axons. Double staining of Mid1 and the indicated markers was performed in polarized neurons at 3 d in vitro (DIV). (C) Mid1 is associated with tubulin in neurons. Coimmunoprecipitation was performed on lysate from cultured cortical neurons at 4 DIV with tubulin antibody. Mouse IgG was used as control. Mid1 and tubulin were detected with specific antibodies. (D) Mid1 is enriched in the axon segment. GFP-transfected neurons were stained for Mid1 at 3 DIV. Relative immunofluorescence intensity of Mid1 in axons and dendrites was quantified. The ratio of the intensity of Mid1 to GFP in dendrites was taken as 1. Results are shown as mean ± SEM. Fourteen neurons were analyzed. ***P < 0.001. Student t test. (Scale bars: A, 200 μm; B and D, 20 μm.)
Previous research reported that Mid1 is a microtubule-associated protein (6). Consistently, we found that ectopic GFP-tagged Mid1 colocalized with tubulin in COS7 cells whereas overexpressing a C terminus truncated form Mid1∆CTD-GFP led to clustered punctate expression in the cytoplasm (Fig. S1D). Furthermore, endogenous Mid1 could be coimmunoprecipitated (co-IP) with tubulin in cultured cortical neurons (Fig. 1C), indicating that Mid1 was also associated with microtubules in neurons. In polarized neurons, the endogenous Mid1 was mainly located in the soma and axons, but was relatively low in the dendrites (Fig. 1B). Normalizing the intensity of Mid1 staining to that of ectopic GFP revealed that the Mid1 staining in axons was about fourfold greater than that in dendrites (Fig. 1D), suggesting a potential role for Mid1 in axonal development.
Depletion of Mid1 Promotes Axon Growth and Branch Formation in Vitro.
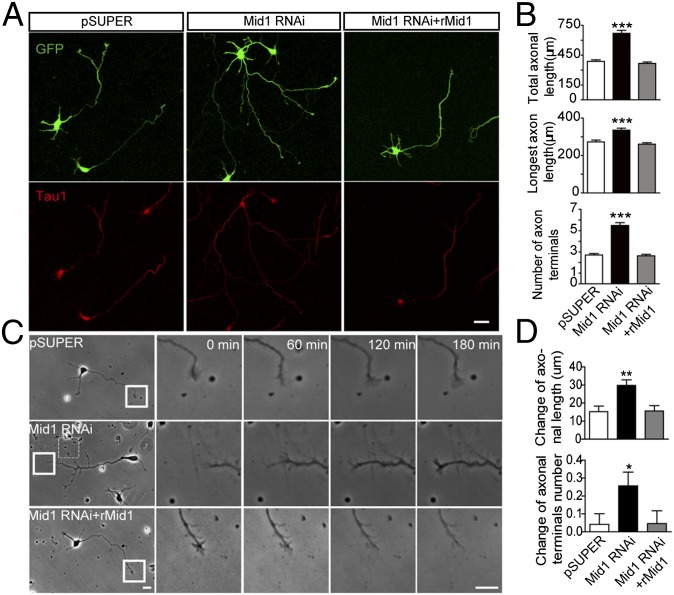
Mutations of MID1 are known to underlie the X-linked form of OS, which is characterized by midline abnormalities, including brain malformations such as agenesis of the corpus callosum (AgCC) (18). To investigate how the loss of Mid1 impacted upon neurons, we down-regulated Mid1 in cultured neurons using RNAi strategy. First, we confirmed that the Mid1 RNAi construct was specific and sufficient to down-regulate endogenous Mid1 protein through Western blotting and immunostainning (Fig. S2 A and B). Neurons transfected with the Mid1 RNAi showed significant increases in total axon length, length of the longest axon, and the number of axon terminals, compared with control cells (Fig. 2 A and B), indicating increased axon growth and branching. Sholl analysis further confirmed this increased axonal complexity (Fig. S2C). However, the establishment of neural polarity appeared to be unaffected (Fig. S2F). To rule out off-target effects, a plasmid encoding the rat Mid1 protein (rMid1) was cotransfected with the Mid1 RNAi to restore Mid1 protein expression (Fig. S2 A and B). As predicted, the axon length and branch number were restored to control levels, demonstrating the specificity of the Mid1 RNAi construct (Fig. 2 A and B). In contrast, the dendritic morphology showed little difference among the three groups (Fig. S2D). To examine whether the axonal phenotype in Mid1–down-regulated neurons was observed only in cortical neurons, we repeated the above experiment in cultured hippocampal neurons. In a similar manner, knocking down Mid1 resulted in increased axonal growth and branching whereas cotransfecting Mid1 rescued this effect (Fig. S2E).
Fig. 2.
