Abstract
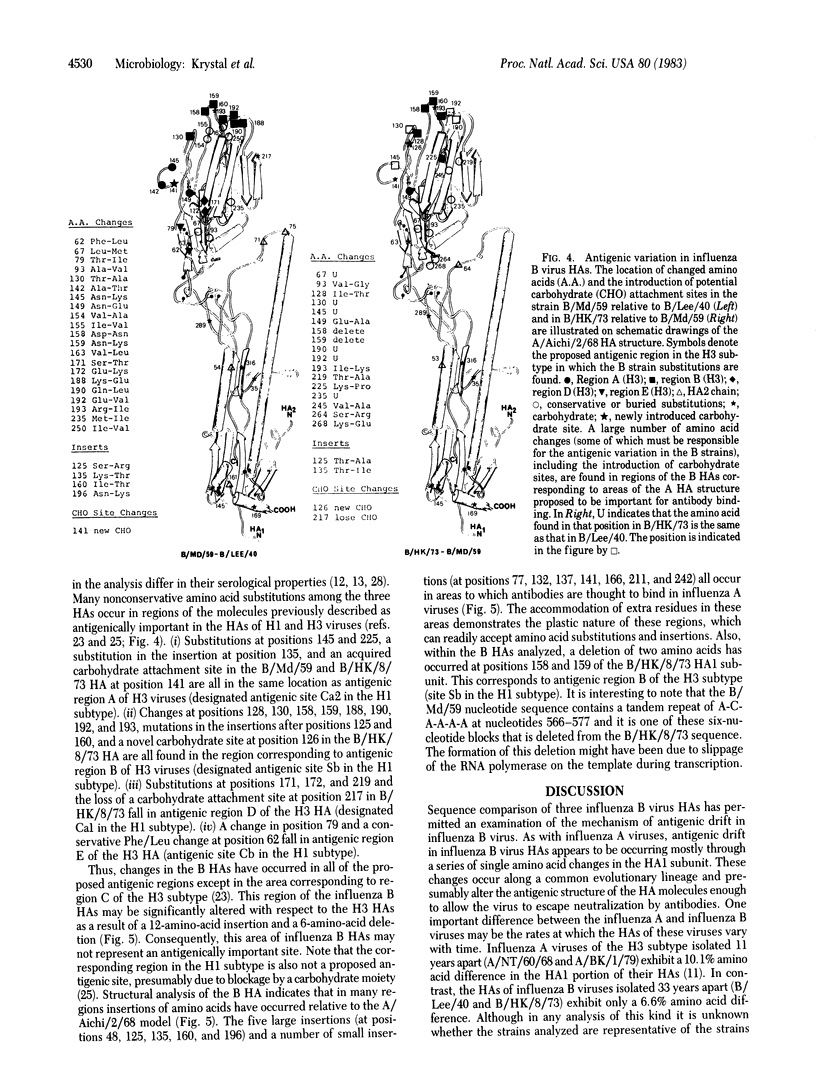
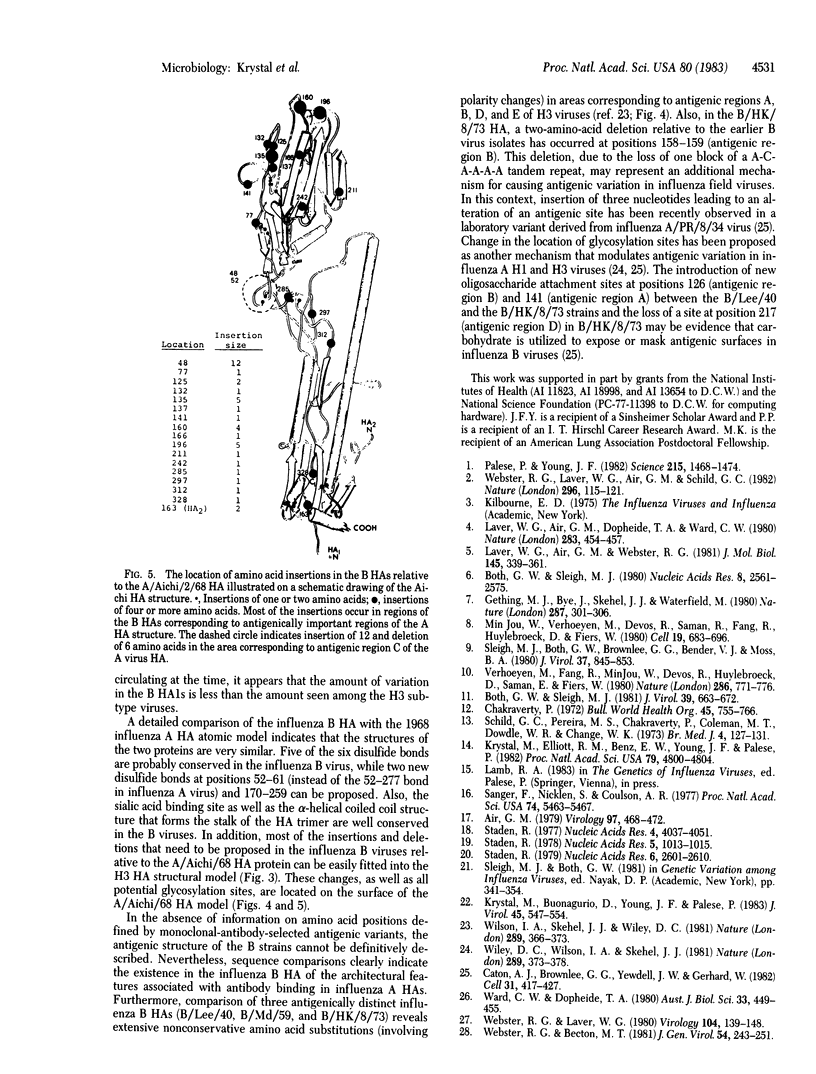
Comparative analysis of the amino acid sequences of hemagglutinins (HAs) of influenza B/Lee/40, B/Md/59, and B/HK/73 viruses has allowed examination of the molecular basis of antigenic variation in type B viruses. As seen with influenza type A viruses, antigenic drift in influenza B viruses proceeds mostly through the accumulation of amino acid substitutions within the HA1 portion of the HA molecule. However, the rate of variation observed among the influenza B virus HAs appears to be significantly lower than the observed rate of variation among influenza A virus HAs. The overall rate of amino acid change in the HA1s of the influenza B viruses studied is 2% per 10 years, whereas the HA1s of H3 influenza A viruses vary by 9.2% per 10 years. The sequences of the influenza B HAs were also examined in relation to the three-dimensional model for the A/Aichi/2/68 HA. When the primary amino acid sequences are compared, it appears that most of the important structural features of the type A HAs--such as the sialic acid binding site, the disulfide linkages, and the stem structure of the trimer--are conserved in the influenza B virus HAs. Regions are also identified where extensive amino acid substitutions have occurred among the three antigenically distinct influenza B virus HAs. The locations of these areas in the B HA structure correspond to antigenic regions proposed for the A virus HAs. In addition, modulation of antigenic regions in B virus HAs may also occur through amino acid deletions and variation in glycosylation sites.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Air G. M. Nucleotide sequence coding for the "signal peptide" and N terminus of the hemagglutinin from an asian (H2N2) strain of influenza virus. Virology. 1979 Sep;97(2):468–472. doi: 10.1016/0042-6822(79)90358-1. [DOI] [PubMed] [Google Scholar]
- Both G. W., Sleigh M. J. Complete nucleotide sequence of the haemagglutinin gene from a human influenza virus of the Hong Kong subtype. Nucleic Acids Res. 1980 Jun 25;8(12):2561–2575. doi: 10.1093/nar/8.12.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Both G. W., Sleigh M. J. Conservation and variation in the hemagglutinins of Hong Kong subtype influenza viruses during antigenic drift. J Virol. 1981 Sep;39(3):663–672. doi: 10.1128/jvi.39.3.663-672.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caton A. J., Brownlee G. G., Yewdell J. W., Gerhard W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell. 1982 Dec;31(2 Pt 1):417–427. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- Chakraverty P. Antigenic relationship between influenza B viruses. Bull World Health Organ. 1971;45(6):755–766. [PMC free article] [PubMed] [Google Scholar]
