Abstract
Vaccinia virus contains ~200 genes classified temporally as early, intermediate or late. We analyzed 53 intermediate promoters to determine whether any have dual late promoter activity. Our strategy involved (i) construction of a cell line that stably expressed the three late transcription factors, (ii) infection with a vaccinia virus mutant that expresses RNA polymerase but neither intermediate nor late transcription factors, and (iii) transfection with plasmids containing a luciferase reporter regulated by an intermediate promoter. After confirming the specificity of the system for late promoters, we found that many intermediate promoters had late promoter activity, the strength of which correlated with a TAAAT at the initiator site and T-content from positions −12 to −8 of the coding strand. In contrast, intermediate promoter activity correlated with the A-content from positions −22 to −14. The sequence correlations were confirmed by altering the specificities of strict intermediate and late promoters.
Keywords: Poxvirus transcription, mRNA synthesis, consensus promoter sequence, regulation of gene expression
Introduction
Poxviruses comprise a large family of DNA viruses that are noted for their medical importance, usefulness as vaccine vectors and ability to replicate entirely in the cytoplasm (Moss, 2013). Replication outside of the nucleus is attributed in large part to poxvirus-encoded proteins for genome replication and transcription. Gene expression is temporally programmed to occur in three consecutive stages: early, intermediate and late (Baldick and Moss, 1993). Transcription is mediated by a multisubunit DNA-dependent RNA polymerase and stage-specific transcription factors that recognize cognate promoters (Moss, 2013). Although characterized primarily for vaccinia virus (VACV), the transcription system is conserved in all chordopoxviruses. Early genes are transcribed with the aid of proteins that are synthesized late in infection and packaged within infectious virus particles (Broyles and Fesler, 1990; Broyles et al., 1988; Gershon and Moss, 1990). Intermediate and late genes are transcribed following genome replication and require the successive synthesis of intermediate and late transcription factors, encoded by early and intermediate genes, respectively (Keck et al., 1990; Sanz and Moss, 1999). Roles for specific cellular proteins in intermediate and late gene expression have also been reported (Katsafanas and Moss, 2004; Knutson et al., 2006; Knutson et al., 2009; Rosales et al., 1994; Wright et al., 2001).
VACV genes that are expressed early are readily recognized by their transcription in the presence of inhibitors of protein and DNA synthesis. Based on these criteria, we classified the VACV transcripts for 118 open reading frames (ORFs) as early and 93 as post-replicative (Yang et al., 2010). Discriminating between intermediate post-replicative and late post-replicative transcripts is difficult because of their nearly concurrent synthesis and read-through of neighboring genes. The most satisfactory method has been to infect cells with VACV in the presence of an inhibitor of DNA replication, transfect candidate intermediate and late genes, and monitor protein synthesis. Only the intermediate genes are expressed because their transcription factors are products of early genes whereas late transcription factors are not made in the absence of DNA replication. Based on this criterion, we identified 53 intermediate and 38 late genes (Yang et al., 2011b). However, for technical reasons gene designations have been based on a triage system. Thus, genes have been designated early even though they might also be expressed at intermediate or late stages. Similarly, genes have been designated intermediate even if they might also be expressed late. Indeed, analyses of individual genes have indicated the existence of tandem early and post-replicative promoters (Cochran et al., 1985; Rosel et al., 1986; Vos and Stunnenberg, 1988) and others have been artificially constructed (Baldick and Moss, 1993). Identifying multistage promoters may help to better understand poxvirus replication and their use may improve expression vectors. Here we show that many intermediate promoters also have substantial late promoter activity.
Results
Strategy to selectively express late genes
During VACV infection the transcription of intermediate genes must precede the transcription of late genes because the late transcription factors are themselves products of intermediate genes. However, in order to distinguish potential dual intermediate/late promoters from intermediate-only promoters we needed a way to selectively prevent intermediate promoter activity while allowing late promoter activity. Our plan was to (i) construct a cell line that stably expresses late transcription factors, (ii) infect such cells with a VACV mutant that is unable to express intermediate or late transcription factors but will express the viral RNA polymerase, and (iii) transfect the cells with a plasmid containing a luciferase (LUC) reporter gene regulated by the putative intermediate/late or intermediate-only promoter (Fig. 1A). Only plasmids with a late promoter element should express LUC. We already had the VACV intermediate-specific transcription factor deletion mutants vA8Δ vA23Δ, which are propagated in the cell line RK13-A8/A23 that expresses the two intermediate transcription factors (Warren et al., 2012). However, we still needed a cell line that expresses the three late transcription factors encoded by the G8R, A1L and A2L ORFs (Keck et al., 1990).
Fig. 1.
