Abstract
Non-coding RNAs (ncRNAs) are important regulatory molecules involving in various physiological cellular processes. Alterations of ncRNAs, particularly microRNAs, play crucial roles in tumorigenesis. Accumulating evidence indicates that small nucleolar RNAs (snoRNAs), another large class of ncRNAs, are gaining prominence and more actively involved in carcinogenesis than previously thought. Some snoRNAs exhibit differential expression patterns in a variety of human cancers and demonstrate capability to affect cell transformation, tumorigenesis, and metastasis. We are beginning to comprehend the functional repercussions of snoRNAs in the development and progression of malignancy. In this review, we will describe current studies that have shed new light on the functions of snoRNAs in carcinogenesis and the potential applications for cancer diagnosis and diagnosis.
Keywords: Cancer, non-coding RNAs, small nucleolar RNAs, diagnosis, therapy
1. Introduction
Cancer is the second most common cause of death in the US, exceeded only by heart disease [1]. Fundamental understanding of the mechanisms underlying tumorigenesis will help develop effective approaches for its early detection and treatments, and hence reduce the mortality. Cancer results from the disrupted regulating cell growth and death, which are maintained by the coordinated function of protein-coding genes [2]. Therefore, protein coding genes have long been considered as important regulatory molecules in tumor initiation and progression.
The human genome encodes approximately 25,000 protein-coding genes, representing only <2% of the total genome sequence [3-5]. However, out of more than 98% of non-protein-coding DNA, at least 90% of the genome is actively transcribed. The transcriptome, which essentially comprises small non-coding (nc) RNAs and long ncRNAs as well, is more complex than a collection of protein-coding genes [5-7]. The small ncRNAs represent a loosely grouped RNA species, including microRNAs (miRNAs), small nucleolar RNAs (snoRNAs), short interfering RNAs (siRNAs), piwi-associated RNAs, small Cajal body-specific RNAs (scaRNAs), snRNAs (small nuclear RNAs). Although ranging in length from 18 to 300 nucleotides (nt), the small ncRNAs have big functions and are of crucial importance in a spectrum of regulatory processes, such as cell development, physiology, and pathogenesis [8,9]. Furthermore, small ncRNAs are emerging as new players in the cancer paradigm and become increasingly important in tumorigenesis. In addition, miRNA-expression profiling of human tumors has identified the signatures that could potentially be used for cancer diagnosis, prognosis, and response to treatments [10-17]. In addition, given their primarily function as post-transcriptional regulators [18, 19], some miRNAs can act as tumor suppressors or oncogenes depending on their target genes [20].
However, other types of small ncRNAs may also plays critical roles in regulating diverse cellular processes, and their dysfunction could consequently contribute to tumorigenesis in previously unanticipated means [21]. Therefore, investigation of dysregulation of other classes of ncRNAs in cancer, and their diagnostic and therapeutic values is of great significance. Especially, small nucleolar RNAs (snoRNAs) or snoRNA dysregulation has recently exhibited important function in tumorigenesis. For instance, several indications of unexpected roles of snoRNAs as cancer genes have emerged from recent studies [22]. The growing knowledge of the role of snoRNAs in tumorigenesis would point towards the potential as novel biomarkers and therapeutic targets for cancer. Therefore, in this review article, we will mainly focus our discussion on the novel functions of snoRNAs in carcinogenesis and the possible applications for cancer diagnosis and treatment in the future.
2. Biogenesis, action mechanisms, and cellular functions of snoRNAs
2.1. Biogenesis
SnoRNAs range in size from 60-300 nt [23]. At least 200 snoRNAs have been identified in mammals, but many more remain to be found [24]. There are two types of snoRNAs: Box C/D or Box H/ACA snoRNAs (Fig. 1A and B). [25-27]. Most of snoRNAs are located within introns of genes transcribed via RNA polyermase II (Fig. 2). However, snoRNAs can also be processed from introns of long non-protein-coding RNAs (lncRNAs) [28, 29]. For example, GAS5, an lncRNA, encodes 9 C/D box snoRNAs (snoRNDs74-81) [30]. After liberation from introns, snoRNAs are processed to remove excess nucleotides from either end via exonuclease activity. Signal sequences within the snoRNAs centered at boxes C and D or H and ACA directs binding of protein interacting partners that represent the functional snoRNP complex.
Figure 1.
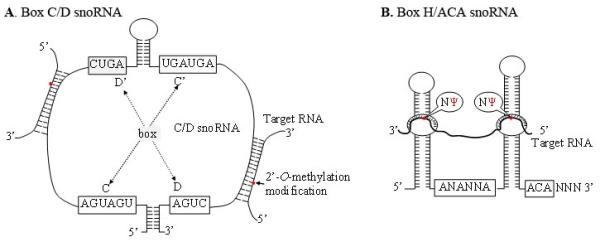
Schematic structures of the C/D-box and H/ACA-box snoRNAs
A. The 2′-O-methylated nucleotides located five nucleotides upstream of the D or D’ box sequences are indicated as red. B. Positions and consensus sequences of the conserved C, C’ box (UGAUGA), and D, D’ boxes (CUGA). The uridine residues selected for pseudouridylation are shown as Ψ.
Figure 2.
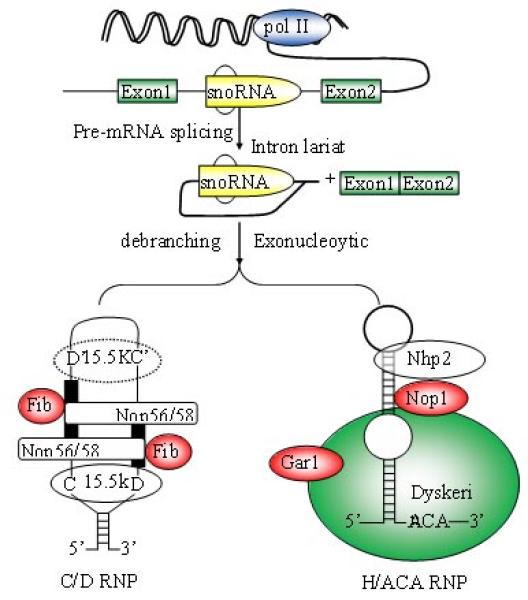
Biogenesis of snoRNA
The great majority of mammalian box C/D and H/ACA snoRNAs are processed from pre-mRNA introns. snoRNA families are not independently transcribed but processed from the pre-mRNA introns, in most cases by exonucleolytic digestion of the debranched lariat. Box C/D snoRNAs contain four evolutionarily conserved, essential proteins, fibrillarin (methyltransferase), Nop56, Nop58, and 15.5kDa. Proteins common to H/ACA snoRNAs include dyskerin (pseudouridine synthase), Gar1, Nhp2, and Nop10p.
