Abstract
IL-9 is a pro-allergic cytokine produced by a newly proposed T helper cell subset TH9. TH9 cells can be generated by treatment of naïve T cells with TGF-β and IL-4 in vitro. But how TGF-β signaling regulates TH9 differentiation is still not clear. Here we demonstrate that Smad2 and Smad4, two transcriptional factors activated by TGF-β signaling, are required for TH9 differentiation in vitro. Deficiency of Smad2 or Smad4 in T cells resulted in impaired IL-9 expression, which was coincident with enrichment of repressive chromatin modification H3K27Me3 and enhanced EZH2 binding to the Il9 locus. Pharmacologic inhibition of EZH2 partially rescued IL-9 production in Smad deficient TH9 cells. Smad proteins may displace EZH2 directly from Il9 locus since Smad2 and Smad4 can bind EZH2. Our data shed light on the molecular mechanisms underlying TH9 cell differentiation, revealing that TGF-β-Smad2/4 signaling pathway regulates IL-9 production through an epigenetic mechanism.
Introduction
IL-9 is a pleiotropic cytokine that plays an important role in asthma induction, parasite expulsion, immune tolerance and anti-tumor response depending on cell types and environmental context (1,2). In addition to mast cells, CD4 helper T cells are major IL-9 producers (1). Even within CD4 T cells, multiple lineages have been reported to express IL-9. IL-9 was first discovered in TH2 cells. Recently it was documented that TH17 and Treg cells can secret this cytokine as well (3,4). However, accumulating evidence suggest that there is a specialized subset of T cells that is dedicated to IL-9 production. This T cell type is called TH9 cells (5,6).
TH9 cells can be generated in vitro from naïve CD4 T cells by TGF-β plus IL-4 treatment (7). These cells are related to TH2 cells because they require IL-4-Stat-6 signaling and GATA-3 for their differentiation. But they have lower expression of TH2 cytokines (5). Several transcriptional factors such as Stat5, Stat6, PU.1 and IRF4 have been identified that may directly regulate IL-9 transcription during TH9 cell differentiation (8,9, 21). The molecular links between cytokine receptor and Il9 transcription during TH9 cell differentiation are still missing. It is clear that IL-4 signaling regulates Il9 transcription either by positive regulation via the induction of IRF4 (10) or by negative regulation through the induction of SOCS protein CIS, which downregulates binding of Stat5 and Stat6 to the Il9 promoter (21). However, how TGF-β signaling contributes to TH9 differentiation has not been thoroughly assessed so far.
TGF-β, by binding to its receptor, induces the phosphorylation of Smad2 and Smad3. Through association with common partner Smad4, phosphorylated Smad2 or Smad3 translocate into the nucleus where they drive the expression of downstream genes (11). In addition, TGF-β triggers Smad-independent cascade (12). Therefore, whether Smad proteins mediate TGF-β signaling during TH9 cell differentiation is still an open question.
In the present study, we have determined the role of both Smad2 and Smad4 during TH9 differentiation and found that both of them are required for IL-9 production. We observed that deletion of Smad2 and Smad4 impaired IL-9 expression, leading to sustained association of repressive H3K27Me3-modification, which was associated with sustained binding of EZH2, a H3K27-specific methylase, to the Il9 locus. Pharmacological inhibition of EZH2 led to partially rescued IL-9 production in Smad2 and Smad4 deficient TH9 cells. Both Smad2 and Smad4 were observed be able to bind EZH2 directly. Our data revealed that TGF-β-Smad signaling regulates IL-9 expression by displacement of inhibitory histone modification enzyme EZH2 from the Il9 locus during TH9 differentiation.
Material and Methods
Mice
Smad2fl/flCD4-Cre and Smad4fl/flCD4-Cre mice were described previously (13,14). All animal experiments were performed following protocols approved by Institutional Animal Care and Use Committee.