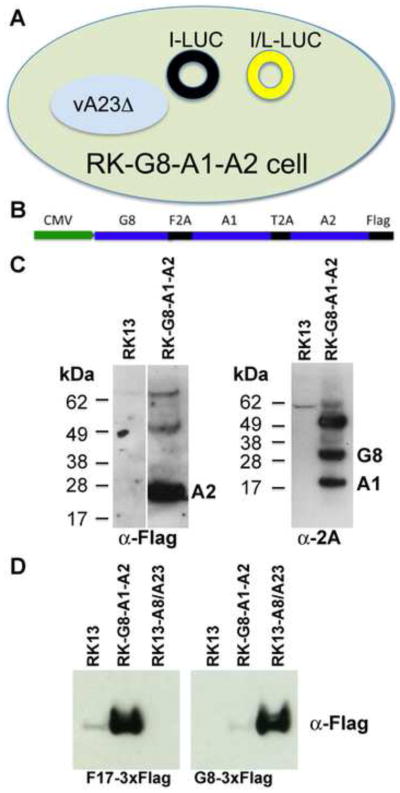
Construction and characterization of the RK-G8-A1-A2Flag cell line. (A) Diagram depicting the assay to determine dual late promoter activity of intermediate promoters. A cell that expresses the three late transcription factors G8, A1 and A2, is infected with a VACV mutant with a deletion of the A23 intermediate transcription factor and is transfected with a plasmid containing LUC regulated by a strict intermediate (I) promoter or by a dual intermediate/late (I/L) promoter. Black circle indicates no LUC expression; yellow circle indicates LUC expression. (B) Diagram of a tricistronic cassette regulated by a CMV promoter. The picornavirus 2A-like sequences F2A and T2A separate the G8R and A1L ORFs and the A1L and A2L ORFs, respectively. The A2L ORF has a C-terminal Flag tag. (C) Western blots showing G8, A1 and A2 expression. A2 was detected with antibody to the Flag epitope; G8 and A1 were detected with antibody to the 2A peptide. The positions of protein markers in kDa are shown on the left and the A2, G8 and A1 bands on the right. (D) Expression of a late and intermediate gene. RK-13, RK-G8-A1-A2Flag, or RK13-A8/A23 cells were infected with vA23Δ and transfected with plasmids encoding the late F17R and intermediate G8R ORFs with 3XFlag tags regulated by their natural promoters. After 18 h, the cells were lysed and analyzed by Western blotting with anti-Flag antibody. The single flag epitope on the A2 protein was not detected in the small amount of extract used for analysis of the triple flag on the F17 and G8 proteins.
To express G8R, A1L and A2L ORFs in similar amounts in the same cell, we made a tricistronic vector containing the three ORFs separated by picornavirus 2A-like CHYSEL peptide sequences that mediate co-translational separation of the nascent chains (de Felipe and Ryan, 2004; Doronina et al., 2008). A Flag-tag was appended to the C-terminus of the A2L ORF and the tricistronic construct was inserted following the CMV promoter in the pcDNA 3.1/Zeo (+) plasmid (Fig. 1B). RK-13 cells were transfected and Zeocin was used for clonal selection of cells. Western blotting was used to detect expression of the VACV A2Flag protein with antibody to the Flag epitope and the VACV A1 and G8 proteins with antibody to the picornavirus 2A peptide in RK-G8-A1-A2Flag cell lysates (Fig. 1C). High molecular weight bands may represent translational read through.
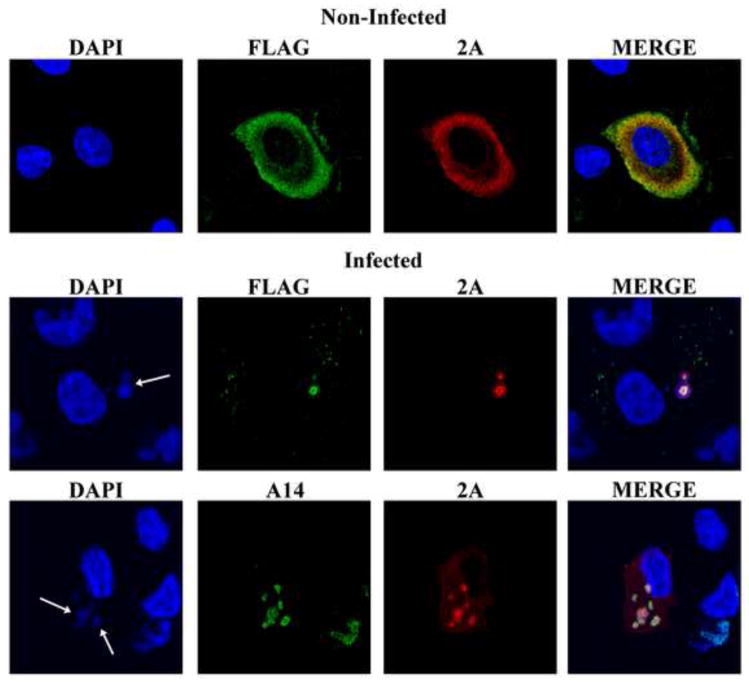
Confocal microscopy was used to determine the intracellular location of the late transcription factors in RK-G8-A1-A2Flag cells. In uninfected cells, the transcription factors were distributed throughout the cytoplasm (Fig. 2, upper panel). However, when the cells were infected with VACV, the transcription factors apparently migrated into virus factories, which were revealed by staining the DNA with DAPI or with antibody to the A14 membrane protein, which was expressed from the viral genome (Fig. 2, lower panels). The re-localization of all three factors was remarkable and taken as an encouraging sign for their ability to function with the viral RNA polymerase, although the mechanism for sequestering the factors in the factory remains to be determined.
Fig. 2.
Intracellular localization of transcription factors in RK-G8-A1-A2Flag cells. Uninfected RK-G8-A1-A2Flag cells (upper panel) and RK-G8-A1-A2 cells infected with VACV for 7 h (lower panels) were fixed and permeabilized. The samples were stained with a rabbit polyclonal primary antibody to the picornavirus 2A peptide, which remains attached to the G8 and A1 proteins, a mouse anti-Flag MAb to detect FLAG attached to the A2 ORF and the mouse MAb to A14, followed by donkey anti rabbit IgG and goat anti-mouse IgG coupled to Alexa Fluor 488 and Alexa Fluor 594, respectively. The nuclei and cytoplasmic viral factories (arrows) were stained with DAPI.
