Abstract
RalA and RalB are small GTPases which support malignant development and progression in experimental models of bladder, prostate and squamous cancer. However, demonstration of their clinical relevance in human tumors remains lacking. Here, we developed tools to evaluate Ral protein expression, activation and transcriptional output and evaluated their association with clinicopathologic parameters in common human tumor types. In order to evaluate the relevance of Ral activation and transcriptional output, we correlated RalA and RalB activation with the mutational status of key human bladder cancer genes. We also identified and evaluated a “transcriptional signature” of genes that correlates with depletion of RalA and RalB in vivo. The Ral transcriptional signature score, but not protein expression as evaluated by immunohistochemistry, predicted disease stage, progression to muscle invasion, and survival in human bladder cancers, and metastatic and stem cell phenotypes in bladder cancer models. In prostate cancer, the Ral transcriptional signature score was associated with seminal vesicle invasion, androgen-independent progression, and reduced survival. In squamous cell carcinoma, this score was decreased in cancer tissues compared with normal mucosa, validating the experimental findings that Ral acts as a tumor-suppressor in this tumor type. Together, our findings demonstrate the clinical relevance of Ral in human cancer and provide a rationale for the development of Ral-directed therapies.
Keywords: bladder cancer, prostate cancer, squamous cell carcinoma, Ral GTPase, gene expression profiling
INTRODUCTION
Ras-like (Ral) GTPases include the homologous paralogs RalA and RalB, which have been implicated in diverse cellular functions (1). Like other small GTPases, Ral GTPases serve as a GDP/GTP conformational switch, with active signaling mediated by the GTP-loaded form, regulated by a family of Ras-dependent and independent GEFs or other factors (2). A large body of literature has implicated these GTPases in key cancer phenotypes such as Ras-mediated transformation (3). This transformation is dependent specifically on RalA (4), and may be further regulated by phosphorylation (5) while other phenotypes such as regulation of cellular migration, invasion, and metastasis (6, 7) are attributed to either RalA or RalB, depending on the model system and cancer type evaluated.
Surprisingly, despite these important in vitro and in vivo findings, there is little evidence supporting the biological relevance of Ral in human cancer. Unlike other GTPases, Ral mutations have not been noted in large or targeted (8) screens of common cancers. In contrast, we have observed overexpression of RalA in a small number of muscle invasive bladder cancers (MIBCs) (8), while others noticed this in advanced prostatic adenocarcinoma (9). Neither of these studies evaluated the tumors by in situ technologies such as immunohistochemistry or in a large enough number of cases to derive sufficiently robust clinical conclusions. Furthermore, while expression of the GTPase itself contributes to the output of the Ral pathway, factors that impact GTPase activation such as microenvironmental stimuli, post-translational modifications including phosphorylation (10), and differential expression of downstream Ral downstream effectors are likely to play significant roles in determining the biological consequences of Ral expression in cancer. Ral GTPases also regulate key transcription factors such as TCF, Jun, NF-κB, Stat3, HSF, E2F, the forkhead family, ZONAB, and RREB1. Targets of these pathways have been demonstrated to include key cancer genes such as cyclin D1(11), VEGFC (12), and CD24 (13), supportive of the important role of Ral-dependent transcription in cancers. Hence using a transcriptional signature associated with Ral expression and/or activity may provide a useful and comprehensive picture of the Ral pathway activity in a cancer.
Here we assess the status and clinical relevance of Ral in several human cancers by establishing and evaluating immunohistochemistry for RalA and RalB in tumor tissues. We also develop gene expression signature based on transcriptional changes induced in response to Ral depletion in cells and determine the utility of this in predicting clinical outcomes in various human cancer types. Our data indicate that only the transcriptional signature of Ral is associated with human tumor characteristics and patient outcomes. In addition, this signature is also associated with experimentally proven Ral phenotypes which validate its relevance as an accurate reporter of Ral-dependent biology. Taken together, this comprehensive approach demonstrates for the first time the broad clinical significance of Ral in human cancer. This work also provides ample justification for the development of therapies to target the Ral pathway.
METHODS
Cell Lines
The BLA-40 cell line panel and its provenance has been detailed before (14). The cells have been tested/validated by gene expression profiling, supporting the bladder cancer origin of the lines, as reported in detail recently (15).
Immunohistochemistry
Immunohistochemistry (IHC) was performed on formalin-fixed paraffin embedded cells and a previously described tissue microarray (TMA) (16) using antibodies specific to RalA (1:1600 dilution, Clone 8, BD Biosciences, San Jose, CA) (17) and RalB (1:400 dilution, R&D Systems, Minneapolis, MN) (5). Specificity workup used pelletted formalin-fixed paraffin embedded UM-UC-3 cells stably expressing FLAG vector or FLAG-tagged RalA or RalB (6) to demonstrate specificity in IHC using standard streptavidin-biotin detection (specific protocols/autostainer settings in Supplementary Information). Staining for RalA or RalB was scored semi-quantitatively as either low (low to moderate intensity, or only focal higher expression in <50% of cells in cores examined) or high (strong, diffuse positive staining in >50% of cells in the TMA cores examined).
Ral Activation Assays and Derivation of the Ral Activation Signature
Ral activation assays, which use the active, GTP:Ral-dependent interaction between the Ral binding domain of RalBP1 conjugated to beads to selectively pull down active RalA or RalB, which may then be quantitated and compared to total RalA or RalB by immunoblotting were as reported (4, 6). Spearman correlation between RalA and RalB percent activation and individual probes and percentage activated RalA or RalB was performed across microarray data generated on the BLA-40 cell line panel (14). False discovery rate was tested by random permutation testing of the Ral activation measurements for the BLA-40 cell lines, in Matlab R2010B (The Mathworks, Natick, MA) by randomly permuting percentage activation numbers and measuring numbers of probes randomly correlated at a range of thresholds and comparing these values to those observed experimentally. The association between distributions of RalA or RalB activation and mutation status was tested by the Mann-Whitney U-test, using publicly available mutation data for cell lines in the BLA-40 panel, recently tabulated by our group (15).
