Abstract
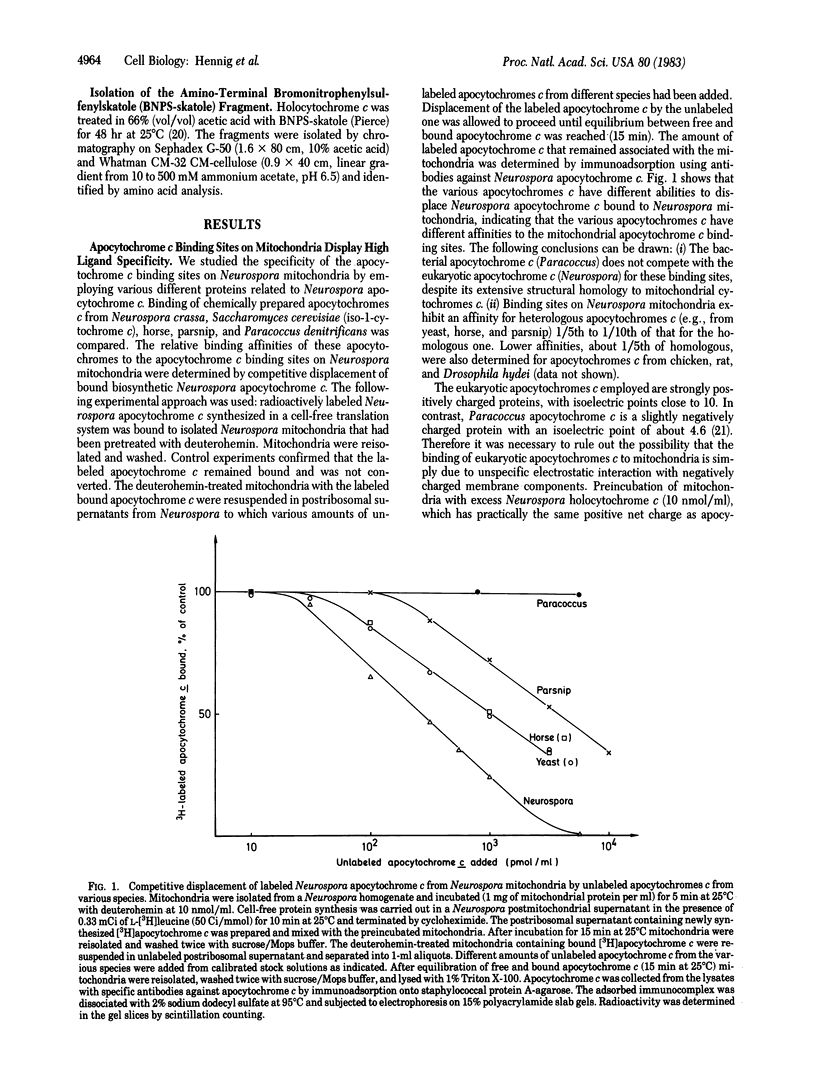
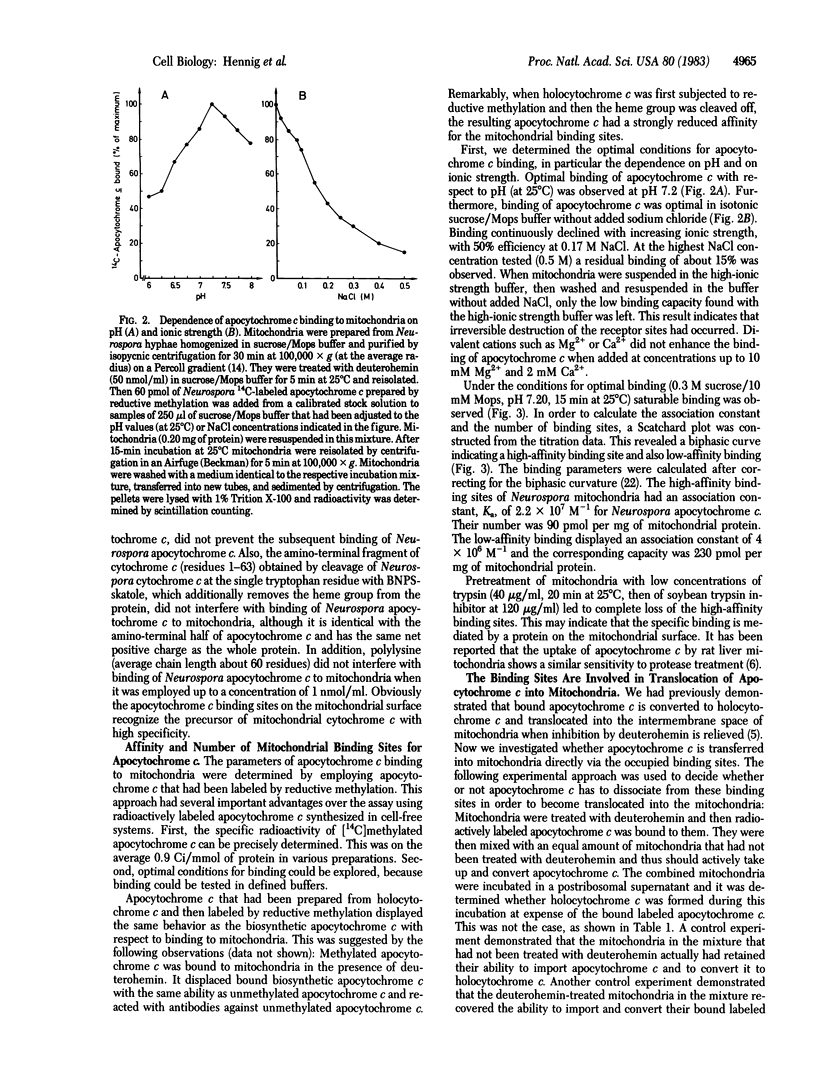
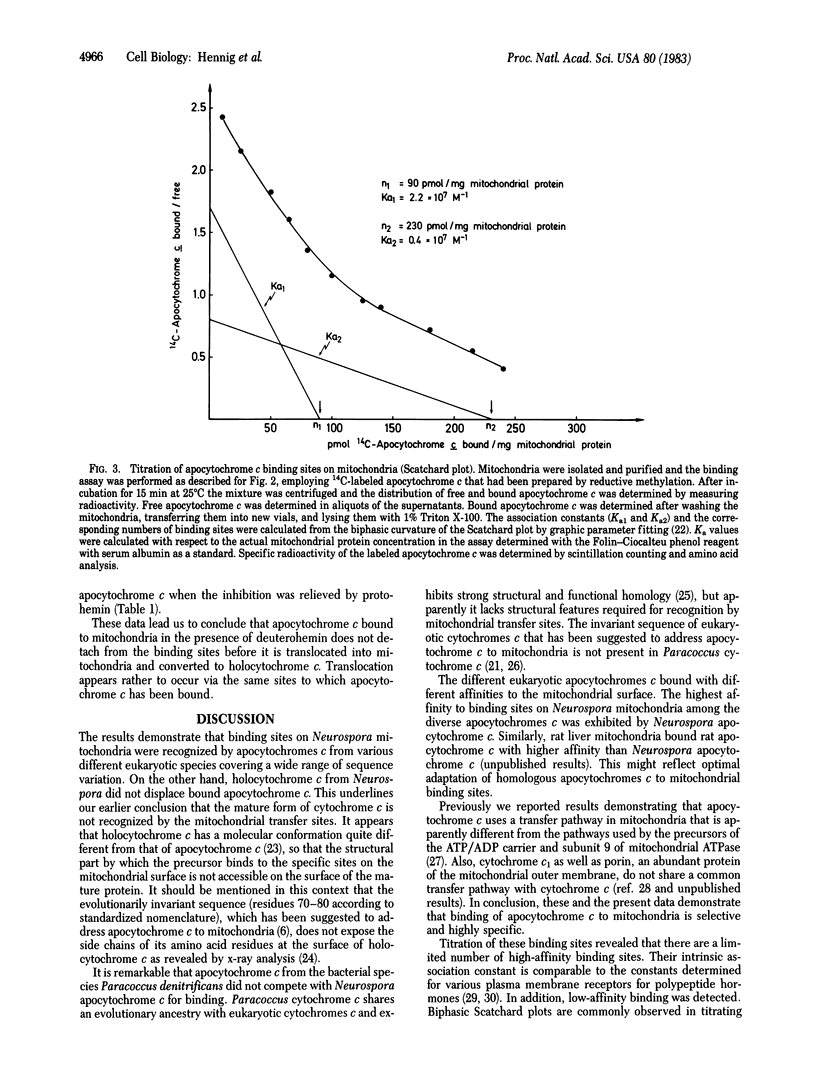
Assembly of cytochrome c involves a series of steps: synthesis of apocytochrome c on free ribosomes, specific binding of apocytochrome c to the mitochondrial surface, transfer across the outer membrane, covalent addition of protoheme, refolding of the polypeptide chain, and association of holocytochrome c with its functional sites at the inner membrane. The binding step of apocytochrome c to Neurospora crassa mitochondria was studied by inhibiting the subsequent transfer steps with the heme analogue deuterohemin. The binding sites are highly specific for mitochondrial apocytochromes c. Bound labeled Neurospora apocytochrome c was competitively displaced by unlabeled apocytochrome c from various species. These exhibited different abilities for displacement. Apocytochrome c from Paracoccus denitrificans, the amino-terminal (heme-binding) fragment of Neurospora apocytochrome c, and Neurospora holocytochrome c did not recognize the binding sites. Polylysine did not interfere with apocytochrome c binding. Apocytochrome c is reversibly bound. The binding sites are present in limited number. High-affinity binding sites were present at about 90 pmol/mg of mitochondrial protein. They displayed an association constant of 2.2 X 10(7) M-1. Apocytochrome c was imported into mitochondria and converted to holocytochrome c directly from the binding sites when inhibition by deuterohemin was relieved. We conclude that the apocytochrome c binding sites on mitochondria represent receptors that function in the recognition and import of this precursor by mitochondria.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alberghina F. A., Trezzi F., Signorini R. C. The biogenesis of mitochondria in Neurospora crassa: ultrastructural changes induced by nutrients. Cell Differ. 1974 Mar;2(6):307–317. doi: 10.1016/0045-6039(74)90009-8. [DOI] [PubMed] [Google Scholar]
