Abstract
The aim of this study was to investigate the effect of high-far-high-energy diet on cloned and non-cloned domestic pigs of both lean and obese phenotype and to evaluate if the lean cloned pigs had a lower inter-individual variation as compared with non-cloned pigs. The microbiota of colon and terminal ileum was investigated in cloned and non-cloned pigs that received a high-far-high-energy diet with either restricted or ad libitum access to feed, resulting in lean and obese phenotypes, respectively. The fecal microbiota of lean pigs was investigated by terminal restriction fragment length polymorphism (T-RFLP). The intestinal microbiota of lean and obese cloned and non-cloned pigs was analyzed by quantitative real time PCR and a novel high-throughput qPCR platform (Fluidigm). Principal component analysis (PCA) of the T-RFLP profiles revealed that lean cloned and non-cloned pigs had a different overall composition of their gut microbiota. The colon of lean cloned pigs contained relatively more bacteria belonging to the phylum Firmicutes and less from the phylum Bacteroidetes than obese cloned pigs as estimated by qPCR. Fluidigm qPCR results revealed differences in specific bacterial groups in the gut microbiota of both lean and obese pigs. Our results suggest that high-far-high-energy diet is associated with changes in the gut microbiota even in the absence of obesity. Overall, the cloned pigs had a different gut microbiota from that of non-cloned pigs. To our knowledge this is the first study to investigate the gut microbiota of cloned domestic pigs of lean and obese phenotype.
Keywords: cloned pigs, gut microbiota, high-fat–high-energy diet, inter-individual variation, obesity
Introduction
Gut microbiota has recently been the target of many investigation regarding different diseases and conditions such as obesity. Microbial changes in the gut has been proposed to be linked to many diseases and implicated to play a role in obesity and obesity related diseases. Animals are frequently used as models for human gut microbiota-obesity-related studies and therefore it is important that these animals have human-like physiology, in order to understand the role of gut microbiota in different diseases and conditions such as obesity. Rodents are commonly used as animal models for many experimental and interventional studies in relation to human diseases, even though pigs have more similarities to humans especially in regard to the physiology of their gastrointestinal track. Pigs are generally considered to be good animal models for studying human diseases, mainly due to their similar physiology.1 However, one factor in such experimental studies is the inter-individual variations between animals. Cloning of animals such as pigs may provide an animal model with smaller inter-individual variation than normally bred siblings, thereby enhancing experimental control and standardization with small number of animals resulting in acceptable statistical power. Cloning of pigs has been performed previously primarily by somatic cell nuclear transfer but this method often results in low numbers of cloned animals due to loss of embryos either during the gestation period or death of the piglets shortly after birth.2,3 It has been suggested that cloning gives rise to altered metabolic characteristics,4 skin abnormalities5,6 and other physiological defects which may limit the use of cloned animals as experimental models. However so far there is only one study that has explored the gut microbiota in obese cloned pigs.7
One of the ground breaking studies regarding the relation between gut microbiota and obesity was performed in germ-free mice8 which have been shown to be protected against diet-induced obesity.9 However, when they are colonized with the gut microbiota of conventionally raised mice, the germ-free mice show an increase in body-fat percent.8 These studies reveal that the composition of the gut microbiota is an important factor that affects energy storage in the body, possibly in part due to an increased energy-harvest by the gut microbiota. The gut microbiota seemingly influences the energy balance and may play a role in development of obesity.10 Furthermore, an obesity-related gut microbiota has been shown to be associated with a reduction in bacterial diversity.11 In high-fat (HF) diet-induced obesity, nutrient load presumably affects the composition of the microbiota as shown in mice,12 humans11 and pigs.7,13 This in turn causes changes in the dominance of one or more bacterial divisions, especially reducing the number of bacteria belonging to the phylum Bacteroidetes and increasing the number of bacteria belonging to the phylum Firmicutes. Both Bacteroidetes and Firmicutes produce short chain fatty acids (SCFA) from digestion of otherwise indigestible dietary compounds which in turn provide their host with extra energy.10,14 Moreover, there seems to be an association between obesity and an increase in Lactobacillus reuteri in humans, however the correlation between specific bacterial species and obesity still remains unclear.15 These studies together suggest that the gut microbiota in obese state extracts energy from the diet more efficiently than the gut microbiota in lean state. In the current study, we aimed to investigate if domestic cloned pigs provide good animal model and their use as an experimental platform in diet-intervention studies. Therefore in this study, the gut microbiota of lean cloned pigs was investigated to evaluate if the cloned pigs had smaller inter-individual variation than lean non-cloned pigs. Furthermore, we investigated the relationship between the intestinal microbiota and high-far-high-energy diet (HF/HE) with diet restriction in lean pigs and in obese pigs on the same diet but fed ad libitum.
Results
Weight and body-fat
All the pigs were fed a standard pig diet (regular diet) after weaning and were weighed just before the start of experimental diet (HF/HE diet). All the pigs were weighed while they received standard pig diet, just before the start of the diet-intervention study (baseline) and the cloned pigs of lean phenotype weighed 65.1 ± 7.4 kg (baseline; age: 22 weeks) and non-cloned pigs (age: 19 weeks) weighed 61.7 ± 1.4 kg. The pigs were subsequently fed a restricted HF/HE diet and were given 60% of the feed consumed by pigs fed ad libitum throughout the diet-intervention period. At the end of the experiment, the cloned pigs (n = 8) and non-cloned pigs (n = 9) on HF/HE restricted diet, weighed 127.1 ± 5.9 kg and 119.1 ± 3.2 kg, respectively.
In the obese group, all the pigs were fed a HF/HE diet ad libitum throughout the diet-intervention period. At the beginning of the experiment while the pigs received standard pig diet (baseline), the cloned pigs (age: 13 weeks) had an average weight of 38 ± 4.1 kg and the non-cloned pigs weighed 38 ± 2.3 kg. By the end of the diet-intervention experiment, the obese cloned pigs (n = 9) had an average weight of 147.5 ± 5.9 kg and obese non-cloned pigs (n = 10) weighed 170.1 ± 5.4 kg.
The weight of lean non-cloned pigs was significantly lower than that of the obese non-cloned pigs (p < 0.0001) and the same was observed for cloned pigs (p < 0.02). CT (CT) scans of the lean and obese pigs showed that the obese pigs both non-cloned and cloned, had a higher percentage of body-fat than the lean non-cloned pigs (p < 0.0004) and lean cloned pigs (p < 0.03) (Fig. 1).