Derivation of the Ral Transcriptional Signature
We have recently reported the biological effects of siRNA-mediated depletion of Ral GTPases in the UM-UC-3 human bladder cell line cell and profiled the transcriptional changes associated with such depletion (18). Original CEL files of these data normalized in RMA (19) as implemented in Matlab, extracting log2 expression values. After calculating fold changes for each of the HG-U133A array probes comparing Ral-intact to Ral-depleted cells, we then defined “Ral Transcriptional Signature” probes as the intersect of those altered >2-fold on average by depletion of both GTPases. A COXEN (CO-eXpression ExtrapolatioN) step was undertaken as reported (20) selecting only those candidates with concordant expression human tumors (COXEN coefficient cutoff, >0). Ral transcriptional signature scores, ranging from 0 to 1, were made from the final probe set using a correlation distance weighted KNN prediction algorithm (WNN) we have reported before (21). This algorithm uses group mean z-score-normalized log2 signature gene expression data from the cells with intact or depleted RalA and RalB as training, then employs Spearman correlation as distance metric to output a distance-weighted posterior probability for each of similarly z-scored normalized clinical test cases. This probability, the Ral signature score, ranges from 0 (like siRal, signature negative cells) to 1 (like siControl, signature positive). These scores were designated Ral signature scores and then compared across relevant clinicopathologic groups. Cross microarray platform comparisons were made by mapping probes by Unigene cluster ID or HUGO gene symbol, specifics for each case detailed in the Supplementary Information. Datasets used for bladder were: (NCBI GEO): GSE88; GSE89; GSE19915; GSE16255; GSE31684; GSE37317; (Array Express): TABM-147; Sanchez-Carbayo et al. data at journal website (22); Dyrskjøt et al. processed data (footnote: http://www.mdl.dk/). Prostate cancer: GSE2443; GSE5803; GSE21887; GSE8702; GSE21034; GSE16560; GSE32269; GSE6956. Squamous cell carcinoma: GSE23400; GSE2944; GSE7803.
Statistical Analysis
Analysis of the association of Ral immunohistochemistry staining with clinicopathologic characteristics in bladder cancer was performed through the chi square test (Matlab) or log rank test (Prism 5.0, GraphPad Software, La Jolla, CA). For comparison of distributions of Ral transcriptional signature scores between groups of patient tumors, values were plotted and tested by the Mann-Whitney U-test, Wilcoxon Matched Pairs test or Kruskal-Wallis test, all in Prism 5.0, while association of Ral transcriptional signature scores with survival was tested with the log rank test (Matlab).
RESULTS
Immunohistochemical Staining for RalA and RalB in Human Bladder Cancer
Given prior observed overexpression by western blot and mRNA of RalA, RalB and their effectors in a small cohort of human bladder cancers (8), we were interested in examining whether Ral protein expression in human tumors was associated with clinicopathologic variables. We developed, optimized (see Supplementary Information, Figures S1A-B, S2A-B) and then performed immunohistochemical (IHC) staining for RalA and RalB, using paralog-specific (5, 6) antibodies. We then stained and scored a tissue microarray of archival human bladder tumor tissues that included urothelial carcinoma (N=110) and other less common histological variants (N=35) (16). We did not find a significant association between level of RalA or RalB staining and patient gender, pathologic stage, lymphovascular space invasion, or presence of carcinoma in situ (Tables S1-S2). Interestingly, tumors of non-urothelial histology (N=35) had significantly different proportions of tumors showing low and high Ral staining compared to urothelial carcinoma (RalA P=0.03, RalB P=0.02, χ2-test, Tables S1-S2).
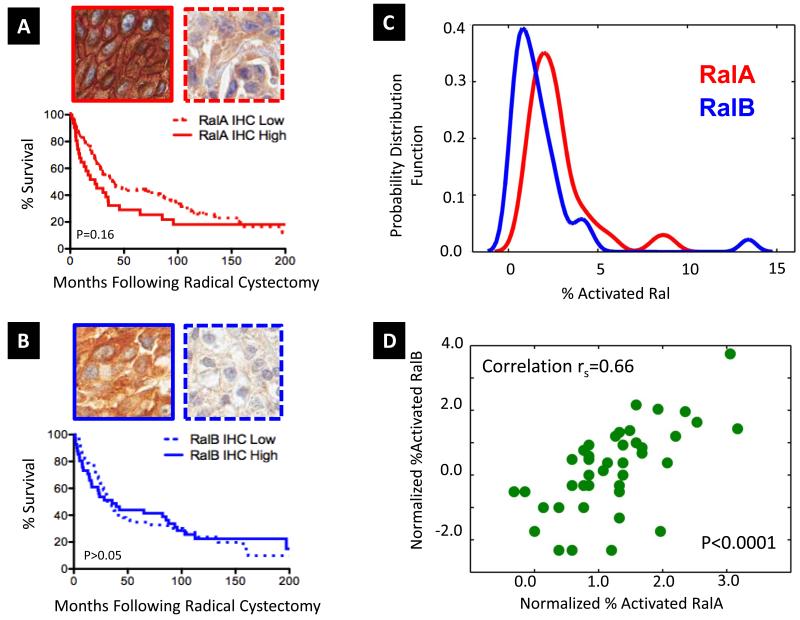
In urothelial tumors (N=110), the most common histology found in bladder cancer, we observed a trend toward decreased overall survival post radical cystectomy in patients with intense staining for RalA (P=0.16, Log Rank, P=0.04 Wilcoxon-Breslow, which weights early events Figure 1A). For RalB, there was no significant association (Figure 1B). Stratification of survival by differing levels of RalA and RalB was inferior to that of RalA alone (P=0.45, Figure S3).
Figure 1. Expression of RalA and RalB by immunohistochemistry in 110 human urothelial bladder tumors in patients treated by radical cystectomy.