As proof of principal for our strategy, we infected normal RK-13 cells, RK13-A8/A23 cells and RK-G8-A1-A2Flag cells with vA23Δ and transfected them with a plasmid encoding the F17 late protein or the G8 intermediate protein under their natural promoters. Upon Western blotting the F17 protein was detected in the RK-G8-A1-A2Flag cells but not in the RK13-A8/A23 cells, whereas the opposite was true for the G8 protein (Fig. 1D), confirming the function and specificity of the transcription factors and validating the system for our purpose.
Intermediate/late promoter screen
The previously classified 53 intermediate ORFs (Yang et al., 2011b) were used as the starting point for the screen. None of these ORFs were expressed in the presence of AraC indicating that they do not have functional early promoters. However, the presence of late promoter elements remained a possibility. Approximately 80 bp preceding each intermediate ORF was cloned adjacent to the Firefly Luc ORF. Both RK-G8-A1-A2Flag cells and RK-13 cells were infected with vA23Δ and after 1 h were transfected with one of the VACV intermediate promoter Firefly LUC plasmids and with a control plasmid encoding Renilla LUC adjacent to the HSV TK promoter for normalization of transfection efficiency. After 16 to 18 h, LUC activities were determined in the cell lysates. The non-specific Firefly LUC values in the RK-13 cells were subtracted from the values for the same plasmids in the RK-G8-A1-A2Flag cells. The LUC activities varied in a continuous fashion from very low to values approaching the strong late F17 promoter (Bertholet et al., 1985), which was used as a positive control (Fig. 3, Table 1). We calculated that 26 of the 53 intermediate promoters had more than 5% of the F17 promoter activity.
Fig. 3.
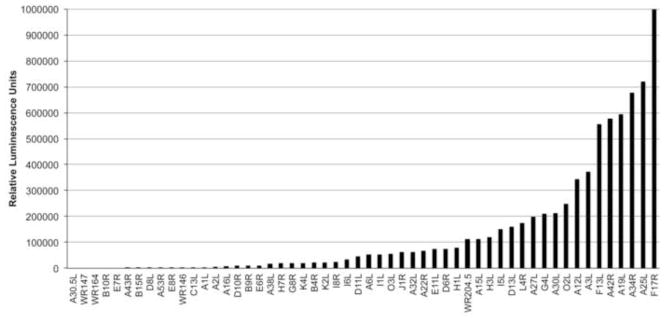
Late activities of intermediate promoters. RK-A8-A1-A2Flag cells were infected with vA23Δ nd transfected with plasmids encoding Firefly LUC regulated by an intermediate promoter. After 16 to 18 h, LUC activity was determined and normalized as described in Methods.
Table 1.
Dual late activities of intermediate promoters
| VACV WR |
VACV COP |
Relativea luminescence | Functional categoriesb |
|---|---|---|---|
| 33 | K2L | 22,623 | Host interaction; EV and WV membrane proteins |
| 35 | K4L | 21,103 | DNA replication, core-associated enzymes and transcriptional factors |
| 52 | F13L | 556,924 | EV and WV membrane proteins |
| 62 | E6R | 10,679 | Required for morphogenesis |
| 63 | E7R | 0 | Unknown |
| 64 | E8R | 826 | Core-associated non-enzymatic proteins |
| 67 | E11L | 74,499 | Core-associated non-enzymatic proteins |
| 69 | O2L | 249,648 | Core-associated enzymes and transcriptional factors |
| 69.5 | O3L | 55,201 | MV membrane-associated proteins; EFC |
| 70 | I1L | 54,568 | Core-associated non-enzymatic proteins; required for morphogenesis; DNA-binding and packing proteins |
| 74 | I5L | 151,740 | MV membrane-associated proteins |
| 75 | I6L | 33,651 | Core-associated non-enzymatic proteins; required for morphogenesis; DNA-binding or packaging proteins |
| 77 | I8R | 25,296 | Core-associated enzymes and transcriptional factors; transcription |
| 81 | G4L | 210,652 | MV membrane-associated proteins; S-S bond formation pathway; required for morphogenesis |
| 86 | G8R | 20,235 | Transcription, late stage |
| 91 | L4R | 174,828 | Core-associated non-enzymatic proteins; DNA-binding proteins; transcription |
| 93 | J1R | 61,879 | Core-associated non-enzymatic proteins |
| 99 | H1L | 79,812 | Core-associated enzymes and transcriptional factors; transcription |
| 101 | H3L | 120,648 | MV membrane-associated proteins |
| 105 | H7R | 19,760 | Contributes to the formation of crescent membrane |
| 111 | D6R | 74,841 | Core-associated enzymes and transcriptional factors; DNA-binding and packing proteins; transcription |
| 113 | D8L | 388 | MV membrane-associated proteins |
| 115 | D10R | 9,701 | Host interaction |
| 116 | D11L | 45,394 | Core-associated enzymes and transcriptional factors; Transcription |
| 118 | D13L | 160,625 | Required for morphogenesis; required for crescent formation |
| 119 | A1L | 4,072 | Transcription, late stage |
| 120 | A2L | 5,502 | Transcription, late stage |
| 122 | A3L | 372,152 | Core-associated non-enzymatic proteins; required for morphogenesis |
| 125 | A6L | 54,180 | Core-associated non-enzymatic proteins; required for morphogenesis |
| 131 | A12L | 343,579 | Core-associated non-enzymatic proteins |
| 135 | A15L | 113,953 | Core-associated non-enzymatic proteins; required for morphogenesis |
| 136 | A16L | 8,392 | MV membrane-associated proteins; EFC |
| 139 | A19L | 593,009 | Unknown |
| 142 | A22R | 67,366 | DNA replication; DNA-binding or packing proteins |
| 146 | no ortholog | 932 | Pseudogene |
| 147 | no ortholog | 0 | Pseudogene |