T cell differentiation
T cell in vitro differentiation was conducted as previously described (13,14) except following conditions were used for TH2 and TH9 cells. FACS-sorted naïve cells (250K) were stimulated in 48 well plates with plate-bound anti-CD3 (1ug/ml;2C11) plus soluble anti-CD28 (1ug/ml;37.51) in the following cytokines or neutralizing antibodies: 4ng/ml TGF-β, 20ng/ml IL-4, 10ug/ml anti-IFN-γ (XMG 1.2) and 30U/ml hIL-2 for TH9; 40ng/ml IL-4, 10ug/ml anti-IFN-γ, 10ug/ml anti-TGB-β (1D11) and 30U/ml hIL-2 for TH2. 2μM of GSK126 (XcessBio) was added in the culture from the start in some experiments. After 4 day stimulation, cells were harvested for chromatin immunoprecipitation (ChIP) and Western Blot analysis or washed and re-stimulated with plate-bound anti-CD3 (1.0ug/ml) for RNA extraction (4hr) or for ELISA (24hr). Cytokine staining was performed as previously described (13,14).
ChIP Assay
Il9 locus definition followed previous study (8). Genomic DNA was extracted from 2∼4 millions of cells by using a commercial kit (Upstate), followed by real-time PCR quantification for Il9 promoter (F-ctcaattggcctcaacttacag, R-ccctttgccatcctccagcag), Il9 CNS (F-aattacagaattttgccccaggtcctg, R-gttaatgcacaattcatgtgccaatcc) and Il4 promoter (F-ctcattttcccttggtttcagc, R-gatttttgtcgcatccgtgg). ChIP-grade antibodies against H3AC, H3K27Me3 and H3K4Me3 were purchased from Millipore. ChIP-grade EZH2 antibody was obtained from active Motif (#39875).
Statistics analysis
Data are presented as mean value ± s.d. Data were analyzed by using Student's t test. A value of p<0.05 was considered significant.
Results and Discussion
Smad2 is required for TH9 differentiation
To study how TGF-β signaling pathway regulates TH9 differentiation, Smad2fl/flCD4-Cre (Smad2 KO) and Smad4fl/flCD4-Cre (Smad4 KO) conditional knockout mice were used to delete Smad2 and Smad4 respectively in T cells (13,14). Age and sex-matched Smad2fl/fl and Smad4fl/fl littermate animals were used as wild-type (WT) controls throughout this study.
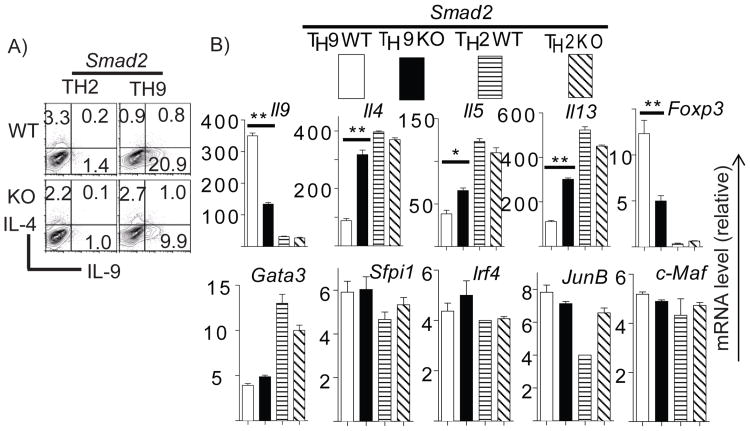
Purified naïve cells from Smad2 KO and WT animals were differentiated under TH9 and TH2 conditions for4 days. Cytokine production was measured by intracellular staining. We found that TH2 cells produced negligible amount of IL-9 but high levels of IL-4. Both IL-4 and IL-9 production, however, were not affected by Smad2 deletion in TH2 cells (Figure 1A). TH9 cells produced higher levels of IL-9 and lower levels of IL-4 compared to TH2 cells. Smad2 deletion significantly reduced IL-9 production but enhanced IL-4 production in TH9 cells (Figure 1A). ELISA analysis of IL-9 and TH2 cytokines from above cells showed the same trend (Supplemental Figure S1A). Consistent with the results at the protein level, mRNA of Il9 was higher in TH9 cells than in TH2 cells and was reduced significantly in TH9 cells by Smad2 deletion. Il4/5/13 mRNA were enhanced in TH9 cells but were not affected in TH2 cells by Smad2 deletion (Figure 1B). These data suggest that Smad2 is required for IL-9 expression and suppression of TH2 cytokine production in TH9 cells while Smad2 is dispensable for IL-4/5/13 production in TH2 cells.