A tissue microarray of bladder carcinomas (16), stages pTa-T4, was stained with antibodies specific for RalA and RalB (see Supplementary Information). A. Representative photomicrographs of strong, diffuse RalA staining (RalA High, solid) and weak RalA staining (RalA Low, dashed). Kaplan-Meier analysis of overall survival in urothelial carcinoma cases (N=110, 109 with adequate staining, Table S1) stratified by expression level of RalA, showing a non-significant trend in favor of poorer overall survival for cases expressing strong RalA (Log Rank P=0.16). Wilcoxon-Breslow testing of these curves, which weights early events, identified a significant difference (P=0.04). B. Similar micrographs to A. but for RalB, showing strong diffuse staining (blue, solid) and weak RalB staining (blue, dashed). Urothelial carcinoma cases (N=110, 104 with adequate staining, Table S2) were evaluated by Kaplan-Meier analysis for RalB expression, finding non-significant difference by Log Rank or Wilcoxon-Breslow methods. C. Activation of RalA and RalB. Smoothed histogram plots of RalA and RalB activation in the BLA-40 cell panel. In vitro biochemical activation assays for RalA and RalB, which employ beads coated with the GTP-Ral dependent interaction domain of the Ral effector, RalBP1 to selectively bind activated GTPase from cell lysates before detection and relative quantitation by immunoblotting, were performed to quantitate percentage active GTP-bound RalA and RalB in matched lysates of bladder cancer cell lines (the BLA-40). RalA red; RalB blue. D. RalA (x-axis) and RalB (y-axis) normalized (for UM-UC-3 levels, see Supplementary Information) percentage activation (log2 scale) scatter plotted across the cell line panel, with indicated Spearman correlation. For full data see Table S3.
The Relationship of RalA and RalB Activation to Common Mutations Found in Human Bladder Cancer
Activation of Ral can occur through various upstream mechanisms. However, the relationship between RalA and RalB activation status and the common pathogenetic mutations in bladder cancer remains unknown, despite the fact that a number of such lesions are known to regulate components of canonical pathways upstream of Ral. To examine this, we performed in vitro GTPase activation assays for RalA and RalB in a large panel of human bladder cancer cell lines, the BLA-40 (14), and examined these findings with respect to cell line mutation status for seven commonly mutated genes in bladder cancer, KRAS, p53, RB, CDKN2a, PTEN, PIK3CA, and FGFR3 (15). In this large panel, activation of RalA and RalB was similarly distributed across the cell lines tested (Figure 1C). Interestingly, RalA and RalB activations were highly correlated across the BLA-40 (rs=0.66, P<0.0001), (Figure 1D, Table S3). Strikingly, neither the activation of RalA, nor of RalB was significantly associated with mutation status for the seven bladder cancer genes (Table S4), suggesting independence of Ral activity from these common molecular lesions.
Development of Gene Expression Signatures of Ral Status
Current biochemical activation assays for RalA and RalB preclude testing of formalin fixed archival bladder cancer tissues, given their requirement for rapid lysis and pull-down of activated GTPase species from fresh cells. Given that long-term clinical data is predominantly available in association with formalin fixed tissues, we sought to overcome these limitations in order to evaluate such patient samples. We first used the Ral activation measurements from the BLA-40 cell lines to identify probes that correlated to Ral activation and thus generate a “Ral activation signature”. Unfortunately, we were unable to uncover any transcripts that correlated to RalA or RalB activation due to an unacceptably high false discovery rate, (Supplementary Information and Figure S4A-B).
Given these findings, we sought to develop a surrogate of Ral pathway status based on the fact that Ral GTPases alter gene expression through various transcription factors (18, 23, 24). Since tumors with the same levels of Ral protein but different levels of GTPase activation or effector interactions may induce such transcription factors to varying levels, which in turn might induce different clinical phenotypes, we hypothesized that Ral-dependent transcriptomic profiles might effectively capture pathway output that will be associated with salient clinicopathologic factors and outcomes. To directly assay for genes dependent on Ral pathway activity, by perturbing it in vivo, we decided to develop a transcriptional signature of Ral pathway status based on profiling cells depleted of RalA or RalB. We used siRNA to deplete RalA or RalB from bladder cancer cells and then profiled the resultant transcriptional changes by microarray (18). Given the significant overlap between RalA and RalB-dependent transcriptional targets and the high degree of correlation between activation of RalA and RalB (Figure 1D), we developed a “core” signature of the transcriptional program common to both RalA and RalB by choosing a union of 60 probesets regulated by RalA and RalB depletion in human bladder cancer cells (minimum 2 fold, >100 microarray expression units difference between closest replicates, Table S5). Importantly, and supportive of the specificity of these probsets to core Ral transcriptional signaling, we observed that this overlap between transcripts regulated by RalA and RalB was highly significant (P<0.0001, X2 test for independence). To this set of 60 probesets we applied the COXEN principle (14, 20) to define a subset of 39 probesets maintaining concordant expression in a published bladder cancer microarray cohort of patients treated by radical cystectomy (N=91) (22). Importantly, as in previous COXEN implementations (20, 25), no clinical outcome or other biological/pathologic information from the patient cohort was used in this step. Table 1 shows these probes, the genes they interrogate, their fold change in Ral intact as compared to Ral depleted cells, and the direction of their differential expression in relevant cancer types.
TABLE 1. Probes in the Ral Signature.
| Probe* | Fold Change^ | Gene Symbol | Bladder Ca# | Squamous Ca# | CaP# |
|---|---|---|---|---|---|
| 222043_at | 4.88 | CLU | (−) | (−) | (−) |
| 203325_s_at | 3.90 | COL5A1 | (−) | (+) | |
| 204396_s_at | 3.73 | GRK5 | |||
| 204584_at | 3.73 | L1CAM | |||
| 212488_at | 3.56 | COL5A1 | (−) | (+) | |
| 212489_at | 3.41 | COL5A1 | (−) | (+) | |
| 213397_x_at | 3.23 | RNASE4 | (−) | (−) | |
| 203845_at | 3.13 | KAT2B | (−) | ||
| 202196_s_at | 3.12 | DKK3 | (−) | (−) | |
| 211071_s_at | 3.10 | MLLT11 | |||
| 206924_at | 3.09 | IL11 | |||
| 205158_at | 2.80 | RNASE4 | (−) | (−) | |
| 218625_at | 2.76 | NRN1 | |||
| 214247_s_at | 2.68 | DKK3 | (−) | (−) | |
| 206117_at | 2.65 | TPM1 | (−) | (+),(−) | (−) |
| 212888_at | 2.51 | DICER1 | |||
| 202952_s_at | 2.42 | ADAM12 | (+) | ||
| 202733_at | 2.40 | P4HA2 | |||
| 213790_at | 2.38 | ADAM12 | (+) | ||
| 202743_at | 2.35 | PIK3R3 | |||
| 213005_s_at | 2.33 | KANK1 | (−) | (−) | |
| 201506_at | 2.33 | TGFBI | (−) | (+) | |
| 221541_at | 2.30 | CRISPLD2 | (−) | (−) | |
| 208792_s_at | 2.30 | CLU | (−) | (−) | (−) |
| 208791_at | 2.28 | CLU | (−) | (−) | (−) |
| 206116_s_at | 2.27 | TPM1 | (−) | (+),(−) | (−) |
| 212099_at | 2.23 | RHOB | |||
| 222062_at | 2.21 | IL27RA | (+) | ||
| 209822_s_at | 2.19 | VLDLR | |||
| 210986_s_at | 2.17 | TPM1 | (−) | (+),(−) | (−) |
| 201505_at | 2.17 | LAMB1 | (+) | (−) | |
| 203871_at | −2.12 | SENP3 | |||
| 218190_s_at | −2.27 | UQCR10 | |||
| 208756_at | −2.31 | EIF3I | (+) | ||
| 215113_s_at | −2.74 | SENP3 | |||
| 221263_s_at | −2.84 | SF3B5 | |||
| 215171_s_at | −3.04 | TIMM17A | (+) | (+) | |
| 201528_at | −3.48 | RPA1 | |||
| 204475_at | −5.05 | MMP1 | (+) | (+) |
Indicated probe from the Affymetrix HG-U133A oligonucleotide microarray platform.