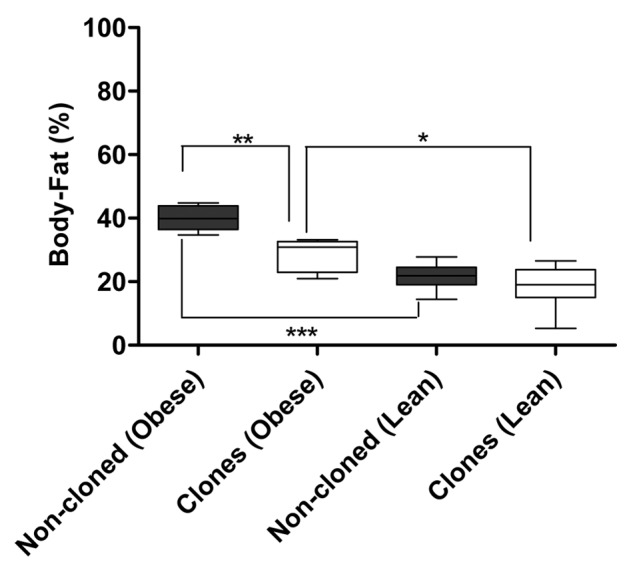
Figure 1. Percent body-fat in lean and obese, cloned and non-cloned pigs (statistics is performed by Mann-Whitney U test, * indicate significance for p < 0.05)
The fecal microbiota of lean cloned pigs and non-cloned pigs
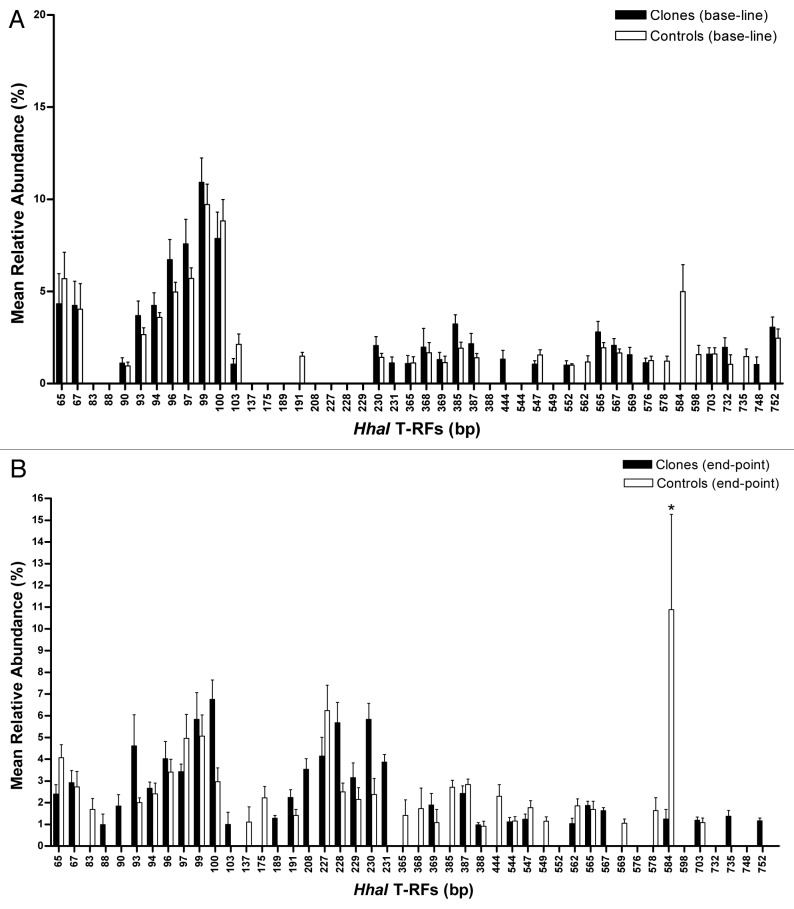
Terminal restriction fragment length polymorphism (T-RFLP) was used to profile the composition of the fecal microbiota and principal component analysis (PCA) of the most predominant terminal restriction fragments (T-RFs) (> 1%) revealed a difference between cloned and non-cloned pigs’ bacterial community in fecal microbiota at endpoint, after being on the HF/HE experimental diet (end of the diet intervention study) (Fig. 2). The bacterial diversity of the fecal microbiota was similar between lean cloned and non-cloned pigs when the pigs were on standard pig diet (baseline) and on HF/HE diet (endpoint), as estimated by the Shannon-Weaver index. Furthermore, there was no change in the diversity of the fecal microbiota determined every four weeks throughout the diet-intervention period in neither the lean cloned pigs nor the non-cloned lean pigs. A comparison of all the T-RFs (bacterial load) between the microbiota of the lean cloned and non-cloned pigs, did not show any significant difference at baseline when the pigs were still on standard pig diet (p < 0.7) (Fig. 3A). At endpoint however, one particular T-RF (584 bp) was significantly higher than all the other T-RFs in the non-cloned pigs (p < 0.03) (Fig. 3B) which according to in silico analysis may belong to the phylum Firmicutes. Based on relative abundance of all the T-RF base pairs, i.e., total bacterial abundance, no difference was observed in lean cloned and non-cloned pigs at the sampling time before the start of diet intervention when the pigs received standard pig diet or at the end of HF/HE diet-intervention period.
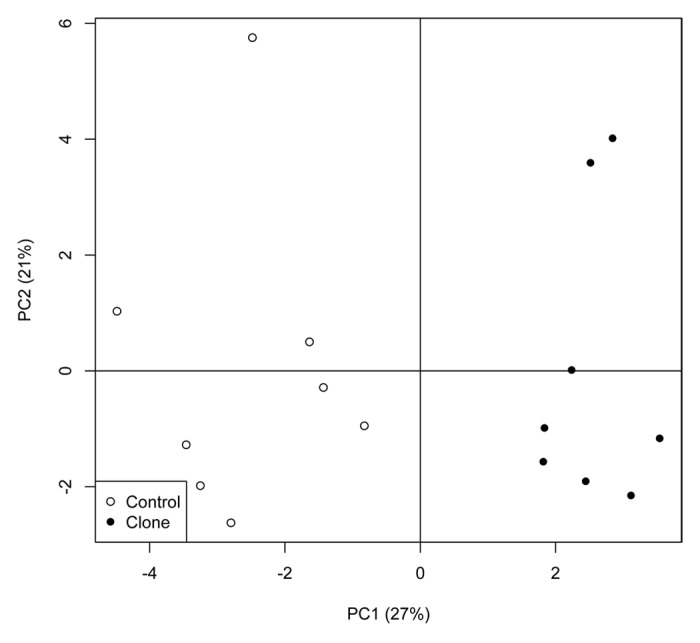
Figure 2. Principal component analysis (PCA) of the most predominant T-RFs in fecal samples from lean cloned pigs (●) and lean non-cloned pigs (controls) (○) at end of diet intervention period as obtained by T-RFLP method. Statistics is performed by Mann-Whitney U test, * indicate significance for p < 0.05 and the error bars represent standard error of mean (SEM).
