Abstract
Cell fate decisions during embryogenesis and adult life govern tissue formation, homeostasis and repair. Two key decisions that must be tightly coordinated are proliferation and differentiation. Overproliferation can lead to hyperplasia or tumor formation while premature differentiation can result in a depletion of proliferating cells and organ failure. Maintaining this balance is especially important in tissues that undergo rapid turnover like skin however, despite recent advances, the genetic mechanisms that balance cell differentiation and proliferation are still unclear. In an unbiased genetic screen to identify genes affecting early development, we identified an essential regulator of the proliferation-differentiation balance in epidermal progenitor cells, the Keratinocyte differentiation factor 1 (Kdf1; 1810019J16Rik) gene. Kdf1 is expressed in epidermal cells from early stages of epidermis formation through adulthood. Specifically, Kdf1 is expressed both in epidermal progenitor cells where it acts to curb the rate of proliferation as well as in their progeny where it is required to block proliferation and promote differentiation. Consequently, Kdf1 mutants display both uncontrolled cell proliferation in the epidermis and failure to develop terminal fates. Our findings reveal a dual role for the novel gene Kdf1 both as a repressive signal for progenitor cell proliferation through its inhibition of p63 and a strong inductive signal for terminal differentiation through its interaction with the cell cycle regulator Stratifin.
Keywords: progenitor cells, epidermis, development, keratinocyte, cell fate decision, mouse, forward genetics
INTRODUCTION
The appropriate balance between cell division and differentiation is crucial for organ formation during development as well as for tissue maintenance postnatally. Excessive proliferation or defective differentiation can lead to tumor formation, while the inverse scenario can result in hypoplastic organs or defects in tissue repair. Stem cells are key to maintaining tissue integrity, acting as a reservoir to produce new progeny cells that will differentiate to replace damaged or lost tissue. Adult mammalian skin is the largest organ in the body and comprises a thin, external epidermis and an underlying, thicker dermis. The epidermis has been an excellent model to define the cellular interactions and genetic factors regulating this delicate balance of stem cell proliferation and differentiation, as this tissue is turned over approximately every 4 to 6 weeks in humans and between 1.5-2 million skin cells are sloughed off every hour (Milstone, 2004). Without replacement, the epidermis and the barrier it provides would rapidly be depleted. Establishment of the epidermis occurs during embryonic development, where epidermal progenitor cells give rise to a fully functional epidermis by birth. Thus, the coordination of proliferation and differentiation within the epidermal progenitor cells is also essential for epidermis formation and function.
During embryonic development, the undifferentiated ectoderm becomes committed to the epithelial lineage by embryonic day (e) 8.5 (Byrne et al., 1994). A few days later, (around e13.5) stratification begins, giving rise to a distal layer of epidermal cells (keratinocytes) called intermediate cells that continue to proliferate but also begin to express the first markers of differentiation (Byrne et al., 1994; Lechler and Fuchs, 2005; Smart, 1970). Intermediate cells are a transient embryonic epidermal population that retain the ability divide, but later differentiate into non-proliferative spinous cells by e15.5 (Koster et al., 2007; Weiss and Zelickson, 1975). From this stage onward, proliferation is restricted to the basal layer of the epidermis (Byrne et al., 2003; Fuchs, 2008; Koster and Roop, 2007). As cells leave the basal layer, they undergo sequential differentiation to produce spinous, granular and cornified layers, which are eventually shed via desquamation (Fuchs and Horsley, 2008; Milstone, 2004). Disruption in the formation of these layers or connections between them can lead to a host of defects including lethal dehydration and microbial infection. Despite recent advances in epidermal biology, our understanding of epidermis formation and homeostasis, particularly in regards to the genes that regulate epidermal progenitor cell proliferation and differentiation is quite limited.
One factor that is essential for normal stratification of the epidermis is the cell cycle regulator Stratifin (14-3-3ψ, Sfn). Although initially studied for its role in DNA damage response (Chan et al., 1999; Chan et al., 2000), Sfn was later shown to promote differentiation of suprabasal cells. Mouse embryos homozygous for a mutation in Sfn develop a thick, hyperplastic epidermis and keratinocytes fail to differentiate into granular or cornified cells (Guenet et al., 1979; Herron et al., 2005; Li et al., 2005). Instead, all suprabasal layers express spinous markers while remaining proliferative, suggesting that these cells are locked into the intermediate fate. A key regulator of Sfn is p63 (Dohn et al., 2001; Westfall et al., 2003), the first transcription factor expressed specifically in epithelial cells that have adopted an epidermal fate. At later stages, p63 is expressed primarily by basal keratinocytes and plays multiple roles during epidermis development (Green et al., 2003; Koster et al., 2004). p63 inhibits the expression of Sfn in basal cells and also promotes their proliferation such that loss of p63 results in a thin but differentiated epidermis (Mills et al., 1999; Yang et al., 1999). However, only a handful of factors that interact with Sfn or p63 to control proliferation and differentiation during embryonic epidermis formation are known.
Forward genetics is a powerful, unbiased tool to identify novel genes with essential roles in biological processes. Using this approach, we uncover a previously unknown but essential regulator of the proliferation-differentiation balance in epidermis, the Keratinocyte Differentiation Factor 1 (Kdf1; 1810019J16Rik) gene. Here we demonstrate that mice carrying mutations in Kdf1 display uncontrolled proliferation of basal progenitor cells and a failure of their progeny to differentiate. This results in a thickened, but non-functional epidermis. Our results further reveal that Kdf1 represses basal cell proliferation by inhibiting p63 and also closely interacts with the cell cycle regulator Sfn to induce terminal differentiation in suprabasal cells. These results identify Kdf1 as a critical negative regulator of keratinocyte proliferation during epidermis development and an essential factor in the differentiation of epidermal progenitor cell progeny.
MATERIALS & METHODS
Mouse Strains
SfnEr (Guenet et al., 1979) mice were kindly provided by M. Karin. The p63Brdm2 line was obtained from JAX. We genotyped mutant alleles as previously described (Herron et al., 2005; Li et al., 2005; Mills et al., 1999) and see Supplementary Figure 5. We obtained ES cells harboring a targeted allele of Kdf1 (1810019J16Riktm1a(EUCOMM)Wtsi) from the International Knockout Mouse Consortium in order to generate the Kdf1 gene trap line (Kdf1GT). This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by Institutional animal care and use committee (IACUC protocol number 2010-11172).
Mutagenesis Screen
C57BL/6J and C3HeB/FeJ mice were obtained from The Jackson Laboratory. ENU mutagenesis was performed essentially as described (Kasarskis et al., 1998). 10 mg/ml ENU in phosphate-citrate buffer was injected intraperitoneally into male mice 9-10 weeks old at the time of the first injection. C57BL/6J males received ENU in three doses of 100 mg/kg body weight administered once a week for 3 weeks. These males were used to establish lines by crossing to C3HeB/FeJ females. Each resulting F1 male was mated to two C3HeB/FeJ females, to generate second-generation (G2) females. Four to ten G2 females per line were mated to the F1 male; lines with two or more litters containing embryos with similar abnormal morphology were considered potentially mutant. G2 males and females from these lines were produced and intercrossed, and litters were screened at 12.5 days post coitum to confirm the heritability of the phenotype and to identify G2 males that carried the mutation. Lines were maintained initially by outcrossing the carrier males to C3HeB/FeJ, intercrossing the resulting progeny, and examining embryos to identify new male carriers. After the mutation responsible for the phenotype was initially mapped, carriers of both sexes were identified by PCR as those carrying the C57BL/6J alleles of markers flanking the induced mutation.
Mapping of shd
The shd mutant was initially identified based on a short limb/snout phenotype at e12.5. Initial mapping of the shd mutation utilized two to five simple-sequence length polymorphism markers per chromosome to identify linkage between the epidermis phenotype and C57BL/6J DNA polymorphisms. Linkage to Chromosome 4 was established and subsequently the shd interval was narrowed to between markers D4Mit204 (132.98 Mb) and D4SKI3 (134.16 Mb; https://mouse.mskcc.org). The shd mutation was crossed >11 generations onto the C3HeB/FeJ background, which removed more than 99.9% of the original mutagenized C57BL/6J background, supporting the idea that the shd phenotype is monogenic. Characterization of the shd phenotype occurred on a mixed C57BL/6J-C3HeB/FeJ background.
Sequence capture
The basic principle of Sequence Capture has been previously described (Hodges et al., 2007; Olson, 2007). In our study, a Nimblegen mouse Sequence Capture 385K array was designed to contain oligos complementary to all exons and splice sites within the shd interval. Genomic DNA was isolated from an e11.5 shd homozygote, and then sheared by sonication and adaptors were ligated to the resulting fragments. The adaptor-ligated templates were fractionated by agarose gel electrophoresis and fragments of the desired size were excised. Extracted DNA was amplified by ligation-mediated PCR, purified, and hybridized to the capture array. The array was washed, and bound DNA was eluted, purified, and amplified by ligation-mediated PCR (similar to methods employed in (Choi et al., 2009)). The capture and sequencing experiments were performed at the W.M. Keck Foundation for Biotechnology Resources at Yale. This array also contained sequences from Chromosomes 11 & 7, unrelated to shd. For details about these sequences, please contact the authors.
Sequencing and mutation analysis
Captured libraries were sequenced on an Illumina Genome Analyzer II as single-end, 75-bp reads, as previously described (Choi et al., 2009). Illumina reads were first trimmed based on their quality scores to remove low-quality regions using the program Btrim (Kong, 2011). A cutoff of 20 for average quality scores within a moving window of size 5-bp was used. Minimum acceptable read length was set to 25-bp. Other parameters of Btrim were set to defaults. The pre-processed reads were then aligned to the mouse genome reference sequence (mm9) using the Burrows-Wheeler Alignment tool(Li and Durbin, 2009). The mapping results were converted into SAMtools pileup format using SAMtools programs (Li et al., 2009). PCR duplicates were removed using the rmdup command from SAMtools. SNVs were called using SAMtools’ pileup command. Further filtering was performed using in-house scripts to exclude those SNV calls that had fewer than 3 reads or a SNP score less than 20. Annotation was added based on the UCSC RefSeq gene model (http://genome.ucsc.edu/) (Pruitt et al., 2009). Following the identification of the shd mutation, we designed primers to discriminate the mutation from wild-type sequence by PCR followed by restriction enzyme digestion (Supplemental Figure 4).