- Gething M. J., Bye J., Skehel J., Waterfield M. Cloning and DNA sequence of double-stranded copies of haemagglutinin genes from H2 and H3 strains elucidates antigenic shift and drift in human influenza virus. Nature. 1980 Sep 25;287(5780):301–306. doi: 10.1038/287301a0. [DOI] [PubMed] [Google Scholar]
- Jou W. M., Verhoeyen M., Devos R., Saman E., Fang R., Huylebroeck D., Fiers W., Threlfall G., Barber C., Carey N. Complete structure of the hemagglutinin gene from the human influenza A/Victoria/3/75 (H3N2) strain as determined from cloned DNA. Cell. 1980 Mar;19(3):683–696. doi: 10.1016/s0092-8674(80)80045-6. [DOI] [PubMed] [Google Scholar]
- Krystal M., Buonagurio D., Young J. F., Palese P. Sequential mutations in the NS genes of influenza virus field strains. J Virol. 1983 Feb;45(2):547–554. doi: 10.1128/jvi.45.2.547-554.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krystal M., Elliott R. M., Benz E. W., Jr, Young J. F., Palese P. Evolution of influenza A and B viruses: conservation of structural features in the hemagglutinin genes. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4800–4804. doi: 10.1073/pnas.79.15.4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laver W. G., Air G. M., Dopheide T. A., Ward C. W. Amino acid sequence changes in the haemagglutinin of A/Hong Kong (H3N2) influenza virus during the period 1968--77. Nature. 1980 Jan 31;283(5746):454–457. doi: 10.1038/283454a0. [DOI] [PubMed] [Google Scholar]
- Laver W. G., Air G. M., Webster R. G. Mechanism of antigenic drift in influenza virus. Amino acid sequence changes in an antigenically active region of Hong Kong (H3N2) influenza virus hemagglutinin. J Mol Biol. 1981 Jan 15;145(2):339–361. doi: 10.1016/0022-2836(81)90209-6. [DOI] [PubMed] [Google Scholar]
- Palese P., Young J. F. Variation of influenza A, B, and C viruses. Science. 1982 Mar 19;215(4539):1468–1474. doi: 10.1126/science.7038875. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild G. C., Pereira M. S., Chakraverty P., Coleman M. T., Dowdle W. R., Chang W. K. Antigenic variants of influenza B virus. Br Med J. 1973 Oct 20;4(5885):127–131. doi: 10.1136/bmj.4.5885.