Knockdown of Mid1 promotes axon growth and branching in vitro. (A) Representative images of neurons transfected with GFP and the constructs indicated. At 4 DIV, the neurons were stained for GFP and Tau1 to visualize the morphology of whole cells and axons. (B) Quantification of total axonal length, longest axon length, and number of axonal terminals in neurons transfected with pSUPER, Mid1 RNAi, and Mid1 RNAi plus rMid1. More than 100 neurons from four independent experiments were analyzed in each group; data are shown as mean ± SEM, ***P < 0.001, Student t test. (C) Dynamics of axonal tips. Neurons transfected with the plasmids indicated were imaged for 180 min at 2 DIV. Right are higher magnification images of the boxed regions at the time points indicated. Magnified images of the dashed box region are shown in Fig. S2F. (D) Quantification of neurite dynamics. The change of axon length and number of axon terminals over 180 min was measured and shown as mean ± SEM, n = 48 in pSUPER group, n = 74 in Mid1 RNAi group, n = 87 in Mid1 RNAi+rMid1 group, *P < 0.05, **P < 0.01, t test. (Scale bars: 20 μm.)
To further dissect how Mid1 affects axon growth, time-lapse imaging was carried out in cultured neurons. We found that knocking down Mid1 promoted axon elongation whereas coexpressing rMid1 restored this phenotype (Fig. 2C and Fig. S2G). By measuring the change of neurite length and branch number over 180 min, we showed that down-regulating Mid1 increased the speed of axonal growth and the rate of axon branching, without affecting dendrites (Fig. 2D and Fig. S2H). Furthermore, replenishing rMid1 restored the growth speed and branching rate of axons to control levels (Fig. 2D).
Silencing Mid1 Accelerates Callosal Axon Growth and Branching.
To determine the role of Mid1 during neuronal development in vivo, we electroporated the Mid1 RNAi and GFP into a subpopulation of neural progenitor cells at E15 and analyzed brain slices at different developmental stages. First, we sorted the GFP-positive cells from electroporated mouse brains at P0 by fluorescent activated cell sorting (FACS). Both the GFP-positive and GFP-negative cells were subjected to Western blotting in control and Mid1 RNAi animals. The result showed that GFP-positive cells in Mid1 RNAi electroporated animals had dramatically decreased Mid1 expression, compared with both GFP-positive/pSUPER cells and GFP-negative cells (Fig. S3A). Although Mid1 mRNA was highly expressed in the VZ/SVZ, the BrdU incorporation experiment showed that down-regulating Mid1 did not affect neural precursor cell proliferation (Fig. S3B). In addition, by examining the brain slices from P3 mice, we found little influence of depleting Mid1 on neuronal migration and dendritic arborization (Fig. S3 C and D). Moreover, Cux1 and Satb2 staining confirmed that the neuronal identity of callosal neurons was not affected by silencing Mid1 (Fig. S3E).
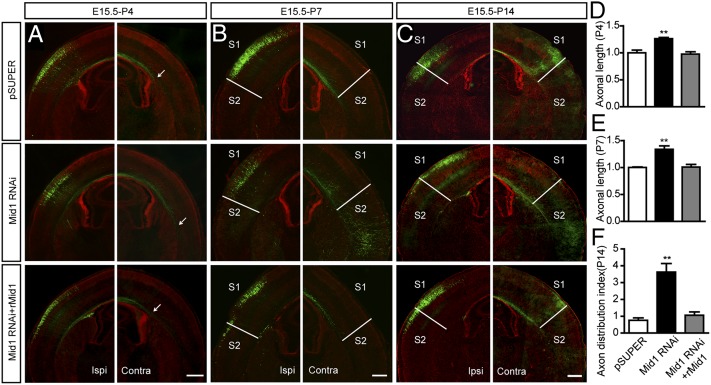
We further assessed callosal axon growth at different postnatal stages. In the control group at P4, callosal axons originating from electroporated neurons had crossed the midline, and axon bundles were restricted to the corpus callosum (CC). In mice of the same age that were transfected with the Mid1 RNAi, the axon terminals were positioned further away from the midline, indicating accelerated axon elongation (Fig. 3A and Fig. S4A). Quantitative analysis revealed that the length of axons in the CC was significantly increased in the Mid1 RNAi group (Fig. 3D). Cotransfecting the rescue construct encoding rMid1 restored this phenotype (Fig. 3 A and D). In P7 mice from the control group, axons terminated predominantly in the area beneath the S1 (primary somatosensory cortex) and S1/S2 (secondary somatosensory cortex) border of the contralateral cortex, with some projections in S2 and the entorhinal cortex (Ect). Suppressing Mid1 expression promoted callosal axon growth to most regions, in particular with more axons invading S2 and Ect (Fig. 3B and Fig. S4B). The axon length was increased by about 30% (Fig. 3E). Replenishing rMid1 also rescued these phenotypes (Fig. 3 B and E). These data suggest that down-regulating Mid1 in callosal projection neurons promotes axon growth and branching in vivo, which is consistent with the in vitro phenotypes.
Fig. 3.