Expression and histological analyses
Hematoxylin/Eosin staining, in situ hybridizations, skeletal staining and immunofluorescence analyses were performed using standard methods (Nagy et al., 2002). Toluidine Blue dye exclusion assays were performed at e18.5 as previously described (Hardman et al., 1998). X-gal staining of genetrap heterozyogotes was performed as previously described (Nagy et al., 2002). Antibodies used for immunostaining included Keratin 10, Keratin 14, Filaggrin, Loricrin (all from Covance), phosphorylated Histone H3 (Millipore), p63 (BD Pharmingen, 559951), Kdf1 (Sigma Prestige Antibody HPA028639; raised against the human ortholog (Uhlen et al., 2005), Rhodamine-Phalloidin (Molecular Probes/Invitrogen) and CD104/β4 integrin (BD biosciences).
RNA extraction and Real-time PCR
The epidermis was isolated from e18.5 embryos and total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA) and RNeasy kit (Quiagen). RNA was treated with DNaseI and RT-PCR was performed on 2 g of total RNA using a Superscript first-strand cDNA synthesis kit (Invitrogen) according to the manufacturer’s instructions. The resulting cDNA was diluted 1:5 in nuclease-free water and stored in aliquots at −80°C until used. Real-time PCR with SYBR green detection (Roche) was performed using a ViiA7 thermocycler (Invitrogen). The following primers were used:
GAPDH Forward: 5′ TGATGACATCAAGAAGGTGGTGAAG 3′,
GAPDH Reverse: 5′ TCCTTGGAGGCCATGTAGGCCAT 3′,
pan p63 Forward: 5′ AGATCCCTGAACAGTTCCGAC 3′,
pan p63 Reverse: 5′ ATCGATCACACGTTCACCAC 3′,
DNp63 Forward: 5′ CTGGAAAACAATGCCCAGAC 3′
DNp63 Reverse: 5′ GAGGAGCCGTTCTGAATCTG 3′,
Sfn Forward: 5′ TCGCACCTCATCAAAGGGG 3′,
Sfn Reverse: 5′ CGGCTAGGTAGCGGTAGTAG 3′.
The experiments were carried out in triplicate for each data point. The relative quantification in gene expression was determined using the 2-Ct method (Livak and Schmittgen, 2001). Using this method, we obtained the fold changes in gene expression normalized to an internal control gene (GAPDH).
Mitotic index analysis
Transverse sections of e12.5 and e16.5 embryos were processed through standard immunostaining protocols. The mitotic index was calculated as the ratio of phosphorylated Histone H3 (PHH3)-positive cells to all nuclei in the basal or suprabasal layer of the lateral epidermis, as appropriate. All data were collected from multiple embryos (e12.5: control n=5, shd−/− n=4; e16.5: control n=4, shd−/−n=5, shd−/−;p63+/−n=5). For quantification of the mitotic index, between 720 and 1100 basal keratinocytes (e12.5), 800 and 2100 basal keratinocytes (e16.5) and between 3700 and 5800 suprabasal cells (e16.5) were scored. The results were expressed as mean ± standard deviation and statistical significance was measured using an unpaired two-tailed student T-test with equal variance.
Isolation of primary keratinocytes and transient transfection
Primary keratinocytes were isolated from e18.5 epidermis and cultured following previously described methods (Nowak and Fuchs, 2009). To test antibody specificity, primary keratinocytes were plated onto glass cover slips and transfected with full length Kdf1 with a C-terminal or N-terminal GFP tag. Both constructs showed the same expression pattern. CMV-GFP and K14-GFP were used as transfection controls. Transient transfection was performed using Lipofectamine LTX (Invitrogen) or Fugene 9 reagent (Promega) according to manufacturer’s instructions. After 24 hours, cover slips were subjected to standard immunostaining procedures and stained for Kdf1 or Rhodamine-phalloidin together with DAPI.
RESULTS
Identification of a recessive, perinatal lethal mouse mutant with epidermal defects
To uncover new factors that regulate cell fate decisions during embryonic development, we performed an unbiased N-ethyl-N-nitrosourea (ENU)-induced mutagenesis screen in mice and scored for morphological defects at embryonic day (e) 12.5. We identified a recessive mutant with a short snout and short limbs (Figure 1A-C), which we designated shorthand (shd). To further examine the phenotype, we analyzed litters at e18.5. At this late stage, shd mutants appeared to lack ears or a mouth opening, exhibited short forelimbs, and showed fusion between the tail, hindlimbs and genitals (Figure 1D). Skeletal analysis revealed the presence of all limbs and the tail in shd mutants (not shown), but the lack of protrusion of the hindlimbs and tail appeared to result from an overgrowth of the epidermis that fused them to the body presenting a smooth, taut surface. Internal organs and muscles were not obviously affected but shd homozygotes died at birth, likely due to the epidermis covering the mouth and nose openings, preventing respiration. shd heterozygous animals were viable, fertile and were phenotypically indistinguishable from wild-type littermates.
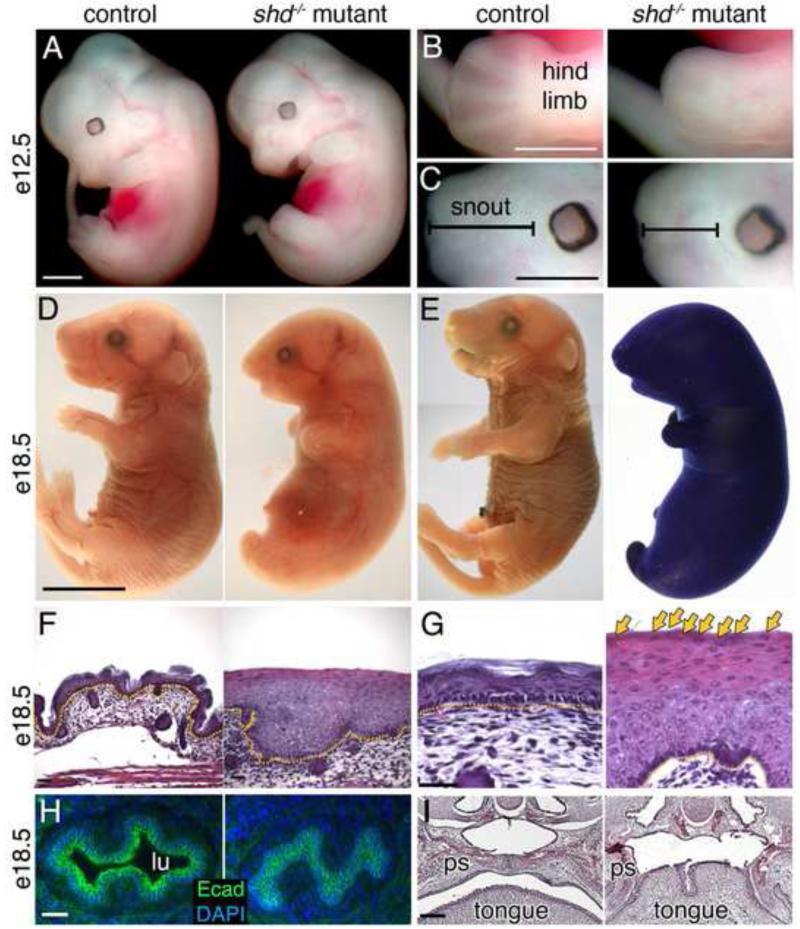
Figure 1. shd mutants display multiple defects linked to keratinocytes.
(A-C) Gross morphological comparison of wild-type and shd mutant embryos at e12.5. shd mutants display shortened and fused limbs (B) and craniofacial malformations, including shortened snout (C). (D) Gross appearance of wild type and shd mutants at e18.5, showing a smooth skin and fusion between hindlimbs and tail. (E) Toluidine Blue exclusion assays of whole embryos show a breakdown in the epidermal barrier in shd homozygotes. (F-G) Hematoxylin and eosin-stained sections of back skin showing a thickened epidermis in shd mutants and retention of nuclei in distal keratinocytes (G, arrows). (H) Sections through the esophagus reveal an absence of a lumen (lu) in shd homozygotes and apparent fusion between the keratinocytes (E-Cadherin+) lining the esophagus. (I) Hematoxylin and eosin-stained sections through heads showed that the palatal shelves (ps) do not fuse along the midline in shd mutants, but instead appear fused to the tongue, resulting in cleft palate. Scale bars: 1mm (A-C) 5mm (D, same for E), 50um (F,G), 20um (H), 100um (I). Yellow dotted lines in F, G mark the dermis (lower) and epidermis (upper) boundary.
shd mutants display keratinocyte defects leading to thickened epidermis and defective epidermal barrier formation
shd mutants possessed a shiny and taut epidermis and, but it was unclear if the epidermis was functional. One measure of epidermal integrity is the formation of an outside-in barrier that keeps the external environment from penetrating the skin. To test if this barrier formed normally in shd mutants, we challenged them with a dye that cannot penetrate mature epidermis. At e18.5, wild-type and heterozygote animals had developed a watertight epidermal barrier, but in shd mutants, we observed global dye penetration indicating a defective epidermal barrier (Figure 1E and not shown). The overall gross morphological phenotype and failure to form a competent barrier prompted us to examine the epidermis in greater detail. Histological analysis of sections through dorsal skin confirmed our initial observation of shd mutant skin being taut and lacking wrinkles. In addition, we observed that the epidermis was 2-10x thicker than in controls (Figure 1F). Moreover, even though an apparent basal layer was present, more differentiated keratinocytes were poorly defined compared to wildtype, which are organized into distinct stratified layers (spinous, granular and cornified). As keratinocytes progress through terminal differentiation, they undergo cornification and lose their nuclei. However, in shd mutants, the most distal keratinocytes still retained their nuclei and the epidermis showed no signs of a cornified layer (Figure 1G). To test if the shd mutation only affected epidermal keratinocytes, we also examined the oral cavity and esophagus, which are lined with keratinocytes. We observed complete fusions of the esophageal epithelium, resulting in no lumen (Figure 1H). In addition, shd mutants developed cleft palate (Figure 1I) due to a fusion of the tongue and palate shelf epithelium, suggesting a widespread defect in keratinocyte development.