Average fold change comparing control siRNA treated UM-UC-3 cells to cells depleted of RalA and RalB.
Direction of gene expression change (+ or −) in bladder carcinoma, squamous carcinoma (of esophagus, cervix, or upper aerodigestive tract), or prostatic adenocarcinoma, compared to normal respective tissue. Direction indicated if the gene were in the top 1% of genes in ≥ 1 study of the relevant tumor type in the Oncomine database (53).
Hoping to use these genes to develop a signature of the Ral pathway to examine across samples and cancer types, next, we wished to test the generality of this signature to another cell type where Ral signaling was manipulated. For this study we employed a recently published (26) dataset of immortalized human embryonic kidney (HEK-HT) cells that had been stably transfected with vector control, G12V mutant HRAS oncogene, or for G12V oncogenic Ras effector loop mutants, which interact with some specificity with the Raf pathway (G12V/T35S), PI3 kinase pathway (G12V/Y40C), or RalGEF-Ral pathway (G12V/E37G).
To test the status of the Ral signature in these cell lines, we employed a weighted KNN (WNN) classifier algorithm, as detailed in Supplementary Information and as reported (21). Briefly, the WNN classifier algorithm uses non-parametric (Spearman) correlation as distance metric to measure similarity of expression of Ral signature genes in a sample to be tested (e.g., one of the HEK-HT Ras samples) to the Ral-depleted or Control cells, outputting a prediction score, which we call the “Ral Signature Score,” ranging from 0 to 1. This score, calculated for each sample solely based on the correlation of its expression of genes in the Ral signature assigns a high signature score to a sample showing strong expression of the Ral transcriptional program (i.e., close to 1) and low score (i.e., close to 0) for a sample without expression of the transcriptional program.
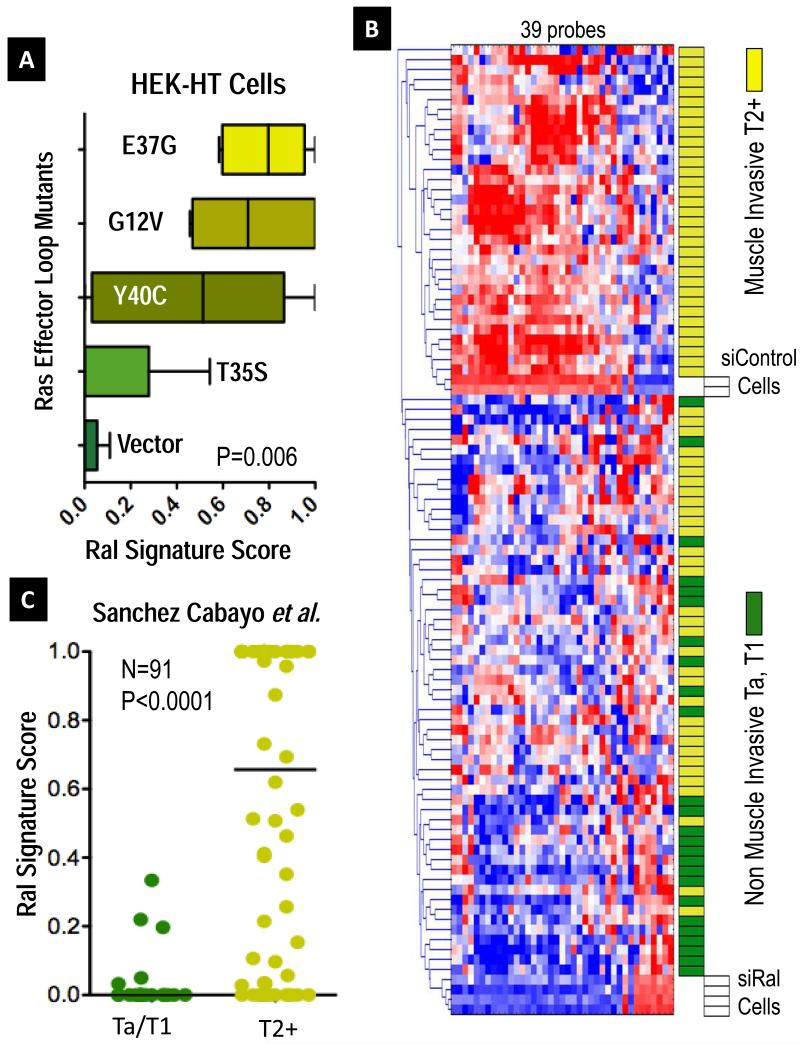
Testing each of the quintuplicate cases of HEK-HT cells (5 each of vector, G12V HRAS, and the three effector loop mutants), we observed the highest Ral signature scores in the G12V/E37G (Ral stimulating) and G12V (stimulating all 3 pathways), compared to the other non-Ral stimulating mutants (see Figure 2A, P=0.006). The G12V/Y40C mutant, which stimulates the PI3K pathway, showed intermediate values, which may correlate recent data showing that PI3 kinase may stimulate the Ral pathway indirectly. We interpreted these data, taken from a different cell type showing Ral perturbation from a gain of function standpoint (instead of loss of function, with our siRNA study) as consistent with the Ral signature being sufficiently general across cell types to allow comparisons and as supportive of the validity of our informatic approach to assay the status of the Ral pathway through expression of the signature.