Silencing Mid1 accelerates callosal axons growth and changes their projection pattern. (A–C) The constructs pSUPER, Mid1 RNAi, or Mid1 RNAi plus rMid1 were coelectroporated with GFP into the ventricles of E15 mice. Animals were killed at different developmental stages. Brain slices at the level of Bregma −1.58 mm were stained with Hoechst and GFP antibodies to visualize the callosal axons. Arrows indicate the location of axon terminals. The S1/S2 border is labeled with white lines. (Scale bar: 500 μm.) Contra, contralateral side to electroporation; Ipsi, ipsilateral side to electroporation; S1, primary somatosensory cortex; S2, second somatosensory cortex. (D–F) Quantitative analysis of callosal axon length and axon distribution index. Results are shown as mean ± SEM. At P4, n = 4 in each group; at P7 and P14, n = 5–7 in each group. **P < 0.01, Student t test.
Depleting Mid1 Affects Callosal Axon Projection Pattern in the Contralateral Hemisphere.
Callosal axons leave the contralateral white matter and start to grow into the cortical plate at P6. This process is completed by P12, when a dense projection of callosal axons forms at the S1/S2 border that arborizes predominantly in layers 5 and 2/3 (19). In our study, a dense collection of callosal axon terminals was observed at the S1/S2 border, with minor axon projections existing in S2 and Ect at P14 in control animals (Fig. 3C and Fig. S4C), consistent with previous report (19). In Mid1-depleted neurons, however, the axon projection to the S1/S2 border was largely decreased, and a dense collection of axon terminals was observed in S2 and Ect (Fig. 3C and Fig. S4C). For quantification, we devised a parameter axon distribution index (ADI) to demonstrate the axon-terminal distribution in the contralateral hemisphere (Fig. S5A). From this analysis, depletion of Mid1 dramatically increased the ADI value (Fig. 3F), indicating a shift of axon-terminal distribution with less in S1 and more in S2 and Ect (Fig. 3C). A similar phenotype was also observed in brain slices from P30 mice, indicating that this phenotype was not transient and was maintained during development (Fig. S5C). Coexpression of rMid1 restored the axon-projection pattern, as the dense region of axon terminals shifted back toward the S1/S2 border (Fig. 3C) and the ADI value returned to a level similar to that of control animals (Fig. 3F).
These changes in axon projection may result from an altered response to axon guidance cues. Thus, we tested whether depleting Mid1 changed the response of growth cones to guidance cues whose roles are well established in callosal axon pathfinding. By performing the growth cone turning assay (20), we found that depleting Mid1 had no obvious effect on the turning response of growth cones to attraction or repulsion molecules Netrin-1 and Wnt5a (Fig. S6 A–C). In addition, Slit2-induced growth-cone collapse was also unaffected (Fig. S6 D and E). We further investigated whether the phenotype was general or specific to the somatosensory cortex by examining coronal sections at Bregma −1.94 mm, where the S2 region becomes the AuV (secondary auditory cortex, ventral). Consistent with what we observed in the S1/S2 region, enhanced axon distribution was also observed in the AuV in the Mid1 RNAi group (Fig. S5D). This result indicated that accelerated axon growth is a general phenotype of Mid1-depleted cells in vivo. Thus, the abnormal projection pattern of callosal axons was probably caused by enhanced axon growth and branching in a cell-autonomous manner.
Accumulation of PP2Ac in Mid1-Depleted Cells Mediates Defects in Axonal Development.
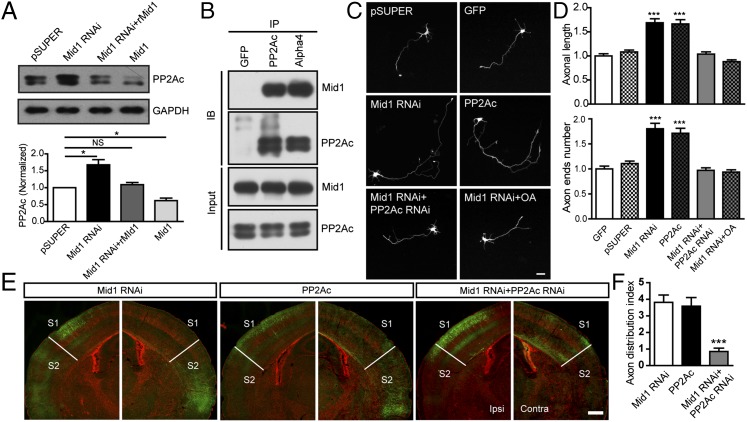
Mid1 has been identified as an E3 ligase of PP2Ac, by binding to its regulatory subunit α4, and a marked accumulation of PP2Ac protein has been observed in fibroblast cells from OS patients (13). Consistent with this work, we found that knocking down Mid1 increased the protein level of PP2Ac, which could be reversed by additional expression of rMid1 (Fig. 4A). We also found that down-regulating Mid1 in cultured cortical neurons led to increasing PP2Ac staining in the axon shaft, compared with adjacently nontransfected cells (Fig. S7B). Simultaneously, the RNAi-transfected cells exhibited increased acetylated-tubulin in axons, with the tyrosinated-tubulin unchanged (Fig. S7C), indicating an altered balance between stabilized and dynamic tubulin, which may contribute to the excessive axon growth. In contrast, overexpression of Mid1 caused an ∼50% reduction of endogenous PP2Ac (Fig. 4A). Consistently, PP2A activity increased in Mid1-depleted neurons whereas overexpressing Mid1 led to its decrease (Fig. S7A). Thus, Mid1 negatively regulates the protein level and catalytic activity of PP2Ac in neurons. To determine whether the up-regulation