The shd gene is required for the differentiation of basal keratinocyte progeny
We decided to focus our efforts on understanding the effects of the shd mutation on the development of epidermal keratinocytes. The epidermis comprises a proliferative basal layer and progressively differentiated spinous, granular and cornified keratinocyte layers. To analyze the differentiation of individual cell types in shd epidermis, we examined markers of different stages of epidermal keratinocyte differentiation. Several lines of evidence indicated a failure of terminal differentiation in shd mutants. First, basal markers Keratins 14 and 5 were expressed throughout shd epidermis in contrast to their restricted expression to basal keratinocytes in controls (Figure 2A and not shown). In addition, although the spinous marker Keratin 10 was induced as cells exited the basal layer, its domain was expanded distally (Figure 2B) and overlapped with basal marker expression in shd mutants, suggesting these cells possessed a mixed identity. Finally, the granular and cornified markers Loricrin and Filaggrin were strongly reduced or absent in shd mutants (Figure 2C,D). Together, these results demonstrate that the gene affected in shd mutants is essential for the normal differentiation of basal progenitor cell progeny. Instead, suprabasal cells in shd mutants express a mix of basal and spinous markers.
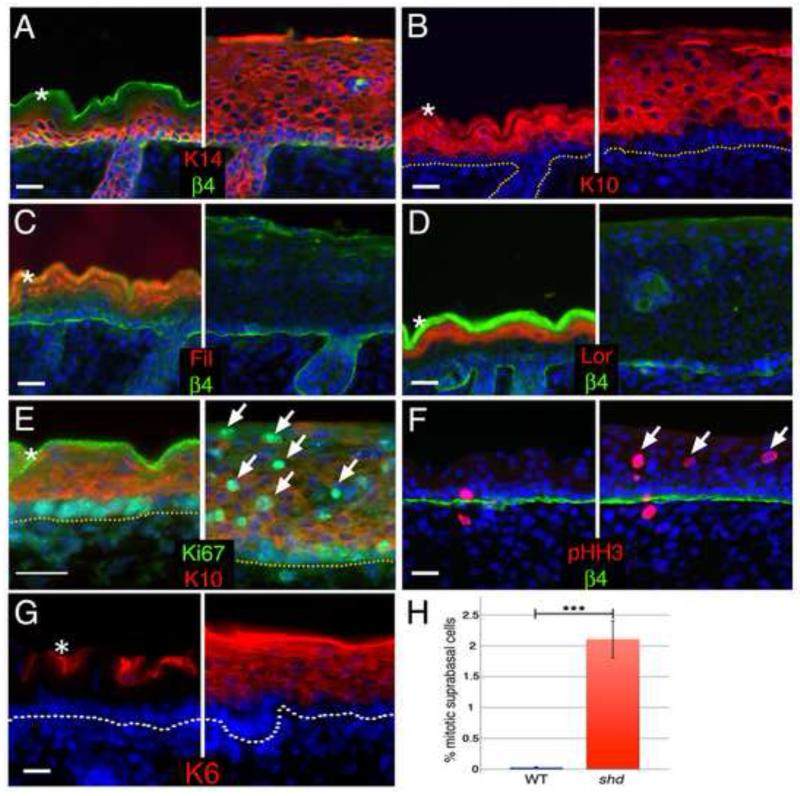
Figure 2. The shd mutation causes hyperproliferation and defective differentiation of the epidermis.
Analysis of e18.5 (A-G) shd epidermis. Basal (A, Keratin 14 in red) and spinous (B, Keratin 10 in red) cell markers are expressed in the distally expanded keratinocytes in shd epidermis. Markers of granular and cornified layers (C, Filaggrin in red; D, Loricrin in red) are largely absent in shd mutants. shd epidermis is hyperproliferative, expressing the mitotic markers Ki-67 (E, green) and phospho-HistoneH3 (F, green) in suprabasal layers (arrows) expressing K10 (E, red), whereas proliferation is restricted to basal layers in controls. Keratin-6 is absent under normal conditions (G) and only induced under hyperproliferative conditions, such as in shd epidermis. (H) Quantification of the mitotic index in the suprabasal cells at e16.5. DAPI is shown in blue (A-G). Asterisks mark unspecific trapping of the secondary antibody in the cornified layer. The dotted lines (B, E, G) or β4 integrin staining (A, C, D, F) mark the border between the epidermis and dermis. Scale bar represents 20um for all panels.
Suprabasal keratinocytes retain proliferative potential in shd mutants
While all epidermal keratinocytes initially have the potential to proliferate, this feature is restricted at later stages to the basal cell layer. As the progeny of basal cells enter the suprabasal layer, they quickly lose the ability to divide and begin to differentiate. The thickened epidermis in shd mutants led us to question whether dysregulated proliferation might also contribute to the skin phenotype. We found that proliferation was no longer restricted to the basal layer and that suprabasal keratinocytes continued to divide (Figure 2E,F). In fact, shd mutant suprabasal cells showed ~70-fold increase in their mitotic index compared to controls (2.11% vs. 0.03%, Figure 2H). Consistent with this observation, shd mutant epidermis showed ectopic Keratin-6 immunoreactivity (Figure 2G), which is observed in hyperproliferative epidermis (Fuchs, 1995; Navarro et al., 1995). Together, our data demonstrate a breakdown in the switch from proliferative basal cells to differentiated, postmitotic progeny in shd mutants. The result is a hyperthickened, poorly-differentiated epidermis that fails to form a normal barrier.
The Stratifin tumor suppressor gene interacts with shd
The cell cycle regulator Stratifin (Sfn; 14-3-3ψ) is a key modulator of cell differentiation in the epidermis (Bhatia et al., 2003; Dellambra et al., 1995). The Sfn gene is mutated in Repeated Epilation (Er) animals, and Er homozygotes phenocopy shd mutants (Guenet et al., 1979; Herron et al., 2005; Li et al., 2005). Like shd mutants, Er homozygotes are perinatal lethal and mutant embryos develop a thickened epidermis that lacks differentiation markers (ex. Loricrin, Filaggrin) but instead shows expanded basal (K14) and spinous (K10) markers. (Guenet et al., 1979; Herron et al., 2005; Li et al., 2005). One possible explanation for the shd phenotype is that Sfn might act downstream of the shd gene to regulate keratinocyte proliferation and differentiation. If so, then we would expect to see reduced levels of Sfn in shd mutant epidermis. In contrast, we found that Sfn is now expressed throughout the epidermis in shd mutants (Figure 3A), and Sfn transcript levels are increased approximately 3 fold (Figure 3B), as assayed by quantitative real-time reverse transcriptase PCR (qRT-PCR). These data argued against the possibility that Sfn acts downstream of shd and instead suggested that Sfn and shd act together to regulate proliferation and differentiation.
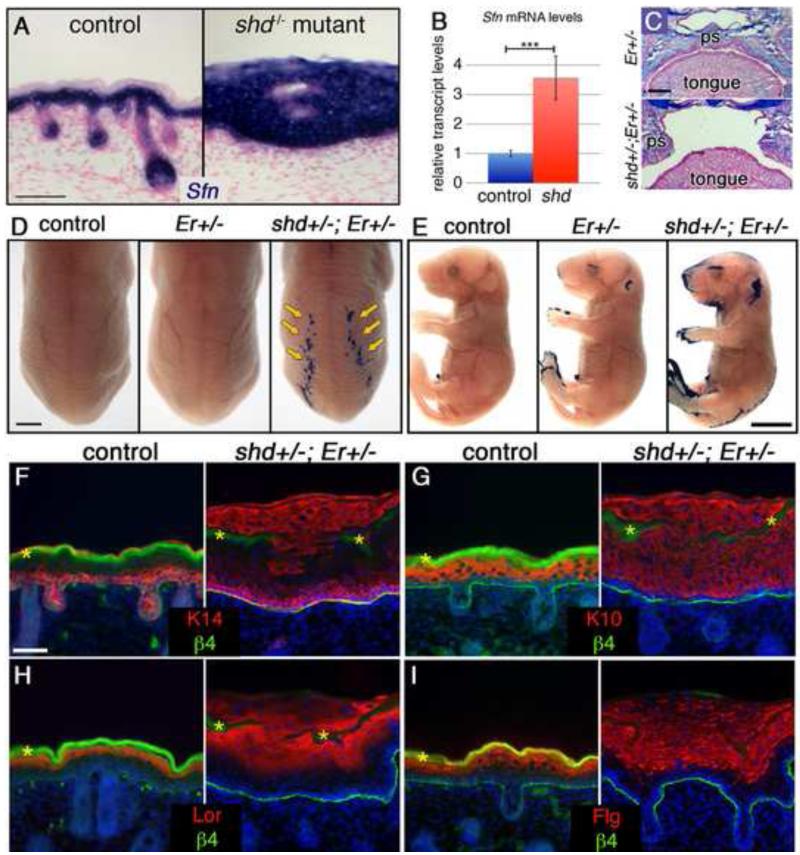
Figure 3. Er-shd double heterozygotes develop cleft palate and epidermal differentiation defects.
(A) in situ hybridization for Sfn (blue) at e18.5 shows expanded expression in shd epidermis. (B) qRT-PCR reveals elevated Sfn transcript levels in shd epidermis. (C) Alcian blue and Fast Red-stained sections through e18.5 heads show that the palatal shelves (ps) elevate and fuse normally in Er heterozygotes but do not fuse along the midline in shd-Er double heterozygotes, resulting in cleft palate. (D,E) Dye exclusion assays show a delay in barrier formation in Er heterozygotes and a more pronounced epidermal barrier defect in shd+/−;Er+/− animals including stripes in caudal back skin (D) and around the eyes, ears, snout, limbs and tail (E). Sections through the barrier-defective dorsal epidermis show that the epidermis is thickened in these regions (F-I) and that basal (F, K14) and spinous (G, K10) markers are abnormally expanded in distal shd+/−;Er+/− keratinocytes. Terminal differentiation markers (E,F) are expressed in shd+/−;Er+/− keratinocytes, but are co-expressed in with K14 and K10. β4 integrin staining marks the border between the epidermis and dermis (F-I). DAPI is shown in blue (F-I). Scale bars represent 50um (A), 250um (C), 2mm (D), 5mm (E), 20um (F-I).
To test for a potential genetic interaction between Sfn and shd we analyzed doubly heterozygous animals (shd+/−;Er+/−). We recovered only a single shd+/−;Er+/− adult out of 51 animals (2% instead of the expected 25%), indicating a postnatal lethal interaction. Focusing on embryonic development, we recovered shd+/−;Er+/− embryos at normal Mendelian ratios (n=7/32; 22%) however, they displayed cleft palate and fusions within the oral cavity, similar to shd or Er homozygotes (Figure 3C). When we tested for epidermal barrier function we observed that shd+/−;Er+/− embryos showed barrier defects in several regions (Figure 3D,E). As mentioned previously, shd+/− carriers did not show barrier defects, however Er+/− embryos displayed abnormal barrier formation around the eyelids, ears, snout and digit/tail tips (Figure 3E). This is likely a delay in barrier formation as Er+/− heterozygotes are viable and appear normal at birth. In shd+/−;Er+/− embryos, while most regions of the epidermis show normal barrier formation, we observed more pronounced barrier defects around the eyelids, ears, snout and digit/tail tips than in Er+/− heterozygotes (Figure 3E), suggesting Sfn and shd cooperate to ensure normal barrier formation.