Figure 2. The Transcriptional Signature of Ral GTPases.
A Ral Transcriptional Signature consisting of 39 probes was developed from genes regulated 2-fold by RalA and RalB expression in UM-UC-3 cells. To validate the association of this signature with Ral across cell types, we used a weighted nearest neighbor (WNN) algorithm (21), which outputs a score from 0, Signature negative, to 1, Signature positive. We classified samples from published data from Chang et al. (26), where quintuplicate preparations of HEK-HT cells stably expressing vector control, G12V oncogenic HRAS, or indicated G12V HRAS effector loop mutants were profiled by oligonucleotide microarrays. A. Boxplots of median and range (whiskers) Ral signature scores for the indicated vector control or Ras mutant, finding higher scores in oncogenic Ras (G12V) and its effector loop mutant (G12V/E37G) stimulating the Ral pathway (P=0.006, Kruskal-Wallis test). B. Hierarchical cluster analysis of gene expression data for 91 bladder cancers (22) and control siRNA treated UM-UC-3 cells (siControl) or RalA and RalB-depleted siRNA duplexes (siRal), showing association of the signature with muscle-invasive tumors (yellow blocks). C. Expression of the Ral signature from cases in B. was again quantitated as before and dotplotted, medians indicated by lines. Differences in score distributions between non-muscle invasive pTa/T1 cases and muscle invasive pT2+ cases were tested by the Mann-Whitney test. Similar significant findings were identified in six additional cohorts comprising over 500 additional patients (see Figure S5A-F).
The Ral Transcriptional Signature Characterizes Invasive Disease in Bladder Cancer
Using the 39 aforementioned Ral signature probes, we clustered the 91 tumors described above (22) with control or Ral-depleted cells and found that non-muscle invasive (stage pTa, pT1) tumors clustered with the Ral-depleted cells, while muscle invasive (stage pT2+) tumors clustered with control treated cells (Figure 2B). To determine quantitatively if there is a relationship between tumor stage in this cohort and expression of the Ral signature, we again used a WNN classifier algorithm to classify the tumors based on similarity to Ral-depleted cells (like siRalA and siRalB) or control cells (like Control cells, expressing Ral and its transcriptional program).
Using this approach, we observed a significant difference in distributions of Ral signature scores between non-muscle invasive bladder cancers and muscle invasive bladder cancers, P<0.0001 (Figure 2C), with non-muscle invasive bladder cancers having lower Ral signature scores than their invasive counterparts. Importantly, we used thousand-fold random permutation testing to examine the significance of our approach, confirming that this degree of difference was only associated with a 0.1% false discovery rate, confirming the importance of Ral signature genes as opposed to global transcriptional differences between non-muscle invasive bladder cancers and muscle invasive bladder cancers (see Supplementary Information). Additional analysis using only data for RalA or RalB was done to assess for any specificity to either GTPase compared to the results of the RalA & RalB “core” signature. This effort found weaker associations of RalA-only or RalB-only signatures with stage, and thus all further implementations used the core signature (see Supplementary Information). Importantly, we applied this core signature to classify tumors of six additional independent cohorts of bladder tumors (27-31) profiled on five different microarray platforms (total additional N=522) and found similar, significant results (Figure S5A-F).
Cells with Metastatic or Stem Cell Characteristics have High Ral Transcriptional Signature Scores
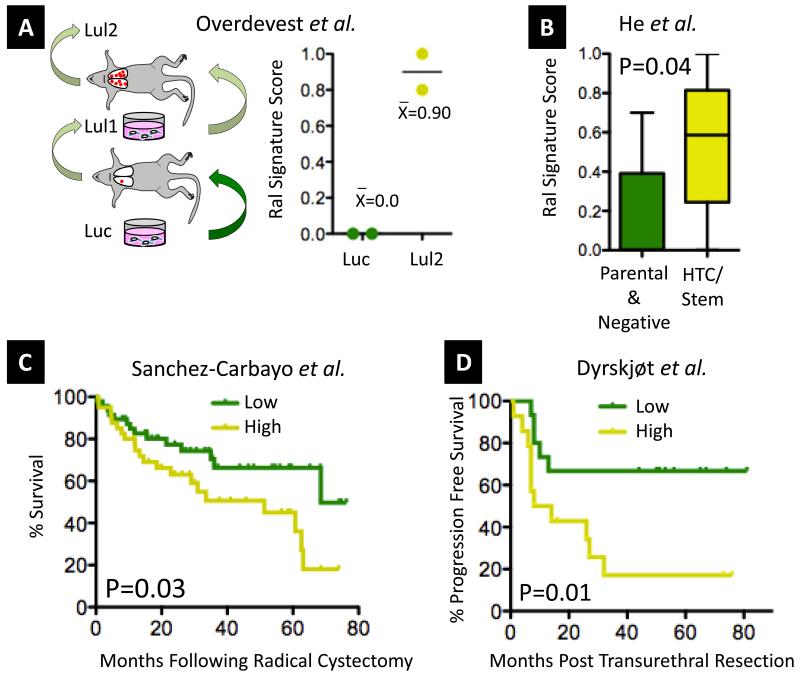
Given the correlation of Ral signature scores with stage in bladder cancer patients, we were interested to determine if this score correlated with development of metastasis after surgery, especially as experimental model systems have demonstrated an important role of RalA and RalB in mediating metastasis in vivo (5). We have recently developed a mouse model of lung metastasis using parental, poorly metastatic UM-UC-3 human bladder cancer cells. UM-UC-3 cells stably expressing firefly luciferase for bioluminescent imaging (Luc), where serially inoculated via tail vein to generate progressively more metastatic variants (Lul1 and Lul2) (Figure 3A) which were then transcriptionally profiled (32). Consistent with the importance of Ral in bladder tumor progression, we found Lul2 had a higher Ral signature score than Luc cells (Figure 3A). Another report used fluorescence activated cell sorting to prospectively isolate an aggressive, highly tumorigenic/stem cell-like population of cells from SW780 bladder cancer cells, which were subsequently profiled by microarray. Suggesting a role for Ral in the stem cell phenotype, the Ral signature score was higher in highly tumorigenic/ stem cell-like SW780 isolates compared to parental and negative-sorted populations (Figure 3B).