of PP2Ac protein after knocking down Mid1 was a consequence of decreased protein turnover by ubiquitin-proteasome system, we treated neurons with the proteasome inhibitor MG132. We found MG132 treatment increased the protein level of endogenous PP2Ac in control neurons. In contrast, this accumulation was not obvious in Mid1-depleted cells (Fig. S7D). Furthermore, we performed IP with PP2Ac antibody and detected the ubiquitinated PP2Ac using poly-ubiquitin antibody. The results showed that down-regulating Mid1 caused a decrease in poly-ubiquitinated PP2Ac (Fig. S7D), supporting the notion that Mid1 acts as an E3 ligase to mediate the ubiquitination and degradation of PP2Ac. To further confirm the relationship between Mid1 and PP2Ac, we performed co-IP in cell lines and cultured cortical neurons. Consistent with previous reports (13, 14, 21), Mid1 and PP2Ac existed in a protein complex when Mid1, α4, and PP2Ac were expressed in HEK293 cells (Fig. S7E). Similarly, PP2Ac and α4 antibody could co-IP endogenous Mid1 and PP2Ac in cultured neurons whereas the control GFP antibody failed (Fig. 4B). These data indicate that Mid1, PP2Ac, and α4 form a complex in neurons and that Mid1 acts as an E3 ligase to regulate the protein turnover of PP2Ac.
Fig. 4.
PP2Ac accumulation mediates the axon abnormality following Mid1 down-regulation. (A) Mid1 negatively regulates the protein level of PP2Ac. Cortical neurons electroporated with the constructs indicated were harvested and subjected to immunoblotting at 4 DIV. For quantification, GAPDH was used as the loading control. n = 3 in each group. *P < 0.05, Student t test. (B) Mid1 and PP2Ac interact in cultured neurons. Cortical neurons were harvested at 4 DIV, and immunoprecipitation was performed with the antibodies indicated. Mid1 and PP2Ac were detected with specific antibodies. (C) Representative images of neurons transfected with different constructs at 4 DIV. GFP was amplified by immunostaining to visualize the cell morphology. (D) Quantitative analysis of the total axonal length and number of axonal terminals. Results are shown as mean ± SEM. More than 100 neurons from four independent experiments were analyzed in each group. ***P < 0.001, Student t test. (E) Representative images of brain slices at the level of Bregma −1.58 mm from P14 animals transfected with indicated constructs. The projection patterns of callosal axons were shown by GFP staining. (F) The axon distribution index of brain slices from P14 mice in different groups. n = 6–7 in each group. ***P < 0.001, compared with Mid1 RNAi, t test. (Scale bar: C, 20 μm; E, 500 μm.)
As a major protein phosphatase, PP2A is required in axonogenesis and involved in axon guidance in C. elegans (22, 23). However, whether Mid1 affects axon growth through PP2A signaling is not known. In cultured neurons, we observed that overexpressing PP2Ac promoted axon outgrowth and branching, similar to the phenotype of Mid1-depleted neurons (Fig. 4 C and D). In addition, cotransfecting Mid1 RNAi and PP2Ac RNAi (Fig. S7F) prevented the excessive axon growth found in the Mid1 RNAi group (Fig. 4 C and D), and chronically inhibiting PP2A activity with okadaic acid (OA) in Mid1-depleted neurons also restored the axonal length and number of axonal terminals to the control levels (Fig. 4 C and D). Similarly, overexpression of PP2Ac in vivo also mimicked the phenotype observed in the Mid1 RNAi-transfected brains (Fig. 4E). And introducing a PP2Ac RNAi construct successfully restored axonal growth and projection pattern, as a dense bundle of axon terminals were observed at the S1/S2 border with few projection to the S2 region (Fig. 4 E and F). Thus, aberrant axon phenotypes that resulted from Mid1 knockdown were predominantly due to the accumulation of PP2Ac in the cells.
Corpus Callosum Development Is Abnormal in Mid1 KO Mice.
Given that knockdown of Mid1 resulted in increased axon outgrowth both in vitro and in vivo, we next sought to determine whether a similar axonal phenotype existed in Mid1 KO mice (24). By comparing the morphology of cortical neurons cultured from WT and KO mice cortices, we found the axon length and branch number increased in KO neurons, which is consistent with the RNAi experiment (Fig. S8A). As Mid1 was undetectable and PP2Ac was increased in KO animals (Fig. S9E), we expressed rMid1 or PP2Ac RNAi in KO neurons and successfully restored the excessive axonal growth and branching (Fig. S8A). Additionally, by performing in utero electroporation, we found that cell proliferation, neuronal migration, and dendritic arborization were unchanged in the Mid1 KO mice cortex (Fig. S9 A–C). Immunostaining with specific cortical neuron markers further confirmed that Mid1 loss-of-function did not affect cell-fate determination or neuronal migration (Fig. S9D).
