Significance
We report that glycolytic genes are synchronously activated in midembryogenesis in developing Drosophila muscles and that muscle-targeted attenuation of their expression leads to reduced muscle size and the presence of unfused myoblasts. Importantly, a “thin muscle” phenotype is also observed in fast muscle fibers of pgam2 zebrafish morphants, strongly suggesting a conserved role of glycolysis in promoting myoblast fusion-based muscle growth. We also show that insulin positively regulates glycolysis and that blocking the insulin pathway phenocopies the loss of function phenotypes of glycolytic genes, leading to myoblast fusion arrest and reduced muscle size. These findings led us to propose that setting metabolism to glycolysis-based high-rate biomass production is part of a core myogenic program that promotes formation of syncytial muscles.
Keywords: morpholino, Danio rerio
Abstract
Muscles ensure locomotion behavior of invertebrate and vertebrate organisms. They are highly specialized and form using conserved developmental programs. To identify new players in muscle development we screened Drosophila and zebrafish gene expression databases for orthologous genes expressed in embryonic muscles. We selected more than 100 candidates. Among them is the glycolysis gene Pglym78/pgam2, the attenuated expression of which results in the formation of thinner muscles in Drosophila embryos. This phenotype is also observed in fast muscle fibers of pgam2 zebrafish morphants, suggesting affected myoblast fusion. Indeed, a detailed analysis of developing muscles in Pglym78 RNAi embryos reveals loss of fusion-associated actin foci and an inefficient Notch decay in fusion competent myoblasts, both known to be required for fusion. In addition to Pglym78, our screen identifies six other genes involved in glycolysis or in pyruvate metabolism (Pfk, Tpi, Gapdh, Pgk, Pyk, and Impl3). They are synchronously activated in embryonic muscles and attenuation of their expression leads to similar muscle phenotypes, which are characterized by fibers with reduced size and the presence of unfused myoblasts. Our data also show that the cell size triggering insulin pathway positively regulates glycolysis in developing muscles and that blocking the insulin or target of rapamycin pathways phenocopies the loss of function phenotypes of glycolytic genes, leading to myoblast fusion arrest and reduced muscle size. Collectively, these data suggest that setting metabolism to glycolysis-stimulated biomass production is part of a core myogenic program that operates in both invertebrate and vertebrate embryos and promotes formation of syncytial muscles.
Intensive research in a variety of models has uncovered many evolutionarily conserved features of making muscle, including initial myoblast specification, the events of their differentiation, and the assembly of the functional musculature (1). However, several gaps still exist in our understanding of muscle development. One of the aspects that remains to be elucidated is the potential involvement of metabolic genes in muscle development and function. A better comprehension of the role of this class of genes seems important to improve our knowledge on metabolic myopathies involving glycolytic genes. For example, phosphoglycerate mutase 2 (pgam2) has been associated with myopathy with exercise intolerance (2). The muscle disruption phenotypes observed in patients with pgam2 mutations, but also with phosphofructokinase (pfk) mutations, suggest a role for glycolytic genes in promoting muscle fiber resistance to contractions (3). This view is supported by the phenotypes observed in mouse mutants for the muscle isoform of pfk (pfkm). Loss of pfkm in mice leads to a reduction of glycolysis in muscles and provokes severe phenotypes ranging from exercise intolerance to death around weaning, which are highly similar to clinical phenotypes of patients carrying pfk mutations (4).
A novel developmental role for glycolytic genes has been reported in a recent study in Drosophila (5). This work provided evidence that glycolytic genes are part of a metabolic switch that supports rapid developmental growth of Drosophila larvae. It is well known that a shift to aerobic glycolysis, known as the Warburg effect, is commonly used by cancer cells (6), but also by rapidly proliferating normal cells (7, 8), to efficiently divert glycolytic intermediates (9) to the synthesis of the macromolecules needed for cell growth. Glycolysis might, therefore, contribute to developmental processes characterized by bursts in cell proliferation and/or by rapid growth. This idea is consistent with the high glycolytic activity observed in early mouse embryos (10) and with the requirement for glycolytic gene function downstream of Drosophila estrogen-related receptor (dERR) signaling in rapidly growing Drosophila larvae (5). Interestingly, dERR was reported to be expressed in developing embryonic muscles and found to activate glycolysis in midembryogenesis (5); however, the role of this transcriptional activation of glycolytic genes in muscle development has not yet been addressed.
Here we applied a large-scale in silico screen for evolutionarily conserved genes expressed in embryonic muscles. Among the identified candidates we found several genes encoding glycolytic enzymes, including the phosphoglyceromutase Pglym78/pgam2 gene involved in human myopathy with exercise intolerance. We show that loss of Pglym78/pgam2 function in developing skeletal muscle results in myoblast fusion defects in both Drosophila and zebrafish embryos, leading to a “thin muscle” phenotype. We also demonstrate that insulin signaling positively regulates glycolysis and together with the target of rapamycin (TOR) pathway is required to promote fusion-based embryonic muscle growth. Thus, glycolytic genes emerge as a part of the evolutionarily conserved myogenic program ensuring proper formation of syncytial muscle. Our findings indicate that metabolic genes contribute to regulatory cascades controlling normal embryonic development.
Results
A Gene Expression-Based Screen Identifies Glycolytic Genes Among Conserved Components of the Myogenic Pathway.