- Almassy R. J., Dickerson R. E. Pseudomonas cytochrome c551 at 2.0 A resolution: enlargement of the cytochrome c family. Proc Natl Acad Sci U S A. 1978 Jun;75(6):2674–2678. doi: 10.1073/pnas.75.6.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambler R. P., Meyer T. E., Kamen M. D., Schichman S. A., Sawyer L. A reassessment of the structure of Paracoccus cytochrome c-550. J Mol Biol. 1981 Apr 5;147(2):351–356. doi: 10.1016/0022-2836(81)90445-9. [DOI] [PubMed] [Google Scholar]
- Basile G., Di Bello C., Taniuchi H. Formation of an iso-1-cytochrome c-like species containing a covalently bonded heme group from the apoprotein by a yeast cell-free system in the presence of hemin. J Biol Chem. 1980 Aug 10;255(15):7181–7191. [PubMed] [Google Scholar]
- Boss J. M., Darrow M. D., Zitomer R. S. Characterization of yeast iso-1-cytochrome c mRNA. J Biol Chem. 1980 Sep 25;255(18):8623–8628. [PubMed] [Google Scholar]
- Chua N. H., Schmidt G. W. Transport of proteins into mitochondria and chloroplasts. J Cell Biol. 1979 Jun;81(3):461–483. doi: 10.1083/jcb.81.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dottavio-Martin D., Ravel J. M. Radiolabeling of proteins by reductive alkylation with [14C]formaldehyde and sodium cyanoborohydride. Anal Biochem. 1978 Jul 1;87(2):562–565. doi: 10.1016/0003-2697(78)90706-6. [DOI] [PubMed] [Google Scholar]
- Fisher W. R., Taniuchi H., Anfinsen C. B. On the role of heme in the formation of the structure of cytochrome c. J Biol Chem. 1973 May 10;248(9):3188–3195. [PubMed] [Google Scholar]
- Fontana A., Vita C., Toniolo C. Selective cleavage of the single tryptophanyl peptide bond in horse heart cytochrome c. FEBS Lett. 1973 May 15;32(1):139–142. doi: 10.1016/0014-5793(73)80757-4. [DOI] [PubMed] [Google Scholar]
- Hennig B. Change of cytochrome c structure during development of the mouse. Eur J Biochem. 1975 Jun 16;55(1):167–183. doi: 10.1111/j.1432-1033.1975.tb02149.x. [DOI] [PubMed] [Google Scholar]
- Hennig B., Neupert W. Assembly of cytochrome c. Apocytochrome c is bound to specific sites on mitochondria before its conversion to holocytochrome c. Eur J Biochem. 1981 Dec;121(1):203–212. doi: 10.1111/j.1432-1033.1981.tb06450.x. [DOI] [PubMed] [Google Scholar]