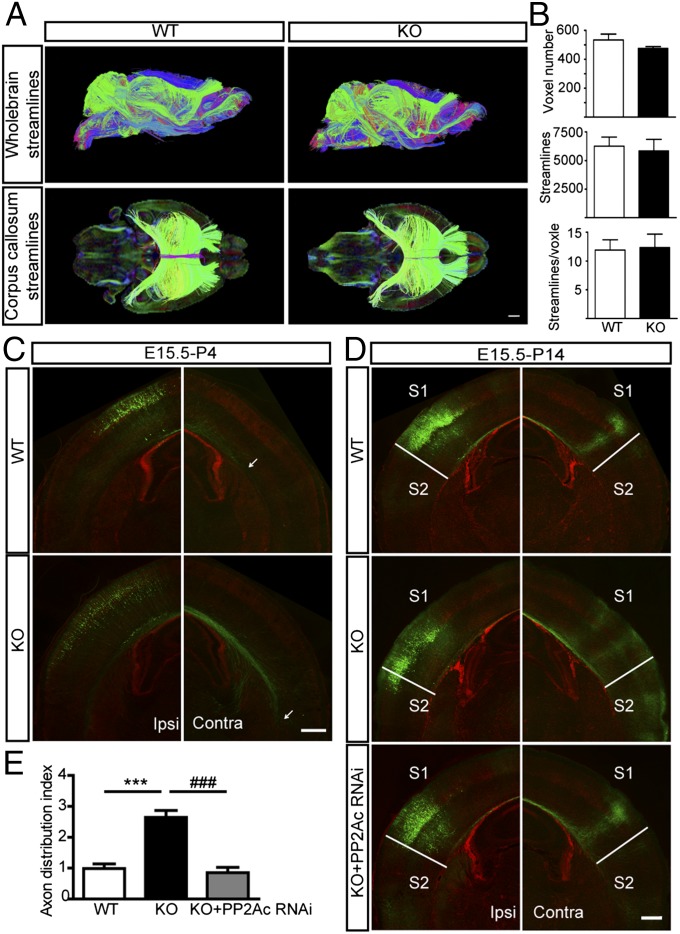
To examine whether genetic ablation of Mid1 resulted in abnormal axon development, we first used diffusion MRI tractography to examine the integrity of the CC in Mid1 KO mice and their WT littermates and found no obvious difference (Fig. 5A). From the whole-brain tractography, streamlines passing through the CC were extracted for comparison, but no morphological differences were observed between the genotypes (Fig. 5A). No difference was observed in the number of voxels, nor the number streamlines alone or when normalized to the CC voxel volume (Fig. 5B). These data suggest that loss of Mid1 does not change gross CC morphology in adult mice. As the diffusion MRI method could not determine the growth rate of developing axons or provide precise information regarding the exact position of axonal projections, we used electroporation to examine callosal axon development in Mid1 KO mice. We found that the callosal axons in KO mice had grown further into the contralateral hemisphere than those in their WT littermates at P4 (Fig. 5C). At P14, compared with the WT littermates, axon distribution was altered in Mid1 KO mice, as a greater number of axon terminals, with a wider distribution, were observed in both the S1 and S2 regions but not restricted at the S1/S2 border (Fig. 5D). Consistently, the ADI of KO mice was significantly increased (Fig. 5E). Meanwhile, we performed anterograde and retrograde tracing experiments to confirm the phenotype. When the retrograde tracer cholera toxin B subunit (CTb) was injected into S2, most of the labeled cell bodies were located in the contralateral S2 in WT animals, with some cell bodies in the contralateral S1 region of KO animals (Fig. S10A), suggesting that S2 also contained axon terminals from callosal projection neurons in the contralateral S1 area. On the other hand, when the anterograde tracer biotinylated dextran amines (BDA) was injected into S1, most axon terminals were seen in the contralateral S1 in WT animals whereas, in KO animals, axon terminals distributed in both S1 and S2 areas (Fig. S10B). Collectively, Mid1 KO caused a broader distribution of the contralateral axon projection pattern, consistent with the knockdown experiment.
Fig. 5.
Corpus callosum development is abnormal in Mid1 KO mice. (A) Comparison of the corpus callosum (CC) between Mid1 KO and WT adult mice using DTI. (Upper) Representative whole-brain streamlines in a lateral-sagittal view of WT and KO mice. (Lower) CC streamlines in a dorsal view of WT and KO mice. Streamline color follows the following orientation code: green, mediolateral; red, rostrocaudal; blue, dorsoventral. (B) The volume of the CC was measured by voxel number, the number of streamlines, and the number of streamlines per voxel. n = 3 in each group. (C) Callosal axon growth is accelerated in Mid1 KO mice. The GFP plasmid was electroporated into neuronal progenitors at E15, and GFP staining was performed in brain slices of P4 animals. Arrows indicate the axon terminals within the corpus callosum. (D) Callosal projections are abnormal in Mid1 KO mice. GFP or PP2Ac RNAi constructs were electroporated at E15. Brain slices (Bregma −1.58 mm) from P14 Mid1 KO mice and their WT littermates were stained for GFP to visualize the callosal projection. (E) Statistical analysis of axon distribution index in D. Results are shown as mean ± SEM, n = 7–8 in each group. ***P < 0.001, compared with WT; ###P < 0.001, compared with KO; Student t test. (Scale bars: A, 1 mm; C and D, 500 μm.)