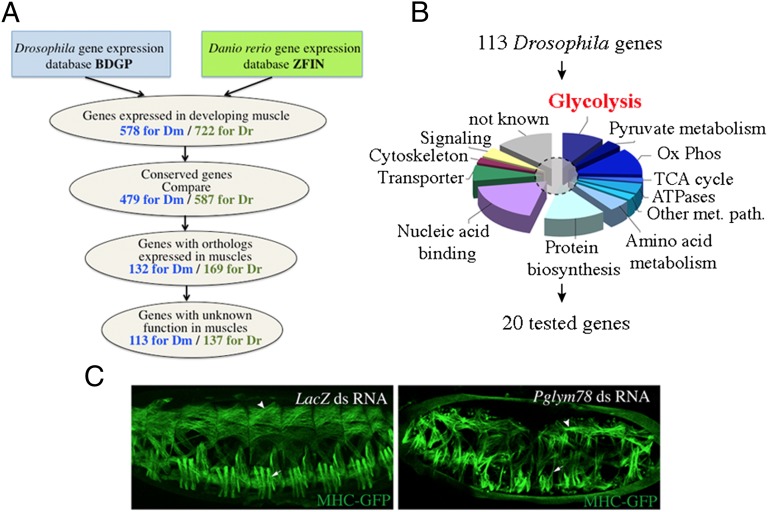
Many developmental processes including formation of different types of muscles are controlled by evolutionarily conserved genes (11, 12). We thus reasoned that by identifying conserved groups of genes expressed in developing muscles we could uncover new developmental regulators. To select conserved muscle genes with so-far-unknown functions we designed a pipeline to screen existing databases (Fig. 1A). We first examined gene expression databases of two evolutionarily distant species, Drosophila melanogaster (BDGP in situ, http://insitu.fruitfly.org) and Danio rerio (ZFIN, www.zfin.org). This led to the identification of 578 somatic/skeletal muscle-expressed genes referenced for Drosophila and 722 for zebrafish. We then applied the multiorganism information system COMPARE (http://compare.ibdml.univ-mrs.fr/) to select only those among the muscle genes that are conserved between Drosophila and zebrafish (479 Drosophila and 587 zebrafish genes) and whose orthologs are expressed in developing muscles in both species (132 Drosophila and 169 zebrafish genes) (Fig. 1A and Table S1). Within the selected pool of conserved muscle genes are some known conserved regulators of muscle development such as Mef2 or FGFR/heartless (htl) (Table S1). However, for the majority of candidates (113 Drosophila and 137 zebrafish genes), myogenic roles remain unknown (Fig. 1A). Gene Ontology analysis of selected conserved muscle genes reveals that more than half of them encode proteins involved in metabolic pathways, including glycolytic enzymes (Fig. 1B and Table S1). In addition to this major gene category we notice that about 20% of candidates code for nucleic acid binding proteins and 10% for transporters (Fig. 1B). To assess whether our screen identified genes that function in muscle development, we performed a functional analysis of randomly chosen genes from different functional classes (Fig. 1B and Table S1) in Drosophila. We used the dsRNA injection-based RNAi technique to attenuate gene expression in MHC-tauGFP transgenic embryos in which somatic muscle formation can be visualized (13) (SI Materials and Methods gives details). Among the tested genes we found that the RNAi-mediated attenuation of Pglym78, encoding one of the glycolytic enzymes, leads to an altered somatic muscle pattern (Fig. 1C). We noticed that the majority of embryonic muscles in the Pglym78 RNAi context have abnormal shapes and appear significantly thinner (arrows and arrowheads in Fig. 1C) compared with the negative control (LacZ dsRNA injections).
Fig. 1.
Gene expression-based screen for new evolutionarily conserved regulators of muscle development. (A) A pipeline applied to screen Drosophila and zebrafish gene expression databases (Table S1 gives the full list of genes). (B) Gene Ontology (GO) classification of 113 conserved Drosophila candidate genes with unknown muscle functions. The dashed circle marks a sample of 20 candidate genes selected for functional analyses by direct dsRNA injection in Drosophila MHC-tauGFP embryos. (C, Left) The muscle pattern of a MHC-tauGFP embryo injected with dsRNA against the yeast LacZ gene (negative control). Ox Phos, oxidative phosphorylation; TCA, tricarboxylic acid cycle; Other met. path., other metabolic pathway. (C, Right) Muscle phenotype observed after injection of dsRNA against Pglym78, encoding phosphoglyceromutase and involved in glycolysis. Notice that after the attenuation of Pglym78 expression muscles appear thinner both in the dorsal (arrowhead) and in the lateral domain (arrows).
Phosphoglycerate Mutase (Pglym78/pgam2) Promotes Fusion-Dependent Embryonic Muscle Growth in Drosophila and in Zebrafish Embryos.