| 148 | A25L | 721,492 | MV membrane-associated proteins; truncated |
| 150 | A27L | 198,933 | MV membrane-associated proteins |
| 153 | A30L | 212,505 | Core-associated non-enzymatic proteins; required for morphogenesis |
| 153.5 | A30.5L | 0 | Unknown |
| 155 | A32L | 62,632 | Core-associated enzymes and transcriptional factors; DNA-binding or packaging proteins |
| 157 | A34R | 676,586 | EV and WV membrane proteins |
| 162 | A38L | 17,456 | CD47-like putative membrane protein |
| 164 | no ortholog | 0 | Pseudogene |
| 167 | A42R | 577,110 | Unknown |
| 168 | A43R | 26 | Host interaction |
| 179 | A53R | 506 | Host interaction |
| 186 | B4R | 21,721 | Unknown |
| 191 | B9R | 10,233 | Unknown |
| 192 | B10R | 0 | Unknown |
| 197 | WR-B15R | 134 | Host interaction |
| 206 | C13L | 2,878 | Unknown |
| 204.5 | no ortholog | 112,865 | Unknown |
Relative to activity (1,000,000 fluorescent units) of F17 late promoter
EFC, Entry-Fusion Complex; EV, enveloped virion; WV, wrapped virion
Correlation of sequence features of intermediate promoters with late promoter activity
Previous studies had pointed out signature features of late promoters: specifically a TAAAT at the RNA start site frequently followed by a G and a T at −10 surrounded by several less conserved Ts in the coding strand (Yang et al., 2011b). In contrast, promoters thought to be primarily or exclusively intermediate frequently lacked the T following the TAAA as well as the T at −10 but contained a more A+T-rich sequence further upstream (Yang et al., 2011b). Here, we identified sequence features of the intermediate promoters that correlated with associated late activity. Most important were the T at +4, which follows the TAAA at the transcription start site, and the T-content between −12 to −8. Individually the sequence features and late promoter activity showed a moderate statistically significant correlation but together the correlation became strong (Table 2). However, neither of those features correlated with intermediate promoter activity (Table 2), which was determined by transfection of cells infected with VACV in the presence of AraC (not shown). Intermediate promoter activity showed a moderate statistically significant correlation with A-content from −22 to −14 but neither A- nor T-content in this region correlated with late transcription activity (Table 2). In addition, there was no significant correlation between the relative magnitudes of intermediate and late promoter activities (not shown).
Table 2.
Correlation coefficient analysis of promoter elements and promoter activities
|
| ||||
|---|---|---|---|---|
| Promoter elementsa | Correlation with late promoter activities | Correlation with intermediate promoter activities | ||
| Spearman rb | P valuec | Spearman rb | P valuec | |
| T following TAAA | 0.56 | <0.0001 | −0.01 | 0.9453 |
| T content (−12 to −8) | 0.45 | 0.001 | −0.01 | 0.9344 |
| T content (−12 to −8) + T following TAAA | 0.69 | <0.0001 | −0.05 | 0.7579 |
| A content (−22 to −14) | −0.11 | 0.4301 | 0.49 | 0.0004 |
Coding strand nucleotides are shown
0.00–0.19, very weak; 0.20–0.39, weak; 0.40–0.59, moderate; 0.60–0.79, strong; 0.80–1.00, very strong.
P<0.05 considered significant.
Mutational confirmation of intermediate and late promoter sequences
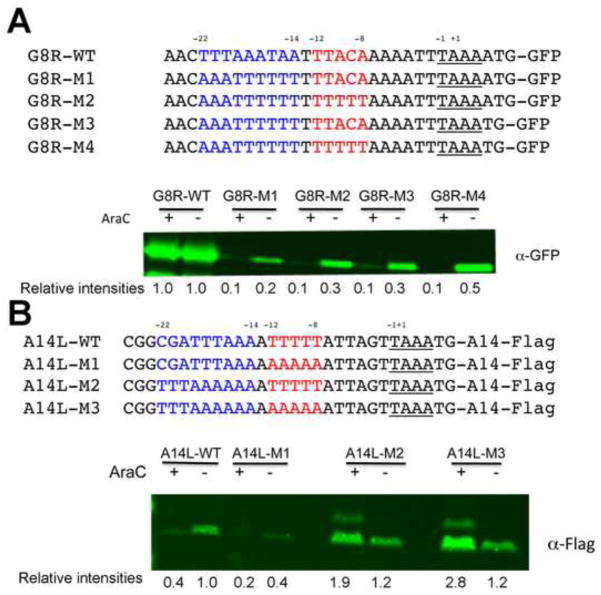
Our analyses suggested that distance from the transcriptional start site and strand specificity for A and T residues, rather than merely A+T DNA content, correlated with intermediate and late promoter activities. To experimentally confirm the sequence correlations, we made mutations in promoters that are exclusively or predominantly intermediate or late to convert them to the opposite specificity. The promoter associated with the G8R ORF was chosen as an example of a relatively strict intermediate promoter (Table 1). Plasmids with green fluorescence protein (GFP) ORF adjacent to the wild type (WT) or mutated G8R promoters were transfected into BS-C-1 cells that had been infected with WT VACV in the presence and absence of AraC. With the WT intermediate promoter there was similar GFP synthesis under both conditions as expected (Fig. 4A). When the sequence between residues −22 to −14 were mostly flipped to the complement (G8R-M1), however, GFP synthesis in the presence and absence of AraC was sharply reduced indicating loss of intermediate promoter function despite retention of the A+T content of the two DNA strands. As shown in Fig. 4A, the late activity was enhanced by increasing the number of T residues between −12 to −8 (G8R-M2) or removing one A in the TAAAAT initiator sequence to make TAAAT (G8R-M3) and even more by combining those mutations (G8R-M4). Nevertheless, these late promoter up mutations did not increase GFP synthesis in the presence of AraC, indicating no effect on intermediate promoter activity.