Figure 1. Smad2 is required for TH9 differentiation.
FACS-sorted naïve cells from Smad2fl/flCD4Cre+ (Smad2 KO) or Smad2fl/flCD4Cre-(Smad2 WT) mice were polarized under TH2 and TH9 conditions for 4 days. A, Cells were then re-stimulated with PMA/Ionomycin in the presence of Golgi Stop for 6hr. IL-4 and IL-9 production was assessed by intracellular staining. B, Cells were re-stimulated with plate-bound anti-CD3 for 4hr prior to Trizol treatment. cDNA was prepared and gene expression was analyzed by real-time RT-PCR. Data were normalized to a reference gene Actb. The expression of each gene in TH0 cells (not shown) was referred as 1. A representative of 4 independent experiments is shown.
TGF-β binding to its receptor triggers the activation of both Smad2 and Smad3 (12). Previous studies have shown that Smad2 and Smad3 have redundant roles in T cells (15). In the present study, although Smad2 deletion resulted in dramatic reduction of IL-9 production (Figure 1), it did not completely abrogate IL-9 production. It is likely that Smad2 and Smad3 play redundant roles during TH9- differentiation.
To elucidate the mechanism underlying impaired IL-9 in Smad2- deficient TH9 cells, we sought to examine the expression of transcription factors regulating IL-9 and IL-4 transcription in T cells. Recently it was reported that PU.1 (Sfpi1) and IRF4 were required for TH9 differentiation (8,9). However, we could not detect any difference of PU.1 or IRF4 expression between Smad2-sufficient and -deficient cells by real-time RT-PCR (Figure1B).
To understand why IL-4/5/13 expression was enhanced in the absence of Smad2 in TH9 cells, we assessed GATA3, JunB and c-Maf expression in these cells. Expression of these TH2 lineage-associated transcription factors was not up-regulated in the absence of Smad2 (Figure1B). Although Foxp3 expression was significantly reduced upon Smad2 deletion (Figure1B), it is not required for TH9 differentiation. This is because retroviral overexpression of Foxp3 failed to rescue IL-9 production in Smad2 deficient TH9 cells (data not shown).
Smad4 is required for TH9 differentiation
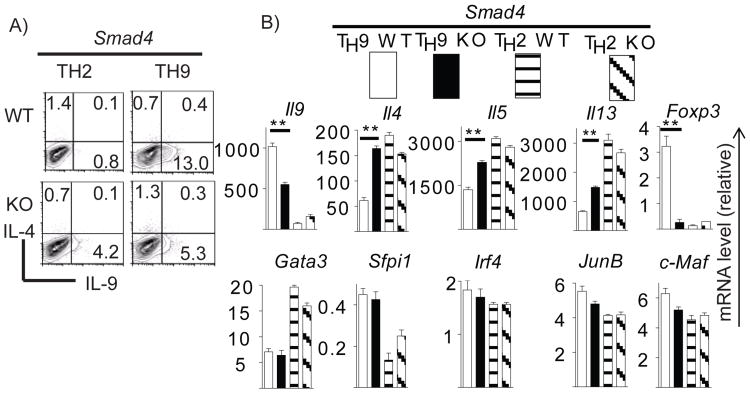
Similarly, naïve T cells from Smad4 KO and WT mice were polarized under TH2 and TH9 conditions. WT TH2 cells produced marginal levels of IL-9. Smad4 deficiency significantly enhanced IL-9 production whereas IL-4 production was slightly reduced in TH2 cells (Figure 2A). Identical to Smad2-deficient TH9 cells, IL-9 production was significantly reduced in Smad4-deficient TH9 cells compared to WT cells. Moreover, IL-4 production in TH9 cells was enhanced in the absence of Smad4 (Figure 2A). ELISA analysis of IL-9 and TH2 cytokines from above cells further confirmed our findings (Supplemental Figure S1B). Measuring cytokinem RNA revealed that Smad4 deletion led to a slight increase of IL-9 production in TH2 cells. However, Smad4 deficiency significantly attenuated IL-9 production but enhanced TH2 cytokines in TH9 cells (Figure 2B). These data indicates that both Smad2 and Smad4 are required for IL-9 expression and suppression of TH2 cytokine production in TH9 cells.