We noticed additional regions with abnormal barrier formation in shd+/−;Er+/− embryos including under the chin as well as in two dorsal stripes (Figure 3D,E). When we sectioned through the dorsal stripes, we found that the barrier defect appeared to be caused by breaks in the cornified layer. Most regions of shd+/−;Er+/− embryos form normal epidermis, including a cornified layer. In the barrier-defective stripe regions, we found the epidermis was greatly thickened and the cornified layer (highlighted by trapping of β4 integrin staining, marked by asterisks) displayed gaps, with keratinocytes spilling out onto the surface of the embryo (see Figure 3F-I). When we assayed the identity of the keratinocytes in the barrier-defective stripes, we found that the epidermis had a clear basal layer (K14+, K10-; Figure 3F,G) but more distal cells were co-expressing basal, suprabasal, and more differentiated markers simultaneously (Figure 3F-I). This phenotype is distinct from shd or Er single homozygotes and shows that in some regions of shd+/−;Er+/− epidermis, differentiation proceeds without turning off basal markers. This suggests that the shd gene and Sfn act together to inhibit basal markers and coordinate the stepwise differentiation of basal cell progeny, at least in a subset of the epidermis.
Basal cell proliferation is increased in shd mutants
shd mutant keratinocytes simultaneously expressed both basal (K14) and spinous (K10) markers in distal layers, while remaining mitotically active. These features are a hallmark of the transient embryonic cell population called intermediate keratinocytes found at e14.5 (Smart, 1970; Weiss and Zelickson, 1975). This led us to hypothesize that shd keratinocytes become trapped in the intermediate cell fate and fail to terminally differentiate. If this were true, we would expect that 1) shd mutants would not show a defect prior to the establishment of intermediate cells, and 2) basal cells would not be affected by the shd mutation. However, we observed premature epidermal thickening by e12.5 in shd mutants (Figure 4A), leading to greatly thickened dorsal epidermis by the time of intermediate identity establishment (Figure 4B). We further observed a significant increase in shd basal keratinocytes that were actively dividing compared to controls (2.82% in control compared to 4.48% in shd mutants) at e12.5 (Figure 4C). These data indicate that the shd gene normally acts within the basal layer as a brake on epidermal progenitor cell proliferation and when disrupted leads to extensive early epidermis overgrowth.
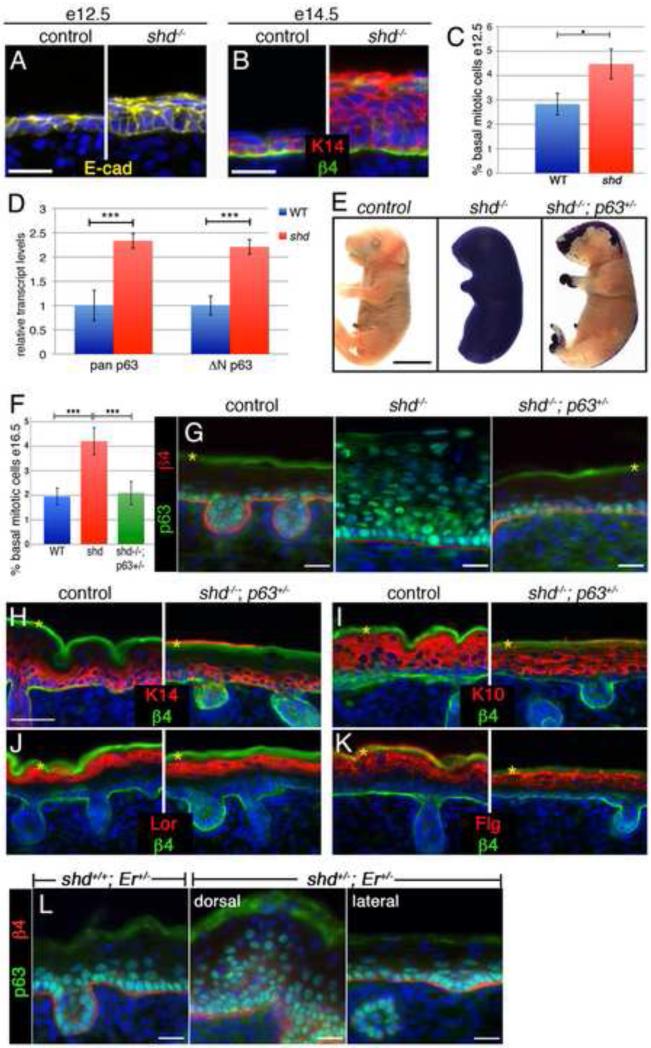
Figure 4. The shd mutation acts through p63 to regulate basal proliferation and epidermal differentiation.
(A) Early epidermis is largely single layered, but shd mutants show extra layers by e12.5. (B) At e14.5, the intermediate keratinocyte layer is largely Keratin14-negative in wild type, but dorsal epidermis in shd mutants is greatly thickened and retains Keratin14 (red) expression. (C) Basal proliferation is already increased at e12.5 in shd mutants (4.48% ± 0.61%) compared to control littermates (2.83% ± 0.44). (D) qRT-PCR shows a ~two-fold upregulation of p63 in shd epidermis, which is mainly due to the ΔNp63 isoform. (E) Dye exclusion assay shows a rescue of epidermal barrier function in shd−/−;p63+/− embryos, especially in lateral regions. (F) Basal progenitor cell proliferation is doubled in shd mutants (4.21% ± 0.55; n=5 embryos) compared to controls (1.95% ±0.34; n=4) at e16.5 and rescued by reducing p63 dosage in shd−/−;p63+/− embryos (2.00% ±0.47; n=5). Error bars: st.dev.; statistical significance was measured using an unpaired two-tailed student T-test with equal variance. ***P < 0.001. (G) At e18.5, p63 protein expression is expanded throughout shd mutant epidermis and restored to wild-type in shd−/−; p63+/− compound mutants. (H-K) Sections of shd−/−;p63+/− lateral back skin show normal epidermis thickness. Basal (H, Keratin 14 in red) and spinous (I, Keratin 10 in red) cell markers are restricted to their normal domains and expression of more differentiated markers is rescued (J, Loricrin in red; K, Filaggrin in red) in shd−/−;p63+/− embryos. (L) p63 expression is greatly expanded in caudal regions of the backskin in shd+/−; Er+/− transheterozygotes which lacked proper barrier function (see Fig. 3E). p63 expression in lateral regions of shd+/−; Er+/− embryos appears similar wild type and to Er+/− animals (not shown). β4 integrin staining marks the border between the epidermis and dermis (B, G-L). Yellow asterisks mark unspecific trapping of secondary antibody in cornified layer. DAPI is shown in blue (A,B, G-L). Control and mutant samples are shown at the same magnification. Scale bars represent 20um (A,B, G-L), 5mm (E).
shd acts upstream of p63 to regulate epidermal proliferation and differentiation
p63 is a member of the p53 tumor suppressor gene family that is expressed primarily by basal keratinocytes and plays multiple roles during epidermis development (Green et al., 2003; Koster et al., 2004). Loss of p63 function results in the opposite phenotype of shd homozygotes, namely a thin but differentiated epidermis (Mills et al., 1999; Yang et al., 1999). This highlights one of the key roles of p63, namely that it promotes proliferation of basal keratinocytes (Truong et al., 2006). Since we observed an increase in the mitotic index of shd keratinocytes at e12.5 (Figure 4C), we tested whether this might be due to an increased level of p63. qRT-PCR assays on e18.5 epidermis showed that p63 transcript levels were increased approximately 2-fold in shd mutants compared to controls (Figure 4D) and that these transcripts are almost exclusively Np63, the main epidermal isoform of p63, which is normally restricted to basal keratinocytes (Romano et al., 2009). Further, using a pan-p63 antibody, we found that p63 was no longer restricted to basal keratinocytes in shd embryos, but was now expressed throughout the epidermis (Figure 4G). These results suggested the possibility that increased levels of p63 might underlie the shd phenotype.
To test the hypothesis that elevated p63 levels drive part of the shd mutant phenotype, we set out to lower p63 gene dosage in the shd background. We first generated shd+/−;p63+/− double heterozygotes. shd+/−;p63+/− animals were viable, fertile and did not display any gross epidermal or hair defects (not shown). The p63 locus is located on Chromosome 16 in the mouse, and we had mapped the shd mutation to Chromosome 4 (see Methods), allowing us to make compound mutants without concerns about linkage. We subsequently crossed these double heterozygotes to shd+/− carriers in order to examine the phenotype of the compound mutant (shd−/−;p63+/−) mice. Strikingly, we found that many aspects of the shd phenotype were rescued by reducing the dosage of p63. The epidermal barrier formation was rescued in most regions of the compound mutants as well as the limb protrusion defects and tail-hindlimb fusion normally observed in shd mutants (Figure 4E). When we examined basal keratinocyte proliferation, we found that the ~2-fold increased mitotic index in shd mutants was rescued in shd−/−;p63+/− embryos (Figure 4F). Further, normal epidermis thickness was restored in lateral (barrier-restored) regions of shd−/−;p63+/− mutants, and p63 was now restricted to basal keratinocytes (Figure 4G). When we examined differentiation markers in the epidermis of shd−/−;p63+/− mutants, we found that early and late differentiation markers were expressed normally (Figure 4H-K).
Sfn has also been shown to inhibit p63 in keratinocytes (Li et al., 2011). We then tested whether Sfn and shd might cooperate to regulate p63. When we examined lateral (i.e phenotypically wild type) regions of shd+/−;Er+/− epidermis we observed normal restriction of p63 to basal keratinocytes. However, we observed a dramatic expansion of p63 throughout the epidermis in the barrier-defective dorsal stripes (Figure 4L). This demonstrates that shd and Sfn can act together to inhibit p63 and allow normal epidermal differentiation. Taken together, these results demonstrate that the shd gene functions to inhibit p63 in the epidermis. Both the block of terminal differentiation as well as the increase in basal progenitor cell proliferation in shd mutants appears to be due to excess p63.