As the PP2Ac protein and PP2A activity were increased in Mid1 KO mice brain lysates (Fig. S9 E and F), to test whether this increase of PP2Ac causes abnormal callosal axonal development, we introduced PP2Ac RNAi into Mid1 KO mice, which successfully restored the axonal projection pattern (Fig. 5D), as well as the ADI value (Fig. 5E).
Interestingly, overexpression of Mid1∆CTD, which is common in OS patients, in Mid1 KO cells led to phenotypes that were different from those generated solely by gene KO. Not only did Mid1∆CTD reverse the effect but it did further decrease axonal length and branching number in vitro (Fig. S8A), impair neuronal migration, and cause an obvious decrease in axon terminals in the contralateral cortex in vivo (Fig. S8B). This result indicated that Mid1∆CTD may cause accumulated toxicity in the cells and further impair neuronal migration and axon development.
Discussion
Various mutations of MID1 have been identified in human patients with X-linked OS (25), and it has been reported as an E3 ligase for PP2Ac (13). However, the role of Mid1 in mammalian neural development and function is largely unknown. Here, we described a unique role for Mid1 in axon development, showing that Mid1-dependent PP2Ac turnover controls axon growth and branching in vitro and regulates axon elongation and projection in vivo.
Correct axonal connections are required for normal neural function (26). It has recently been shown that depleting FGF13, another mental retardation (MR) candidate gene, increases callosal axon branching in the rodent cortex (27), indicating that abnormal axonal branching and targeting may underlie some MR. Depleting Mid1 causes a dramatic axon overgrowth both in cultured cells and in vivo, indicating that Mid1 may act as an inhibitory factor to regulate axon growth to ensure precise structural and functional patterning, which is crucial for proper circuit formation.
As OS patients often have defects associated with midline development, including agenesis of the corpus callosum (18), we initially expected Mid1 KO mice to display midline abnormalities of the CC. However, DTI analysis showed that the gross structure of CC was normal (Fig. 5 A and B). Furthermore, no obvious midline defects were observed in these Mid1-deficient mice, nor in a Mid1 KO line generated by another group (28). This discrepancy between human symptoms and animal phenotypes may result from different regulatory mechanisms in different species. Specifically, Mid2, a close homolog of Mid1 (29, 30), may play a redundant role and compensate for the effect of Mid1 deletion (11, 31). In addition, many mutations identified in OS patients are located in the C-terminal region of the protein (8), which causes Mid1 to dissociate from the microtubules and form clusters (6, 14) (Fig. S1D). Interestingly, overexpressing Mid1∆CTD could not rescue the phenotypes caused by deleting Mid1 but resulted in different defects in vitro and in vivo (Fig. S8 A and B). We speculate that the reversed effect of Mid1∆CTD in cultured neurons was not due to its ability to rescue the phenotype; instead, Mid1∆CTD possibly has a gain-of-function consequence that causes accumulated toxicity in the cells, which further impairs neuronal migration and axon development in vivo.
Increasing evidence has demonstrated that remodeling of the cytoskeleton plays important roles in various aspects of brain development (32). Several genes associated with MR have also been found to be involved in cytoskeletal processes (27, 33), indicating that a common pathological mechanism may underlie different human disorders. As PP2A is a major phosphatase and microtubule dynamics are widely regulated by protein phosphorylation and dephosphorylation (34), it is reasonable to hypothesize that Mid1 plays a role in neural development by modulating the cytoskeleton. In individuals with OS, most mutations in the MID1 gene result in disruption of the normal distribution of protein along the microtubules, indicating that functional MID1 is required along the length of the cytoskeleton (18). Indeed, subsequent studies have shown that in OS the normal regulated turnover of PP2Ac along the microtubules is perturbed, leading to other microtubule-associated proteins being caught in an abnormal phosphorylation state (13), which may affect microtubule polymerization and stability. Recently, mTORC1 signaling, which is also involved in cytoskeletal remodeling, was identified as a downstream component of the Mid1-PP2Ac pathway (35). Further investigation should therefore be focused on elucidating the downstream molecular events triggered by Mid1-directed PP2Ac regulation.
On the other hand, Madd-2, the homolog of Mid1 in C. elegans, has recently been shown to play a role in axon guidance through the Netrin/DCC pathway (9, 10). Although the response of growth cone to Netrin-1, Wnt5a, and Slit2 exhibits no significant difference between pSUPER and Mid1 RNAi transfected neurons (Fig. S6), it would be interesting to explore whether Mid1 regulates mammalian axon development via other guidance cues.
Materials and Methods
Mid1 mutant mice, a kind gift from Timothy Cox, were genotyped as described previously (24). We also used female ICR and C57BL/6 mice in in utero electroporation experiments. The morning of vaginal plug was considered to be E0.5. All experimental procedures using animals were approved by the animal care and use committee of the Institute of Neuroscience, Chinese Academy of Sciences.