- Korb H., Neupert W. Biogenesis of cytochrome c in Neurospora crassa. Synthesis of apocytochrome c, transfer to mitochondria and conversion to Holocytochrome c. Eur J Biochem. 1978 Nov 15;91(2):609–620. doi: 10.1111/j.1432-1033.1978.tb12714.x. [DOI] [PubMed] [Google Scholar]
- Matner R. R., Sherman F. Differential accumulation of two apo-iso-cytochromes c in processing mutants of yeast. J Biol Chem. 1982 Aug 25;257(16):9811–9821. [PubMed] [Google Scholar]
- Matsuura S., Arpin M., Hannum C., Margoliash E., Sabatini D. D., Morimoto T. In vitro synthesis and posttranslational uptake of cytochrome c into isolated mitochondria: role of a specific addressing signal in the apocytochrome. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4368–4372. doi: 10.1073/pnas.78.7.4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery D. L., Leung D. W., Smith M., Shalit P., Faye G., Hall B. D. Isolation and sequence of the gene for iso-2-cytochrome c in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1980 Jan;77(1):541–545. doi: 10.1073/pnas.77.1.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollet R. J., Standaert M. L., Haase B. A. Insulin binding to the human lymphocyte receptor. Evaluation of the negative cooperativity model. J Biol Chem. 1977 Aug 25;252(16):5828–5834. [PubMed] [Google Scholar]
- Powell-Jones C. H., Thomas C. G., Jr, Nayfeh S. N. Thyrotropin receptors in normal human thyroid. Nonclassical binding kinetics not explained by the negative cooperativity model. J Biol Chem. 1980 May 10;255(9):4001–4010. [PubMed] [Google Scholar]
- Sabatini D. D., Kreibich G., Morimoto T., Adesnik M. Mechanisms for the incorporation of proteins in membranes and organelles. J Cell Biol. 1982 Jan;92(1):1–22. doi: 10.1083/jcb.92.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla R. C., Agne K. M., Wu R. Isolation and structure of a rat cytochrome c gene. J Biol Chem. 1981 Jun 25;256(12):6480–6486. [PubMed] [Google Scholar]
- Scholes P. B., McLain G., Smith L. Purification and properties of a c-type cytochrome from Micrococcus denitrificans. Biochemistry. 1971 May 25;10(11):2072–2076. doi: 10.1021/bi00787a017. [DOI] [PubMed] [Google Scholar]
- Sherman F., Taber H., Campbell W. Genetic determination of iso-cytochromes c in yeast. J Mol Biol. 1965 Aug;13(1):21–39. doi: 10.1016/s0022-2836(65)80077-8. [DOI] [PubMed] [Google Scholar]