Figure 3. Mean relative abundance of the most predominant T-RFs (> 1%) obtained by T-RFLP method in the fecal samples at baseline (A) and endpoint (B) in cloned pigs (■) and non-cloned pigs (□). The error bars represent standard deviation.
Several T-RFs’ with mean relative abundances of more than one percent were present in lean cloned pigs at baseline but absent at endpoint. There was a clear distinction between the individual T-RFs at baseline (on standard pig diet) (Fig. 3A) and endpoint (on HF/HE diet) in both lean cloned and lean non-cloned pigs (Fig. 3B). The relative abundance (> 1%) of T-RFs 93 bp to 100 bp at baseline was higher than their relative abundance at endpoint in both lean cloned pigs and lean non-cloned pigs. The subsequent in silico analysis of these T-RFs indicated that some of these T-RFs probably represent the bacteria belonging to the phylum Bacteroidetes.
Relative abundance of the phyla Firmicutes and Bacteroidetes varies between lean and obese pigs
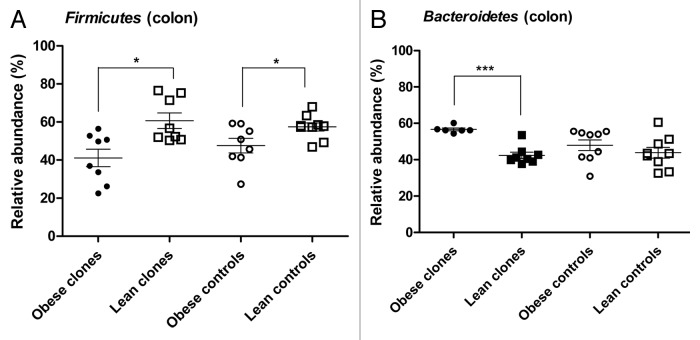
The relative abundance of Firmicutes and Bacteroidetes as obtained by quantitative real-time PCR (qPCR) performed on the microbiota of colon and terminal ileum at the end of the diet intervention study was different between lean and obese cloned and non-cloned pigs. The qPCR analysis of the microbiota in colon revealed that the lean cloned pigs had a higher relative abundance of Firmicutes (p < 0.01) (Fig. 4A) and a lower relative abundance of Bacteroidetes (p < 0.0007) than the obese cloned pigs (Fig. 4B). The lean non-cloned pigs had a higher relative abundance of Firmicutes in their colon than the obese non-cloned pigs (p < 0.02) (Fig. 4A). There was no difference in relative abundance of the phylum Bacteroidetes between lean and obese non-cloned pigs (Fig. 4B).
Figure 4. Relative abundance of Firmicutes (A) and Bacteroidetes (B) in colon of lean and obese, cloned pigs and non-cloned (control) pigs by qPCR. Obese cloned pigs (●), lean cloned pigs (■), obese non-cloned pigs (○) and lean non-cloned pigs (□). Statistics is performed by Mann-Whitney U test and significant differences are indicated by * for p < 0.05.
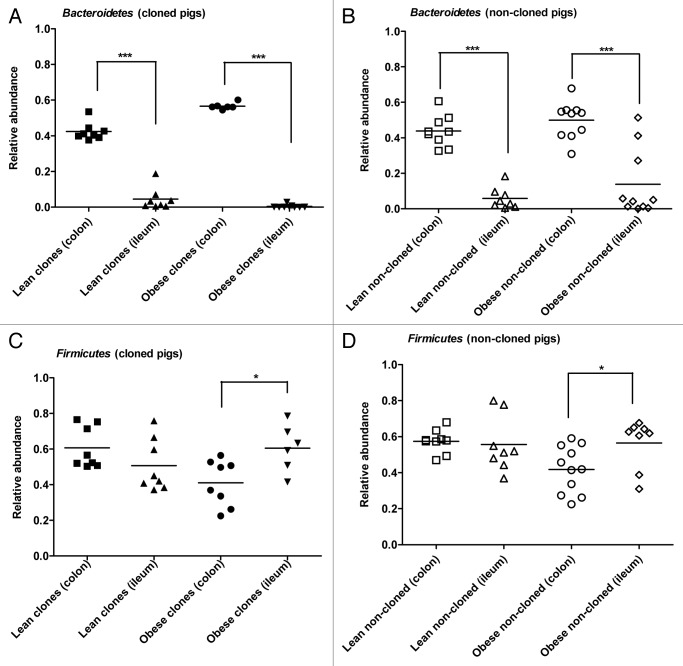
When comparing the microbiota of colon and terminal ileum, the relative abundance of Bacteroidetes in colon was significantly higher than in terminal ileum in both lean cloned pigs (p < 0.0002) and obese cloned pigs (p < 0.0004) (Fig. 5A). The same was observed in both lean non-cloned (p < 0.0001) and obese non-cloned pigs (p < 0.0003) (Fig. 5B).
Figure 5. Comparison of relative abundance of Firmicutes and Bacteroidetes between colon and terminal ileum of lean and obese pigs obtained by qPCR. Relative abundance of Bacteroidetes in cloned pigs (A) and non-cloned pigs (B). Relative abundance of Firmicutes in cloned pigs (C) and non-cloned pigs (D). Statistics performed by Mann-Whitney U test and significant differences are indicated by * for p < 0.05. (Lean clones’ colon [■], lean clones’ ileum [▲], obese clones’ colon [●], obese clones’ ileum [▼], lean non-cloned pigs’ colon [□], lean non-cloned pigs’ ileum (Δ), obese non-cloned pigs’ colon [○] and obese non-cloned pigs’ ileum [◊]).
There was no difference in the abundance of Firmicutes between colon and ileum microbiota of lean cloned pigs (Fig. 5C) and lean non-cloned pigs (Fig. 5D). In the obese cloned and obese non-cloned pigs, a higher abundance of Firmicutes was observed in terminal ileum (p < 0.03) than in colon (p < 0.02) (Fig. 5C-D).