2. 2. Action mechanisms
The snoRNAs have two basic action mechanisms: 2′-O-methylation and pseudouridylation. 2′-O-methylation is carried out by the box C/D snoRNA family, which contains two short sequence motifs, C and D (Fig. 1A). The 4-5 nucleotides at both termini of the snoRNA form a terminal stem-box required for snoRNA biogenesis and nucleolar localization [31]. The conserved C, C’, D and D’ box elements are in the central portion of the snoRNA, where RNA-protein interactions occur to direct the proper assembly of the functional ribonucleoprotein complexes [32]. Upstream of the box D or D’ elements are either one or two antisense elements, which are complementary to a specific site of rRNA, allowing for proper alignment and methylation of the appropriate nucleotide. The snoRNA component directs the snoRNP complex to the appropriate rRNA location, where the methyltransferase, fibrillarin, catalyzes the methylation reaction [31-33]. Pseudouridylation is accomplished by the H/ACA box snoRNAs family that is composed of two hairpins, which form a hairpin-hinge-hairpin-tail secondary structure (Fig. 1B and Fig 2) [32-34]. The H box is in the hinge region, while the ACA motif is in the tail region. Guide sequences that direct the snoRNAs to the appropriate rRNA sequence are in one or both of the hairpin loop domains. After targeting the snoRNP complex to the appropriate site, the substrate rRNA uridine locates at the base of the upper stem [35]. There is 14-16 nt distance between the box motifs, and this site is modified by the pseudouridine synthase, dyskerin, [32].
2.3. Cellular functions
The central functions of snoRNAs have long been believed to modify, mature, and stable rRNAs. These post-transcriptional modifications are important for the production of efficient and accurate ribosomes [36]. However, snoRNAs’ cellular functions continue to expand. For example, snoRNAs can modify small nuclear or snRNAs that mediate mRNA splicing. A brain specific box C/D snoRNA (HBII-52) that contains a 18 nt conserved target recognition element is 100% complimentary to the serotonin receptor 5-HT (2C) mRNA [37]. The snoRNA HBII-52 controls the processing of mRNA expression of the serotonin receptor 2C by regulating the alternative splicing, and hence contributes to the Prader-Willi syndrome [37,38]. Furthermore, snoRNA transcripts serve as the precursors of miRNA-like small RNAs and the regulators of alternative splicing [39, 40]. In addition, the processed RNAs could hold the key to some of the newly found effects of snoRNAs [22]. For example, Ender et al. [39] found that the processing of H/ACA box snoRNA ACA45 produced 20-25 nt-long RNAs that associated with Argonaut proteins and targeted specific mRNAs, including CDK11. Ono et al. [41] found that a subset of miRNAs shares functional H/ACA box snoRNA characteristics, and thus suggested that these miRNAs might have evolved from snoRNAs. Moreover, some snoRNAs could be processed to produce small RNAs, of which, some functioned like miRNAs. Such processing could be of crucial importance, because miRNAs have essential roles in a spectrum of regulatory processes, such as the control of cell survival and proliferation. Recently, Brameier et al. [42] denote the small RNAs that are produced from snoRNAs as sno-miRNAs. The sno-miRNAs could have dual functions: the same transcript could produce both to a snoRNA and through subsequent nucleolytic processing steps to a miRNA [43]. Therefore, snoRNAs, which arose early in evolution, may have given rise to the first miRNAs in early metazoan cells [22]. Additionally, vertebrate telomerase RNAs contain a typical H/ACA domain that function as the template for telomere DNA synthesis. Because the box H/ACA motif of the human telomerase reverse transcriptase (hTER) is required for its association with four proteins (dyskerin, NHP2, NOP10, and GAR1) that are common in snoRNAs, snoRNAs might involve in function of telomere [44-46].
With new discoveries for cellular RNA functions increasing, the novel roles for snoRNA-mediated nucleotide modifications will continually be discovered. Importantly, several independent lines of evidence have strongly indicated that alterations of snoRNAs play important functions in cancer development and progression [47-53]. Therefore, further indetifying new functions of snoRNAs and in-deep understanding the roles of their dysregulations in malignancy will help comprehend tumorigenesis, and hence provides potential biomarkers and therapeutic targets for the disease.
3. Evidence for snoRNA aberrations involved in tumorigenesis
The first report linking snoRNA molecules to cancer was demonstrated by Chang et al. who found that h5sn2, a box H/ACA snoRNA, was significantly downregulated in human meningiomas compared with normal brain tissues [50] (Table 1.) Subsequently, Donsante et al. [51] observed that normal mice and mice with mucopolysaccharidosis VII could develop hepatocellular carcinoma (HCC) after neonatal injection of an adeno-associated viral (AAV) vector expressing β-glucuronidase. The vector was isolated from tumor tissue specimens and located within a 6-kilobase region of chromosome 12. Interestingly, this locus encodes several imprinted transcripts, including snoRNAs. Furthermore, the locus includes Rian gene, which also encodes nine snoRNAs and microRNAs. The findings implicate that these snoRNAs could play an important role in the development of HCC. Furthermore, to identify prostate-associated genes in chromosome 6q14-q22, whose deletion is common in multiple human cancers, Dong et al. narrowed the common region of deletion to a 2.5 Mb interval at 6q14-15. Of the 11 genes located in this minimal deletion region, only snoRNA U50 was discovered to be mutated in prostate cancer cells [52]. Furthermore, a homozygous 2 bp (TT) deletion in the snoRNA U50 was found in two of 30 prostate cancer cell lines/xenografts and nine of 89 prostate cancers. In addition, the heterozygous genotype of the same deletion also occurred in 8 of 31 (26%) breast cancer cell lines tested [53]. Coincidently, chromosome 6q14-15 where snoRNA U50 located at is a breakpoint of chromosomal translocation t(3;6)(q27;q15) for human-B cell lymphoma [54]. Altogether, the studies imply that the snoRNA gene could involve in tumorigenesis of a variety of cancers. Mourtada-Maarabouni et al. found that growth arrest-specific transcript 5 (GAS5) transcript levels were substantially reduced in breast tumors relative to adjacent normal breast epithelial tissues [55]. GAS5 can control mammalian apoptosis and cell population growth. Intriguingly, GAS5 has no significant protein-coding potential, but encodes nine box C/D snoRNAs in its introns. The observations suggest that the snoRNAs form a novel family of genes that could control oncogenesis and sensitivity to therapy in breast cancer. Moreover, Nakamura et al. found that GAS5 could be a novel partner of the BCL6 in a patient with diffuse large B-cell lymphoma, who had chromosomal translocation t(1;3)(q25;q27) [56]. In this case, the chromosome 1 breakpoint (1q25) was located within the intronic snoRNA sequence of GAS5 and the chromosome 3 breakpoint (3q27) at 4 kb upstream of BCL6 exon 1a. As the result of the chromosomal translocation, the GAS5-BCL6 chimeric transcripts were expressed, in which the 5′-terminal oligopyrimidine (5′TOP) sequence of GAS5 was fused to the whole coding sequence of BCL6. Therefore, the snoRNAs enclosed in GAS5 also contribute to human lymphoma due to chromosomal translations or breakages. By profiling ncRNA signatures in 22 NSCLC tissues and matched noncancerous lung tissues, we identified six snoRNAs that displayed higher expressions in lung tumor tissues compared with their normal counterparts (Table 1) [57]. The data imply the close association of the snoRNA alterations with tumorigenesis of lung cancer. Furthermore, like miRNAs, some snoRNAs are located at chromosomal amplified regions that frequently involve in human carcinogenesis [58-62]. Notably, five of the six snoRNAs displaying dysregulations in lung tumor specimens were located in commonly frequent genomic amplified regions in human solid cancers [57, 58]. For instance, SNORD33 is located in chromosome 19q13.3 that contain potential oncogenes in malignancies, including lung cancer [57, 58], while SNORD66 and SNORD76 are situated in chromosomal regions 3q27.1 and 1q25.1, respectively. 3q27.1 and 1q25.1 are two of the most frequently amplified chromosomal segments in human solid tumors [58-62]. Recently, Gee et al. used four snoRNAs, RNU44, RNU48, RNU43 and RNU6B, as internal control genes to analyze cancer-related miRNAs in two patient series: 219 breast cancer and 46 head and neck squamous cell carcinomas [63]. Surprisingly, low expressions of RNU43, RNU44, and RNU48 in the tumors were significantly associated with a poor prognosis of the cancer patients. Taken together, differential snoRNA aberrations found in a variety of cancer types suggest that snoRNA dysfunctions are truly involved in important functions in regulating cellular homeostasis and tumor biology. Therefore, investigating new functions of snoRNAs in carcinogenesis is imperative in the cancer research field.