Figure 2. Smad4 is necessary for TH9 differentiation.
FACS-sorted naïve cells from Smad4fl/fl CD4Cre+(Smad4 KO) or Smad4fl/fl CD4Cre-(Smad4 WT) mice were differentiated under TH2 and TH9 conditions for 4 days. Intracellular staining of IL-4 and IL-9 (A) and real-time RT-PCR analysis of gene expression (B) was performed as Figure 1.
Since Smad4 is co-factor of Smad2 and Smad3 for their binding and subsequent nuclear translocation, Smad4 deletion will impair both Smad2- and Smad3–mediated signaling. However, despite pronounced reduction of IL-9 in Smad4-deficient TH9 cells, these cells still have residual IL-9 production (Figure 2A). Therefore, Smad-independent TGF-β signaling probably is also involved in IL-9 production in T cells.
Similar to Smad2-deficient TH9 cells, the gene expression of potential TH9 lineage transcriptional factors (IRF4 and PU.1 (Sfpi1)) and TH2 lineage transcriptional factors (GATA3, c-Maf, JunB) were not affected in TH9 cells in the absence of Smad4 (Figure 2B). This suggests that TGF-β-Smad signaling regulates IL-9 expression not through these factors.
Smad2 and Smad4 regulates histone modification at the Il9 locus
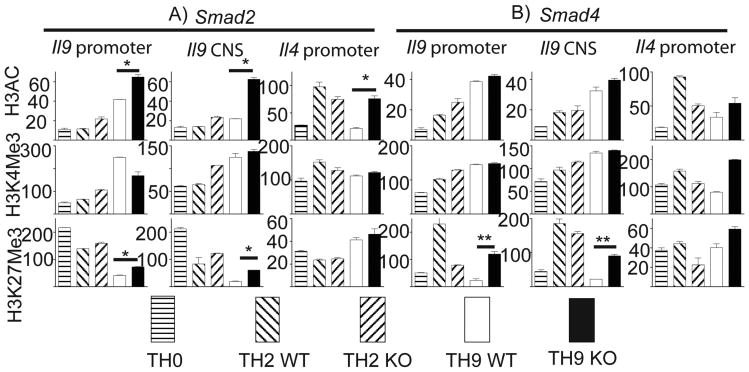
Epigenetic regulation particularly through chromatin modification is an important mechanism involved in gene regulation and T cell differentiation (16,17). We speculate that TGF-β-Smad signaling pathway may directly regulate IL-9 production via epigenetic regulation. To address this question, naïve T cells from Smad2 KO and WT mice were polarized under TH9 conditions with TH2 and TH0 cells included as controls. We used ChIP assay to examine both permissive chromatin modification, including total H3 acetylation (H3AC) and H3K4 trimethylation (H3K4Me3), and repressive chromatin modification such as H3K27 trimethylation (H3K27Me3). Both H3AC and H3K4Me3 at either promoter region or conserved non-coding sequence (CNS) region of the Il9 locus were higher in TH9 cells than that in TH0 and TH2 cells whereas H3K27Me3association were significantly reduced accompanying TH9 differentiation (Figure 3). However, Il4 promoter in TH9 cells was enriched with both permissive and repressive histone modification (Figure 3), a feature shared by bivalent domain possessing low levels of transcription (17). Therefore, chromatin modifications at the Il9 and Il4 loci are compatible with robust IL-9 and modest IL-4 production in TH9 cells.