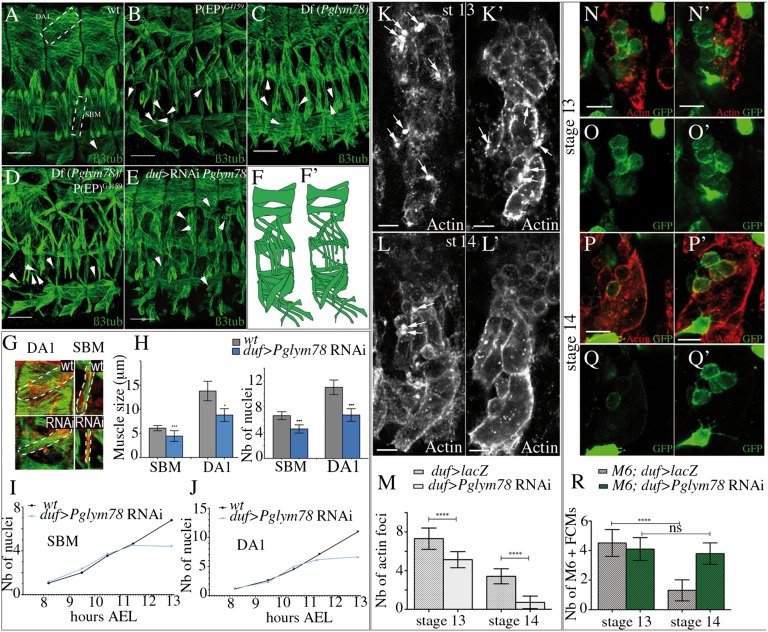
The muscle phenotype observed after attenuation of Pglym78 expression suggested a so-far-unknown developmental function of this gene in muscle formation and prompted us to further characterize effects of its loss of function. We first analyzed embryos carrying the P(EP)G4159 element insertion within the Pglym78 gene (Fig. S1A) and found that all somatic muscles appear thinner (compare Fig. 2A with Fig. 2B). Similarly, embryos homozygous for the Pglym78 deficiency [Df(Pglym78)] generated by flippase recognition target (FRT)-mediated recombination (Fig. S1A and Fig. 2C) or those transheterozygous for Df(Pglym78) and P(EP)G4159 (Fig. 2D) showed highly reminiscent muscle phenotypes with reduced muscle size and the presence of unfused myoblasts (arrowheads in Fig. 2 B–D and scheme in Fig. 2 F and F′), suggesting that myoblast fusion defects contribute to the observed phenotype. To test whether Pglym78 function is specifically required in the growing fiber, we crossed a UAS-Pglym78 RNAi line with the muscle-specific duf-Gal4 (Fig. 2E) and Mef2-Gal4 (Fig. S1B) driver lines. Indeed, muscle-specific attenuation of Pglym78 also leads to apparent fusion defects (arrowheads in Fig. 2E) and to the formation of thinner muscles. To quantify the observed phenotypes we measured the width of two muscles, the laterally located segmental border muscle (SBM) and the dorsally located dorsal acute 1 (DA1) and counted the number of nuclei they contain (Fig. 2G). The quantifications were done in wild-type and in muscle-specific Pglym78 RNAi contexts at the end of embryonic stage 15 [13 h after egg laying (AEL)], when myoblast fusion is completed. We found that Pglym78-deficient muscles (in both duf-Gal4 and Mef2-Gal4 contexts) display a significantly reduced size and contain fewer nuclei compared with the control (Fig. 2H and Fig. S1B and Table S2A), supporting the view that myoblast fusion is affected. Because the duf-Gal4 driver allows targeting of Pglym78 in the time window more accurately corresponding to its expression in embryonic muscles, we chose to use duf-Gal4 in further analyses. The reduced number of nuclei in the muscles may be due to a decreased rate of fusion or to a premature arrest of the fusion process. To answer this question we counted nuclei in SBM and in DA1 muscles at five time points between 8 and 13 h AEL (stages 12 to 15) in wild-type and in duf > Pglym78 RNAi embryos. Our data demonstrate that in both muscles examined attenuation of Pglym78 leads to a premature fusion arrest at 11–11.5 h AEL (Fig. 2 I and J and Table S2B). This effect temporally coincides with the previously reported burst of glycolytic gene expression (5), but we cannot exclude that the maternal load of Pglym78 ensures the initial round of fusion. Altogether these data indicate a requirement of Pglym78 function for fusion-based growth of Drosophila body-wall muscles. Consistent with this view, attenuation of Pglym78 in the cardiac muscle cells that do not undergo fusion or in other cell types such as epidermal or neural cells has no effect on their developmental pattern (Fig. S1 C–E).
Fig. 2.
Loss of Pglym78 function leads to a myoblast fusion defect and reduced muscles size. (A–D) Muscle pattern in wild-type (A), in P(EP)G4159 (B), in Df(Pglym78) (C), and in transheterozygous Df(Pglym78)/P(EP)G4159 (D) mutant embryos. (E) Muscle phenotype observed upon the muscle-specific Pglym78 RNAi knockdown. Arrowheads point to unfused myoblasts. In all Pglym78 mutant contexts, muscles appear thinner than in wildtype. Lateral views of three abdominal hemisegments are shown, anterior to the left, dorsal up. (Scale bar: 30 µm.) (F, F′) Schematic of the observed muscular defects. (G) Enlarged view of DA1 and SBM muscles in wildtype and in the duf > Pglym78 RNAi context. Nuclear staining of DA1 and SBM corresponds to Eve and Lb staining, respectively (in red). Positions of DA1 and SBM muscles are shown in A (dashed rectangles). (H) Muscle size and number of nuclei of SBM and DA1 in wildtype and in the Pglym78 RNAi context. Bar graphs show the mean number of nuclei or the mean muscle size and error bars correspond to the SD. (I and J) Kinetics of fusion in SBM and DA1 in wild-type and in Pglym78 RNAi embryos. Mean values are used to visualize the number of nuclei according to time AEL. (K–L′) Appearance of actin foci (arrows) in developing muscles in ventro-lateral domain of stage-13 (9.5 h AEL) (K and K′) and stage-14 (11 h AEL) (L and L′) embryos. Representative hemisegments of control (duf > LacZ) (K and L) and mutant (duf > Pglym78 RNAi) (K′ and L′) embryos are shown for each developmental stage. (M) Bar graph showing the mean number of actin foci counted in 30 hemisegments in ventro-lateral area of control and Pglym78 RNAi contexts in stage-13 (9.5 h AEL) and stage-14 (11 h AEL) embryos. (N–Q′) Monitoring of M6-gapGFP–positive FCMs in stage-13 (9.5 h AEL) (N–O′) and stage-14 (11 h AEL) (P–Q′) embryos. Ventral area of a representative hemisegment from control (M6-gapGFP;duf > LacZ) (N–Q) and mutant (M6-gapGFP;duf > Pglym78 RNAi) (N′, O′, P′, and Q′) contexts are shown for each developmental stage. (R) The mean number of M6-gapGFP–positive FCMs counted in 30 hemisegments in ventral area of control and Pglym78 RNAi contexts are represented as a graph. Refer also to Table S3 for the mean values. Asterisks indicate significance of changes compared with the wildtype. ***P < 0.0001; **0.001 < P < 0.01; *0.01 < P < 0.05.