Table 1.
Representative snoRNAs involved in cancer and their proposed roles in tumorigenesis.
| SnoRNAs | Class | Changes | Proposed role | Cancer type | References |
|---|---|---|---|---|---|
| U50 | C/D box | Decreased | TS | Breast & Prostate cancer | [53, 54] |
| h5sn2 | H/ACA | Decreased | TS | Meningioma | [51] |
| RNU43 | C/D box | Decreased | TS | Breast cancer & HNSCC | [64] |
| RNU44 | C/D box | Decreased | TS | Breast cancer & HNSCC | [64] |
| snoRD33 | C/D box | Increased | OG | NSCLC | [58] |
| snoRD66 | C/D box | Increased | OG | NSCLC | [58] |
| snoRD76 | C/D box | Increased | OG | NSCLC | [58] |
| snoRA42 | H/ACA box | Increased | OG | NSCLC | [58,104] |
| snoRD44 | C/D box | Increased | OG | Breast cancer | [110] |
TS, tumor suppressor; OG, oncogene
4. Emerging role of snoRNAs in tumorigenesis
4.1. Imprinting snoRNAs in cancers
The loss of the imprinting genes and their associated aberrant gene expressions are key features of cancer [64,65]. Imprinting snoRNAs have been reported to be associated with certain cancers [51, 66]. For instance, integration of an AAV vector containing a cytomegalovirus enhancer and human β-glucuronidase gene into normal newborn mouse genome could produce HCC. The AAV-HCC locus on chromosome 12 harbors multiple imprinted genes, including Rian [67-69]. The Rian encodes at least nine imprinting snoRNAs. Interestingly, the imprinted snoRNAs encoded in Rian were overexpressed by 9- to 539-fold in tumor tissues as compared with normal tissues. Therefore, the oncogenic effect of the vector integration might be due to the overexpressions of the snoRNAs [51]. Another example is MEG3, a maternally expressed imprinted gene. It possesses tumor suppressor activities and its down-regulation inhibits cancer cell proliferation by both p53-dependent and p53-independent pathways [66]. Interestingly, MEG3 harbors a couple of snoRNAs, including SNORD112, SNORD113, and SNORD114 and tumor suppressor miRNAs [66,70,71]. Furthermore, MEG8 is another imprinting gene, which is an lncRNA and locates in chromosome 14q32 region. Dysfunction of MEG8 is implicated in several diseases including Prader-Willi/Angelman syndromes [72]. Interestingly, the chromosomal locus 14q32 has been proposed to have tumor suppressor function [73]. Like MEG3, MEG8 RNA contains repeats of two intronic snoRNAs: SNORD113 (9 copies) and SNORD114 (31 copies) (73). Altogether, the imprinting snoRNAs could have important functions in carcinogenesis. Nevertheless, research toward a better understanding of the precise biological role of the enclosed imprinting snoRNAs and determination of whether the snoRNAs have independent function from the host genes in cancer initiation and progression is required [22].
4.2. Human telomerase RNA (hTR)-associated H/ACA snoRNAs in cancer
hTR shares H/ACA box characteristics with snoRNAs, which localizes to Cajal body in nucleus of cells. Telomerase is a reverse transcriptase that carries its own RNA molecule [74-77]. hTR contains 11 nucleotides (5′-CUAACCCUAAC), which are complementary to the telomere sequence (TTAGGG) repeats [74-77]. The telomere sequence repeats act as a template for DNA synthesis by adding telomeres to the ends of chromosomes [74-77]. Therefore, hTR contributes to protection of chromosomes against a variety of challenges. The deregulation of telomerase RNA involves in tumorigenesis (78-82). Interestingly, hTR H/ACA domain is responsible for pre-RNP formation and nucleolar localization of the telomerase RNP itself. Importantly, the H/ACA snoRNP has been implicated in the X-linked genetic disorder dyskeratosis congenita (DKC) due to its affiliation with human telomerase [83]. Patients with DKC are predisposed to a variety of cancers, including hematological malignancies, melanomas, prostate cancer, and breast cancer [84-86]. Therefore, dysfunction of hTR-associated H/ACA snoRNAs may also play a crucial role in the development and progression of cancer [86]. However, the detailed biological functional of the hTR-associated H/ACA snoRNAs in carcinogenesis remains to be investigated.
4.3. SnoRNA-associated ribosomopathies in cancer
The snoRNA components of snoRNPs are essential in the modifying, processing, and dynamic folding rRNAs for the creation of efficient and accurate ribosomes [87]. Dysregulations of ribosome biogenesis and associated ribosomopathies have been frequently documented in a variety of cancers [88-90]. For instance, abnormal ribosome biogenesis results in chromosomal instability, a common feature in human tumors [91]. Furthermore, ribosome biogenesis requires numerous proteins, which are linked to DNA replication [92]. Therefore, deregulation of one or more of these proteins will promote cancer intiaition and progression through an aberrant protein synthesis and altered chromosomal segregation [90]. For example, H/ACA box snoRNAs guide specific rRNA sites for the modification and assist dyskerin to catalyze pseouridylation. Dyskerin is involved in all basic cellular events such as protein translation, cell growth, and proliferation. Dyskerin is also responsible for stabilization of telomerase RNA component and proper function of telomerase enzymatic complex. Mutations in dyskerin cause ribosomopathies that lead to dyskeratosis congenita. Interestingly, dyskeratosis congenita is characterized by increased susceptibility to cancers, such as skin, breast, and prostate tumors [84-86]. Furthermore, the gene associated with retinoid-interferon-induced mortality-1 (GRIM-1) can inhibit rRNA maturation by suppressing H/ACA box snoRNA expressions and rRNA processing [93]. GRIM-1 inhibits cell growth by sequestering NAF1, which in turn causes a loss of box H/ACA RNAs and mature rRNA levels [93]. Interestingly, GRIM-1 expression was suppressed in human prostate cancer, indicating gain of ribosome function can result in high synthetic capacities. Moreover, elevated expression of a nucleolar Nop5/Sik, which is a snoRNA binding protein, is actively involved in ribosome biogenesis through augmenting the activities of nucleolus in metastatic melanoma cells [94]. It has also been shown that reduced ribosome biogenesis is correlated with malignant transformation in zebrafish [95]. Recently, Michel et al. [96] identified the 60S ribosomal protein rpL13a gene as a critical factor for palmitate-induced metabolic stress and cell death. Promoter trap mutation could disturb expression of rpL13-encoded box C/D snoRNAs U32a, U33, and U35a [96]. Knockdown of U32a, U33, and U35a would protect cells from palmitate induced oxidative stress including ER stress, and subsequent cell death [96]. This protective effect was independent of 2′-O-ribose methylation of rRNA targets, which is the primary function that is associated with box C/D snoRNAs. Therefore, such alteration of snoRNAs could induce ribosomal stress, which in turn may act as oncogenic stress [22, 96]. Altogether, the snoRNA-associated ribosomopaties could contribute to carcinogenesis.