The shd mutation affects a previously unknown epidermal regulator, 1810019J16Rik
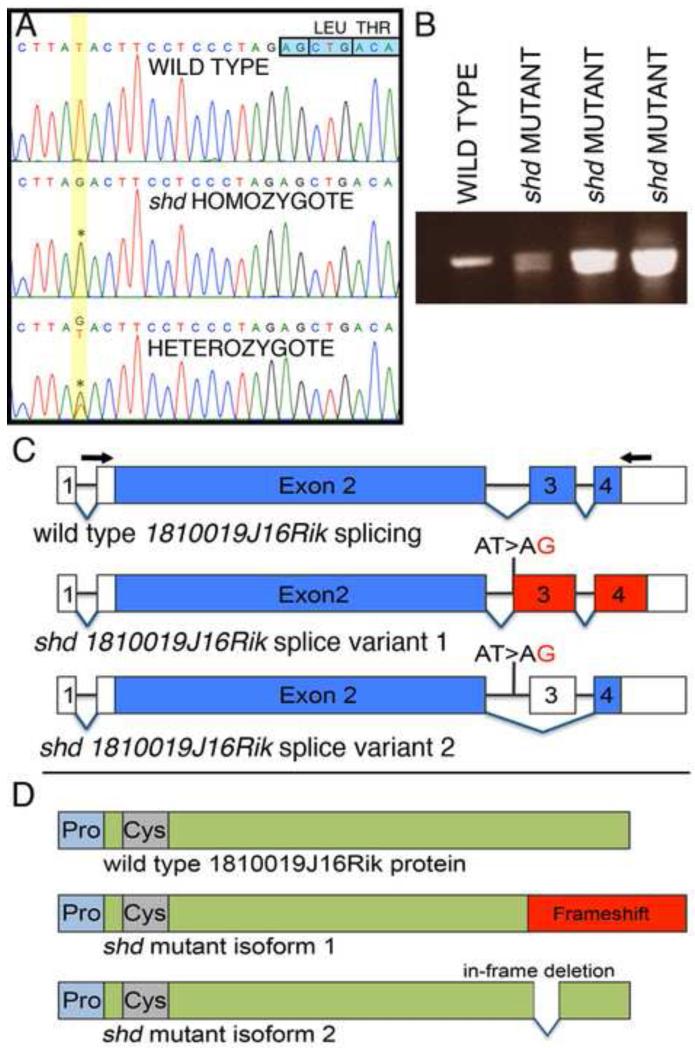
To identify the gene affected by the shd mutation, we initially mapped the shd mutation to a region of mouse Chromosome 4 that is syntenic to human Chromosome 1p36, which has been intensely studied due to developmental phenotypes and skin cancers associated with haploinsufficiency in this region (Bagchi and Mills, 2008; Campeau et al., 2008; Gajecka et al., 2007). Using a targeted capture and sequencing approach, we identified a homozygous mutation in shd animals within the previously uncharacterized 1810019J16Rik gene, which we have renamed Keratinocyte differentiation factor 1 (Kdf1). shd mutants contain a T>G transversion within intron 2 of Kdf1, 14 bases upstream of exon 3 (Figure 5A). This changed an AT site to AG, raising the possibility that this might be recognized by the splicing machinery as a splice acceptor. To test this, we performed RT-PCR and found that the shd mutation eliminates the wild-type transcript by altering splicing and results in two shd-specific splice variants. (Figure 5B). One splice variant utilizes the mutation as a new splice acceptor site, resulting in an out of frame, but longer transcript. The other splice variant skips exon 3 altogether, resulting in an in-frame deletion (Figure 5C).
Figure 5. Identification of a point mutation within the 1810019j16Rik locus, causing splicing defects in shd mutants.
(A) Sequence traces from wild type, heterozygous and homozygous shd embryos. In shd mutants and carriers, there is a T>G transversion (asterisks) 14 bases upstream of the start of exon 3 (indicated by the boxed nucleotides, top right) of the Kdf1 (1810019J16Rik) transcript. (B) RT-PCR across exons 2-4 of the Kdf1 transcript showing a single band in wild type and two bands in shd homozygotes. (C) Representation of the splice variants in wild type and shd mutant. The Kdf1 mRNA spans four exons (exon 2-4 are coding exons). The shd mutation, a T>G transversion in intron 2, results in either a frame shift (splice variant 1) or skipping of exon 3 altogether (splice variant 2). Blue: coding exons; red: frame shift regions; white: untranslated regions. Arrows indicate the position of primers used for RT-PCR in (B). (D) Schematic representation of the Kdf1 protein. The predicted protein of Kdf1 does not contain conserved protein domains, except slightly proline-rich (light blue) and cysteine-rich (grey) regions in the N-terminus of the protein. The frame shift in the shd mutant isoform1 will result in a longer protein with the C-terminal being out of frame (red). Alternatively, skipping of exon 3 will give rise to a truncated protein with a 24 amino acid in-frame deletion.
The Kdf1 gene is unique within the mouse genome but lacks any recognizable protein domains, with the exception of proline and cysteine-rich domains near the N-terminus (Figure 5D). The shd-specific transcripts should give rise to two protein variants, one with a C-terminal frameshift and one with an internal deletion (Figure 5D, Supplemental Figure 1). The Kdf1 protein shares 90% identity with the human homolog (C1ORF172) and is highly conserved across mammals (Supplemental Figure 2). This conservation falls off outside of mammals, but orthologs of Kdf1 have been isolated in multiple vertebrate lineages including birds, amphibians and fish (Supplemental Figure 3). The C-terminal portion of the protein, which is affected in shd mutants, is particularly well conserved in mammals suggesting this region is vital for gene function.
A complementation test confirms the identity of the shd gene
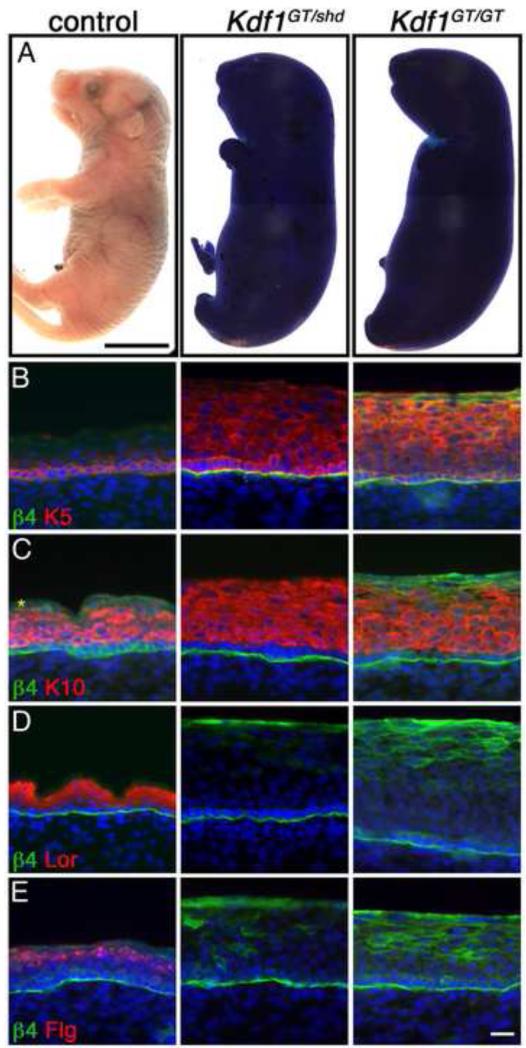
We validated the Kdf1 mutation by standard Sanger sequencing in multiple shd mutants, however it was possible that this mutation was merely linked to the shd phenotype instead of being causative. To verify that loss of function of the Kdf1 gene underlies the shd phenotype, we generated mice carrying a targeted gene trap allele of Kdf1 from embryonic stem cells (1810019J16Riktm1a(EUCOMM)Wtsi; Kdf1GT). Kdf1GT/+ heterozygotes were viable and non-phenotypic. Our characterization of Kdf1GT/GT homozygotes showed epidermal barrier defects (Figure 6) and abnormal differentiation similar to shd mutants. This suggested that the two recessive mutations might affect the same gene, however these two mutations could produce the same phenotype but affect different genes. To discriminate between these possibilities, we performed a complementation test. Briefly, if the Kdf1GT allele was able to complement the shd mutant allele when placed in trans and produce wild-type embryos, this would indicate that the mutations affect different genes (Griffiths et al., 2000). However, when we crossed Kdf1GT/+ heterozygotes with shd/+ animals they failed to complement, with Kdf1GT/shd transheterozygotes exhibiting the same phenotype as shd homozygotes or Kdf1GT homozygotes, This included increased thickness of the epidermis, a breakdown in the epidermal barrier and an abnormal differentiation program in Kdf1GT/shd epidermis (Figure 6A-E). Together these data demonstrate that the shd mutant phenotype is caused by a mutation in Kdf1.
Figure 6. A second Kdf1 allele fails to complement the shd phenotype verifying that Kdf1 is the affected gene in shd mutants.
(A) Dye exclusion assay at e18.5 demonstrating the failure of Kdf1 gene trap homozygotes (Kdf1GT/GT) and shd-Kdf1 gene trap transheterozygotes (Kdf1GT/shd) to establish a functional epidermal barrier. Basal (B, Keratin 5 in red) and spinous (C, Keratin 10 in red) cell markers are expanded in keratinocytes in Kdf1GT homozygote embryos as well as in Kdf1GT/shd transheterozygotes. Markers of granular and cornified layers (D, Loricrin in red; E, Filaggrin in red) are not induced in Kdf1GT homozygote embryos and in shd-Kdf1GT transheterozygotes. This failure of the two alleles to complement demonstrates that the shd phenotype derives from a mutation in the Kdf1 gene. β4 integrin staining marks the border between the epidermis and dermis (B-E). DAPI is shown in blue (B-E). Asterisks marks unspecific antibody trapping. Control and mutant samples are shown at the same magnification. Scale bar represents 5mm (A), 20um (for B-E; shown in E).