To get further insight into the role of Pglym78 in myoblast fusion we monitored the appearance of actin foci that are formed in the fusion-competent myoblasts (FCMs) and in muscle precursors to which FCMs are going to fuse and that are thought to be an obligatory step in myoblast fusion (14, 15). We first analyzed stage-13 embryos (9.5 h AEL) and found that in both control (duf > LacZ) and duf > Pglym78 RNAi conditions the FCMs get in contact with myotubes and that actin foci are present (arrows in Fig. 2 K and K′). However, at stage 14 (11 h AEL), at the time fusion arrest occurs in duf > Pglym78 RNAi embryos, the capacity to form actin foci is severely affected (Fig. 2 L, L′, and M and Table S3). Because actin accumulation is particularly prominent on the side of adhering FCMs (15) the loss of actin foci indicates a nonautonomous effect of glycolytic gene knockdown. It has been proposed by Gildor et al. (16) that a signal emitted by the myotubes might be responsible for Notch decay in FCMs, making them competent to form actin foci and fuse. We thus used a M6-gapGFP sensor line (17) to follow Notch activity in FCMs. At embryonic stage 13 (9.5 h AEL), the number of M6-gapGFP–positive FCMs monitored in the ventral region was similar in control (duf > LacZ) and in mutant (duf > Pglym78 RNAi) contexts (Fig. 2 N–O′and R and Table S3). In contrast, at stage 14 (11 h AEL) the number of M6-gapGFP–positive FCMs goes down in control embryos, whereas in Pglym78 RNAi embryos it remains similar to the number observed at stage 13 (9.5 h AEL). Altogether, this suggests that the fusion arrest in Pglym78 RNAi condition is due to loss of actin foci formation, which in FCMs can be correlated with inefficient Notch decay. This negative influence on Notch activity is consistent with previously reported data (18) but also suggests an altered emission of a so-far-unidentified signal by glycolysis deficient myotubes. To examine one candidate signal, we tested whether glycolytic gene knockdown leads to activation of Delta in myotubes, which would then maintain Notch in FCMs. We found that membrane-associated Delta is only present in unfused FCMs but not in myotubes (Fig. S2B). We also observed that myotube-targeted Delta expression does not influence the fusion process (Fig. S2C). Thus, it is unlikely that muscle-derived Delta is responsible for inefficient Notch decay in FCMs in a glycolysis-deficient context. Testing the influence of glycolysis on other signaling pathways that are required for muscle development and are able to negatively regulate Notch would provide further insight into the genetic cascade linking glycolysis and myoblast fusion. For example, the JAK/STAT pathway could be one promising candidate pathway that may contribute to this link (19, 20).
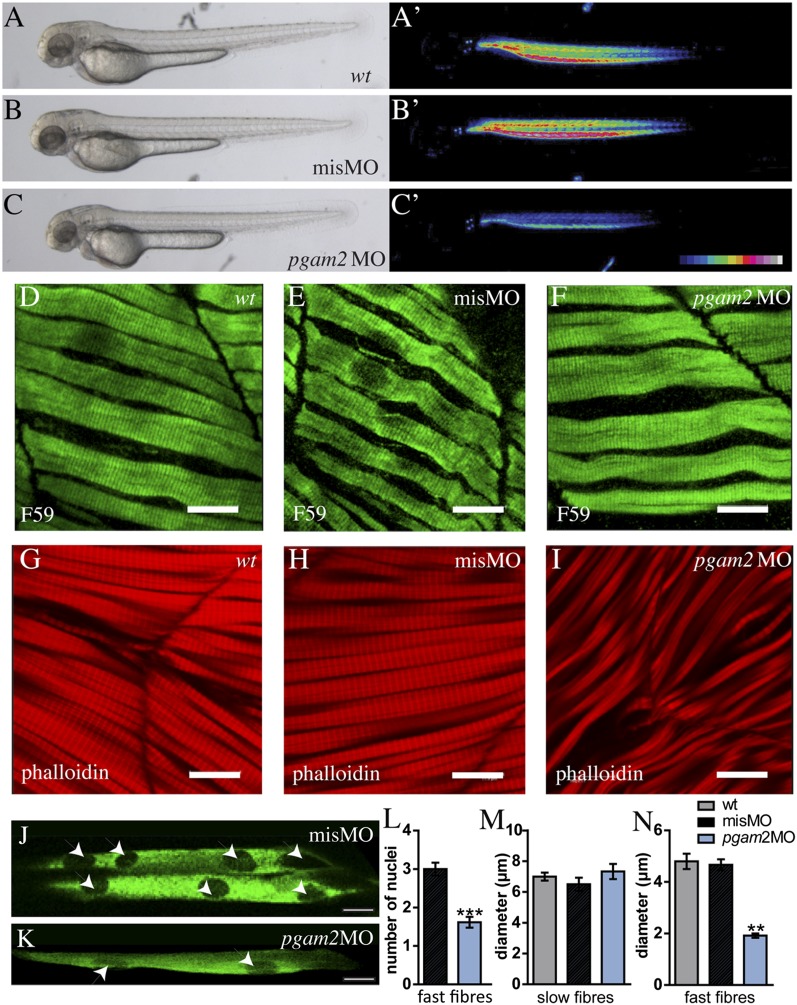
The identification of Pglym78 in our screen for conserved muscle genes prompted us to test whether its vertebrate ortholog, pgam2, plays a similar role in zebrafish muscle development. We attenuated expression of pgam2 by morpholino-mediated knockdown and found that, compared with control morpholino-injected embryos, pgam2 morphants exhibit strongly reduced birefringence (21), indicating a perturbation of muscle fiber structure or arrangement (Fig. 3 C and C′ compared with Fig. 3 B and B′ and Fig. S3A). Interestingly, the slow and fast muscle fibers were differentially affected. Phalloidin staining of syncytial fast muscle fibers revealed large gaps between the fibers and a reduction of their width (Fig. 3 G–I and N), associated with a reduced number of nuclei per fiber (Fig. 3 J–L). The overall number of fast fibers as well as of Pax7-positive myoblasts was significantly higher in pgam2 knockdown context compared with control condition (Fig. S3), indicating that the developmental muscle defects are related to inefficient fusion. In contrast, slow muscle fibers, which do not undergo fusion and contain only one nucleus per fiber (22), appear normal in the morphants (Fig. 3 D–F and M). Furthermore, pgam2 knockdown did not affect heart muscle, which does not form syncytia either, and heartbeat in the morphants was normal (Fig. S3). Together, these findings are consistent with a perturbation of myoblast fusion by attenuated pgam2 expression also in zebrafish embryos and suggest a conserved role for Pglym78/pgam2 in muscle development over large evolutionary distances.