4.4. SnoRNAs have oncogenic and tumor suppressor roles in tumorigenesis
Similar to protein-coding oncogenes, some snoRNAs have exhibited evidence in promoting cellular pathways that lead to tumorigenesis. One example of such an oncogenic snoRNA is snoRNA42, an H/ACA snoRNA. We have found that snoRA42 is one of the most commonly overexpressed snoRNAs in lung tumors [97]. Gene amplification is a major mechanism allowing for increased expression of oncogenes that contribute to cancer development and progression [98]. Interestingly, snoRA42 is located in 1q22, a frequent genomic amplified region observed in a variety of solid tumors, including non-small cell lung cancer (NSCLC) [60, 61. 99]. Furthermore, SNORA42 resides in intron 10 of KIAA0907. Therefore, both SNORA42 and its host gene (KIAA0907) might be targets for the genomic amplicon. Interestingly, SNORA42 rather than its host gene displayed a pattern of high expression similar to that of its increased genomic dosage in all cancer cell lines. However, although exhibiting high genomic dosage in the cancer cell lines tested, KIAA0907 was overexpressed in only three cancer cell lines. SNORA42 overexpression is, therefore, activated by its genomic amplification. Furthermore, SNORA42 knockdown in NSCLC cells inhibited the in vitro and in vivo tumorigenicity, whereas enforced SNORA42 expression in normal bronchial epitheliums increased cell growth and colony formation. In addition, down-regulation of SNORA42 could initiate caspase-3-dependent apoptosis. Intriguingly, upon suppression of SNORA42, the cancer cell lines displaying apoptosis were p53 wild-type cells. However, the cell lines that did not exhibit apoptosis are either p53 null cells or cells with mutated p53. Moreover, the p53 wild type cancer cell lines after p53 was knockdown did not exhibit apoptosis when SNORA42 was reduced. Finally, p53 was down-regulated in the cells with enforced SNORA42 expression, whereas p53 was up-regulated in the cells with SNORA42 knockdown. P53 is a tumor suppressor and has functions to mediate cell growth, apoptosis, and tumorigenesis. Importantly, p53 is the most frequent target of genetic inactivation in human cancer and regulation of p53 is central to normal cell growth and tumor suppression [100,101]. Our findings indicate that the pleiotropy of SNORA42 suppression is achieved through increased apoptosis of NSCLC cells in a p53-dependent manner. Furthermore, activation of SNORA42 in cancer cells could be due to its genomic amplification. Therefore, down-regulation of SNORA42 can inhibit cancer cell growth and proliferation, providing evidence that the snoRNA has oncognic function in lung tumorigenesis. Nevertheless, the mechanism of regulation of p53 by SNORA42 remains to be investigated.
Recently, Xiao et al. found that an H/ACA box snoRNA-derived miRNA50, miR-605 (a sno-miRNA), could play a crucial role in stress-induced stabilization of the p53 [102]. Because p53 transcriptionally activates its negative regulator, MDM2, in addition to miR-605, the sno-miRNA could respond MDM2 through post-transcriptional repression. This sno-miRNA might offset the MDM2 negative-feedback loop, thus generating a positive-feedback loop to enable the rapid accumulation of p53. However, study towards determination of whether this regulation of p53 by the sno-miRNAs is relevant to cancer biology should be carried out. Taken together, these independent studies from diffident groups points to the possibility of ‘oncogenic snoRNA’ that upon dysfunction could silence p53 and induce the expression of oncogenes priming the cell for transformation.
Several recent studies have illuminated several examples of ‘tumor-suppressor snoRNA’. The first one is snoRNA U50. Using a deletion mapping approach to analyze 30 prostate tumors, Dong et al. [52] localized the tumor suppressor candidates to 2.5 Mb at 6q14-15. Examining the expression of all the candidates in this minimal region of deletion, they defined four genes for further consideration. The genes include three protein-encoding genes (LOC441164, NT5E, and SYNCRIP) and one snoRNA, U50. Comprehensively studying the candidates in the prostate cancer samples for cancer-specific mutations identified the snoRNA U50, rather than the protein-coding genes, as a tumor suppressor, because a homozygous 2-bp deletion was detected in the multiple samples. Importantly, tumor suppressor role of the snoRNA U50 was confirmed by a functional assay, in which wild-type but not mutant U50 inhibited cell proliferation or survival in the colony formation assay [52].
Consistent with the findings in prostate cancer, the U50 snoRNA gene displayed frequent copy number loss and transcriptional downregulation in primary breast tumors [53]. Furthermore, a 2-bp deletion occurred both somatically and in germline, leading to increased incidence of homozygosity for the deletion in the breast cancer cells. In addition, heterozygous genotype of the deletion was more frequent in blood specimens of women with breast cancer than those without cancer. Importantly, re-expressing the U50 snoRNA in breast cancer cell lines could dramatically decrease colony formation. Taken together, the snoRNA U50 could be a tumor suppressor gene, because it had mutations, was down-regulated, and reduced colony numbers in prostate and breast cancers. However, it is remains to determine the mechanism by which the snoRNA down-regulations inhibit cell survival and cell proliferation, and prevent oncogenesis.
5. Use of snoRNAs as potential biomarkers for diagnosis and prognosis of cancer
Given their vital role in diverse cellular processes, defining differential expression patterns of cancer type-specific snoRNAs should exploit for the development of novel cancer biomarkers. Indeed, we and others have demonstrated that snoRNAs are present in a stable form and consistently measurable in blood plasma, sputum, and urine samples [206-210]. Therefore, snoRNAs possess the potential as fluid-based biomarkers for cancers. We have recently identified a panel of three plasma snoRNAs (SNORD33, SNORD66 and SNORD76) that produce 81.1% sensitivity and 95.8% specificity in distinguishing NSCLC patients from both normal individuals and patients with chronic obstructive pulmonary disease. Therefore, measuring plasma snoRNAs would serve as a potential noninvasive approach to improve diagnosis of NSCLC [57]. Furthermore, we investigated clinical significance of SNORA42 dysregulation on frozen surgically resected lung tumor tissues of 64 patients with stage I NSCLC. High SNORA42 expression in tumor tissues was a predictive of shorter survival time versus low SNORA42 expression (P<0.01) [97]. The potential for the development of snoRNAs as biomarkers is also supported by a recent report from Gee et al [63]. They find that expression levels of C/D box SnoRNAs RNU44 and RNU43 are associated with poor prognosis in head and neck squamous cell carcinomas and breast cancers [63]. Taken together, snoRNAs may provide possible biomarkers for both diagnosis and prognosis of malignancies. Futures use of comprehensive and high-throughput techniques (e.g., second generation sequencing techniques) would identify more informative snoRNA-biomarkers that can be used in conjunction with miRNAs and protein-coding genes for cancer diagnosis and prognosis with higher accuracy.