Kdf1 is expressed throughout epidermal development
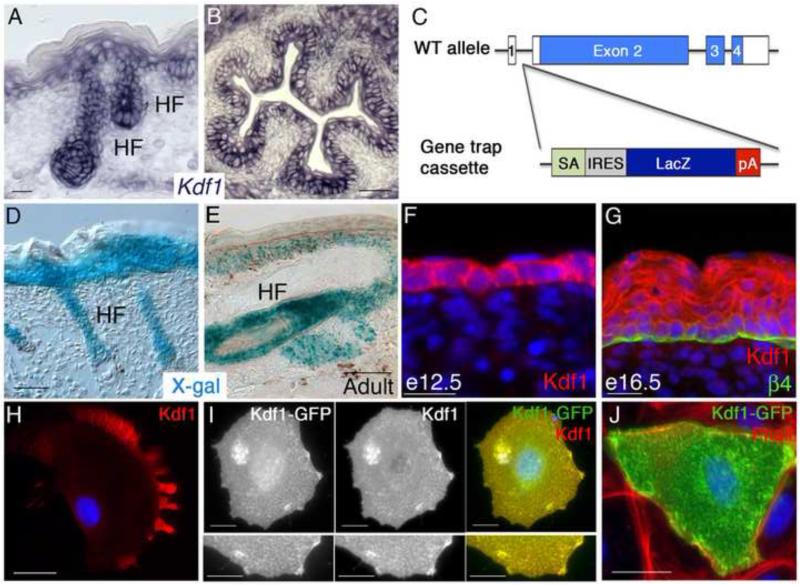
In mature e18.5 mouse skin, we observed strongest expression of Kdf1 transcripts in the basal and suprabasal layers of the epidermis as well as within the developing hair follicles (Figure 7A). Kdf1 transcripts are also found in additional keratinocyte populations, like those lining the lumen of the esophagus (Figure 7B), consistent with the defects observed in shd mutants. We confirmed Kdf1 expression using the LacZ gene within the genetrap cassette in Kdf1GT carriers (Figure 7C). The genetrap is expressed in epidermis and hair follicles at e18.5 (Figure 7D) and maintains a similar pattern in adults (Figure 7E) suggesting that it may also function postnatally in maintenance or regeneration of the epidermis.
Figure 7. Kdf1 is expressed in keratinocytes throughout epidermal development.
(A) in situ hybridization shows Kdf1 transcripts in e18.5 epidermis with strongest expression in the basal layer, including hair follicles (HF) and decreasing levels in differentiated layers. (B) Kdf1 transcripts are also found in other keratinocyte populations including the esophagus at e18.5. (C) Simplified schematic of the genetrap inserted into the Kdf1 locus. The genetrap cassette lies between exons 1 (non-coding) and 2 (first coding exon) and is under the control of the endogenous Kdf1 promoter. The genetrap cassette contains a strong splice acceptor (SA), which is separated from a downstream LacZ cassette by an IRES site. pA = polyadenylation sequence. (D) X-Gal staining of e18.5 Kdf1GT heterozygous skin reveals a similar pattern as the Kdf1 transcript, with strong expression in basal and suprabasal epidermal layers and hair follicles. (E) X-Gal staining adult Kdf1GT heterozygous tail skin shows continued Kdf1 locus expression in epidermis and hair follicles. Kdf1 protein localizes to the cytoplasm and cell borders in single-layered epidermis at e12.5 (F) and retains this pattern in basal and spinous cells at e16.5 but is more diffusely localized in the granular layer (G). Endogenous Kdf1 protein in cultured primary keratinocytes appears to be enriched at the leading edge and cell protrusions (H). Overexpression of Kdf1-GFP in primary keratinocytes and co-staining with Kdf1 antibody supports the Kdf1 antibody specificity (I). Overexpression Kdf1-GFP co-localizes with actin structures, recognized by Rhodamine-Phalloidin (J). β4 integrin staining marks the border between the epidermis and dermis (G). DAPI is shown in blue (F-H). Scale bar represents 20um (A-D, H-J), 50um (E,D).
To test where Kdf1 resides within cells, we used a commercial antibody and analyzed Kdf1 localization in developing mouse epidermis. In single layered keratinocytes (e12.5), the Kdf1 protein is found throughout the epidermis in the cytoplasm and appeared enriched at cell borders (Figure 7F). After the onset of stratification, Kdf1 showed two distinct expression patterns, an enrichment at cell borders in the basal and spinous layer, but becoming more diffuse in more differentiated layers (Figure 7G). This pattern is disrupted in shd mutant epidermis and in Kdf1GT homozygous animals (Supplemental Figure 4). We also examined the localization of endogenous Kdf1 in primary cultured mouse keratinocytes. We found that Kdf1 is expressed at low levels, but strongest localization occurred near the cell membrane, especially in cellular protrusions (Figure 7H). To verify that the antibody recognizes Kdf1, we transfected keratinocytes with a GFP-tagged version of Kdf1 and found co-localization using the Kdf1 antibody and GFP (Figure 7I), although the pattern of high levels of overexpressed Kdf1 was distinct from endogenous Kdf1. Cell protrusions had highest levels of Kdf1 and using fluorescently labeled phalloidin to visualize F-actin we found that those Kdf1-positive protrusions contained actin (Figure 7J).
DISCUSSION
The coordination of proliferation and differentiation is a fundamental process in all organisms. In multicellular organisms, proliferation during early embryonic stages is critical to expand the embryo while differentiation is needed to subfunctionalize tissues and organs. Thus the balance between proliferation and differentiation must be tightly coordinated to ensure stable size and function of organs and tissues. Defining the factors that control the rate of cell division and those that drive the decision of mitotically active cells to differentiate is key to our understanding not only of organ formation but also of tissue homeostasis and for defining the mechanisms underlying human disease.
Forward genetics provides a critical, unbiased, yet focused approach to identify factors essential for the regulation of proliferation and differentiation as well as other key cell fate decisions. Human genetics studies have been greatly aided in recent years by the use of targeted enrichment and next-generation sequencing technologies to identify mutations underlying disease syndromes. This and other recent studies demonstrate the potential for these techniques to transform model organism studies. Selective, large-scale sequencing of mutants results in a rapid progression from phenotype to affected gene, and facilitates mutagenesis screens in mice as a tractable avenue for diagnostic gene discovery.
In the present study, we used a forward genetic screen coupled with sequence capture to identify a recessive mouse mutant (shd) containing a splice site mutation in the previously unstudied Kdf1 (1810019J16Rik) gene (Figure 5). We showed that Kdf1 is a key inhibitor of cell proliferation and promoter of differentiation in the epidermis (Figure 2). We generated a gene trap allele of Kdf1 (Kdf1GT), which displayed a similar phenotype to shd mutants, suggesting that shd is a loss of function allele of Kdf1. We then verified that the splicing mutation in Kdf1 underlies the shd phenotype by performing a complementation test with the Kdf1GT allele (Figure 6). We observed that Kdf1 is expressed in the epidermis from early stages of embryonic development through adulthood, and appears to localize to the cytoplasm of keratinocytes, with a close association to the membrane (Figure 7). We further show that Kdf1 acts, at least in part, through the negative regulation of p63, which is upregulated and expanded throughout the epidermis in shd mutants instead of being restricted to the basal layer. The increased p63 expression underlies many of the shd epidermal defects including increased basal keratinoctye proliferation and failure of distal epidermal cells to differentiate (Figure 4).
Although the factors acting downstream of Kdf1 are unknown, our genetic interaction data place Kdf1 upstream of p63 in epidermal development, as reducing p63 gene dosage rescues many of the epidermal defects in shd mutants. The divergent subcellular localizations of p63 (nucleus) and Kdf1 (cytoplasmic) suggest that Kdf1 acts indirectly to regulate p63 levels. One attractive candidate for Kdf1 to act through is the Notch pathway, due to the location of Notch receptors at the cell membrane. Notch signaling antagonizes p63 and inhibits proliferation while promoting differentiation (Estrach et al., 2008; Nguyen et al., 2006; Tadeu and Horsley, 2013). However, the phenotypes of mice lacking Notch signaling (Blanpain et al., 2006; Moriyama et al., 2008) are much milder than we observed in either shd mutants or Kdf1GT homozygotes, suggesting that if Kdf1 affects Notch signaling, it must have other functions as well.
We further show that Kdf1 and Sfn genetically interact to regulate the stepwise differentiation of keratinocytes (Figure 3). In animals doubly heterozygous for Kdf1 and Sfn mutations (shd+/−;Er+/−), some regions of the epidermis showed expansion of p63 and distal keratinocytes retained basal markers in addition to more differentiated markers, resulting in a breakdown in epidermal barrier formation. As Sfn is a key regulator of the cell cycle, it will be important to determine if Kdf1 also functions by regulating cell cycle progression in epidermal progenitor cells. Intriguingly, mice with mutations in Ikkα or Irf6 phenocopy shd and Er homozygotes. This raises the possibility that these four genes might act in a common pathway to regulate stem cell proliferation and differentiation (Hu et al., 1999; Ingraham et al., 2006; Li et al., 1999; Richardson et al., 2006; Takeda et al., 1999). Notably, human mutations in Irf6 and Ikka underlie the keratinocyte-related van der Woude syndrome, Popliteal Pterygium syndrome, and Cocoon syndrome (Kondo et al., 2002; Lahtela et al., 2010), highlighting the importance of future efforts to define the organization of this pathway for human health.
The human ortholog (C1Orf172) is also expressed in adult epidermis (Uhlen et al., 2005) which suggests that Kdf1 has a conserved function in mammalian keratinocytes and may continue to function in adult epidermal maintenance. Together, our data suggest a model where the shd gene product normally downregulates p63 in basal stem cells to maintain a specific proliferation rate. As cells exit the basal layer, the shd gene product acts with Sfn, a known negative regulator of p63, to further downregulate p63 levels and promote differentiation and repression of basal characteristics.
The shd mutant was first identified at e12.5 based on the observation that the snout and limbs were shorter (Figure 1 A-C). At that early stage, the epidermis in wild-type animals is primarily single-layered (Byrne et al., 1994; Lechler and Fuchs, 2005; Smart, 1970), while in shd mutants, the epidermis displays premature formation of additional layers (Figure 4A). However, the thickening of the epidermis is not visible by wholemount examination of shd mutants. This raises the question of whether the limb, snout and epidermal phenotypes have the same root cause or reflect distinct functions of the Kdf1 gene. It is unclear what causes the short snout phenotype, however it is known that digit length is regulated by a stratified epithelia signaling center called the apical ectodermal ridge (AER) that forms at the distal edge of the limb (Saunders, 1948; Summerbell, 1974; Sun et al., 2002). We have observed defects in the organization of the (AER) in shd mutants (Lee & Weatherbee, unpublished results), suggesting that Kdf1 acts in multiple stratified epithelia including the epidermis and AER. Interestingly, p63 also functions in the AER and is required for normal limb development in mice (Mills et al., 1999; Yang et al., 1999) and humans (Amiel et al., 2001; Celli et al., 1999; Duijf et al., 2002; Ianakiev et al., 2000; McGrath et al., 2001; van Bokhoven et al., 2001). Although beyond the scope of this study, it will be important to determine whether Kdf1 also functions in the AER to regulate p63, or if Kdf1 acts through a different mechanism in the limb. These studies could uncover a role for Kdf1 as a modifier of human syndromes associated with p63 including Split Hand-Foot Malformation, Ectrodactyly, Ectodermal dysplasia and Cleft lip/palate syndrome, Limb mammary syndrome, Ankyloblepharon-Ectodermal defects-Clefting syndrome and Acro-Dermato-Ungual-Lacrimal-Tooth syndrome
Our findings provide the first evidence that the novel gene Kdf1 (1810019J16Rik) functions as a critical regulator of proliferation and differentiation in epidermal progenitor cells and thus has potential broad implications for the understanding of tissue morphogenesis, homeostasis and cancer. While both the shd and gene trap alleles of Kdf1 are perinatal lethal, it will be useful to determine whether Kdf1 is required postnatally for epidermal maintenance. The continued expression of Kdf1 in adult epidermis (Figure 7E), suggests that Kdf1 may play a similar role in controlling adult epidermal cells proliferation and differentiation.