Fig. 3.
Knockdown of pgam2 specifically affects developing fast muscle fibers in zebrafish embryos. (A–C′) Morpholino mediated pgam2 knockdown (pgam2 MO) leads to a decrease in muscle birefringence. (A–C) Bright-field images; A′–C′ are the same embryos as in A–C under polarized light. Intensity of birefringence is color-coded. misMO, mismatch control morpholino injected embryos. (D–I) A muscle phenotype characterized by disturbance of fiber arrangement and reduced fiber diameters is only observed in fast muscle (G–I) of zebrafish pgam2 morphants. (D–F) Slow muscle fibers stained with the F59 antibody (green). (G–I) Fast muscle stained with phalloidin (red). (Scale bars: 11.9 µm.) (J and K) Fast fibers stained with Unc45-b-GFP (green) to reveal position of nuclei (arrowheads). (L) The number of nuclei per fast muscle fiber is significantly reduced in pgam2 morphants (***P = 0.0001). (M and N) The diameter (micrometers) of fast fibers (N), but not of slow fibers (M), is significantly reduced in pgam2 morphants (**P = 0.0027). A–I, M, and N: 48 hours postfertilization (hpf) embryos; J–L: 72 hpf embryos.
Glycolytic Genes Are Synchronously Activated in Embryonic Drosophila Muscles and Required for Their Proper Formation.
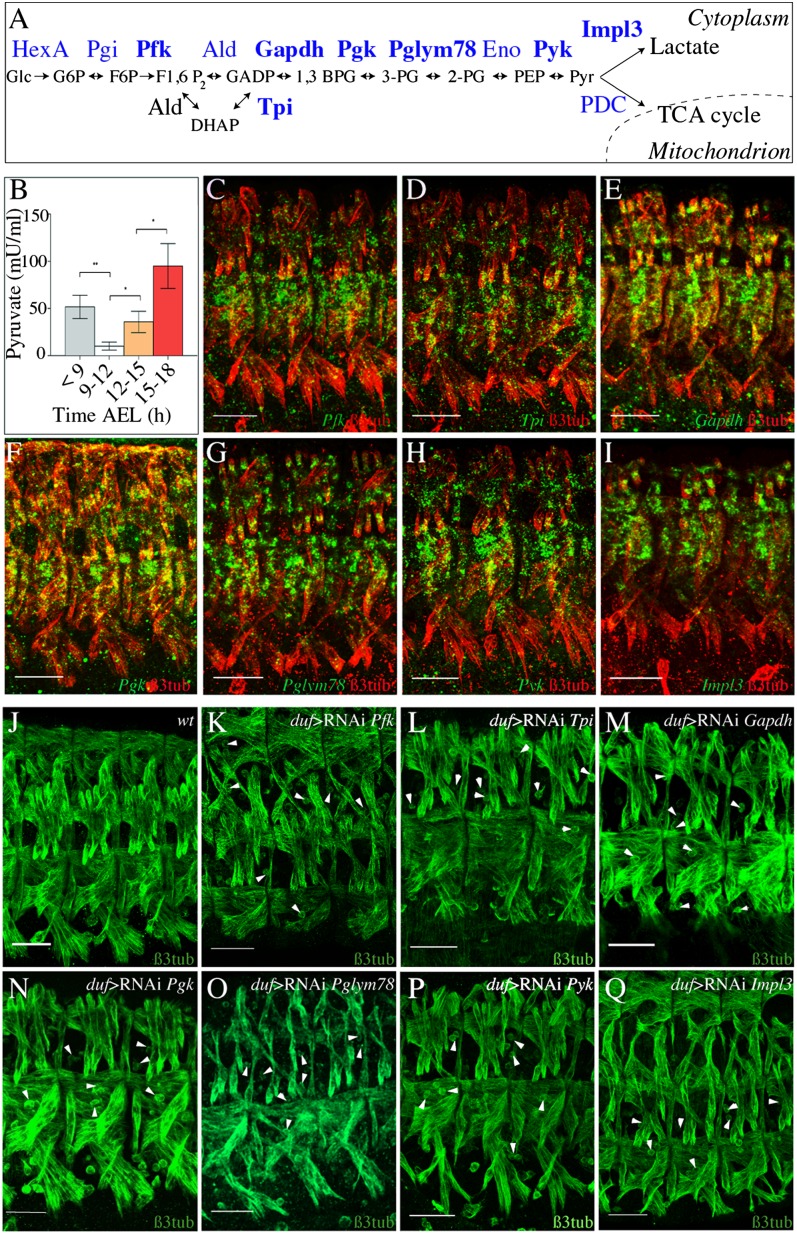
Among the candidates identified by our screen and involved in metabolism we found 14 other genes, in addition to Pglym78, that encode glucose or pyruvate metabolism enzymes (Table S1), strongly suggesting that these pathways may contribute to embryonic muscle development. To investigate this possibility we first analyzed the embryonic expression patterns of six of the glycolytic genes and of the Impl3 gene, which catalyzes the reduction of pyruvate into lactate (see scheme, Fig. 4A). We found that the transcripts of Pfk, Tpi, Gapdh1, Pgk, Pglym78, and PyK as well as that of Imlp3 accumulate specifically and synchronously in developing muscles starting from the embryonic stage 13 (9.5 h AEL) and that the expression of all these genes becomes predominantly muscular at the beginning of embryonic stage 15 (12 h AEL) (Fig. 4 C–I and Fig. S4). Consistent with this, enzymatic activity of PyK, the last enzyme in the glycolytic pathway (Fig. 4A), progressively increases in stage-15 to -16 embryos (12–15 h AEL) (Fig. 4B), providing further evidence that glycolysis is active in developing muscles. To examine the function of this muscle-specific expression of glycolytic genes in midembryogenesis we analyzed effects of loss of function of Pfk, Tpi, and Pyk (Fig. S5 A–C), as well as muscle-specific attenuation of Pfk, Tpi, Gapdh1, Pgk, and PyK (Fig. 4 J–P). In all cases analyzed we observed a moderately affected muscle pattern characterized by the presence of thinner muscles and unfused myoblasts—a phenotype highly reminiscent of that of Pglym78 RNAi embryos (compare with Fig. 4O and Fig. 2 B–E). Furthermore, attenuation of Impl3 (Fig. 4Q) induced similar muscle phenotypes, suggesting that the conversion of the glycolysis-derived pyruvate to lactate is one important metabolic pathway branch that promotes embryonic muscle development. In contrast, analyses of mutant alleles and RNAi knockdown contexts of the pyruvate dehydrogenase complex components CG11876 and CG5261 and of the respiratory chain component CoVa suggested that down-regulation of these genes does not affect the muscle pattern (Fig. S5 D–G′). Altogether, these findings indicate that the myogenic functions of the glycolytic pathway are uncoupled from the oxidative phosphorylation pathway.