6. The potential use of snoRNAs in cancer therapies
Although our understanding of the molecular mechanisms of snoRNA function in tumorigenesis is still limited, some features of snoRNAs would make them ideal candidates for therapeutic intervention. For instance, snoRNAs that mediate transcriptional gene silencing pathways could be of high therapeutic benefit. Furthermore, the progress in the use of RNAi-mediated gene silencing for cancer treatment is encouraging and could be applied to selectively silence oncogenic snoRNAs. In addition, some snoRNAs appear to have protein-binding or functional potential that is dependent on secondary structure, providing a means of therapeutic intervention. For example, snoRNA42 is strongly expressed in lung tumor tissue specimens. As such, we developed siRNAs-based system to exploit the tumor-specific expression of snoRNA42, primarily tested in treating lung cancer cell lines [97]. Cancer cells transfected with snoRA42-siRNA exhibited a substantial loss of snoRA42 expression [97]. Importantly, snoRA42 knockdown had significant anti-proliferation and viability activities in NSCLC cells. Furthermore, cancer cells with snoRA42 downregulation displayed much smaller number and size of colonies compared to cancer cells with scrambled siRNA and mock control. Therefore, suppression of snoRA42 diminished in vitro tumorigenicity of NSCLC cells. Moreover, inhibition of the in vivo tumorigenicity by snoRA42 knockdown was found in both ectopic and orthotopic xenograft mouse models. Collectively, the findings provide strong evidence to indicate the potential in developing snoRNA-mediated therapies. However, many technical challenges need to be overcome for use of therapeutic siRNAs to knockdown the oncogenic snoRNA. These include the development of reliable delivery systems, dosage regimes, and techniques to improve siRNA off target effects. When the technical limitations are overcome, snoRNAs could be potential targets for therapy due to their high turnover rate as well as their direct and specific regulatory functions.
7. Conclusion and future directions
SnoRNAs represent critical regulators of cellular processes, including proliferation, differentiation and survival. SnoRNA dysfunctions are critically associated with the development and progression of cancer. However, the functional role in carcinogenesis for the vast majority of these unique genes is still in question. Research for comprehensively understanding mechanisms by which aberrant snoRNAs contribute to the development and progression of cancer is required. With expanded understanding of snoRNAs’ new functions, the snoRNA-based studies may change the landscape of cancer biology and genetics and uncover new pathways that drive tumorigenesis. Furthermore, profiling snoRNA expression patterns specific to different type of cancers will offer not only novel diagnostic and prognostic biomarkers, but also effective therapeutic strategies to eventually cure cancer.
Acknowledgements
The authors’ work cited in this review was funded by grants from National Cancer Institute (NCI) grants CA161837, CA135382, CA137742, and CA133956), American Cancer Society Research Scholar Grant, an clinical innovator award from Flight Attendant Medical Research Institute to F. J.
Footnotes
Conflict of interest The authors declare that they have no conflict of interest.
References
- [1].American Cancer Society Cancer Facts & Figures. 2011 [Google Scholar]
- [2].Croce CM. Oncogenes and cancer. N. Engl. J. Med. 2008;358(5):502–511. doi: 10.1056/NEJMra072367. [DOI] [PubMed] [Google Scholar]
- [3].Stein LD. Human Genome: End of the Beginning. Nature. 2004;431:915–916. doi: 10.1038/431915a. [DOI] [PubMed] [Google Scholar]
- [4].Pennisi E. A Low Number wins the GeneSweep pool. Science. 2003;300:1484. doi: 10.1126/science.300.5625.1484b. [DOI] [PubMed] [Google Scholar]
- [5].Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, Sementchenko V, Piccolboni A, Bekiranov S, Bailey DK, Ganesh M, Ghosh S, Bell I, Gerhard DS, Gingeras TR. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- [6].Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P, Boyle PJ, Cao H, Carter NP, Clelland GK, Davis S, Day N, Dhami P, Dillon SC, Dorschner MO, Fiegler H. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15(1):R17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- [8].Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol. 2010;220(2):126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- [9].Will CL, Luhrmann R. In: The RNA World. 3rd edition Gesteland RF, Cech R, Atkins AJ, editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2006. pp. 369–400. [Google Scholar]
- [10].Nana-Sinkam P, Croce CM. MicroRNAs in diagnosis and prognosis in cancer: what does the future hold? Pharmacogenomics. 2010;11(5):667–669. doi: 10.2217/pgs.10.57. [DOI] [PubMed] [Google Scholar]
- [11].Qu H, Xu W, Huang Y, Yang S. Circulating miRNAs: promising biomarkers of human cancer. Asian Pac J Cancer Prev. 2011;12(5):1117–1125. [PubMed] [Google Scholar]
- [12].Duttagupta R, Jiang R, Gollub J, Getts RC, Jones KW. Impact of cellular miRNAs on circulating miRNA biomarker signatures. PLoS One. 2011;6(6):e20769. doi: 10.1371/journal.pone.0020769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Shen J, Todd NW, Zhang H, Yu L, Lingxiao X, Mei Y, Guarnera M, Liao J, Chou A, Lu CL, Jiang Z, Fang H, Katz RL, Jiang F. Plasma microRNAs as potential biomarkers for non-small-cell lung cancer. Lab Invest. 2011;91(4):579–587. doi: 10.1038/labinvest.2010.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Xie Y, Todd NW, Liu Z, Zhan M, Fang H, Peng H, Alattar M, Deepak J, Stass SA, Jiang F. Altered miRNA expression in sputum for diagnosis of non-small cell lung cancer. Lung Cancer. 2010;67(2):170–176. doi: 10.1016/j.lungcan.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Saito AJ, Schetter M, Mollerup S, Kohno T, Skaug V, Bowman ED, Mathé EA, Takenoshita S, Yokota J, Haugen A, Harris CC. The association of microRNA expression with prognosis and progression in early-stage, non-small cell lung adenocarcinoma, A retrospective analysis of three cohorts. Clin Cancer Res. 2011;17(7):1875–1882. doi: 10.1158/1078-0432.CCR-10-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids—the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8(8):467–477. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale. strategies and challenges, Nat Rev Drug Discov. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- [19].Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- [20].Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302(1):1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- [21].Galasso M, Elena SM, Volinia S. Non-coding RNAs: a key to future personalized molecular therapy? Genome Med. 2010;2(2):12–21. doi: 10.1186/gm133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Williams GT, Farzaneh F. Are snoRNAs and snoRNA host genes new players in cancer? Nat Rev Cancer. 2012;12:84–88. doi: 10.1038/nrc3195. [DOI] [PubMed] [Google Scholar]