For details, see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. Mu-ming Poo and Tara Walker for insightful comments on the manuscript. This work was supported by 973 Program Grant 2011CBA00400; NSFC Grants 30925016, 31021063, and 31123002; and Program of Shanghai Subject Chief Scientist Grant 12XD1405500. L.J.R. is supported by a National Health and Medical Research Council, Australia, Principal Research Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1303687110/-/DCSupplemental.
References
- 1.Robin NH, Opitz JM, Muenke M. Opitz G/BBB syndrome: Clinical comparisons of families linked to Xp22 and 22q, and a review of the literature. Am J Med Genet. 1996;62(3):305–317. doi: 10.1002/(SICI)1096-8628(19960329)62:3<305::AID-AJMG20>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 2.Quaderi NA, et al. Opitz G/BBB syndrome, a defect of midline development, is due to mutations in a new RING finger gene on Xp22. Nat Genet. 1997;17(3):285–291. doi: 10.1038/ng1197-285. [DOI] [PubMed] [Google Scholar]
- 3.Han X, Du H, Massiah MA. Detection and characterization of the in vitro e3 ligase activity of the human MID1 protein. J Mol Biol. 2011;407(4):505–520. doi: 10.1016/j.jmb.2011.01.048. [DOI] [PubMed] [Google Scholar]
- 4.Dal Zotto L, et al. The mouse Mid1 gene: Implications for the pathogenesis of Opitz syndrome and the evolution of the mammalian pseudoautosomal region. Hum Mol Genet. 1998;7(3):489–499. doi: 10.1093/hmg/7.3.489. [DOI] [PubMed] [Google Scholar]
- 5.Pinson L, et al. Embryonic expression of the human MID1 gene and its mutations in Opitz syndrome. J Med Genet. 2004;41(5):381–386. doi: 10.1136/jmg.2003.014829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schweiger S, et al. The Opitz syndrome gene product, MID1, associates with microtubules. Proc Natl Acad Sci USA. 1999;96(6):2794–2799. doi: 10.1073/pnas.96.6.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cainarca S, Messali S, Ballabio A, Meroni G. Functional characterization of the Opitz syndrome gene product (midin): Evidence for homodimerization and association with microtubules throughout the cell cycle. Hum Mol Genet. 1999;8(8):1387–1396. doi: 10.1093/hmg/8.8.1387. [DOI] [PubMed] [Google Scholar]
- 8.Gaudenz K, et al. Opitz G/BBB syndrome in Xp22: Mutations in the MID1 gene cluster in the carboxy-terminal domain. Am J Hum Genet. 1998;63(3):703–710. doi: 10.1086/302010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hao JC, et al. The tripartite motif protein MADD-2 functions with the receptor UNC-40 (DCC) in Netrin-mediated axon attraction and branching. Dev Cell. 2010;18(6):950–960. doi: 10.1016/j.devcel.2010.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alexander M, et al. MADD-2, a homolog of the Opitz syndrome protein MID1, regulates guidance to the midline through UNC-40 in Caenorhabditis elegans. Dev Cell. 2010;18(6):961–972. doi: 10.1016/j.devcel.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 11.Suzuki M, Hara Y, Takagi C, Yamamoto TS, Ueno N. MID1 and MID2 are required for Xenopus neural tube closure through the regulation of microtubule organization. Development. 2010;137(14):2329–2339. doi: 10.1242/dev.048769. [DOI] [PubMed] [Google Scholar]
- 12.Granata A, Quaderi NA. The Opitz syndrome gene MID1 is essential for establishing asymmetric gene expression in Hensen’s node. Dev Biol. 2003;258(2):397–405. doi: 10.1016/s0012-1606(03)00131-3. [DOI] [PubMed] [Google Scholar]
- 13.Trockenbacher A, et al. MID1, mutated in Opitz syndrome, encodes an ubiquitin ligase that targets phosphatase 2A for degradation. Nat Genet. 2001;29(3):287–294. doi: 10.1038/ng762. [DOI] [PubMed] [Google Scholar]
- 14.Short KM, Hopwood B, Yi Z, Cox TC. MID1 and MID2 homo- and heterodimerise to tether the rapamycin-sensitive PP2A regulatory subunit, alpha 4, to microtubules: Implications for the clinical variability of X-linked Opitz GBBB syndrome and other developmental disorders. BMC Cell Biol. 2002;3:1. doi: 10.1186/1471-2121-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berti C, Fontanella B, Ferrentino R, Meroni G. Mig12, a novel Opitz syndrome gene product partner, is expressed in the embryonic ventral midline and co-operates with Mid1 to bundle and stabilize microtubules. BMC Cell Biol. 2004;5:9. doi: 10.1186/1471-2121-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collison A, et al. The E3 ubiquitin ligase midline 1 promotes allergen and rhinovirus-induced asthma by inhibiting protein phosphatase 2A activity. Nat Med. 2013;19(2):232–237. doi: 10.1038/nm.3049. [DOI] [PubMed] [Google Scholar]