The microbiota in terminal ileum and colon of lean and obese pigs obtained by high throughput qPCR
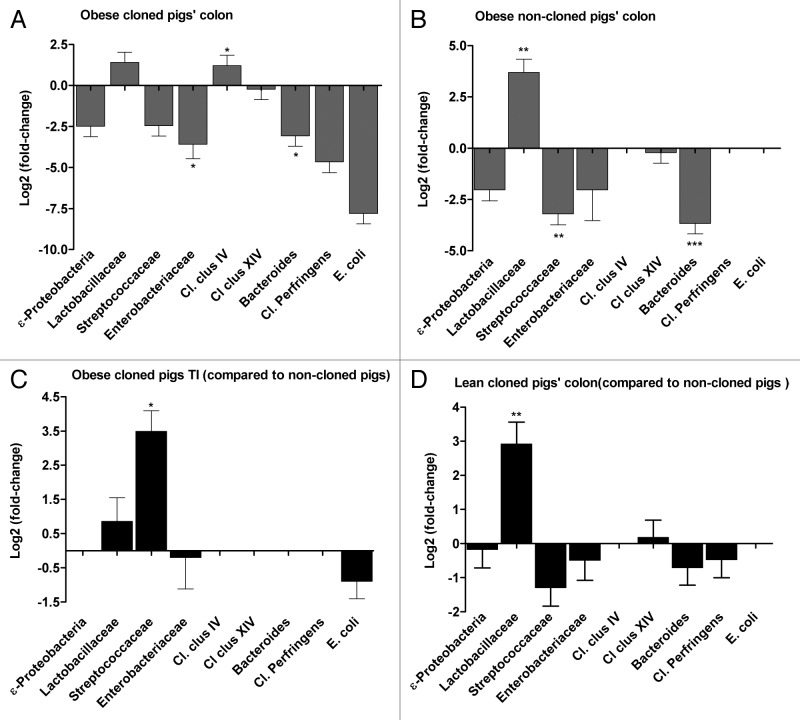
In colon samples from obese cloned pigs the relative abundance of Lactobacillaceae and Clostridium clusters IV (Cl. clus IV) were respectively 2.6- and 2.3-fold higher compared with lean cloned pigs, however only Cl. clus IV was significantly higher (p < 0.03) (Fig. 6A). Overall the relative abundances of ε-Proteobacteria, Streptococcaceae, Enterobacteriaceae (p < 0.04), Bacteroides (p < 0.03), Cl. perfringens and E. coli were lower in colon of obese cloned pigs as compared with colon of lean cloned pigs (Fig. 6A).
Figure 6. Comparison of selected bacterial groups in microbiota of cloned and non-cloned pigs by calculating the ratio (fold-differences) of the 16S rRNA gene of different bacteria at different phylogenetic levels. Obese vs. lean: obese cloned pigs’ colon (A), obese non-cloned pigs’ colon (B) (the fold-differences are compared against the lean animals in each group). Cloned vs. non-cloned pigs: obese cloned vs. obese non-cloned pigs’ terminal ileum (C), lean cloned vs. non-cloned pigs’ colon (D) (the fold-differences are compared against the non-cloned pigs). Statistics performed by Mann-Whitney U test and significant differences are indicated by * for p < 0.05, the error bars represent the standard deviation.
In colon of obese non-cloned pigs the relative abundance of Lactobacillaceae was 12.9-fold higher than lean non-cloned pigs (p < 0.001) (Fig. 6B). The relative abundance of Streptococcaceae (p < 0.003) and Bacteroides (p < 0.0007) were lower in colon of obese non-cloned pigs compared with the lean non-cloned pigs (Fig. 6B).
In terminal ileum of obese cloned pigs, the relative abundances of Streptococcaceae, Enterobacteriaceae and E. coli were respectively 6.3, 1.3 and 1.4 fold higher than in terminal ileum of lean cloned pigs. In terminal ileum of obese non-cloned pigs, the relative abundance of ε-Proteobacteria, Enterobacteriaceae and Cl. cluster XIV were 1.3, 1.5 and 2 fold higher than the lean non-cloned pigs, however these fold differences were not significant.
The relative abundance of Lactobacillaceae, Cl. Cluster IV, Cl. Cluster XIV and Cl. perfringens were 1.5, 1.4, 1.2, and 3.1 fold higher in the colon of obese cloned pigs as compared with the obese non-cloned pigs however these fold differences were not significant (data not shown). The relative abundance of Streptococcaceae was 11-fold higher (p < 0.04) in terminal ileum of obese cloned pigs compared with obese non-cloned pigs (Fig. 6C). The relative abundance of Lactobacillaceae was 3-fold higher (p < 0.008) in colon of lean cloned pigs as compared with lean colon of non-cloned pigs (Fig. 6D).
Discussion
In this study cloned pigs were evaluated for their use as experimental animal model for diet intervention studies and obesity. Pigs were cloned in order to reduce inter-individual variation between the pigs and to further elucidate the relation between HF/HE diet and the composition of the fecal microbiota of lean pigs. We found that the lean cloned pigs had a different overall fecal microbiota than the lean non-cloned pigs while they were on standard pig diet (baseline) and the same was observed at the end of the diet intervention study after being on HF/HE restricted-diet regiment. Based on the microbial profiles of the lean cloned and non-cloned pigs we did not find any evidence of smaller inter-individual variation among the cloned pigs as compared with non-cloned pigs which was also found in our previous study performed on the obese cloned and non-cloned pigs.7 One explanation for this could be that in the process of cloning, a single somatic cell nucleus is introduced to an enucleated oocyte containing the maternal mitochondria and therefore maternal mitochondrial effect could cause the inter-individual variation between the cloned pigs. Other studies have shown that the cloned animals have different phenotypic characters as compared with the non-cloned animals, such as skin abnormalities5 and other physiological defects.6,16 Therefore optimization of the cloning procedure is needed in order to establish the cloned pig as an ideal animal model.
Characterization of the gut microbiota in this study revealed that the bacterial diversity did not change from when the pigs received standard pig diet and throughout the HF/HE diet-intervention period in both cloned and non-cloned pigs of lean phenotype. This has been shown in a previous study where the overall bacterial diversity did not change over time while the individual bacterial divisions may change.17 The pig gut microbiota on normal diet has shown to be dominated by Firmicutes and Bacteroidetes, while Clostridiales, Bacteroidales, Spirochaetales, unclassified gammaproteobacteria and Lactobacillales were the most abundant bacterial orders.18 In a previous study we showed a lower relative abundance of Fimicutes in fecal microbiota of both cloned and non-cloned pigs on standard pig diet when compared with HF/HE diet,7 indicating an effect of diet on the gut microbial community. In the present study we observed differences in the bacterial community from the time the animals were on standard pig diet (baseline) compared with the end of the study, after the animals had received HF/HE diet, which indicates the effect of diet on gut microbiota.