- [23].Viltteri W. Mining the transcrptome-methods and applications. Royal Institute of Technology, School of Biotechnology; Stockholm: 2006. [Google Scholar]
- [24].Gardner PP, Bateman A, Poole AM. SnoPatrol: how many snoRNA genes are there? J. Biol. 2010;9:4–7. doi: 10.1186/jbiol211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lafontaine DL, Tollervey D. Birth of the snoRNPs: the evolution of the modification-guide snoRNAs. Trends Biochem Sci. 1998;23(10):383–388. doi: 10.1016/s0968-0004(98)01260-2. [DOI] [PubMed] [Google Scholar]
- [26].Terns MP, Terns RM. Small nucleolar RNAs: versatile trans-acting molecules of ancient evolutionary origin. Gene Expr. 2002;10(1-2):17–39. [PMC free article] [PubMed] [Google Scholar]
- [27].Weinstein LB, Steitz JA. Guided tours: from precursor snoRNA to functional snoRNP. Curr Opin Cell Biol. 1999;11(3):378–384. doi: 10.1016/S0955-0674(99)80053-2. [DOI] [PubMed] [Google Scholar]
- [28].Smith CM, Steitz JA. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- [29].Bortolin ML, Kiss T. Human U19 intron-encoded snoRNA is processed from a long primary transcript that possesses little potential for protein coding. RNA. 1998;4:445–454. [PMC free article] [PubMed] [Google Scholar]
- [30].Smith CM, Steitz JA. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5′-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol Cell Biol. 1998;18(12):6897–6909. doi: 10.1128/mcb.18.12.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Cavaillé J, Bachellerie JP. SnoRNA-guided ribose methylation of rRNA: structural features of the guide RNA duplex influencing the extent of the reaction. Nucleic Acids Res. 1998;26(7):1576–1587. doi: 10.1093/nar/26.7.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kiss T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 2001;20(14):3617–3622. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bachellerie JP, Cavaillé J, Hüttenhofer A. The expanding snoRNA world. Biochimie. 2002;84(8):775–90. doi: 10.1016/s0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- [34].Kiss AM, Jády BE, Bertrand E, Kiss T. Human Box H/ACA Pseudouridylation Guide RNA Machinery. Mol. Cell. Biol. 2004;24(13):5797–5807. doi: 10.1128/MCB.24.13.5797-5807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ganot P, Caizergues-Ferrer M, Kiss T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 1997;11(7):941–956. doi: 10.1101/gad.11.7.941. [DOI] [PubMed] [Google Scholar]
- [36].Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem Sci. 2002;27(7):344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- [37].Kishore S, Stamm S. The snoRNA HBII-52 Regulates Alternative Splicing of the Serotonin Receptor 2C. Science. 2006;Vol.311(no. 5758):230–232. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- [38].Cavaillé J, Buiting K, Kiefmann M, Lalande M, Brannan CI, Horsthemke B, Bachellerie JP, Brosius J, Hüttenhofer A. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc Natl Acad Sci U S A. 2000;97(26):14311–14316. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ender C, Krek A, Friedländer MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32(4):519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- [40].Kishore S, Khanna A, Zhang Z, Hui J, Balwierz PJ, Stefan M, Beach C, Nicholls RD, Zavolan M, Stamm S. The snoRNA MBII-52 (SNORD 115) is processed into smaller RNAs and regulates alternative splicing. Hum Mol Genet. 2010;19(7):1153–1164. doi: 10.1093/hmg/ddp585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ono M, Scott MS, Yamada K, Avolio F, Barton GJ, Lamond AI. Identification of human miRNA precursors that resemble box C/D snoRNAs. Nucleic Acids Research. 2011;39(9):3879–3891. doi: 10.1093/nar/gkq1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Brameier M, Herwig A, Reinhardt R, Walter L, Gruber J. Human box C/D snoRNAs with miRNA like functions: expanding the range of regulatory RNAs. Nucleic Acids Res. 2011;39(2):675–686. doi: 10.1093/nar/gkq776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Scott MS, Ono M. From snoRNA to miRNA: Dual function regulatory non-coding RNAs. Biochimie. 2011;93:1987–1992. doi: 10.1016/j.biochi.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Mitchell JR, Cheng J, Collins K. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol. Cell. Biol. 1999;19:567–576. doi: 10.1128/mcb.19.1.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Pogacic V, Dragon F, Filipowicz W. Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol. Cell. Biol. 2000;20:9028–9040. doi: 10.1128/mcb.20.23.9028-9040.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Dez C, Henras A, Faucon B, Lafontaine D, Caizergues-Ferrer M, Henry Y. Stable expression in yeast of the mature form of human telomerase RNA depends on its association with the box H/ACA small nucleolar RNP proteins Cbf5p, Nhp2p and Nop10p, Nucl. Acids Res. 2001;29:598–603. doi: 10.1093/nar/29.3.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Skryabin BV, Gubar LV, Seeger B, Pfeiffer J, Handel S, Robeck T, Karpova E, Rozhdestvensky TS, Brosius J. Deletion of the MBII-85 snoRNA gene cluster in mice results in postnatal growth retardation. PLoS Genet. 2007;3(12):e235. doi: 10.1371/journal.pgen.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Sahoo T, del Gaudio D, German JR, Shinawi M, Peters SU, Person RE, Garnica A, Cheung SW, Beaudet AL. Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat Genet. 2008;40(6):719–721. doi: 10.1038/ng.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Ding F, Li HH, Zhang S, Solomon NM, Camper SA, Cohen P, Francke U. SnoRNA Snord116 (Pwcr1/MBII-85) deletion causes growth deficiency and hyperphagia in mice. PLoS ONE. 2008;3(3):e1709. doi: 10.1371/journal.pone.0001709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Chang LS, Lin SY, Lieu AS, Wu TL. Differential expression of human 5S snoRNA genes. Biochem Biophys Res Commun. 2002;299:196–200. doi: 10.1016/s0006-291x(02)02623-2. [DOI] [PubMed] [Google Scholar]