Fig. 4.
Mutational analyses of an intermediate and late promoter. (A) Intermediate promoter mutations. The sequences of the WT G8R promoter and mutated G8R promoters attached to GFP are shown. Plasmids containing these constructs were transfected into cells that had been infected with VACV in the absence (−) or presence (+) of AraC. After 16 h, the cells were lysed and analyzed by Western blotting with fluorescent antibody to GFP. Relative band intensities shown below the blot were determined using ImageJ. (B) Late promoter mutations. The WT A14L promoter and mutated A14L promoters were attached to the A14L ORF with a Flag tag. The sequences of the promoters are shown. Infection, transfection and Western blotting were similar to that of panel A except that antibody to the Flag epitope was used.
The promoter associated with the late A14L ORF was used to represent a strict late promoter. Plasmids with Flag-tagged A14L ORF adjacent to WT or mutated promoters were transfected into infected HeLa cells in the presence or absence of AraC. Under these conditions, the WT A14L promoter showed faint activity in the presence of AraC and strong activity in the absence of the drug, consistent with a predominantly late promoter (Fig. 4B). When the five Ts in the −12 to −8 region were flipped to As (A14L-M1), there was a dramatic decrease in A14 synthesis (Fig. 4B), again showing the importance of strand specificity of the T and A residues. When we modified the −22 to −14 region to make it similar to the WT G8R promoter but left the original T-run from −12 to −8 (A14L-M2), synthesis increased in the presence of AraC (Fig. 4B), indicating acquisition of intermediate promoter function. Changing the T-run from −12 to −8 (A14L-M3) did not decrease and appeared to slightly increase the intermediate promoter activity as measured by A14 synthesis in the presence of AraC. Thus, the sequences that correlated with late and intermediate promoter activities were confirmed experimentally.
Discussion
Poxviruses regulate gene expression by packaging the early transcription system within the infectious virus particle and sequentially synthesizing virus-encoded intermediate and late transcription factors that collaborate with the virus-encoded multisubunit DNA-dependent RNA polymerase and corresponding promoter elements. Analysis of selected early, intermediate and late transcripts demonstrated a strict regulation (Baldick and Moss, 1993). However, we had noted that some intermediate genes were expressed relatively more strongly in the absence than in the presence of AraC, suggesting dual intermediate/late promoters (Yang et al., 2011b). Here, we succeeded in dissecting the promoter elements of hybrid promoters by employing a mutant virus with a deletion of an intermediate transcription factor gene and a stable cell line that expressed the three late transcription factors. The dual late function of each intermediate promoter was assessed by measuring LUC activity of a transfected recombinant plasmid as an indirect measure of transcription. The LUC activities varied in a continuous fashion from very low to values approaching the strong late F17 promoter. Of 53 intermediate promoters analyzed, we found that 26 had 5% or more of the late activity of the strong F17R late promoter and some were nearly as strong. However, we could not compare the relative intermediate and late activities of individual dual promoters because of the different cell lines and viruses used for the assays.
An important aspect of our work was the establishment of a cell line that expressed all three late transcription factors. This was accomplished using picornavirus 2A-like CHYSEL peptide sequences that mediate co-translational separation of the nascent chains (de Felipe and Ryan, 2004; Doronina et al., 2008). The ability to simultaneously express three or more proteins from a single transfected vector is an important advantage of this system. Although short peptides remained at the C-termini of the G8 and A1 proteins, they did not prevent functional activity and furthermore were used as epitope tags with specific antibody. A remarkable feature of the RK-G8-A1-A2 cells was the relocation of the transcription factors upon VACV infection. In uninfected cells, the factors were distributed throughout the cytoplasm. However, upon infection, they localized within the viral factories. Since this phenomenon was not the focus of our study, we can only speculate as to how this might occur. Previous studies demonstrated that the late transcription factors interact with each other and with the H5 viral protein (Dellis et al., 2004; McCraith et al., 2000). The interaction suggests that the H5 protein might be involved in localization of the late transcription factors. H5 is a constitutively expressed multifunctional nucleic acid binding protein involved in post-replicative transcription and mRNA processing (Cresawn and Condit, 2007; D’Costa et al., 2008; Kovacs and Moss, 1996), genome replication (D’Costa et al., 2010) and virion morphogenesis (DeMasi and Traktman, 2000). Moreover, biochemical and biophysical properties suggest that the H5 protein is intrinsically disordered and may be a “hub” protein (Kay et al., 2013). Although the late transcription factor cell line was successfully used to express reporter genes with late promoters, we have been unable to use it for isolation of a VACV late transcription factor deletion mutant. Whether the failure is due to insufficient amounts of the transcription factors or to other reasons is unknown at this time.