Figure 3. Effects of Smad2 and Smad4 deficiency on histone modification at the Il9 locus in TH9 cells.
Naïve T cells from Smad2 (A) or Smad4 (B) mice as in Figure 1 and Figure 2 were differentiated under TH2 and TH9 conditions for 4 days and harvested for ChIP analysis of histone modification at the Il9 locus. WT TH0 cells served as a control for TH9 cells. H3AC, acetylation of histone H3. H3K4Me3, trimethylation of histone H3; H3K27Me3, trimethylation of Histone H3 K27. Control IgG was used as negative control for anti-H3AC, anti-H3K4Me3 and anti-H3K27Me3 (not shown). Total input DNA before IP was used for normalization of data. Histone modification was determined by a quantitative PCR. 3 independent experiments were performed with similar results. * P<0.05, ** P<0.001
Smad2 deletion increased H3AC modification (Figure 3A top row) while having no effect on H3K4Me3 modification at the Il9 locus in TH9 cells (Figure 3A, middle row). Considering impaired production of IL-9 in Smad2-deficienct TH9 cells, permissive histone modification at Il9 locus is not likely the causative mechanism that is targeted by Smad2 during TH9 differentiation. Interestingly, H3K27Me3 was significantly enhanced in all tested regions at the Il9 locus in Smad2-deficient cells compared to WT cells (Figure 3A bottom row). This suggests that Smad2 was involved in demethylation of histone 3 at lysine 27 at the Il9 locus in T cells. Next we sought to determine whether Smad4 also utilizes the same mechanism to regulate IL-9 expression. Ablation of Smad4 had no effect on both H3Ac and H3K4Me3 modifications at the Il9 locus (Figure 3B top and middle rows) whereas it strongly enhanced H3K27Me3 at the same locus (Figure 3B bottom row). These data demonstrated that both Smad2 and Smad4 are required for histone H3K27 demethylation at the Il9 locus during TH9 differentiation.
Smad2 and Smad4 are required to displace EZH2 from the Il9 locus during TH9 differentiation
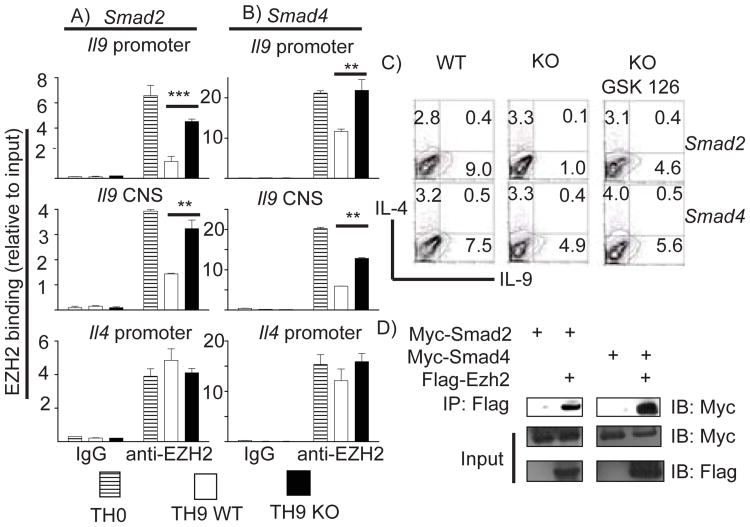
H3K27me3 demethylation is associated with gene activation. Maintaining H3K27me3 is enforced by polycomb repressor complex 2 (PRC2) and represents an important mechanism for gene silencing. EZH2, a component of PRC2 complex, is responsible for catalyzing and maintaining H3K27me3 modification (18). Since deficiency of either Smad2 or Smad4 led to enrichment of H3K27me3 histone modification at the Il9 locus during TH9 differentiation, we hypothesized that enrichment of H3K27me3 modification in Smad2/4-deficient T cells is the result of impaired de-association of EZH2 from the Il9 locus during TH9 differentiation. ChIP analysis of EZH2 binding to the Il9 locus was conducted to assess this. EZH2 constitutively bound to all tested sites of the Il9 locus in TH0 cells (Figure 4A, 4B). This was consistent with no IL-9 production in those cells (data not shown). In contrast, EZH2 binding to the Il9 locus was significantly reduced in WT TH9 cells (Figure 4A, 4B, top and middle rows, open bars). Ablation of either Smad2 or Smad4, however, led to defective de-association of EZH2 from the Il9 locus (Figure 