- Smith M., Leung D. W., Gillam S., Astell C. R., Montgomery D. L., Hall B. D. Sequence of the gene for iso-1-cytochrome c in Saccharomyces cerevisiae. Cell. 1979 Apr;16(4):753–761. doi: 10.1016/0092-8674(79)90091-6. [DOI] [PubMed] [Google Scholar]
- Takano T., Dickerson R. E. Conformation change of cytochrome c. II. Ferricytochrome c refinement at 1.8 A and comparison with the ferrocytochrome structure. J Mol Biol. 1981 Nov 25;153(1):95–115. doi: 10.1016/0022-2836(81)90529-5. [DOI] [PubMed] [Google Scholar]
- Teintze M., Slaughter M., Weiss H., Neupert W. Biogenesis of mitochondrial ubiquinol:cytochrome c reductase (cytochrome bc1 complex). Precursor proteins and their transfer into mitochondria. J Biol Chem. 1982 Sep 10;257(17):10364–10371. [PubMed] [Google Scholar]
- Timkovich R., Dickerson R. E., Margoliash E. Amino acid sequence of Paracoccus denitrificans cytochrome c550. J Biol Chem. 1976 Apr 25;251(8):2197–2206. [PubMed] [Google Scholar]
- Veloso D., Basile G., Taniuchi H. Formation of a cytochrome c-like species from horse apoprotein and hemin catalyzed by yeast mitochondrial cytochrome c synthetase. J Biol Chem. 1981 Aug 25;256(16):8646–8651. [PubMed] [Google Scholar]
- Weder H. G., Schildknecht J., Lutz R. A., Kesselring P. Determination of binding parameters from Scatchard plots. Theoretical and practical considerations. Eur J Biochem. 1974 Mar 1;42(2):475–481. doi: 10.1111/j.1432-1033.1974.tb03361.x. [DOI] [PubMed] [Google Scholar]
- Weiss H., von Jagow G., Klingenberg M., Bücher T. Characterization of Neurospora crassa mitochondria prepared with a grind-mill. Eur J Biochem. 1970 May 1;14(1):75–82. doi: 10.1111/j.1432-1033.1970.tb00263.x. [DOI] [PubMed] [Google Scholar]
- Zimmermann R., Hennig B., Neupert W. Different transport pathways of individual precursor proteins in mitochondria. Eur J Biochem. 1981 Jun 1;116(3):455–460. doi: 10.1111/j.1432-1033.1981.tb05357.x. [DOI] [PubMed] [Google Scholar]
- Zimmermann R., Paluch U., Neupert W. Cell-free synthesis of cytochrome c. FEBS Lett. 1979 Dec 1;108(1):141–146. doi: 10.1016/0014-5793(79)81196-5. [DOI] [PubMed] [Google Scholar]