Some features of VACV intermediate and late promoters were previously recognized by sequence comparisons (Davison and Moss, 1989; Yang et al., 2011a; Yang et al., 2011b) and mutational analyses (Baldick et al., 1992; Davison and Moss, 1989; Hirschmann et al., 1990; Knutson et al., 2006). These studies led to the idea that VACV promoters have stage specific core and initiator sequences separated by a spacer. By correlating the late transcription activities of the intermediate promoters, we found two statistically significant features for late activity: a T following TAAA at +4 in the initiator region and T residues between −12 to −8 in the core region of the coding DNA strand. Neither of these features correlated with intermediate transcription activity. On the other hand, the frequency of A residues from −22 to −14 correlated with intermediate transcription activity but not with late transcription activity. The importance of the T-residue at +4, to form TAAAT, and the greater role of T-residues compared to A-residues at −12 to −8 of late promoters had previously been recognized (Davison and Moss, 1989), although at the time the distinction between intermediate and late promoters had not been discovered. A previous mutational study of G8R promoter also suggested the importance of A-residues from −22 to −14 for intermediate promoters (Baldick et al., 1992). In addition, interconversion of intermediate and late promoters with TAAAT initiator sequences could be achieved to some extent by changing the spacing between the core and initiator sequences (Knutson et al., 2006). Here we demonstrated the importance of the A-content of the coding strand from −22 to −14 for intermediate promoter activity and the T-content of the coding strand between −12 to −8 by transforming a strict intermediate promoter into a late promoter and vice versa.
We also noted that some promoters classified as early based on expression in the presence of a DNA synthesis inhibitor, have a TAAA or TAAAT sequence near the start of the ORF suggesting intermediate or late stage activity. Transfection studies with a representative sample of such promoters suggested that many have weak intermediate or late promoter activity but additional work is needed to confirm this (our unpublished data).
In principal, the utilization of three stages of transcription as well as hybrid promoters enables poxviruses to fine-tune the level and duration of expression of individual genes. Previously, we compared the biological roles of genes with early, intermediate and late promoters. We now extended this analysis by comparing the intermediate promoters with relatively low and high late promoter activity. Since the late activities varied continuously (Fig. 3, Table 1), we used an arbitrary value of 5% of the strong F17 late promoter to divide the intermediate promoters into two groups. The rationale for 5% was that half of the promoters were lower and half higher. The intermediate genes encoding virion core and membrane proteins and proteins involved in virion morphogenesis tended to have high late promoter activity, whereas those involved in transcription and DNA replication tended to have low or no late promoter activity (Fig. 5).
Fig. 5.
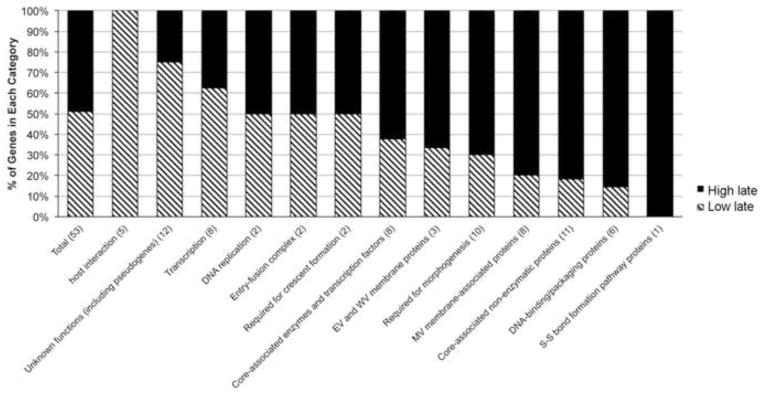
Gene functions of intermediate promoters with low and high late promoter activities. Intermediate promoters were divided into those with low <5% (striped) and high >5% (black) of the activity of the late F17R promoter in the late promoter-specific transfection assay. The numbers of genes in each category are indicated.
Materials and Methods
Cells and viruses
RK-13 cells were grown in Dulbecco’s minimum essential medium, supplemented with 10% fetal bovine serum (FBS), 100 units of penicillin and 100 μg of streptomycin per ml (Quality Biologicals, Gaithersburg, MD). The medium for RK-G8-A1-A2Flag cells was supplemented with 300 μg/ml Zeocin. The VACV deletion mutant vA23Δ was propagated in a RK-13 cell line expressing the A8 and A23 intermediate transcription factors as previously described (Warren et al., 2012).
Construction of the RK-G8-A1-A2Flag cell line
A G8-A1-A2Flag cassette separated by picornavirus 2A-like CHYSEL peptide sequences was cloned in pcDNA 3.1/Zeo (+) plasmid (Life Technologies, Grand Island, NY) as shown in Fig. 1B. The plasmid was transfected into RK-13 cells using Lipofectamine 2000 (Life Technologies) following the manufacturer’s instructions. After 48 h, the transfected cells were distributed to new flasks at approximately 25% confluence with fresh medium containing 750 μg/ml Zeocin. The cells were fed with selective medium every 3 days until cell foci were identified on day 10. The individual colonies were isolated with cloning discs (Sigma Aldrich) and transferred to 96 well plates and screened for Flag-epitope synthesis by Western blotting. The positive colonies were put through a second phase of selection with 750 μg/ml Zeocin. The established recombinant RK-G8-A1-A2Flag cell line was grown as described above and supplemented with 300 μg/ml Zeocin to maintain the selection pressure.