4A, 4B, top and middle rows, filled bars), which was not due to altered EZH2 expression in these cells (Supplemental Figure S2). This implicated that both Smad2 and Smad4 were required for displacement of EZH2 from the Il9 locus during TH9 differentiation. To corroborate that EZH2 is downstream of Smad proteins and mediates TGF-β regulated IL-9 production, WT and Smad2/4 deficient cells were polarized under TH9 condition with GSK126, a specific EZH2 inhibitor (19). GSK126 significantly reduced global H3K27Me3 modification (Supplemental Figure S2). Strikingly, IL-9 production in both Smad2 and Smad4 deficient TH9 cells were partially restored by GSK126 treatment to levels close to that in WT cells (Figure 4C). We also detected that both Smad2 and Smad4 directly bound to EZH2 in 293T cells (Figure 4D). These data strongly suggested that Smad2 or Smad4 were involved in displacement of EZH2 from the Il9 locus via direct interaction during TH9 differentiation. Complementary to two recent studies in which Smad prote in were shown to form complex with either Notch or IRF4 to bind Il9 promoter and drive transcription of IL-9 (20, 22), our data support the concept that TGF-β signaling regulates IL-9 production by multiple mechanism.
Figure 4. Deficiency of Smad2 and Smad4 results in sustained EZH2 binding to the Il9 locus in TH9 cells.
A and B, Naïve T cells from Smad2 (A) or Smad4 (B) mice were differentiated under TH9 condition for 4 days and harvested for ChIP analysis of EZH2 binding at the Il9 locus. WT TH0 cells served as a positive control for EZH2 binding at the Il9 locus in TH9 cells. Control IgG was used as non-specific binding to the Il9 and the Il4 locus. EZH2 binding was determined by a quantitative PCR. This is a representative of two experiments. (* P<0.05, ** P<0.001). C, Naïve T cells from indicated mice were differentiated under TH9 condition with or without addition of 2uM of GSK126 for 4 days. D, Smad2, Smad4 and EZH2 encoding plasmids were transfected into 293T cells. 24hr later, cells were harvested, lysed, and immunoreprecipted with anti-Flag antibody followed by detection with anti-Myc antibody.
We also examined EZH2 binding to the Il4 promoter in TH9 cells. EZH2 was found persistently bound to the Il4 promoter in these cells during TH9 differentiation, which was in contrast to reduced levels of EZH2 binding to this locus in TH2 differentiation (data not shown). Smad2 deletion had no effect on EZH2 binding to Il4 promoter in TH9 cells while ablation of Smad4 led to a slight enhancement of EZH2 binding to Il4 promoter (Figure 4A and 4B, bottom row). Since both Smad2 and Smad4 deficiency increased IL-4 production in TH9 cells (Figure 1 and 2), H3K27me3 modification and EZH2 binding at the Il4 promoter region may not be directly involved in IL-4 expression in these T cells. Mechanism underlying reciprocal up-regulation of TH2 cytokine in Smad2 and Smad4-deficient TH9 cells is not clear so far. It is possible that attenuated expression of Foxp3 de-represses IL4/5/13 expression during TH9 differentiation in the absence of Smad2 and Smad4.