When comparing the intestinal microbiota of lean and obese pigs in our study, we found that the lean cloned pigs had a higher relative abundance of the phylum Firmicutes and correspondingly less relative abundance of the phylum Bacteroidetes. The same observations were made in the lean non-cloned pigs as compared with obese non-cloned pigs. These findings are in contrast with the findings in mice,19 pigs20 and humans,21 where the lean individuals had higher abundance of Bacteroidetes compared with their obese counterparts. The lean animals in our study received HF/HE diet in restricted amounts and since they remained lean, the HF/HE diet could have affected the gut microbial community even in the absence of obesity and could explain the higher abundance of Firmicutes and lower abundance of Bacteroidetes. Furthermore, the T-RFLP data indicates that the gut microbiotas of both lean cloned and non-cloned pigs were different at baseline, when the pigs were on standard pig diet, as compared with the microbiota at endpoint when the pigs had received HF/HE diet. These results collectively suggest that HF/HE diet may affect the composition of the gut microbiota independent of weight or body-fat composition. Alterations in the gut microbial community upon switch to a HF diet in absence of obesity have been shown before22 and our observations are in agreement with these findings.
Many factors may explain the conflicting reports on the relative proportions of Firmicutes and Bacteroidetes in obese and lean state found between different studies. One factor may be the difference in test subject or animal models used in each study and/or different methodological approaches used in different studies to quantify bacterial proportions. In our study the composition of the gut microbiota in lean cloned pigs suggest a gut microbial profile that resembles that of the obese phenotype in regard to high relative abundance of Firmicutes and lower relative abundance of Bacteroidetes, perhaps due to the HF/HE diet. However since we did not have a control group that only received standard pig diet throughout the study period, further investigations are needed to support these findings.
When we investigated the microbiota of colon and terminal ileum in lean and obese, cloned and non-cloned pigs, we found an increase in representation of Bacteroidetes in colon as compared with terminal ileum in the obese cloned and obese non-cloned pigs, suggesting that terminal ileum has generally a lower abundance of Bacteroidetes than colon. However, the abundance of Firmicutes was higher in terminal ileum than colon in the obese pigs while this difference was not observed in the lean pigs, suggesting that HF/HE diet may result in shifts of the bacterial population along the intestinal tract.
The analysis of bacteria in lower phylogenetic levels revealed community differences in the gut microbiota between obese and lean pigs. The most noteworthy finding was the higher abundance of the Lactobacillaceae in obese non-cloned pigs with a 12.9-fold higher relative abundance compared with lean non-cloned pigs. There have been reports on high abundance of Lactobacillaceae in fecal microbiota of obese humans and our results are in agreement with these findings.15,23
We also observed higher relative abundances of the gram-negative bacteria; ε-Proteobacteria and Enterobacteriaceae in terminal ileum of obese pigs. Lipopolysaccharide of gram-negative bacteria is implicated in playing a role in a low grade inflammation observed in obese mice, called metabolic endotoxemia.24 The hypothesis is that the HF diet modulates the gut microbial community resulting in activation of the inflammatory cascade by the gut bacteria, causing metabolic endotoxemia.25,26 The fold increase in the mentioned gram-negative group of bacteria observed in obese pigs is an interesting finding and needs further investigation.
Taken together our results suggest that HF/HE diet results in changes in gut microbiota in several different phylogenetic groups of bacteria. Although many studies have revealed great insights into the obesity related gut microbiota, profiling of the obese and lean gut microbiota in different bacterial phylogenetic groups may provide the information that can be used to manipulate the gut microbial community in obese subjects that perhaps may reverse the metabolic disorders observed in obese subjects.
In conclusion, the overall composition but not the diversity of the fecal microbiota differed between lean cloned and lean non-cloned pigs. Based on gut microbial profiles of lean cloned pigs, we did not observe smaller biological variation among the lean cloned pigs as compared with lean non-cloned pigs. Together our results suggest that cloned pigs do not provide a better model for gut microbiota related studies due to similar levels of inter-individual variations and higher cost as compared with non-cloned pigs. Gut microbial analyses of the lean pigs however seems to support the hypothesis that HF/HE diet is associated with changes in the gut microbiota even in the absence of obesity. Finally, diet induced obesity caused changes in different phylogenetic groups of bacteria in the gut, specially an increase in Lactobacillaceae and several gram-negative bacteria in pigs.
Methods
Animals
The cloning experiments were performed as described previously27using donor cells obtained from cultured ear fibroblasts from a Danish Landrace (L) x Yorkshire (Y) (65% x 35%) sow. The cloned embryos were subsequently transferred surgically to surrogate sows (recipients) five to six days after cloning.4 Five surrogate sows gave birth to a total of 17 female cloned pigs, during a period of three years. The non-cloned pigs (n = 19) (75% L x 25% Y) were obtained from 6 sows after standard artificial insemination. The pigs were reared in the experimental stables at University of Aarhus (Tjele, Denmark) and the studies were approved by the Danish Animal Experimental Committee.
Experimental set up
Lean group
The cloned pigs (n = 8) and non-cloned pigs (n = 9) that were allocated as the lean group were delivered vaginally and the pigs were subsequently nursed by the sows for four weeks. After weaning all the pigs received a standard pig diet with an energy distribution of 18.5% protein, 7.9% fat, 72.4% carbohydrate, and 1.2% fiber, until the cloned pigs were 22 weeks old and the non-cloned pigs were 19 weeks old. After this period, the pigs were housed individually and fed a wheat-based high-far-high-energy (HF/HE) experimental diet consisting of 19.5% protein, 27% fat, 53% carbohydrates and 0.5% fiber,28 in restricted amounts for approximately 21 weeks. The lean group of pigs received the same diet as the pigs in the obese group (ad libitum) but restricted to 60% of the feed consumed by the ad libitum group.
Obese group
The pigs that were allocated as the obese group, consisted of cloned pigs (n = 9) and non-cloned pigs (n = 10). In this group, four cloned pigs and four non-cloned pigs were vaginally delivered while the remaining pigs were delivered by caesarean section. All pigs were nursed for four weeks and subsequently received a standard pig diet for the next nine weeks. The pigs were then housed individually and received a HF/HE diet and fed ad libitum for the following 20 weeks. All the pigs were euthanized after 24 h of fasting. The cloned pigs were euthanized at 39 weeks of age and the non-cloned pigs were euthanized at 40 weeks of age. At the end of the experiment, the body-fat composition of the pigs was measured by CT scanning.
Sampling and tissue collection
Baseline fecal samples were collected while the pigs were still on standard pig diet (one day before the pigs started on the experimental diet). After the start of diet intervention (HF/HE diet), samples were collected biweekly until the pigs were euthanized. The fecal samples taken at the end of the study are mentioned as endpoint samples throughout the manuscript. Tissue samples from terminal ileum and distal colon with content (digesta) were also collected at the end of the study when the animals were sacrificed. The samples were immediately frozen in liquid nitrogen and stored at -80°C for later analyses.