Figure 3. Association of the Ral Transcriptional Signature Score with experimental and patient outcomes.
A. Using a recently developed metastasis model that we have analyzed by microarray (32), we found significantly higher Ral signature scores in metastatic Lul2 cells compared to parental Luc cells. B. Using microarray data from a recent publication where cells with stem cell-like properties were isolated from bladder cancer cells by cell sorting (51), we found significantly higher Ral signature scores in these cells compared to parental or negative sorted cells (Mann-Whitney, plot shows median plus 95% CI). C. In the Sanchez Carbayo et al. cohort of 91 bladder cancers used in Figure 2A-B, the Ral signature score was associated with overall survival (Kaplan-Meier plot showing signature score >0.5 high versus <0.5 low, Log Rank test). D. In non-muscle invasive tumors, expression of the Ral signature is significantly associated with subsequent progression to muscle invasion in a previously published cohort (N=29) (33) (Kaplan-Meier plot and Log Rank test to C.).
Prognostic value of the Ral Transcriptional Signature Score in Human Bladder Cancer
Next, we examined the status of the Ral signature in tumors with respects to survival in the Sanchez-Carbayo et al. cohort (Figure 3C). Using a Ral transcriptional signature score cutoff of >0.5 or <0.5 to classify as signature high or low, respectively, we found that the signature score significantly stratified cases by survival, with signature high cases showing significantly worse survival (P=0.03, Log Rank), though this difference was not independent of the association of scores with stage in multivariate Cox models (P=0.57). Furthermore, several groups have reported that non-muscle invasive (Ta and T1 stage) tumors that subsequently progress to muscle invasion following transurethral resection exhibit, a priori, the molecular characteristics of muscle invasive tumors (27, 29). Based on these observations and our findings above showing the Ral transcriptional signature is associated with more aggressive tumor behavior, we hypothesized that the Ral transcriptional signature might be prognostic of subsequent progression in such cases. Using two published microarray cohorts of non-muscle invasive disease where progression during follow-up was documented (29, 33), we evaluated the Ral transcriptional signature score with respect to progression to muscle invasive stage disease. We found that the score significantly stratified progression free survival in a series (N=29) by Dyrskjøt et al. (Figure 3D, P=0.01). Though time to progression data were not available for a second series reported by Lindgren et al. (N=97, only binary, +/− progression among non-muscle invasive vases), we again observed significantly higher Ral signature scores in cases demonstrating progression (P=0.04, Figure S5B).
Human Squamous Cell Carcinoma has a Lower Ral Transcriptional Signature Score than Normal Mucosa
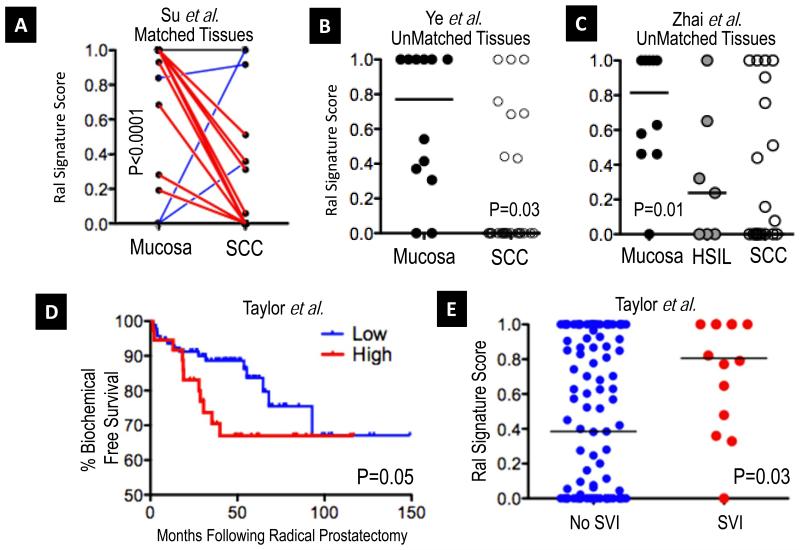
Recent reports suggest that Ral may play a tumor suppressor role in squamous cell carcinoma (SCC) (34). Hence, we reasoned that if these data have clinical significance, the Ral signature score should be lower in SCCs as compared to normal squamous mucosa. We evaluated the signature in a published cohort of matched SCCs and histologically normal adjacent mucosae of the esophagus (N=53 matched tissues) evaluated by microarray (35). Strikingly, we found significantly lower Ral signature scores in SCCs compared to normal mucosae (Figure 4A, P<0.0001). The significance of this difference over background differences in gene expression was tested by random permutation testing, observing a false discovery rate of 0.5%, supportive of the importance of the Ral transcriptional signature. The signature was then tested in a second, smaller cohort of oral SCCs (N=26) as compared to normal mucosae (N=12), profiled on a different microarray platform (36), finding significant difference in signature score distributions between normal and cancer (Figure 4B, P=0.03) consistent in direction with the first set where SCC had lower scores than normal mucosae. To further extend these findings to an additional organ system, we tested the signature on a third cohort of squamous cell carcinomas (N=21), high-grade squamous intraepithelial lesions (HSIL, N=7), and normal mucosae of the uterine cervix (N=10) reported by Zhai et al. (37). We observed the same pattern as in squamous malignancy of the esophagus and oropharynx: higher signature scores in the normal mucosae as compared to the HSIL and SCC (P=0.01, Figure 4C). Notably the biologically intermediate group of HSILs showed intermediate signature scores between normal and cancer.
Figure 4. Ral Transcriptional Signature scores in squamous malignancy.