Supplementary Material
HIGHLIGHTS.
The shd mutant shows keratinocyte defects including fusions and epidermal overgrowth
Epidermal differentiation is blocked in shd mutants and proliferation increases
The shd gene functions together with Stratifin to promote epidermal differentiation.
The shd gene acts upstream of p63; reduced p63 gene dose rescues the shd phenotype
Splicing of the novel Kdf1-1810019J16Rik gene is affected by the shd mutation
ACKNOWLEDGEMENTS
We would like to thank Valentina Greco, Antonio Giraldez, Kasey Christopher, Emily Mis and Anita Fernandez for helpful discussions and comments on the manuscript, and William Horton and Stephanie Andrade for technical assistance. We also thank the Yale Center for Genome Analysis for help with sequence capture and the Yale Animal Genomics Service for blastocyst injection and chimera generation of the gene trap allele.
FUNDING
This work was supported by pilot project funding through ACS IRG 58-012-51 (S.D. Weatherbee), institutional startup funds from the Yale Genetics Department (S.D. Weatherbee) and NIH grants R01AR059687 to Scott Weatherbee, R01NS044385 to Kathryn Anderson, R01HD32427 to Lee Niswander, and U01HD43478 to Kathryn Anderson and Lee Niswander.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Amiel J, Bougeard G, Francannet C, Raclin V, Munnich A, Lyonnet S, Frebourg T. TP63 gene mutation in ADULT syndrome. Eur J Hum Genet. 2001;9:642–645. doi: 10.1038/sj.ejhg.5200676. [DOI] [PubMed] [Google Scholar]
- Bagchi A, Mills AA. The quest for the 1p36 tumor suppressor. Cancer research. 2008;68:2551–2556. doi: 10.1158/0008-5472.CAN-07-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia K, Siraj AK, Hussain A, Bu R, Gutierrez MI. The tumor suppressor gene 14-3-3 sigma is commonly methylated in normal and malignant lymphoid cells. Cancer Epidemiol Biomarkers Prev. 2003;12:165–169. [PubMed] [Google Scholar]
- Blanpain C, Lowry WE, Pasolli HA, Fuchs E. Canonical notch signaling functions as a commitment switch in the epidermal lineage. Genes & development. 2006;20:3022–3035. doi: 10.1101/gad.1477606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne C, Hardman M, Nield K. Covering the limb--formation of the integument. Journal of anatomy. 2003;202:113–123. doi: 10.1046/j.1469-7580.2003.00142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne C, Tainsky M, Fuchs E. Programming gene expression in developing epidermis. Development (Cambridge, England) 1994;120:2369–2383. doi: 10.1242/dev.120.9.2369. [DOI] [PubMed] [Google Scholar]
- Campeau PM, Ah Mew N, Cartier L, Mackay KL, Shaffer LG, Der Kaloustian VM, Thomas MA. Prenatal diagnosis of monosomy 1p36: a focus on brain abnormalities and a review of the literature. American journal of medical genetics. 2008;146A:3062–3069. doi: 10.1002/ajmg.a.32563. [DOI] [PubMed] [Google Scholar]
- Celli J, Duijf P, Hamel BC, Bamshad M, Kramer B, Smits AP, Newbury-Ecob R, Hennekam RC, Van Buggenhout G, van Haeringen A, Woods CG, van Essen AJ, de Waal R, Vriend G, Haber DA, Yang A, McKeon F, Brunner HG, van Bokhoven H. Heterozygous germline mutations in the p53 homolog p63 are the cause of EEC syndrome. Cell. 1999;99:143–153. doi: 10.1016/s0092-8674(00)81646-3. [DOI] [PubMed] [Google Scholar]
- Chan TA, Hermeking H, Lengauer C, Kinzler KW, Vogelstein B. 14-3-3Sigma is required to prevent mitotic catastrophe after DNA damage. Nature. 1999;401:616–620. doi: 10.1038/44188. [DOI] [PubMed] [Google Scholar]
- Chan TA, Hwang PM, Hermeking H, Kinzler KW, Vogelstein B. Cooperative effects of genes controlling the G(2)/M checkpoint. Genes & development. 2000;14:1584–1588. [PMC free article] [PubMed] [Google Scholar]
- Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, Nayir A, Bakkaloglu A, Ozen S, Sanjad S, Nelson-Williams C, Farhi A, Mane S, Lifton RP. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellambra E, Patrone M, Sparatore B, Negri A, Ceciliani F, Bondanza S, Molina F, Cancedda FD, De Luca M. Stratifin, a keratinocyte specific 14-3-3 protein, harbors a pleckstrin homology (PH) domain and enhances protein kinase C activity. Journal of cell science. 1995;108(Pt 11):3569–3579. doi: 10.1242/jcs.108.11.3569. [DOI] [PubMed] [Google Scholar]
- Dohn M, Zhang S, Chen X. p63alpha and DeltaNp63alpha can induce cell cycle arrest and apoptosis and differentially regulate p53 target genes. Oncogene. 2001;20:3193–3205. doi: 10.1038/sj.onc.1204427. [DOI] [PubMed] [Google Scholar]
- Duijf PH, Vanmolkot KR, Propping P, Friedl W, Krieger E, McKeon F, Dotsch V, Brunner HG, van Bokhoven H. Gain-of-function mutation in ADULT syndrome reveals the presence of a second transactivation domain in p63. Human molecular genetics. 2002;11:799–804. doi: 10.1093/hmg/11.7.799. [DOI] [PubMed] [Google Scholar]
- Estrach S, Cordes R, Hozumi K, Gossler A, Watt FM. Role of the Notch ligand Delta1 in embryonic and adult mouse epidermis. The Journal of investigative dermatology. 2008;128:825–832. doi: 10.1038/sj.jid.5701113. [DOI] [PubMed] [Google Scholar]
- Fuchs E. Keratins and the skin. Annual review of cell and developmental biology. 1995;11:123–153. doi: 10.1146/annurev.cb.11.110195.001011. [DOI] [PubMed] [Google Scholar]
- Fuchs E. Skin stem cells: rising to the surface. The Journal of cell biology. 2008;180:273–284. doi: 10.1083/jcb.200708185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E, Horsley V. More than one way to skin. Genes & development. 2008;22:976–985. doi: 10.1101/gad.1645908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajecka M, Mackay KL, Shaffer LG. Monosomy 1p36 deletion syndrome. Am J Med Genet C Semin Med Genet. 2007;145C:346–356. doi: 10.1002/ajmg.c.30154. [DOI] [PubMed] [Google Scholar]
- Green H, Easley K, Iuchi S. Marker succession during the development of keratinocytes from cultured human embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:15625–15630. doi: 10.1073/pnas.0307226100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths AJ, Miller JH, Suzuki DT, Lewontin RC, Gelbart WM. An Introduction to Genetic Analysis. 7th ed W.H. Freeman; New York: 2000. [Google Scholar]
- Guenet JL, Salzgeber B, Tassin MT. Repeated epilation: a genetic epidermal syndrome in mice. The Journal of heredity. 1979;70:90–94. doi: 10.1093/oxfordjournals.jhered.a109223. [DOI] [PubMed] [Google Scholar]
- Hardman MJ, Sisi P, Banbury DN, Byrne C. Patterned acquisition of skin barrier function during development. Development (Cambridge, England) 1998;125:1541–1552. doi: 10.1242/dev.125.8.1541. [DOI] [PubMed] [Google Scholar]
- Herron BJ, Liddell RA, Parker A, Grant S, Kinne J, Fisher JK, Siracusa LD. A mutation in stratifin is responsible for the repeated epilation (Er) phenotype in mice. Nature genetics. 2005;37:1210–1212. doi: 10.1038/ng1652. [DOI] [PubMed] [Google Scholar]
- Hodges E, Xuan Z, Balija V, Kramer M, Molla MN, Smith SW, Middle CM, Rodesch MJ, Albert TJ, Hannon GJ, McCombie WR. Genome-wide in situ exon capture for selective resequencing. Nature genetics. 2007;39:1522–1527. doi: 10.1038/ng.2007.42. [DOI] [PubMed] [Google Scholar]
- Hu Y, Baud V, Delhase M, Zhang P, Deerinck T, Ellisman M, Johnson R, Karin M. Abnormal morphogenesis but intact IKK activation in mice lacking the IKKalpha subunit of IkappaB kinase. Science (New York, N.Y. 1999;284:316–320. doi: 10.1126/science.284.5412.316. [DOI] [PubMed] [Google Scholar]
- Ianakiev P, Kilpatrick MW, Toudjarska I, Basel D, Beighton P, Tsipouras P. Split-hand/split-foot malformation is caused by mutations in the p63 gene on 3q27. American journal of human genetics. 2000;67:59–66. doi: 10.1086/302972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingraham CR, Kinoshita A, Kondo S, Yang B, Sajan S, Trout KJ, Malik MI, Dunnwald M, Goudy SL, Lovett M, Murray JC, Schutte BC. Abnormal skin, limb and craniofacial morphogenesis in mice deficient for interferon regulatory factor 6 (Irf6) Nature genetics. 2006;38:1335–1340. doi: 10.1083/ng1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasarskis A, Manova K, Anderson KV. A phenotype-based screen for embryonic lethal mutations in the mouse. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:7485–7490. doi: 10.1073/pnas.95.13.7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Schutte BC, Richardson RJ, Bjork BC, Knight AS, Watanabe Y, Howard E, de Lima RL, Daack-Hirsch S, Sander A, McDonald-McGinn DM, Zackai EH, Lammer EJ, Aylsworth AS, Ardinger HH, Lidral AC, Pober BR, Moreno L, Arcos-Burgos M, Valencia C, Houdayer C, Bahuau M, Moretti-Ferreira D, Richieri-Costa A, Dixon MJ, Murray JC. Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nature genetics. 2002;32:285–289. doi: 10.1038/ng985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y. Btrim: A fast, lightweight adapter and quality trimming program for next-generation sequencing technologies. Genomics. 2011;98:152–153. doi: 10.1016/j.ygeno.2011.05.009. [DOI] [PubMed] [Google Scholar]