Plasmids, Transfection, Antibodies and Western blotting
Recombinant plasmids were constructed by cloning PCR-amplified target DNA fragments into Zero blunt TOPO vector (Life Technologies). The inserted DNA was verified by sequencing. BS-C-1 cells were transfected with plasmids and Lipofectamine 2000 (Life Technologies) according to the manufacturer’s instructions. The cells were lysed at 16 to 18 h after transfection. For Western blotting, proteins in cell lysates were resolved by SDS polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes using an iBlot apparatus (Life Technologies). The membranes were blocked with 5% nonfat milk in Tris-buffered saline with 0.05% Tween-20 for 1 h, incubated with primary antibody for 1 to 2 h at room temperature or overnight at 4°C, washed with Tris-buffered saline with 0.05% Tween-20, incubated with horseradish peroxidase-conjugated secondary antibody, washed with Tris-buffered saline with 0.05% Tween-20 and developed using chemiluminescent substrate (Pierce, Rockford, IL). Alternatively, a fluorescent secondary antibody was used and the signal was detected with an Odyssey imaging system (LiCor). The band intensities were determined with ImageJ (Wayne Rasband, Research Services Branch, National Institute of Mental Health, Bethesda, MD). Rabbit anti-2A and mouse anti-Flag M2 antibodies were purchased from Millipore (Billerica, MA) and Agilent Technologies (Santa Clara, CA), respectively. Mouse anti-A14 MAb was a gift from Dr. Yan Xiang (University of Texas Health Science Center, TX).
LUC assays
Firefly and Renilla LUC activities were measured simultaneously with a dual LUC assay system (Promega, Madison, WI) according to the manufacturer’s instruction. The transfection efficiency for each experiment was normalized by expression of a co-transfected Renilla LUC plasmid under HSV-TK promoter as the internal control. Data were averaged from the results of transfections performed in at least two independent experiments. Intermediate and late LUC activities from different batches of experiments were normalized by G8R and F17R promoter activity, respectively.
Confocal microscopy
RK-G8-L1-L2Flag cells grown on coverslips were uninfected or infected for 7 h and fixed with 4% paraformaldehyde in phosphate buffered saline (PBS) for 15 min at room temperature (RT) and washed with PBS. The cells were permeabilized for 15 min with 0.1% Triton X-100 in PBS at RT and blocked with 10% FBS for 30 min. After blocking the cells were incubated with the primary antibody in PBS containing 10% FBS for 1 h at RT. Cells were washed and incubated with the secondary antibody conjugated to dye (Molecular Probes, Eugene, OR) for 1 h. The coverslips were washed and mounted on a glass slide by using prolong gold (Life Technologies). Micrographs were acquired with a Leica TCS SP5 confocal inverted-base microscope with a 63x oil objective.
A cell line that expresses vaccinia virus late transcription factors was constructed
Selective expression of late activities of intermediate promoters was measured
Numerous intermediate promoters were shown to have dual late activities
Late activities correlated with TAAAT at initiator site and Ts from −12 to −8
Acknowledgments
We thank Catherine Cotter for maintenance of cell cultures, Yan Xiang for antibody to the A14 protein, members of our laboratory for helpful discussions, and the NIAID Biological Imaging Section for help with confocal microscopy. Research support was provided by the Division of Intramural Research, NIAID, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baldick CJ, Jr, Moss B. Characterization and temporal regulation of mRNAs encoded by vaccinia virus intermediate stage genes. J Virol. 1993;67:3515–3527. doi: 10.1128/jvi.67.6.3515-3527.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldick CJ, Keck JG, Moss B. Mutational analysis of the core, spacer and initiator regions of vaccinia virus intermediate class promoters. J Virol. 1992;66:4710–4719. doi: 10.1128/jvi.66.8.4710-4719.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertholet C, Drillien R, Wittek R. One hundred base pairs of 5′ flanking sequence of a vaccinia virus late gene are sufficient to temporally regulate late transcription. Proc Natl Acad Sci USA. 1985;82:2096–2100. doi: 10.1073/pnas.82.7.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broyles SS, Fesler BS. Vaccinia virus gene encoding a component of the viral early transcription factor. J Virol. 1990;64:1523–1529. doi: 10.1128/jvi.64.4.1523-1529.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broyles SS, Yuen L, Shuman S, Moss B. Purification of a factor required for transcription of vaccinia virus early genes. J Biol Chem. 1988;263:10754–10760. [PubMed] [Google Scholar]
- Cochran MA, Puckett C, Moss B. In vitro mutagenesis of the promoter region for a vaccinia virus gene: evidence for tandem early and late regulatory signals. J Virol. 1985;54:30–37. doi: 10.1128/jvi.54.1.30-37.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresawn SG, Condit RC. A targeted approach to identification of vaccinia virus postreplicative transcription elongation factors: genetic evidence for a role of the H5R gene in vaccinia transcription. Virology. 2007;363:333–341. doi: 10.1016/j.virol.2007.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Costa SM, Bainbridge TW, Condit RC. Purification and properties of the vaccinia virus mRNA processing factor. J Biol Chem. 2008;283:5267–5275. doi: 10.1074/jbc.M709258200. [DOI] [PubMed] [Google Scholar]
- D’Costa SM, Bainbridge TW, Kato SE, Prins C, Kelley K, Condit RC. Vaccinia H5 is a multifunctional protein involved in viral DNA replication, postreplicative gene transcription, and virion morphogenesis. Virology. 2010;401:49–60. doi: 10.1016/j.virol.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison AJ, Moss B. The structure of vaccinia virus late promoters. J Mol Biol. 1989;210:771–784. doi: 10.1016/0022-2836(89)90108-3. [DOI] [PubMed] [Google Scholar]