In summary, we provided genetic evidence that Smad2 and Smad4 are required for TH9 differentiation. We demonstrated that Smad2 and Smad4 were not involved in the regulation of PU.1 and IRF4 expression during TH9 differentiation. We found, instead, that TGF-β signaling regulated IL-9 production through displacement of EZH2 and removal of suppressive H3K27 histone modification at the Il9 locus. This will not only provide further insight into our understanding molecular mechanism underlying TH9 differentiation, but also help us to design therapeutic strategies to manipulate TH9 cells in vivo in future.
Supplementary Material
Acknowledgments
We thank Drs. Chrisopher Wilson for the CD4-Cre mice, Liz Robertson for Smad2 and Smad4 conditional mice, the flow cytometry core at the MD Anderson Cancer Center for help on cell sorting, Dionne Prescod for help maintaining mouse colony and our lab members for technical support and assistance.
This work is supported by research grants from NIH (to C.D.)
The abbreviations used in this paper
- IL-9
interleukin-9
- H3AC
Histone 3 acetylation
- H3K4Me3
Histone 3 K4 trimethylation
- H3K27Me3
Hisotne 3 K27 trimethylation
- ChIP
chromatin immunoprecipitation
Footnotes
Disclosures: The authors have no financial conflicts of interest
References
- 1.Noelle RJ, Nowak EC. Cellular sources and immune functions of interleukin-9. Nat Rev Immunol. 2010;10:683–687. doi: 10.1038/nri2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Purwar R, Schlapbach C, Xiao S, Kang HS, Elyaman W, Jiang X, Jetten AM, Khoury SJ, Fuhlbrigge RC, Kuchroo VK, Clark RA, Kupper TS. Robust tumor immunity to melanoma mediated by interleukin-9-producing T cells. Nat Med. 2012;18:1248–1253. doi: 10.1038/nm.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nowak EC, Weaver CT, Turner H, Begum-Haque S, Becher B, Schreiner B, Coyle AJ, Kasper LH, Noelle RJ. IL-9 as a mediator of Th17-deriven inflammatory disease. J Exp Med. 2009;206:1653–1660. doi: 10.1084/jem.20090246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu LF, Lind EF, Gondek DC, Bennett KA, Gleeson MW, Pino-Lagos K, Scott ZA, Coyle AJ, Reed JL, Van Snick J, Strom TB, Zheng XX, Noelle RJ. Mast cells are essential intermediates in regulatory T-cell tolerance. Nature. 2006;442:997–1002. doi: 10.1038/nature05010. [DOI] [PubMed] [Google Scholar]
- 5.Dardalhon V, Awasthi A, Kwon H, Galileos G, Gao W, Sobel RA, Mitsdoerffer M, Strom TB, Elyaman W, Ho IC, Khoury S, Oukka M, Kuchroo VK. IL-4 inhibits TGF-beta-induced Foxp3+ T cells and, together with TGF-beta, generates IL-9+IL-10+Foxp3- effector T cells. Nat Immunol. 2008;9:1347–1355. doi: 10.1038/ni.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Veldhoen M, Uyttenhove C, Van Snick J, Helmby H, Westendorf A, Buer J, Martin B, Wilhelm C, Stockinger B. Transforming growth factor-beta reprograms the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nat Immunol. 2008;9:1341–1346. doi: 10.1038/ni.1659. [DOI] [PubMed] [Google Scholar]
- 7.Schmitt E, Germann T, Goedert S, Hoehn P, Huels C, Koelsch S, Kuhn R, Muller W, Palm N, Rude E. IL-9 production of naives CD4+ T cells depends on IL-2, is synergistically enhanced by combination of TGF-beta and IL-4, and is inhibited by IFN-gama. J Immunol. 1994;153:3989–3996. [PubMed] [Google Scholar]