DNA extraction
DNA was extracted from the fecal samples taken every four week from lean cloned (n = 8) and non-cloned pigs (n = 9) starting at 22 weeks of age (baseline) until they were euthanized after the diet-intervention period (endpoint). The same DNA extraction method was used to extract DNA from digesta of distal colon and terminal ileum obtained at the end of the study from all the cloned pigs (n = 17) and the non-cloned pigs (n = 19).
The DNA was extracted from 200 mg fecal and digesta samples using the QIAamp DNA Stool Mini Kit (QIAamp DNA Stool Mini Kit, Qiagen) according to manufacturer’s instructions. An additional bead beating step for 2 min was added in order to disrupt the cell wall of Gram-positive bacteria. DNA concentrations were measured by spectrophotometer (NanoDrop Technologies).
The analysis of fecal microbiota by T-RFLP
The microbiota in fecal samples from cloned pigs (n = 8) and non-cloned pigs (n = 9) in the lean group was characterized by terminal restriction fragment length polymorphism (T-RFLP) as described previously.29 The extracted DNA samples were diluted in nuclease-free water to obtain a final concentration of 5 ng µl−1 and a PCR was performed in duplicates as described previously.29 The primer set that targeted most bacterial 16S rRNA gene was Eub-8fm (5′- AGAGTTTGATCMTGGCTCAG- 3′) labeled with 5′FAM and Eub-926r (5-’CCGTCAATTCCTTTRAGTTT- 3′) (DNA Technology, Aarhus, Denmark).29 The PCR mix of a total of 50 µl per sample consisted of 5 µl Fermentas Taq-buffer, 4 µl MgCl2 (25 mM), 2.0 µl (10 µM) deoxyribonucleotide triphosphate (dNTP), 0.5 µl Fermentas Taq polymerase (2.5 µl−1), 0.5 µl of each primer (20 µM), 35.5 µl nuclease-free water and 2 µl DNA sample. The cycling conditions were: an initial denaturing step at 94°C for 6 min followed by 32 denaturing cycles at 94°C for 45 s, annealing at 56°C for 45 s, an extension step at 72°C for 2 min, and a final extension at 72°C for 10 min. The PCR products were then verified by gel electrophoresis adding 10 µl sample per well. The PCR products were purified using the High Pure PCR Purification Kit (Roche Applied Sciences) and 200 ng of this PCR product was digested with 2.0 µl of the restriction enzyme HhaI (Promega Corporation) at 37°C for 3 h. A mixture of the digested PCR products (2 µl), formamide (10 µl) and 0.50 µl Megabase ET900-R size standard (GE Health Care, Buckinghamshire, UK) was subsequently run in duplicates on a capillary electrophoresis genetic analyzer (Applied Biosystems Genetic Analyzer 3130/3130xl).
The terminal restriction fragments (T-RFs) were analyzed using the BioNumerics software (Applied Maths). The T-RF profiles were aligned against an internal standard. T-RF fragments with difference less than two base pairs were considered identical. The bands present in both duplicates were accepted as bacterial fragments and the duplicate with the best intensity was chosen for microbial profiling. T-RF profiles were subsequently identified in silico using the MiCA online software30 and 16S rRNA gene sequences were assigned to their taxonomic names on the Ribosomal Database project.31 All the data was transferred to Microsoft Excel for further analysis. In Microsoft Excel, the intensities less than 50 were removed and the relative intensity of a given T-RF in the range of 60–800 bp, was calculated by relating the sum of the intensity of a given T-RF in the entire group of pigs to that of the total intensity of all T-RFs in the samples. For both graphical presentations and principal component analysis (PCA), only the predominant T-RFs with a mean relative intensity above 1% were considered.
Statistical analysis of T-RF profiles
The T-RFs between 60 and 800 bp were imported into the statistical software programs Stata 11.0 (StataCorp) and Unscrambler version 9.8 (CAMO). Comparisons of group differences in the overall microbial communities between cloned and non-cloned pigs at the different sampling points were investigated with PCA. In these models, the T-RFs were standardized (centered and 1/SD) prior to the modeling phase to ensure that all the T-RFs equally influence the models, and possible outliers were inspected visually and with Hotelling T2. The Shannon-Weaver index of diversity (H’) was used to estimate the diversity of the bacterial fragments as described previously32 and was calculated based on all of the initial T-RFs. At each of the sampling points, comparisons of cloned and non-cloned pigs with respect to this index were evaluated using the Mann Whitney U-test. Group differences were considered statistically significant when p < 0.05.
Relative quantification of Bacteroidetes and Firmicutes
Quantitative real time PCR (qPCR) was performed on 16S rRNA gene DNA extracted from terminal ileum and distal colon content for quantification of the relative abundance of the phyla Bacteroidetes and Firmicutes. The annealing temperature and MgCl2 were optimized for the thermocycler used in this study (Rotor-Gene Q Real Time PCR cycler [Qiagene]).The Pure cultures of three bacteria; Clostridium perfringens (NCTC 8449), Odoribacter splanchnicus (isolate DJF_B089) and Escherichia coli (ATCC 25922) were used to establish standard curves for each primer set. 10-fold serial dilutions of all DNA samples were made from 2.5 × 102 ng µL−1 to 2.5 × 10−6 ng µL−1. Furthermore, standard curves were constructed from 10-fold serial dilutions of DNA extracted from two random samples obtained from the colon and terminal ileum content of cloned and non-cloned pigs to verify if PCR inhibitors were present in extracted DNA.
qPCR primers and conditions
For qPCR the phyla specific primers by Bacchetti De Gregoris et al. (2011)33 were used. A universal primer set with an amplicon length of 147 bp (S-D-Bact-0907-a-S-20 5′-AAACTCAAAGGAATTGACGG-3′; S-D-Bact-1054-a-A-20 5-’ ACGAGCTGACGACAGCCATG-3′)34 was used to detect all bacteria. The specific primer sets were: Bacteroidetes primer set with an amplicon length of 240 bp (798cfbF 5′ CRAACAGGATTAGATACCCT’3 and cfb967R 5′ GGTAAGGTTCCTCGCGTAT ‘3) and Firmicutes primer set with an amplicon length of 200 bp (928F-Firm 5′ TGAAACTYAAAGGAATTGACG ‘3; 1040firmR, 5′ ACCATGCACCACCTGTC ‘3).33 The qPCR reactions contained 12.5 µl of SYBR® Green JumpStart™ Taq ReadyMix™ without MgCl2 (Sigma-Aldrich, Saint Louis, 3050 MO, USA), 0.3 µmol l−1 of each primer and 5 µl template DNA adjusted to 5 ng µL−1. MgCl2 concentration was optimized and a final concentration of 2.5 mM MgCl2 per reaction was chosen. All PCR conditions were subsequently verified by conventional PCR and gel electrophoresis. A non template control (NTC) was included in all runs to detect contamination and DNA artifacts. At the end of each run a melting curve analysis was performed for each individual sample to detect presence of primer-dimers.
qPCR was performed with an initial denaturing step of 10 min at 95°C, 95°C for 30 s, 35 cycles of 56°C for 20 s and an elongation step of 72°C for 20 s. The threshold cycle (Ct) and the bacterial enumeration (DNA copies [µl−1]) were determined automatically by Rotor Gene software (Rotor-Gene Q 2.0.2 (Qiagene)).