Fig. 4.
Glycolytic genes are specifically expressed in developing muscles and required for proper muscle development. (A) Diagram representing the 10 steps of glycolysis. Genes in bold were functionally analyzed in this study. (B) Developmental profile of PyK activity during Drosophila embryogenesis. Error bars show SD. Asterisks indicate significance of changes between stages: *0.01 < P value< 0.05. (C–I) Expression profiles of glycolytic (C–H) and Impl3 (I) genes. Lateral views of three abdominal segments from stage-15 (13 h AEL) embryos double-stained for glycolytic gene transcripts (green) and for β-3 tubulin to reveal muscles (red). Note that all glycolytic genes and Impl3 show predominantly muscular expression. (J) Wild-type muscle pattern in three abdominal hemisegments as revealed by anti–β-3 tubulin staining on stage-15 (13 h AEL) embryo. (K–Q) Effects of muscle-specific RNAi knockdown of glycolytic genes and Impl3. Representative views of three abdominal hemisegments are shown. (Scale bars: 30 µm.) Arrowheads point to unfused myoblasts.
Insulin Signaling Positively Regulates Glycolysis and Together with the TOR Pathway Promotes Myoblast Fusion and Embryonic Muscle Growth.
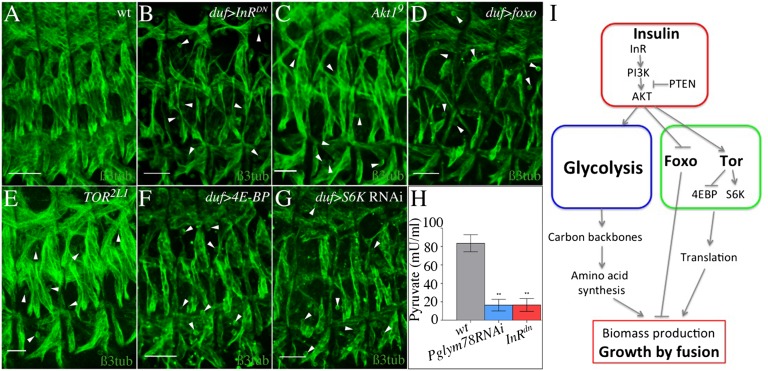
The observation that the attenuation of glycolytic genes results in reduced muscle size prompted us to ask whether their action is coordinated with pathways controlling cell size. The insulin pathway is the major cell-size regulator and is known to control glucose metabolism in vertebrates (23). We thus tested whether it also regulates glycolysis and controls the size of embryonic muscles in Drosophila. PyK activity is decreased in embryos expressing a dominant negative form of the insulin receptor (InRDN) in muscles (Fig. 5H), providing evidence for a role of the insulin pathway in promoting glycolysis. Moreover, the muscle-specific expression of InRDN results in a phenotype similar to those of loss of glycolytic genes (compare Fig. 5B with Fig. 4 K–P). The same is true for loss of Akt1 function (Fig. 5C) and for the muscle-specific expression of the negative regulator of the insulin pathway Pten (Fig. S6E), as well as for muscle-specific expression of the translational repressor foxo, an insulin target (Fig. 5D). The InRDN-induced patterning defects seem specific to somatic muscles and are not seen in other tissues composed of mononucleated cells such as cardiac, epidermal, and neural cells (Fig. S1 C–E). In addition, similar to Pglym78 RNAi, the duf > InRDN embryos show loss of actin foci and disturbed Notch decay in FCMs (Fig. S6 K–P). In contrast, muscle-targeted overexpression of positive regulators of the insulin pathway (namely, the wild-type insulin receptor form InRwt, Akt1, and PI3K) or loss of foxo function do not seem to influence muscle pattern (Fig. S6 B–D and H–J). Insulin is also known to control muscle growth via the regulation of TOR (24). Consistent with this, loss of function of TOR, muscle-specific gain of function of the negative TOR effector 4E-BP, or muscle-specific knockdown of S6K lead to InRDN-like thin muscle phenotypes (Fig. 5 E–G and Fig. S7 A–F and Table S2), whereas gain of function of TOR does not lead to muscular defects (Fig. S6F). Moreover, mutants of TORC2 component rictor have no muscle defects (Fig. S6 G and I), indicating that the phenotypes observed when TOR is attenuated specifically depend on TORC1 activity. These phenotypes do not seem to be a secondary consequence of energy depletion, because neither loss nor gain of function of the ATP sensor AMPK, which deactivates TOR signaling under energy stress, affects muscle development (Fig. S7 G–J). Examining the kinetics of myoblast fusion in InRDN, UAS-foxo, UAS-4E-BP, and UAS-S6K RNAi contexts (Fig. S7 E and F and Table S2) reveals, in all cases, a premature fusion arrest as previously observed in Pglym78 RNAi embryos (Fig. 2 I and J). The arrest temporally coincides with the muscle-specific burst of zygotic expression of glycolytic genes. Altogether, this suggests that both the glycolytic pathway and the TOR pathway, acting downstream of InR signaling, are required to complete the fusion programs in developing muscles (Fig. 5I).