- 8.Chang HC, Sehra S, Goswami R, Yao W, Yu Q, Stritesky GL, Jabeen R, McKinley C, Ahyi AN, Han L, Nguyen ET, Robertson MJ, Perumal NB, Tepper RS, Nutt SL, Kaplan MH. The transcription factor PU.1 is required for the development of IL-9-producing T cells and allergic inflammation. Nat Immunol. 2010;11:527–534. doi: 10.1038/ni.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Staudt V, Bothur E, Klein M, Lingnau K, Reuter S, Grebe N, Gerlitzki B, Hoffmann M, Ulges A, Taube C, Dehzad N, Becker M, Stassen M, Steinborn A, Lohoff M, Schild H, Schmitt E, Bopp T. Interferon-regulatory factor 4 is essential for the development program of T helper 9 cells. Immunity. 2010;33:192–202. doi: 10.1016/j.immuni.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 10.Goswami R, Jabeen R, Yagi R, Pham D, Zhu J, Goenka S, Kaplan MH. Stat6-dependent regulation of Th9 development. J Immunol. 2012;188:968–975. doi: 10.4049/jimmunol.1102840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Massague J, Chen YG. Controlling TGFβ signaling. Gene and development. Genes Dev. 2000;14:627–644. [PubMed] [Google Scholar]
- 12.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 13.Martinez GJ, Zhang Z, Reynolds JM, Tanaka S, Chung Y, Liu T, Robertson E, Lin X, Feng XH, Dong C. Smad2 positively regulates the generation of Th17 cells. J Biol Chem. 2010;285:29039–29043. doi: 10.1074/jbc.C110.155820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang XO, Nurieva R, Martinez GJ, Kang HS, Chung Y, Pappu BP, Shah B, Chang SH, Schluns KS, Watowich SS, Feng XH, Jetten AM, Dong C. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity. 2008;29:44–56. doi: 10.1016/j.immuni.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takimoto T, Wakabayashi Y, Sekiya T, Inoue N, Morita R, Ichiyama K, Takahashi R, Asakawa M, Muto G, Mori T, Hasegawa E, Saika S, Hara T, Nomura M, Yoshimura A. Smad2 and Smad3 are redundantly essential for the TGF-beta-mediated regulation of regulatory T plasticity and Th1 development. J Immunol. 2010;185:842–855. doi: 10.4049/jimmunol.0904100. [DOI] [PubMed] [Google Scholar]
- 16.Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 17.Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, Schones DE, Peng W, Sun HW, Paul WE, O'Shea JJ, Zhao K. Global Mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiation CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A, Diaz E, LaFrance LV, Mellinger M, Duquenne C, Tian X, Kruger RG, McHugh CF, Brandt M, Miller WH, Dhanak D, Verma SK, Tumminoand PJ, Creasy CL. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–12. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 20.Elyaman W, Bassil R, Bradshaw EM, Orent W, Lahoud Y, Zhu B, Radtke F, Yagita H, Khoury SJ. Notch receptors and Smad3 signaling cooperate in the induction of interleukin-9-producing T cells. Immunity. 2012;36:623–34. doi: 10.1016/j.immuni.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang XO, Zhang H, Kim BS, Niu X, Peng J, Chen Y, Kerketta R, Lee H, Chang SH, Corry DB, Wang D, Watowich SS, Dong C. The signaling suppressor CIS controls proallergic T cell development and allergic airway inflammation. Nat Immunol. 2013;14:732–740. doi: 10.1038/ni.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamiya T, Ichiyama K, Kotani H, Fukaya T, Sekiya T, Shichita T, Honma K, Yuri K, Matsuyama T, Nakao T, Fukuyama S, Inoue H, Nomura M, Yoshimura A. Smad2/3 and IRF4 play a cooperative role in IL-9-producing T cell induction. J Immunol. 2013;191:2360–2371. doi: 10.4049/jimmunol.1301276. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.