- 17.Krauss S, et al. Translation of HTT mRNA with expanded CAG repeats is regulated by the MID1-PP2A protein complex. Nat Commun. 2013;4:1511. doi: 10.1038/ncomms2514. [DOI] [PubMed] [Google Scholar]
- 18.Cox TC, et al. New mutations in MID1 provide support for loss of function as the cause of X-linked Opitz syndrome. Hum Mol Genet. 2000;9(17):2553–2562. doi: 10.1093/hmg/9.17.2553. [DOI] [PubMed] [Google Scholar]
- 19.Wang CL, et al. Activity-dependent development of callosal projections in the somatosensory cortex. J Neurosci. 2007;27(42):11334–11342. doi: 10.1523/JNEUROSCI.3380-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng JQ, Felder M, Connor JA, Poo MM. Turning of nerve growth cones induced by neurotransmitters. Nature. 1994;368(6467):140–144. doi: 10.1038/368140a0. [DOI] [PubMed] [Google Scholar]
- 21.Liu J, Prickett TD, Elliott E, Meroni G, Brautigan DL. Phosphorylation and microtubule association of the Opitz syndrome protein mid-1 is regulated by protein phosphatase 2A via binding to the regulatory subunit alpha 4. Proc Natl Acad Sci USA. 2001;98(12):6650–6655. doi: 10.1073/pnas.111154698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu LQ, et al. Protein phosphatase 2A facilitates axonogenesis by dephosphorylating CRMP2. J Neurosci. 2010;30(10):3839–3848. doi: 10.1523/JNEUROSCI.5174-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogura K, et al. Protein phosphatase 2A cooperates with the autophagy-related kinase UNC-51 to regulate axon guidance in Caenorhabditis elegans. Development. 2010;137(10):1657–1667. doi: 10.1242/dev.050708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perry J, Palmer S, Gabriel A, Ashworth A. A short pseudoautosomal region in laboratory mice. Genome Res. 2001;11(11):1826–1832. doi: 10.1101/gr.203001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fontanella B, Russolillo G, Meroni G. MID1 mutations in patients with X-linked Opitz G/BBB syndrome. Hum Mutat. 2008;29(5):584–594. doi: 10.1002/humu.20706. [DOI] [PubMed] [Google Scholar]
- 26.Gazzaniga MS. Principles of human brain organization derived from split-brain studies. Neuron. 1995;14(2):217–228. doi: 10.1016/0896-6273(95)90280-5. [DOI] [PubMed] [Google Scholar]
- 27.Wu QF, et al. Fibroblast growth factor 13 is a microtubule-stabilizing protein regulating neuronal polarization and migration. Cell. 2012;149(7):1549–1564. doi: 10.1016/j.cell.2012.04.046. [DOI] [PubMed] [Google Scholar]
- 28.Lancioni A, et al. Lack of Mid1, the mouse ortholog of the Opitz syndrome gene, causes abnormal development of the anterior cerebellar vermis. J Neurosci. 2010;30(8):2880–2887. doi: 10.1523/JNEUROSCI.4196-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buchner G, et al. MID2, a homologue of the Opitz syndrome gene MID1: Similarities in subcellular localization and differences in expression during development. Hum Mol Genet. 1999;8(8):1397–1407. doi: 10.1093/hmg/8.8.1397. [DOI] [PubMed] [Google Scholar]
- 30.Perry J, et al. FXY2/MID2, a gene related to the X-linked Opitz syndrome gene FXY/MID1, maps to Xq22 and encodes a FNIII domain-containing protein that associates with microtubules. Genomics. 1999;62(3):385–394. doi: 10.1006/geno.1999.6043. [DOI] [PubMed] [Google Scholar]
- 31.Granata A, et al. Evidence of functional redundancy between MID proteins: Implications for the presentation of Opitz syndrome. Dev Biol. 2005;277(2):417–424. doi: 10.1016/j.ydbio.2004.09.036. [DOI] [PubMed] [Google Scholar]
- 32.Heng JI, Chariot A, Nguyen L. Molecular layers underlying cytoskeletal remodelling during cortical development. Trends Neurosci. 2010;33(1):38–47. doi: 10.1016/j.tins.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 33.Robertson SP, et al. OPD-Spectrum Disorders Clinical Collaborative Group Localized mutations in the gene encoding the cytoskeletal protein filamin A cause diverse malformations in humans. Nat Genet. 2003;33(4):487–491. doi: 10.1038/ng1119. [DOI] [PubMed] [Google Scholar]
- 34.Avila J, Domínguez J, Díaz-Nido J. Regulation of microtubule dynamics by microtubule-associated protein expression and phosphorylation during neuronal development. Int J Dev Biol. 1994;38(1):13–25. [PubMed] [Google Scholar]
- 35.Liu E, Knutzen CA, Krauss S, Schweiger S, Chiang GG. Control of mTORC1 signaling by the Opitz syndrome protein MID1. Proc Natl Acad Sci USA. 2011;108(21):8680–8685. doi: 10.1073/pnas.1100131108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.