- de Felipe P, Ryan MD. Targeting of proteins derived from self-processing polyproteins containing multiple signal sequences. Traffic. 2004;5:616–626. doi: 10.1111/j.1398-9219.2004.00205.x. [DOI] [PubMed] [Google Scholar]
- Dellis S, Strickland KC, McCrary WJ, Patel A, Stocum E, Wright CF. Protein interactions among the vaccinia virus late transcription factors. Virology. 2004;329:328–336. doi: 10.1016/j.virol.2004.08.017. [DOI] [PubMed] [Google Scholar]
- DeMasi J, Traktman P. Clustered charge-to-alanine mutagenesis of the vaccinia virus H5 gene: isolation of a dominant, temperature-sensitive mutant with a profound defect in morphogenesis. J Virol. 2000;74:2393–2405. doi: 10.1128/jvi.74.5.2393-2405.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doronina VA, de Felipe P, Wu C, Sharma P, Sachs MS, Ryan MD, Brown JD. Dissection of a co-translational nascent chain separation event. Biochem Soc Trans. 2008;36:712–716. doi: 10.1042/BST0360712. [DOI] [PubMed] [Google Scholar]
- Gershon PD, Moss B. Early transcription factor subunits are encoded by vaccinia virus late genes. Proc Natl Acad Sci USA. 1990;87:4401–4405. doi: 10.1073/pnas.87.11.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschmann P, Vos JC, Stunnenberg HG. Mutational analysis of a vaccinia virus intermediate promoter in vivo and in vitro. J Virol. 1990;64:6063–6069. doi: 10.1128/jvi.64.12.6063-6069.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsafanas GC, Moss B. Vaccinia virus intermediate stage transcription is complemented by Ras-GTPase-activating protein SH3 domain-binding protein (G3BP) and cytoplasmic activation/proliferation-associated protein (p137) individually or as a heterodimer. J Biol Chem. 2004;279:52210–52217. doi: 10.1074/jbc.M411033200. [DOI] [PubMed] [Google Scholar]
- Kay NE, Bainbridge TW, Condit RC, Bubb MR, Judd RE, Venkatakrishnan B, McKenna R, D’Costa SM. Biochemical and biophysical properties of a putative hub protein expressed by vaccinia virus. J Biol Chem. 2013;288:11470–11481. doi: 10.1074/jbc.M112.442012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck JG, Baldick CJ, Moss B. Role of DNA replication in vaccinia virus gene expression: A naked template is required for transcription of three late transactivator genes. Cell. 1990;61:801–809. doi: 10.1016/0092-8674(90)90190-p. [DOI] [PubMed] [Google Scholar]
- Knutson BA, Liu X, Oh J, Broyles SS. Vaccinia virus intermediate and late promoter elements are targeted by the TATA-binding protein. J Virol. 2006;80:6784–6793. doi: 10.1128/JVI.02705-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson BA, Oh J, Broyles SS. Downregulation of vaccinia virus intermediate and late promoters by host transcription factor YY1. J Gen Virol. 2009;90:1592–1599. doi: 10.1099/vir.0.006924-0. [DOI] [PubMed] [Google Scholar]
- Kovacs GR, Moss B. The vaccinia virus H5R gene encodes late gene transcription factor 4: purification, cloning and overexpression. J Virol. 1996;70:6796–6802. doi: 10.1128/jvi.70.10.6796-6802.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCraith S, Holtzman T, Moss B, Fields S. Genome-wide analysis of vaccinia virus protein-protein interactions. Proc Natl Acad Sci USA. 2000;97:4879–4884. doi: 10.1073/pnas.080078197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss B. Poxviridae. In: Knipe DM, Howley PM, editors. Fields Virology. 6. Vol. 2. Vol. 2. Wolters Kluwer/Lippincott Williams & Wilkins; Philadelphia: 2013. pp. 2129–2159. [Google Scholar]
- Rosales R, Sutter G, Moss B. A cellular factor is required for transcription of vaccinia viral intermediate stage genes. Proc Natl Acad Sci USA. 1994;91:3794–3798. doi: 10.1073/pnas.91.9.3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosel JL, Earl PL, Weir JP, Moss B. Conserved TAAATG sequence at the transcriptional and translational initiation sites of vaccinia virus late genes deduced by structural and functional analysis of the HindIII H genome fragment. J Virol. 1986;60:436–439. doi: 10.1128/jvi.60.2.436-449.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz P, Moss B. Identification of a transcription factor, encoded by two vaccinia virus early genes, that regulates the intermediate stage of viral gene expression. Proc Natl Acad Sci USA. 1999;96:2692–2697. doi: 10.1073/pnas.96.6.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos JC, Stunnenberg HG. Derepression of a novel class of vaccinia virus genes upon DNA replication. EMBO J. 1988;7:3487–3492. doi: 10.1002/j.1460-2075.1988.tb03224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren RD, Cotter C, Moss B. Reverse genetic analysis of poxvirus intermediate transcription factors. J Virol. 2012;86:9514–9519. doi: 10.1128/JVI.06902-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright CF, Oswald BW, Dellis S. Vaccinia virus late transcription is activated in vitro by cellular heterogeneous nuclear ribonucleoproteins. J Biol Chem. 2001;276:40680–40686. doi: 10.1074/jbc.M102399200. [DOI] [PubMed] [Google Scholar]
- Yang Z, Bruno DP, Martens CA, Porcella SF, Moss B. Simultaneous high-resolution analysis of vaccinia virus and host cell transcriptomes by deep RNA sequencing. Proc Natl Acad Sci USA. 2010;107:11513–11518. doi: 10.1073/pnas.1006594107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Bruno DP, Martens CA, Porcella SF, Moss B. Genome-wide analysis of the 5′ and 3′ ends of vaccinia virus early mRNAs delineates regulatory sequences of annotated and anomalous transcripts. J Virol. 2011a;85:5897–5909. doi: 10.1128/JVI.00428-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Reynolds SE, Martens CA, Bruno DP, Porcella SF, Moss B. Expression profiling of the intermediate and late stages of poxvirus replication. J Virol. 2011b;85:9899–9908. doi: 10.1128/JVI.05446-11. [DOI] [PMC free article] [PubMed] [Google Scholar]