A. Using data from a published cohort of 53 patient-matched esophageal squamous cell carcinomas (SCC) and adjacent normal mucosae (35), we observed a significant trend toward lower Ral signature score in cancer (P<0.0001, Wilcoxon matched pairs test). Plot shows matched pairs of mucosa and cancer, with decreases in signature score plotted red (N=37), similar scores plotted black (N=13) and increased scores plotted blue (N=3). B. In a second, unmatched cohort of 12 mucosae and 26 oropharyngeal SCCs (36), a similar pattern was identified (Mann-Whitney test, signature scores plotted and medians per group indicated, black lines) that was significant. C. In a third cohort of SCCs of the uterine cervix (N=21), high grade squamous intraepithelial lesions (HSIL, (N=7), and normal cervical mucosae (N=10), we observed significantly lower scores in the neoplastic tissues as compared to the mucosae (Mann-Whitney test, Mucose vs. HSIL & SCC; scores plotted as in B), with biologically intermediate HSIL lesions showing scores between those of normal mucosae and SCC. The Ral Transcriptional Signature score and prostate cancer disease aggression. D. Using data from a recently published cohort (N=131) of prostatic adenocarcinomas from Taylor et al. (40) we found that Ral signature scores could significantly stratify biochemical recurrence free survival. (Kaplan-Meier plot, Ral signature classes plotted at optimal discriminating point for survival, Log Rank Test). E. In the Taylor et al. cohort, a significant difference was observed between cases that did or did not evince seminal vesicle invasion (SVI) at prostatectomy (Mann-Whitney test, signature scores plotted and medians indicated per group, black lines).
The Ral Signature in Progression of Prostatic Adenocarcinoma
In animal models of prostate cancer, RalA and/or RalB have been associated with metastasis and androgen independence (7, 12, 38). We thus examined the status of the Ral signature with respects to important clinicopathologic surrogates of tumor aggressiveness in two recently published, large patient cohorts (39, 40). In patients treated by radical prostatectomy (N=131, Taylor et al. (40)), we did not observe significant correlations between the Ral signature scores and Gleason grade at biopsy (r=0.11, P=0.19) or prostatectomy (r=0.05, P=0.53), or with pathologic stage (P=0.86). However, Ral signature scores could risk stratify patients as a function of biochemical recurrence (P=0.05, Figure 4D). Analogous to the results described regarding invasion in bladder cancer, Ral signature scores were significantly higher in cases showing seminal vesicle invasion (SVI), a poor prognostic factor (P=0.028, Figure 4E). We extended and generalized these findings by evaluating the Ral signature score on data from the Swedish Watchful Waiting Cohort (N=281) (39). In this cohort, cases were incidentally diagnosed on transurethral resection (clinical T1a-b), and managed with observation only over a 10-year period. Here Ral signature score was significantly correlated with Gleason score (r=0.13, P=0.03) and could stratify patients by disease specific survival (P=0.03, Figure S6). Ral signature scores were not significantly associated with the TMPRSS-ERG fusion (41) in this cohort (P=0.77). Finally, given the discrepancy noted in correlation between Ral signature and Gleason signature scores between these two cohorts, we analyzed one additional cohort (N=69) from Wallace et al. (42) to further test for association with Gleason grade. As in the Taylor et al. cohort, we observed a no significant correlation (r=−0.11, P = 0.37).
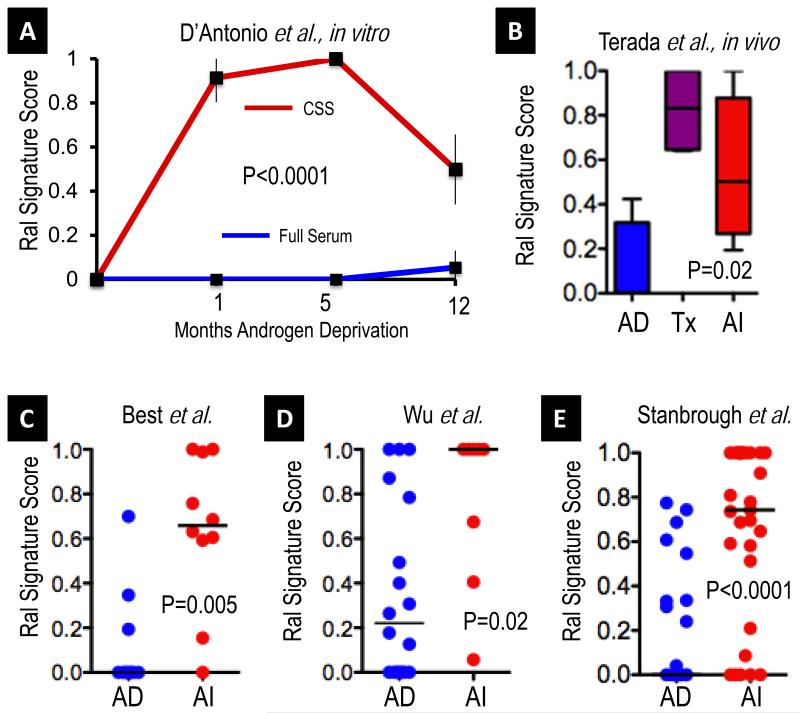
A clinically important dimension of prostate cancer biology is the issue of androgen dependence of disease. A recent report has functionally implicated RalA through induction of VEGFC upon androgen withdrawal (12). To examine whether androgen withdrawal was associated with changes in the Ral signature score through long-term androgen withdrawal, as occurs during therapy, we used a published gene expression study of longitudinal (1 year) in vitro androgen deprivation of LNCAP cells (43). Comparing the Ral signature scores of replicate androgen deprived cells to control cells over time, we observed an induction of the Ral signature scores over time (Figure 5A, P<0.0001). Next, we examined an explanted tumor xenograft model of androgen independent progression of prostate cancer, KUCaP-2, which has been transcriptionally profiled at baseline, at their growth nadir upon castration, and upon androgen independent regrowth (44). We found an induction of the Ral signature score over time that paralleled that observed in the LNCAP in vitro model (Figure 5B).
Figure 5. The Ral Transcriptional Signature score is sensitive to androgen status in prostate cancer.