A standard curve was constructed for each primer set from serial dilution of extracted DNA from specific bacteria: C. perfringens, O. splanchnicus and E. coli. The relative abundance of the 16S rRNA gene from the bacteria belonging to phylum Bacteroidetes and the phylum Firmicutes was calculated against the 16S rRNA gene obtained from all bacteria and the relative abundance of bacteria from the two phyla in each pig was subsequently calculated. Group differences were investigated with Mann Whitney U-test and the correlation analyses were performed by the Spearman rank-order correlation in GraphPad prism version 5.00 for Windows (GraphPad software). A statistical significance was considered when p < 0.05.
Ileal and colonic microbial identification by 48.48 dynamic array
High throughput quantitative real time PCR (qPCR) was performed using a 48.48 Dynamic Array Integrated Fluidic Circuits (Fluidigm, San Francisco, CA USA) which combines 24 primers (in duplicates) with 48 samples to run 2304 simultaneous qPCR reactions. This Fluidigm chip was designed by Hermann-Bank et al. 2013 (manuscript in publication*) in which the primer sets target the small subunit rRNA genes (16S or 23S) of the mammalian intestinal microbiota at various taxonomic levels (Domain, Phyla, class, Family, Genus and Species level, respectively) and normalizes each group according to a universal primer set. Phyla included are: Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria. After each qPCR run, Fluidigm Real-Time PCR Analysis software (Fluidigm) generates a heatmap of Ct values constructed as 48 × 48 small squares presenting all the 2304 primer sample combinations, for comparison of bacterial profiles across the samples included in the present study. High primer specificity of the Microbiotassay has been confirmed first by in silico analysis and second by sequencing of purified sample amplicons from each of the 24 primer assays (a 454 GS FLX Titanium Sequencer (Roche). Serial dilutions of DNA extracted from a wide range of reference bacteria has previously been used to confirm a wide dynamic range as well as a high specificity of the gut microbiotassay (manuscript in publication). The samples consisted of 20 µM forward and reverse primers, 1× assay loading reagent (Fluidigm, PN85000746), 1x low EDTA TE buffer (VWR, APLIA8569.0500) and master mix consisting of: 20× DNA binding dye sample loading reagent (Fluidigm, PN 100–0388), 20× EvaGreen DNA binding dye (Biotium, PN 31000) and 2× Taqman master mix (Applied Biosystems). The concentrations of 16S rRNA gene were optimized and subsequently samples were adjusted to a concentration of 50 ng µl−1. In each chip a non template control (NTC) was included to detect any contamination or non-specific amplification. For more details on qPCR reaction mix and cycling conditions as well as all 24 primer sets we refer to (manuscript in publication*).The obtained Ct values were subsequently exported to Microsoft Excel for further analysis. The relative proportion of bacteria representing each taxon was calculated based on the Livak method.35
Hence, the relative quantifications of the qPCR signal of the target 16S rRNA gene in the obese pigs was related to that of the lean pigs which were considered to harbor the reference composition of the gut microbiota. Similarly, in comparisons between cloned and non-cloned pigs, the latter were considered to be the reference. Fold differences in the different bacterial groups were subsequently calculated by 2-ΔΔCt and log2-transformed for visual appraisal in the figures. All the statistical analysis and figure arts were performed on GraphPad prism version 5.00 for Windows (GraphPad software).
Acknowledgments
We would like to thank Sophia Rasmussen and Thomas Pihl for their excellent technical assistance. This work was supported by a grant from the Danish Strategic Research Council (FØSU 2101–06–0034).
Authors’ contributions
RP performed the experimental work, data and statistical analysis and wrote the manuscript. MB contributed in designing the experimental procedures. ADA performed the statistical analysis on the T-RFLP (Shannon-Weaver and PCA) and JS designed and conducted the diet-intervention experiments. RP, MB, ADA and JS contributed in editing the manuscript and approved the final manuscript. ML provided us with the microfluidic dynamic Chip primer set up.
Submitted
04/11/2013
Revised
08/06/2013
Accepted
08/10/2013
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/gutmicrobes/article/26108
References
- 1.Litten-Brown JC, Corson AM, Clarke L. Porcine models for the metabolic syndrome, digestive and bone disorders: a general overview. Animal. 2010;4:899–920. doi: 10.1017/S1751731110000200. [DOI] [PubMed] [Google Scholar]
- 2.Whitworth KM, Li R, Spate LD, Wax DM, Rieke A, Whyte JJ, Manandhar G, Sutovsky M, Green JA, Sutovsky P, et al. Method of oocyte activation affects cloning efficiency in pigs. Mol Reprod Dev. 2009;76:490–500. doi: 10.1002/mrd.20987. [DOI] [PubMed] [Google Scholar]
- 3.Park MR, Cho SK, Lee SY, Choi YJ, Park JY, Kwon DN, Son WJ, Paik SS, Kim T, Han YM, et al. A rare and often unrecognized cerebromeningitis and hemodynamic disorder: a major cause of sudden death in somatic cell cloned piglets. Proteomics. 2005;5:1928–39. doi: 10.1002/pmic.200401079. [DOI] [PubMed] [Google Scholar]
- 4.Clausen MR, Christensen KL, Hedemann MS, Liu Y, Purup S, Schmidt M, Callesen H, Stagsted J, Bertram HC. Metabolomic phenotyping of a cloned pig model. BMC Physiol. 2011;11:14. doi: 10.1186/1472-6793-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hwang KC, Cho SK, Lee SH, Park JY, Kwon DN, Choi YJ, Park C, Kim JH, Park KK, Hwang S, et al. Depigmentation of skin and hair color in the somatic cell cloned pig. Dev Dyn. 2009;238:1701–8. doi: 10.1002/dvdy.21986. [DOI] [PubMed] [Google Scholar]
- 6.Tian XC, Park J, Bruno R, French R, Jiang L, Prather RS. Altered gene expression in cloned piglets. Reprod Fertil Dev. 2009;21:60–6. doi: 10.1071/RD08214. [DOI] [PubMed] [Google Scholar]