- Koster MI, Dai D, Marinari B, Sano Y, Costanzo A, Karin M, Roop DR. p63 induces key target genes required for epidermal morphogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:3255–3260. doi: 10.1073/pnas.0611376104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster MI, Kim S, Mills AA, DeMayo FJ, Roop DR. p63 is the molecular switch for initiation of an epithelial stratification program. Genes & development. 2004;18:126–131. doi: 10.1101/gad.1165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster MI, Roop DR. Mechanisms regulating epithelial stratification. Annual review of cell and developmental biology. 2007;23:93–113. doi: 10.1146/annurev.cellbio.23.090506.123357. [DOI] [PubMed] [Google Scholar]
- Lahtela J, Nousiainen HO, Stefanovic V, Tallila J, Viskari H, Karikoski R, Gentile M, Saloranta C, Varilo T, Salonen R, Kestila M. Mutant CHUK and severe fetal encasement malformation. The New England journal of medicine. 2010;363:1631–1637. doi: 10.1056/NEJMoa0911698. [DOI] [PubMed] [Google Scholar]
- Lechler T, Fuchs E. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature. 2005;437:275–280. doi: 10.1038/nature03922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England) 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics (Oxford, England) 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Lu Q, Estepa G, Verma IM. Identification of 14-3-3{sigma} mutation causing cutaneous abnormality in repeated-epilation mutant mouse. Proceedings of the National Academy of Sciences of the United States of America. 2005 doi: 10.1073/pnas.0508310102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Lu Q, Hwang JY, Buscher D, Lee KF, Izpisua-Belmonte JC, Verma IM. IKK1-deficient mice exhibit abnormal development of skin and skeleton. Genes & development. 1999;13:1322–1328. doi: 10.1101/gad.13.10.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Sambandam SA, Lu HJ, Thomson A, Kim SH, Lu H, Xin Y, Lu Q. 14-3-3{sigma} and p63 play opposing roles in epidermal tumorigenesis. Carcinogenesis. 2011 doi: 10.1093/carcin/bgr207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- McGrath JA, Duijf PH, Doetsch V, Irvine AD, de Waal R, Vanmolkot KR, Wessagowit V, Kelly A, Atherton DJ, Griffiths WA, Orlow SJ, van Haeringen A, Ausems MG, Yang A, McKeon F, Bamshad MA, Brunner HG, Hamel BC, van Bokhoven H. Hay-Wells syndrome is caused by heterozygous missense mutations in the SAM domain of p63. Human molecular genetics. 2001;10:221–229. doi: 10.1093/hmg/10.3.221. [DOI] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- Milstone LM. Epidermal desquamation. Journal of dermatological science. 2004;36:131–140. doi: 10.1016/j.jdermsci.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Moriyama M, Durham AD, Moriyama H, Hasegawa K, Nishikawa S, Radtke F, Osawa M. Multiple roles of Notch signaling in the regulation of epidermal development. Developmental cell. 2008;14:594–604. doi: 10.1016/j.devcel.2008.01.017. [DOI] [PubMed] [Google Scholar]
- Nagy A, Gertsenstain M, Vintersten K, Behringer RR. Manipulating the Mouse Embryo: A Laboratory Manual. 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2002. [Google Scholar]
- Navarro JM, Casatorres J, Jorcano JL. Elements controlling the expression and induction of the skin hyperproliferation-associated keratin K6. The Journal of biological chemistry. 1995;270:21362–21367. doi: 10.1074/jbc.270.36.21362. [DOI] [PubMed] [Google Scholar]
- Nguyen BC, Lefort K, Mandinova A, Antonini D, Devgan V, Della Gatta G, Koster MI, Zhang Z, Wang J, Tommasi di Vignano A, Kitajewski J, Chiorino G, Roop DR, Missero C, Dotto GP. Cross-regulation between Notch and p63 in keratinocyte commitment to differentiation. Genes & development. 2006;20:1028–1042. doi: 10.1101/gad.1406006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak JA, Fuchs E. Isolation and culture of epithelial stem cells. Methods in molecular biology (Clifton, N.J. 2009;482:215–232. doi: 10.1007/978-1-59745-060-7_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson M. Enrichment of super-sized resequencing targets from the human genome. Nature methods. 2007;4:891–892. doi: 10.1038/nmeth1107-891. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Harrow J, Harte RA, Wallin C, Diekhans M, Maglott DR, Searle S, Farrell CM, Loveland JE, Ruef BJ, Hart E, Suner MM, Landrum MJ, Aken B, Ayling S, Baertsch R, Fernandez-Banet J, Cherry JL, Curwen V, Dicuccio M, Kellis M, Lee J, Lin MF, Schuster M, Shkeda A, Amid C, Brown G, Dukhanina O, Frankish A, Hart J, Maidak BL, Mudge J, Murphy MR, Murphy T, Rajan J, Rajput B, Riddick LD, Snow C, Steward C, Webb D, Weber JA, Wilming L, Wu W, Birney E, Haussler D, Hubbard T, Ostell J, Durbin R, Lipman D. The consensus coding sequence (CCDS) project: Identifying a common protein-coding gene set for the human and mouse genomes. Genome research. 2009;19:1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson RJ, Dixon J, Malhotra S, Hardman MJ, Knowles L, Boot-Handford RP, Shore P, Whitmarsh A, Dixon MJ. Irf6 is a key determinant of the keratinocyte proliferation-differentiation switch. Nature genetics. 2006;38:1329–1334. doi: 10.1038/ng1894. [DOI] [PubMed] [Google Scholar]
- Romano RA, Ortt K, Birkaya B, Smalley K, Sinha S. An active role of the DeltaN isoform of p63 in regulating basal keratin genes K5 and K14 and directing epidermal cell fate. PloS one. 2009;4:e5623. doi: 10.1371/journal.pone.0005623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders JW., Jr. The proximo-distal sequence of origin of the parts of the chick wing and the role of the ectoderm. The Journal of experimental zoology. 1948;108:363–403. doi: 10.1002/jez.1401080304. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular systems biology. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart IH. Variation in the plane of cell cleavage during the process of stratification in the mouse epidermis. The British journal of dermatology. 1970;82:276–282. doi: 10.1111/j.1365-2133.1970.tb12437.x. [DOI] [PubMed] [Google Scholar]
- Summerbell D. A quantitative analysis of the effect of excision of the AER from the chick limb-bud. Journal of embryology and experimental morphology. 1974;32:651–660. [PubMed] [Google Scholar]
- Sun X, Mariani FV, Martin GR. Functions of FGF signalling from the apical ectodermal ridge in limb development. Nature. 2002;418:501–508. doi: 10.1038/nature00902. [DOI] [PubMed] [Google Scholar]
- Tadeu AM, Horsley V. Notch signaling represses p63 expression in the developing surface ectoderm. Development (Cambridge, England) 2013 doi: 10.1242/dev.093948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, Takeuchi O, Tsujimura T, Itami S, Adachi O, Kawai T, Sanjo H, Yoshikawa K, Terada N, Akira S. Limb and skin abnormalities in mice lacking IKKalpha. Science (New York, N.Y. 1999;284:313–316. doi: 10.1126/science.284.5412.313. [DOI] [PubMed] [Google Scholar]
- Truong AB, Kretz M, Ridky TW, Kimmel R, Khavari PA. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes & development. 2006;20:3185–3197. doi: 10.1101/gad.1463206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M, Bjorling E, Agaton C, Szigyarto CA, Amini B, Andersen E, Andersson AC, Angelidou P, Asplund A, Asplund C, Berglund L, Bergstrom K, Brumer H, Cerjan D, Ekstrom M, Elobeid A, Eriksson C, Fagerberg L, Falk R, Fall J, Forsberg M, Bjorklund MG, Gumbel K, Halimi A, Hallin I, Hamsten C, Hansson M, Hedhammar M, Hercules G, Kampf C, Larsson K, Lindskog M, Lodewyckx W, Lund J, Lundeberg J, Magnusson K, Malm E, Nilsson P, Odling J, Oksvold P, Olsson I, Oster E, Ottosson J, Paavilainen L, Persson A, Rimini R, Rockberg J, Runeson M, Sivertsson A, Skollermo A, Steen J, Stenvall M, Sterky F, Stromberg S, Sundberg M, Tegel H, Tourle S, Wahlund E, Walden A, Wan J, Wernerus H, Westberg J, Wester K, Wrethagen U, Xu LL, Hober S, Ponten F. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics. 2005;4:1920–1932. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- van Bokhoven H, Hamel BC, Bamshad M, Sangiorgi E, Gurrieri F, Duijf PH, Vanmolkot KR, van Beusekom E, van Beersum SE, Celli J, Merkx GF, Tenconi R, Fryns JP, Verloes A, Newbury-Ecob RA, Raas-Rotschild A, Majewski F, Beemer FA, Janecke A, Chitayat D, Crisponi G, Kayserili H, Yates JR, Neri G, Brunner HG. p63 Gene mutations in eec syndrome, limb-mammary syndrome, and isolated split hand-split foot malformation suggest a genotype-phenotype correlation. American journal of human genetics. 2001;69:481–492. doi: 10.1086/323123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LW, Zelickson AS. Embryology of the epidermis: ultrastructural aspects. II. Period of differentiation in the mouse with mammalian comparisons. Acta dermato-venereologica. 1975;55:321–329. [PubMed] [Google Scholar]
- Westfall MD, Mays DJ, Sniezek JC, Pietenpol JA. The Delta Np63 alpha phosphoprotein binds the p21 and 14-3-3 sigma promoters in vivo and has transcriptional repressor activity that is reduced by Hay-Wells syndrome-derived mutations. Molecular and cellular biology. 2003;23:2264–2276. doi: 10.1128/MCB.23.7.2264-2276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, McKeon F. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.