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleigh M. J., Both G. W., Underwood P. A., Bender V. J. Antigenic drift in the hemagglutinin of the Hong Kong influenza subtype: correlation of amino acid changes with alterations in viral antigenicity. J Virol. 1981 Mar;37(3):845–853. doi: 10.1128/jvi.37.3.845-853.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. A strategy of DNA sequencing employing computer programs. Nucleic Acids Res. 1979 Jun 11;6(7):2601–2610. doi: 10.1093/nar/6.7.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Further procedures for sequence analysis by computer. Nucleic Acids Res. 1978 Mar;5(3):1013–1016. doi: 10.1093/nar/5.3.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Sequence data handling by computer. Nucleic Acids Res. 1977 Nov;4(11):4037–4051. doi: 10.1093/nar/4.11.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson R. G., Hooley R. D., Findlay J. K., Hopkins P. S. Effects of heat stress on the lactation performance of ewes accustomed to tropical conditions and the total fluid intake of their lambs. Aust J Biol Sci. 1980 Aug;33(4):449–456. doi: 10.1071/bi9800449. [DOI] [PubMed] [Google Scholar]
- Verhoeyen M., Fang R., Jou W. M., Devos R., Huylebroeck D., Saman E., Fiers W. Antigenic drift between the haemagglutinin of the Hong Kong influenza strains A/Aichi/2/68 and A/Victoria/3/75. Nature. 1980 Aug 21;286(5775):771–776. doi: 10.1038/286771a0. [DOI] [PubMed] [Google Scholar]
- Webster R. G., Berton M. T. Analysis of antigenic drift in the haemagglutinin molecule of influenza B virus with monoclonal antibodies. J Gen Virol. 1981 Jun;54(Pt 2):243–251. doi: 10.1099/0022-1317-54-2-243. [DOI] [PubMed] [Google Scholar]
- Webster R. G., Laver W. G., Air G. M., Schild G. C. Molecular mechanisms of variation in influenza viruses. Nature. 1982 Mar 11;296(5853):115–121. doi: 10.1038/296115a0. [DOI] [PubMed] [Google Scholar]
- Webster R. G., Laver W. G. Determination of the number of nonoverlapping antigenic areas on Hong Kong (H3N2) influenza virus hemagglutinin with monoclonal antibodies and the selection of variants with potential epidemiological significance. Virology. 1980 Jul 15;104(1):139–148. doi: 10.1016/0042-6822(80)90372-4. [DOI] [PubMed] [Google Scholar]
- Wiley D. C., Wilson I. A., Skehel J. J. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981 Jan 29;289(5796):373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- Wilson I. A., Skehel J. J., Wiley D. C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981 Jan 29;289(5796):366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]