- 7.Pedersen R, Andersen AD, Mølbak L, Stagsted J, Boye M. Changes in the gut microbiota of cloned and non-cloned control pigs during development of obesity: gut microbiota during development of obesity in cloned pigs. BMC Microbiol. 2013;13:30. doi: 10.1186/1471-2180-13-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004;101:15718–23. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bäckhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci U S A. 2007;104:979–84. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jumpertz R, Le DS, Turnbaugh PJ, Trinidad C, Bogardus C, Gordon JI, Krakoff J. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am J Clin Nutr. 2011;94:58–65. doi: 10.3945/ajcn.110.010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–4. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102:11070–5. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pedersen R, Ingerslev HC, Sturek M, Alloosh M, Cirera S, Christoffersen BØ, Moesgaard SG, Larsen N, Boye M. Characterisation of gut microbiota in Ossabaw and Göttingen minipigs as models of obesity and metabolic syndrome. PLoS One. 2013;8:e56612. doi: 10.1371/journal.pone.0056612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwiertz A, Taras D, Schäfer K, Beijer S, Bos NA, Donus C, Hardt PD. Microbiota and SCFA in lean and overweight healthy subjects. Obesity (Silver Spring) 2010;18:190–5. doi: 10.1038/oby.2009.167. [DOI] [PubMed] [Google Scholar]
- 15.Million M, Maraninchi M, Henry M, Armougom F, Richet H, Carrieri P, Valero R, Raccah D, Vialettes B, Raoult D. Obesity-associated gut microbiota is enriched in Lactobacillus reuteri and depleted in Bifidobacterium animalis and Methanobrevibacter smithii. Int J Obes (Lond) 2012;36:817–25. doi: 10.1038/ijo.2011.153. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Archer GS, Dindot S, Friend TH, Walker S, Zaunbrecher G, Lawhorn B, Piedrahita JA. Hierarchical phenotypic and epigenetic variation in cloned swine. Biol Reprod. 2003;69:430–6. doi: 10.1095/biolreprod.103.016147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–3. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 18.Lamendella R, Domingo JW, Ghosh S, Martinson J, Oerther DB. Comparative fecal metagenomics unveils unique functional capacity of the swine gut. BMC Microbiol. 2011;11:103. doi: 10.1186/1471-2180-11-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy EF, Cotter PD, Healy S, Marques TM, O’Sullivan O, Fouhy F, Clarke SF, O’Toole PW, Quigley EM, Stanton C, et al. Composition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models. Gut. 2010;59:1635–42. doi: 10.1136/gut.2010.215665. [DOI] [PubMed] [Google Scholar]
- 20.Guo X, Xia X, Tang R, Zhou J, Zhao H, Wang K. Development of a real-time PCR method for Firmicutes and Bacteroidetes in faeces and its application to quantify intestinal population of obese and lean pigs. Lett Appl Microbiol. 2008;47:367–73. doi: 10.1111/j.1472-765X.2008.02408.x. [DOI] [PubMed] [Google Scholar]
- 21.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hildebrandt MA, Hoffmann C, Sherrill-Mix SA, Keilbaugh SA, Hamady M, Chen Y-Y, Knight R, Ahima RS, Bushman F, Wu GD. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137:1716–24, e1-2. doi: 10.1053/j.gastro.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Armougom F, Henry M, Vialettes B, Raccah D, Raoult D. Monitoring bacterial community of human gut microbiota reveals an increase in Lactobacillus in obese patients and Methanogens in anorexic patients. PLoS One. 2009;4:e7125. doi: 10.1371/journal.pone.0007125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761–72. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 25.Creely SJ, McTernan PG, Kusminski CM, Fisher M, Da Silva NF, Khanolkar M, Evans M, Harte AL, Kumar S. Lipopolysaccharide activates an innate immune system response in human adipose tissue in obesity and type 2 diabetes. Am J Physiol Endocrinol Metab. 2007;292:E740–7. doi: 10.1152/ajpendo.00302.2006. [DOI] [PubMed] [Google Scholar]
- 26.Cani PD, Delzenne NM, Amar J, Burcelin R. Role of gut microflora in the development of obesity and insulin resistance following high-fat diet feeding. Pathol Biol (Paris) 2008;56:305–9. doi: 10.1016/j.patbio.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 27.Kragh PM, Vajta G, Corydon TJ, Purup S, Bolund L, Callesen H. Production of transgenic porcine blastocysts by hand-made cloning. Reprod Fertil Dev. 2004;16:315–8. doi: 10.1071/RD04007. [DOI] [PubMed] [Google Scholar]
- 28.Christensen KL, Hedemann MS, Jørgensen H, Stagsted J, Knudsen KE. Liquid chromatography-mass spectrometry based metabolomics study of cloned versus normal pigs fed either restricted or ad libitum high-energy diets. J Proteome Res. 2012;11:3573–80. doi: 10.1021/pr201253h. [DOI] [PubMed] [Google Scholar]
- 29.Mølbak L, Johnsen K, Boye M, Jensen TK, Johansen M, Møller K, Leser TD. The microbiota of pigs influenced by diet texture and severity of Lawsonia intracellularis infection. Vet Microbiol. 2008;128:96–107. doi: 10.1016/j.vetmic.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 30.Shyu C, Soule T, Bent SJ, Foster JA, Forney LJ. MiCA: a web-based tool for the analysis of microbial communities based on terminal-restriction fragment length polymorphisms of 16S and 18S rRNA genes. Microb Ecol. 2007;53:562–70. doi: 10.1007/s00248-006-9106-0. [DOI] [PubMed] [Google Scholar]
- 31.Maidak BL, Cole JR, Lilburn TG, Parker CT, Jr., Saxman PR, Farris RJ, Garrity GM, Olsen GJ, Schmidt TM, Tiedje JM. The RDP-II (Ribosomal Database Project) Nucleic Acids Res. 2001;29:173–4. doi: 10.1093/nar/29.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andersen AD, Mølbak L, Michaelsen KF, Lauritzen L. Molecular fingerprints of the human fecal microbiota from 9 to 18 months old and the effect of fish oil supplementation. J Pediatr Gastroenterol Nutr. 2011;53:303–9. doi: 10.1097/MPG.0b013e31821d298f. [DOI] [PubMed] [Google Scholar]
- 33.Bacchetti De Gregoris T, Aldred N, Clare AS, Burgess JG. Improvement of phylum- and class-specific primers for real-time PCR quantification of bacterial taxa. J Microbiol Methods. 2011;86:351–6. doi: 10.1016/j.mimet.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 34.Leser TD, Amenuvor JZ, Jensen TK, Lindecrona RH, Boye M, Møller K. Culture-independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl Environ Microbiol. 2002;68:673–90. doi: 10.1128/AEM.68.2.673-690.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]