- [51].Donsante A, Miller DG, Li Y, Vogler C, Brunt EM, Russell DW, Sands MS. AAV vector integration sites in mouse hepatocellular carcinoma. Science. 2007;317:477–477. doi: 10.1126/science.1142658. [DOI] [PubMed] [Google Scholar]
- [52].Dong XY, Rodriguez C, Guo P, Sun X, Talbot JT, Zhou W, Petros J, Li Q, Vessella RL, Kibel AS, Stevens VL, Calle EE, Dong JT. SnoRNA U50 is a candidate tumor-suppressor gene at 6q14.3 with a mutation associated with clinically significant prostate cancer. Hum Mol Genet. 2008;17(7):1031–1042. doi: 10.1093/hmg/ddm375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Dong XY, Guo P, Boyd J, Sun X, Li Q, Zhou W, Dong JT. Implication of snoRNA U50 in human breast cancer. J Genet Genomics. 2009;8:447–454. doi: 10.1016/S1673-8527(08)60134-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Tanaka R, Satoh H, Moriyama M, Satoh K, Morishita Y, Yoshida S, Watanabe T, Nakamura Y, Mori S. Intronic U50 small-nucleolar-RNA (snoRNA) host gene of no protein-coding potential is mapped at the chromosome breakpoint t(3;6)(q27;q15) of human B-cell lymphoma. Genes Cells. 2000;5(4):277–287. doi: 10.1046/j.1365-2443.2000.00325.x. [DOI] [PubMed] [Google Scholar]
- [55].Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28:195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- [56].Nakamura Y, Takahashi N, Kakegawa E, Yoshida K, Ito Y, Kayano H, Niitsu N, Jinnai I, Bessho M. The GAS5 (growth arrest-specific transcript 5) gene fuses to BCL6 as a result of t(1;3)(q25;q27) in a patient with B-cell lymphoma. Cancer Genet Cytogenet. 2008;182(2):144–149. doi: 10.1016/j.cancergencyto.2008.01.013. [DOI] [PubMed] [Google Scholar]
- [57].Liao J, Yu L, Mei Y, Guarnera M, Shen J, Li R, Liu Z, Jiang F. Small nucleolar RNA signatures as biomarkers for non-small-cell lung cancer. Mol Cancer. 2010;27:198–207. doi: 10.1186/1476-4598-9-198. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Li R, Wang H, Bekele BN, Yin Z, Caraway NP, Katz RL, Stass SA, Jiang F. Identification of putative oncogenes in lung adenocarcinoma by a comprehensive functional genomic approach. Oncogene. 2006;18:2628–2635. doi: 10.1038/sj.onc.1209289. [DOI] [PubMed] [Google Scholar]
- [59].Jiang F, Yin Z, Caraway NP, Li R, Katz RL. Genomic profiles in stage I primary non small cell lung cancer using comparative genomic hybridization analysis of cDNA microarrays. Neoplasia. 2004;6:623–635. doi: 10.1593/neo.04142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Gebhart E. Double minutes, cytogenetic equivalents of gene amplification, in human neoplasia - a review. Clin Transl Oncol. 2005;7:477–485. doi: 10.1007/BF02717000. [DOI] [PubMed] [Google Scholar]
- [61].Schwab M. Oncogene amplification in solid tumors. Semin Cancer Biol. 1999;9:319–325. doi: 10.1006/scbi.1999.0126. [DOI] [PubMed] [Google Scholar]
- [62].Bell DW. Our changing view of the genomic landscape of cancer. J Pathol. 2010;220:231–243. doi: 10.1002/path.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Gee HE, Buffa FM, Camps C, Ramachandran A, Leek R, Taylor M, Patil M, Sheldon H, Betts G, Homer J, West C, Ragoussis J, Harris AL. The small-nucleolar RNAs commonly used for microRNA normalisation correlate with tumour pathology and prognosis. Br J Cancer. 2011;104:1168–1177. doi: 10.1038/sj.bjc.6606076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Jelinic P, Shaw P. Loss of imprinting and cancer. J Pathol. 2007;211(3):261–268. doi: 10.1002/path.2116. [DOI] [PubMed] [Google Scholar]
- [65].Jirtle RL. Genomic Imprinting and Cancer. Exp Cell Res. 1994;248:18–24. doi: 10.1006/excr.1999.4453. [DOI] [PubMed] [Google Scholar]
- [66].Benetatos L, Vartholomatos G, Hatzimichael E. MEG3 imprinted gene contribution in tumorigenesis. Int J Cancer. 2011;129:773–779. doi: 10.1002/ijc.26052. [DOI] [PubMed] [Google Scholar]
- [67].Seitz H, Royo H, Bortolin ML, Lin SP, Ferguson-Smith AC, Cavaille J. A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome Res. 2004;14:1741–1748. doi: 10.1101/gr.2743304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Lin SP, Youngson N, Takada S, Seitz H, Reik W, Paulsen M, Cavaille J, Ferguson-Smith AC. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat Genet. 2003;35:97–102. doi: 10.1038/ng1233. [DOI] [PubMed] [Google Scholar]
- [69].Shimoda M, Morita S, Obata Y, Sotomaru Y, Kono T, Hatada I. Imprinting of a small nucleolar RNA gene on mouse chromosome 12. Genomics. 2002;79:483–486. doi: 10.1006/geno.2002.6727. [DOI] [PubMed] [Google Scholar]
- [70].da Rocha ST, Edwards CA, Ito M, Ogata T, Ferguson-Smith AC. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet. 2008;24:306–316. doi: 10.1016/j.tig.2008.03.011. [DOI] [PubMed] [Google Scholar]
- [71].Kagami M, Sekita Y, Nishimura G, Irie M, Kato F, Okada M, Yamamori S, Kishimoto H, Nakayama M, Tanaka Y, Matsuoka K, Takahashi T, Noguchi M, Tanaka Y, Masumoto K, Utsunomiya T, Kouzan H, Komatsu Y, Ohashi H, Kurosawa K, Kosaki K, Ferguson-Smith AC, Ishino F, Ogata T. Deletions and epimutations affecting the human 14q32.2 imprinted region in individuals with paternal and maternal upd(14)-like phenotypes. Nat Genet. 2008;40:237–2. 10. doi: 10.1038/ng.2007.56. [DOI] [PubMed] [Google Scholar]
- [72].Cavaillé J, Seitz H, Paulsen M, Ferguson-Smith AC, Bachellerie JP. Identification of tandemly-repeated C/D snoRNA genes at the imprinted human 14q32 domain reminiscent of those at the Prader-Willi/Angelman syndrome region. Hum Mol Genet. 2002;11(13):1527–1538. doi: 10.1093/hmg/11.13.1527. [DOI] [PubMed] [Google Scholar]
- [73].Ko JM, Yau WL, Chan PL, Lung HL, Yang L, Lo PH, Tang JC, Srivastava G, Stanbridge EJ, Lung ML. Functional evidence of decreased tumorigenicity associated with monochromosome transfer of chromosome 14 in esophageal cancer and the mapping of tumor-suppressive regions to 14q32. Genes Chromosomes Cancer. 2005;43(3):284–293. doi: 10.1002/gcc.20190. [DOI] [PubMed] [Google Scholar]
- [74].Blackburn EH. Telomerases. Annu Rev Biochem. 1992;61:113–129. doi: 10.1146/annurev.bi.61.070192.000553. [DOI] [PubMed] [Google Scholar]