Fig. 5.
The insulin pathway regulates glycolysis and is required for myoblast fusion. (A–G) Inhibition of the insulin pathway by expressing a dominant negative form of InR (InRDN) (B) or by mutating Akt1 (C) leads to a thin muscle phenotype. Muscles of reduced size are also detected in embryos with overexpression of foxo in muscles (D) or affected TOR pathway (E–G). Lateral views of three hemisegments from stage-15 (13 h AEL) embryos stained for β-3 tubulin are shown. (Scale bars: 30 µm.) Arrowheads point to unfused myoblasts. (H) PyK activity decreases strongly in embryos expressing InRDN (red) and in the Pglym78 RNAi context (blue). Error bars show SD. **0.001 < P value < 0.01. (I) Model: Insulin acts upstream of Glycolysis and Tor pathway to stimulate biomass production and muscle growth by fusion.
Discussion
One property that makes muscle cells different from other cell types is their syncytial character. In midembryogenesis, muscle precursors increase their size by fusing with surrounding fusion-competent myoblasts, but the metabolic requirements of rapidly growing muscle precursors during the fusion process have not yet been analyzed. Here we identify glycolytic genes as conserved embryonic muscle genes and report that they promote myoblast fusion in both Drosophila and zebrafish embryos.
The shift to glycolysis in developing muscles during midembryogenesis is reminiscent of the switch to aerobic glycolysis in rapidly proliferating cancer cells known as the Warburg effect (6). Even though developing muscle cells do not proliferate at that time, the fusion increases the number of nuclei and, thereby, the number of transcriptionally active genes per cell, which results in a higher demand for biomass production. The stimulation of both glycolysis (to provide building blocks such as amino acids or nucleotides) and the TOR pathway (to stimulate protein biosynthesis) together with the repression of Foxo (to reduce protein degradation) meets this demand (Fig. 5I). In embryos deficient for Pglym78, only the first fusion events occur. It is thus conceivable that a nutritional checkpoint exists in developing muscles, which leads to transcriptional activation of glycolytic genes and thereby promotes fusion. Importantly, this function of glycolysis seems conserved in vertebrates because in zebrafish embryos deficient for the Pglym78 ortholog the size of syncytial fast muscle fibers is reduced. Interestingly, the size of the mononucleated, nonfusing slow fibers remains normal, indicating that the metabolic requirements of muscle precursors that undergo fusion are different from those of mononucleated cells.
How the metabolic and developmental processes are interconnected in muscle precursors remains poorly understood. A recent screen for Notch regulators in Drosophila revealed that glycolysis negatively regulates the Notch pathway (18). Furthermore, decreasing Notch signaling in fusion competent myoblasts (FCMs) is a key event in making them competent to fuse (16), thereby linking the Notch pathway to myoblast fusion. Here, we provide evidence that targeted attenuation of the glycolytic gene Pglym78 in developing muscles leads to fusion arrest accompanied by loss of actin foci and inefficient Notch decay in FCMs. It is tempting to speculate that the metabolic state of developing muscles and Notch signaling are connected via the hypoxia-inducible factor-1α (Hif-1α), a key factor induced in metabolic stress conditions and able to intensify Notch signaling (25). However, whether the Hif-1α level increases in embryos deficient for glycolytic genes and whether it interacts with Notch in myoblasts remains to be investigated. Also, the myogenic functions of the JAK/STAT pathway (19) and its capacity to negatively regulate Notch (20) suggest it might be involved in linking glycolysis with myoblast fusion.
Regardless of the mechanism connecting metabolic and developmental pathways, our finding that Pglym78/pgam2 is required to ensure fusion-based muscle growth sheds light on the pathogenesis of pgam2-involving human myopathy (2). Our data suggest that the exercise intolerance observed in patients could result not only from the perturbed energy metabolism, as has been thought so far, but also from a perturbation of the fusion process that may affect the repair of muscle damage occurring upon exercise.
Altogether, our data provide evidence that setting metabolism to glycolysis-based, high-rate biomass production is part of the core myogenic program that promotes formation of syncytial muscles during development.
Materials and Methods
Detailed information about the fly stocks and zebrafish morpholino design and injections can be found in SI Materials and Methods. This section also includes all information on experimental procedures including dsRNA injection, in situ hybridization, immunostaining, pyruvate kinase activity measurements, staging embryos, nuclei counting, and muscle size measurement. Myoblast fusion events have been analyzed by visualizing actin foci.
Supplementary Material
Acknowledgments
We thank collaborating teams from the Myores network, S. Schiaffino (University of Padova), A. Vincent (Centre de Biologie du Développement, Toulouse), and B. D. Weger (Institute of Toxicology and Genetics, Karlsruhe) for stimulating discussions, and N. Allegre for technical assistance. This work was supported by a European FP6 grant to network of excellence Myores, by L’Agence Nationale de la Recherche Grant MYO-ID, la Fondation pour la Recherche Médicale (FRM) Grant Equipe FRM and Association Française Contre les Myopathies (AFM) grants (to K.J.) and by FP7 IP ZF-HEALTH and COST BM0804 EUFishBioMed (to U.S.). L.B. was supported by the FRM and AFM Grant MUSCLE-ID-PROPA. We are also thankful for support by the Helmholtz Society, the Deutsche Forschungsgemeinschaft, the AFM (T.D.) and the Studienstiftung des deutschen Volkes (M.W.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1301262110/-/DCSupplemental.