- [75].de Lange T. Stringent sequence requirements for the formation of human telomeres. Proc Natl Acad Sci USA. 1994;91:2882–2885. doi: 10.1073/pnas.91.19.8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Feng J, Funk WD, Wang S, Weinrich SL, Avilion AA, Chiu C, Adams RR, Chang E, Allsopp RC, Yu J, Le S, West MD, Harley CB, Andrews WH, Greider CW, Villeponteau B. The RNA component of human telomerase. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- [77].Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- [78].Meyerson M, Counter CM, Eaton EN, Ellisen LW, Steiner P, Caddle SD, Ziaugra L, Beijersbergern RL, Davidoff MJ, Liu Q, Bacchetti S, Haber DA, Weinberg RA. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- [79].Harley CB. Telomere loss: mitotic clock or genetic time bomb? Mutat Res. 1991;256:271–282. doi: 10.1016/0921-8734(91)90018-7. [DOI] [PubMed] [Google Scholar]
- [80].Sharma HW, Maltese JY, Zhu X, Kaiser HE, Narayanan R. Telomeres, telomerase and cancer: is the magic bullet real? Anticancer Res. 1996;16:511–515. [PubMed] [Google Scholar]
- [81].Nakano K, Watney E, McDougall JK. Telomerase activity and expression of telomerase RNA component and telomerase catalytic subunit gene in cervical cancer. Am J Pathol. 1998;153(3):857–864. doi: 10.1016/S0002-9440(10)65627-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Yan P, Coindre JM, Benhattar J, Bosman FT, Guillou L. Telomerase activity and human telomerase reverse transcriptase mRNA expression in soft tissue tumors: correlation with grade, histology, and proliferative activity. Cancer Res. 1999;59:3166–3170. [PubMed] [Google Scholar]
- [83].Trahan C, Dragon F. Dyskeratosis congenita mutations in the H/ACA domain of human telomerase RNA affect its assembly into a pre-RNP. RNA. 2009;15(2):235–243. doi: 10.1261/rna.1354009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Alter BP, Giri N, Savage SA, Rosenberg PS. Cancer in dyskeratosis congenital. Blood. 2009;113(26):6549–6557. doi: 10.1182/blood-2008-12-192880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Sieron P, Hader C, Hatina J, Engers R, Wlazlinski A, Müller M, Schulz WA. DKC1 overexpression associated with prostate cancer progression. British Journal of Cancer. 2009;101:1410–1416. doi: 10.1038/sj.bjc.6605299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Montanaro L, Bigotti M, Clohessy J, Barbieri S, Ceccarelli C, Santini D, Taufurelli M, Calienti M, Teruya-Feldstein J, Trere D, Pandolfi PP, Derenzini M. Dyskerin expression influences the level of ribosomal RNA pseudo-uridylation and telomerase RNA component in human breast cancer. J Pathol. 2006;210:10–18. doi: 10.1002/path.2023. [DOI] [PubMed] [Google Scholar]
- [87].Watkins NJ, Bohnsack MT. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Mol Biosyst. 2010;6(3):481–493. doi: 10.1002/wrna.117. [DOI] [PubMed] [Google Scholar]
- [88].Ruggero D, Pandolf PP. Does the ribosome translate cancer? Nat Rev Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- [89].Ruggero D, Grisendi S, Piazza F, Rego E, Mari F, Rao PH, Cordon-Cardo C, Pandolfi PP. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science. 2003;299:259–262. doi: 10.1126/science.1079447. [DOI] [PubMed] [Google Scholar]
- [90].Montanaro L. Dyskerin and cancer: more than telomerase. The defect in mRNA translation helps in explaining how a proliferative defect leads to cancer. J. Pathol. 2010;222:345–349. doi: 10.1002/path.2777. [DOI] [PubMed] [Google Scholar]
- [91].Killian A, Le Meur N, Sesboue R, Bourguignon J, Bougeard G, Gautherot J, Bastard C, Frébourg T, Flaman JM. Inactivation of the RRB1-Pescadillo pathway involved in ribosome biogenesis induces chromosomal instability. Oncogene. 2004;23:8597–8602. doi: 10.1038/sj.onc.1207845. [DOI] [PubMed] [Google Scholar]
- [92].Berthon J, Fujikane R, Forterre P. When DNA replication and protein synthesis come together. Trends Biochem Sci. 2009;34(9):429–434. doi: 10.1016/j.tibs.2009.05.004. [DOI] [PubMed] [Google Scholar]
- [93].Nallar SC, Lin L, Srivastava V, Gade P, Hofmann ER, Ahmed H, Reddy SP, Kalvakolanu DV. GRIM-1, a Novel Growth Suppressor, Inhibits rRNA Maturation by Suppressing Small Nucleolar RNAs. PLoS ONE. 2011;6(9):e24082. doi: 10.1371/journal.pone.0024082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Nakamoto K, Ito A, Watabe K, Koma Y, Asada H, Yoshikawa K, Shinomura Y, Matsuzawa Y, Nojima H, Kitamura Y. Increased expression of a nucleolar Nop5/Sik family member in metastatic melanoma cells: evidence for its role in nucleolar sizing and function. Am J Pathol. 2001;159(4):1363–1374. doi: 10.1016/s0002-9440(10)62523-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Amsterdam A, Sadler KC, Lai K, Farrington S, Bronson RT, Lees JA, Hopkins N. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol. 2004;2(5):E139. doi: 10.1371/journal.pbio.0020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Michel CI, Holley CL, Scruggs BS, Sidhu R, Brookheart RT, Listenberger LL, Behlke MA, Ory DS, Schaffer JE. Small nucleolar RNAs U32a, U33, and U35a are critical mediators of metabolic stress. Cell Metab. 2011;14(1):33–44. doi: 10.1016/j.cmet.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Mei YP, Liao JP, Shen JP, Yu L, Liu BL, Liu L, Li RY, Ji L, Dorsey SG, Jiang ZR, Katz RL, Wang JY, Jiang F. Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis. Oncogene. 2011;449:1–11. doi: 10.1038/onc.2011.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Albertson DG, Ylstra B, Segraves R, Collins C, Dairkee SH, Kowbel D, Kuo WL, Gray JW, Pinkel D. Quantitative mapping of amplicon structure by array CGH identifies CYP24 as a candidate oncogene. Nat Genet. 2000;25(2):144–146. doi: 10.1038/75985. [DOI] [PubMed] [Google Scholar]
- [99].Taguchi T, Cheng GZ, Bell DW, Balsara B, Liu Z, Siegfried JM, Testa JR. Combined chromosome microdissection and comparative genomic hybridization detect multiple sites of amplification DNA in a human lung carcinoma cell line. Genes Chromosomes Cancer. 1997;20(2):208–212. doi: 10.1002/(sici)1098-2264(199710)20:2<208::aid-gcc13>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- [100].Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- [101].Vousden KH. Activation of the p53 tumor suppressor protein. Biochim Biophys Acta. 2002;1602:47–59. doi: 10.1016/s0304-419x(02)00035-5. [DOI] [PubMed] [Google Scholar]
- [102].Xiao J, Lin H, Luo X, Luo X, Wang Z. miR-605 joins p53 network to form a p53:miR-605:Mdm2 positive feedback loop in response to stress. EMBO J. 2011;30(3):524–532. doi: 10.1038/emboj.2010.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Appaiah HN, Goswami CP, Mina LA, Badve S, Sledge GW, Jr, Liu Y, Nakshatri H. Persistent upregulation of U6:SNORD44 small RNA ratio in the serum of breast cancer patients. Breast Cancer Res. 2011;13(5):R86. doi: 10.1186/bcr2943. [DOI] [PMC free article] [PubMed] [Google Scholar]