References
- 1. Taylor M (2006) Comparison of muscle development in Drosophila and vertebrates. Muscle Development in Drosophila, Intelligence Unit Series, ed Sink H (Landes Bioscience, Austin, TX), pp 169–190.
- 2.DiMauro S, Miranda AF, Olarte M, Friedman R, Hays AP. Muscle phosphoglycerate mutase deficiency. Neurology. 1982;32(6):584–591. doi: 10.1212/wnl.32.6.584. [DOI] [PubMed] [Google Scholar]
- 3.Servidei S, et al. Fatal infantile form of muscle phosphofructokinase deficiency. Neurology. 1986;36(11):1465–1470. doi: 10.1212/wnl.36.11.1465. [DOI] [PubMed] [Google Scholar]
- 4.García M, et al. Phosphofructo-1-kinase deficiency leads to a severe cardiac and hematological disorder in addition to skeletal muscle glycogenosis. PLoS Genet. 2009;5(8):e1000615. doi: 10.1371/journal.pgen.1000615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tennessen JM, Baker KD, Lam G, Evans J, Thummel CS. The Drosophila estrogen-related receptor directs a metabolic switch that supports developmental growth. Cell Metab. 2011;13(2):139–148. doi: 10.1016/j.cmet.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agathocleous M, et al. Metabolic differentiation in the embryonic retina. Nat Cell Biol. 2012;14(8):859–864. doi: 10.1038/ncb2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang T, Marquardt C, Foker J. Aerobic glycolysis during lymphocyte proliferation. Nature. 1976;261(5562):702–705. doi: 10.1038/261702a0. [DOI] [PubMed] [Google Scholar]
- 9.Lunt SY, Vander Heiden MG. Aerobic glycolysis: Meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 10.Johnson MT, Mahmood S, Patel MS. Intermediary metabolism and energetics during murine early embryogenesis. J Biol Chem. 2003;278(34):31457–31460. doi: 10.1074/jbc.R300002200. [DOI] [PubMed] [Google Scholar]
- 11.de Joussineau C, Bataillé L, Jagla T, Jagla K. Diversification of muscle types in Drosophila: Upstream and downstream of identity genes. Curr Top Dev Biol. 2012;98:277–301. doi: 10.1016/B978-0-12-386499-4.00011-2. [DOI] [PubMed] [Google Scholar]
- 12.Harvey RP. NK-2 homeobox genes and heart development. Dev Biol. 1996;178(2):203–216. doi: 10.1006/dbio.1996.0212. [DOI] [PubMed] [Google Scholar]
- 13.Kennerdell JR, Carthew RW. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell. 1998;95(7):1017–1026. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- 14.Haralalka S, et al. Asymmetric Mbc, active Rac1 and F-actin foci in the fusion-competent myoblasts during myoblast fusion in Drosophila. Development. 2011;138(8):1551–1562. doi: 10.1242/dev.057653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Onel SF, Renkawitz-Pohl R. FuRMAS: Triggering myoblast fusion in Drosophila. Dev Dyn. 2009;238(6):1513–1525. doi: 10.1002/dvdy.21961. [DOI] [PubMed] [Google Scholar]
- 16.Gildor B, Schejter ED, Shilo BZ. Bidirectional Notch activation represses fusion competence in swarming adult Drosophila myoblasts. Development. 2012;139(21):4040–4050. doi: 10.1242/dev.077495. [DOI] [PubMed] [Google Scholar]
- 17.Rebeiz M, Reeves NL, Posakony JW. SCORE: A computational approach to the identification of cis-regulatory modules and target genes in whole-genome sequence data. Site clustering over random expectation. Proc Natl Acad Sci USA. 2002;99(15):9888–9893. doi: 10.1073/pnas.152320899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saj A, et al. A combined ex vivo and in vivo RNAi screen for notch regulators in Drosophila reveals an extensive notch interaction network. Dev Cell. 2010;18(5):862–876. doi: 10.1016/j.devcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 19.Liu YH, et al. A systematic analysis of Tinman function reveals Eya and JAK-STAT signaling as essential regulators of muscle development. Dev Cell. 2009;16(2):280–291. doi: 10.1016/j.devcel.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 20.Flaherty MS, Zavadil J, Ekas LA, Bach EA. Genome-wide expression profiling in the Drosophila eye reveals unexpected repression of notch signaling by the JAK/STAT pathway. Dev Dyn. 2009;238(9):2235–2253. doi: 10.1002/dvdy.21989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Felsenfeld AL, Walker C, Westerfield M, Kimmel C, Streisinger G. Mutations affecting skeletal muscle myofibril structure in the zebrafish. Development. 1990;108(3):443–459. doi: 10.1242/dev.108.3.443. [DOI] [PubMed] [Google Scholar]
- 22.Simionescu A, Pavlath GK. Molecular mechanisms of myoblast fusion across species. Adv Exp Med Biol. 2011;713:113–135. doi: 10.1007/978-94-007-0763-4_8. [DOI] [PubMed] [Google Scholar]
- 23.Dimitriadis G, Mitrou P, Lambadiari V, Maratou E, Raptis SA. Insulin effects in muscle and adipose tissue. Diabetes Res Clin Pract. 2011;93(Suppl 1):S52–S59. doi: 10.1016/S0168-8227(11)70014-6. [DOI] [PubMed] [Google Scholar]
- 24.Manning BD, Cantley LC. AKT/PKB signaling: Navigating downstream. Cell. 2007;129(7):1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gustafsson MV, et al. Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev Cell. 2005;9(5):617–628. doi: 10.1016/j.devcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.