A. Using published expression profiling data for LNCAP cells treated with control or charcoal-stripped (steroid hormone free) medium over a time course of 12 months (43), we observed significant and durable induction of the Ral signature over time in androgen deprived (charcoal stripped serum (CSS) red) as compared to control full serum treated cells (full serum, blue), Mann-Whitney test. B. Quadruplicate KuCAP-2 (44) xenografts were analyzed at androgen-dependent baseline (AD), at castration induced growth nadir (Tx), and during androgen independent (AI) regrowth. A significantly higher Ral signature score was seen in treated and androgen independent tumors (Mann-Whitney test of all treated versus baseline replicates, plot shows median plus 95% CI). C. Consistent with the in vitro and in vivo results in A and B, we observed significantly higher Ral signature scores in a published cohort (45) of androgen independent tumors (N=10) as compared to androgen dependent (N=10) cases (Mann Whitney test, signature scores plotted, and medians indicated per group, black lines). D. We also observed a similar higher Ral signature score in androgen independent cases in a second cohort (46) of androgen independent (N=8) as compared to androgen dependent cases (N=18). E. We observed similar higher Ral signature score in androgen independent metastatic tissues of a third cohort (47) of androgen independent (N=29) as compared to androgen naïve primary cases (N=22).
To determine whether such a mechanism operated in human tumors, we examined the Ral signature score in a dataset of microarray profiled, microdissected androgen dependent (N=10) and androgen independent (N=10) prostate tumors (45). We observed that the Ral signature score distributions differed significantly, with higher scores in androgen independent disease (Figure 5C, P=0.005). Random permutation testing suggested that the observed degree of difference between androgen dependent and independent cases was specific to the Ral transcriptional signature rather than global differences in transcription (false discovery rate 1%). We further tested this approach on a second cohort (46) of androgen dependent (N=18) and androgen independent cases (N=18), profiled on a different, custom microarray platform (Figure 5D, P=0.02), thus generalizing these observations. For further validation of the Ral signature in the setting of androgen independent metastatic prostate cancer, we used an additional dataset, reported by Stanbrough et al (47), where primary, hormone naïve tissues (N=22) and androgen independent bone metastatic cases (N=29) were profiled by microarray. We observed again a highly significant difference in Ral signature scores, with increased scores in androgen independent cases (P<0.0001, Figure 5E), similar to the two other cohorts. Again, as in the Swedish Watchful Waiting cohort above, Ral signature scores were not associated with the TMPRSS-ERG fusion status of the cases (P=0.32).
DISCUSSION
RalA and RalB have been implicated in transformation, regulation of survival, migration and metastasis. However, though ample dysregulation of pathways upstream of Ral signaling, including mutation of Ras paralogs, have been described in human tumors, mutations of RalA and RalB have not been found. In addition, despite observations of differential expression or activation of RalA or RalB in small cohorts of bladder (8), prostate (9), squamous (34), and pancreatic cancers, there is no evidence supporting the relevance of this pathway on clinical outcome in large patient series that include multiple cancer types.
To our knowledge, our findings provide the first evidence supporting a role of Ral in mediating clinically meaningful phenotypes in human cancer. First, we demonstrated that a Ral signature, derived from comparing bladder cancer cells with and without depletion of RalA and RalB (loss of function) was associated, in the proper, opposite direction, with Ras mutant activity, including specifically the E37G Ras effector loop mutant stimulating RalGEF pathway, (both gain of function) in another cell type (human embryonic kidney cells) (26). These findings argue strongly for the generality of this signature and support our downstream studies comparing the signature in different tissue types. Also, findings of our novel Ral transcriptional signature closely parallel experimentally demonstrated roles of Ral in model systems. This is perhaps most striking for squamous cell carcinoma (SCC), where Ral was shown to act as a tumor suppressor in experimental systems, in contrast to its role in other models (34). This relevance of this role in human cancer was supported by our findings using the Ral transcriptional signature score, which was lower in tumors compared to normal mucosa. This finding also speaks to the specificity of the Ral signature to Ral biology. For example, if the score were simply a surrogate of a global phenotype such as transformation, we would not expect lower signature scores in SCC compared to normal mucosa.
One key aspect of Ral biology is the ability of RalA and RalB to regulate transcription, through a number of transcriptional pathways including TCF, Jun, NF-κB, Stat3, HSF, forkhead family members (2). We recently added the metastasis and stem-cell associated gene, CD24, to this list (13, 32), and have also implicated the RREB1 transcription factor pathway therein (18, 24). However, it bears consideration that a number of the Ral signature genes (Table 1) have been demonstrated to play important roles in bladder and other cancers. Clusterin has substantial literature in bladder cancer as an antiapoptotic protein, both as a biomarker for disease aggression and, when inhibited, as a chemosensitizing agent to the key chemotherapeutic drug, gemcitabine; in prostate cancer, an inhibitor of clusterin, custirsen (OGX-011) has shown promise as a chemosensitizing agent in clinical trials castration resistant disease (48). Overexpression of replication protein A1 (RPA1), which we found to be lower in Ral signature positive cells, has been recently demonstrated to be a positive prognostic factor in bladder cancer (49), which correlates our signature findings. The L1CAM adhesion molecule, higher in signature positive cells, has an extensive literature in invasion and metastasis (50) and may be involved in cooperative signaling with the aforementioned CD24. Taken together these and prior findings suggest that Ral may coordinately regulate genes involved in aggression and metastasis.
Interestingly, based on this foundation and observations in this manuscript on a cohort of bladder cancer cell isolates stratified for their stem cell-like properties (51), we implicate for the first time Ral biology with this key phenotype. This is particularly interesting given the finding that human pancreatic cancer stem cells (CSCs) express high levels of CD24 (52) and pancreatic cancer is a K-Ras/Ral driven cancer (4). Since Ral regulates CD24 (13), the data presented here implicating Ral in the CSC phenotype is consistent with the literature. These findings suggest Ral might have a role in regulating this key cellular subpopulation thought to play a therapeutically central role in patients. Hence targeting the Ral GTPase would help delete this population of cells, reducing drug resistance with consequently beneficial clinical results.
The core signature of Ral-dependent transcription shared by RalA and RalB is a pervasive feature of muscle-invasive bladder cancer, all the more striking given its consistency across a large number of cohorts from different institutions, geographical locations, and profiled on different microarray platforms. In the case of one cohort by Sanchez-Carbayo et al. where survival data were available, we found that the signature was associated with survival, consistent with the role of Ral in experimental metastasis (5) as well as our observation herein that the Ral signature is associated with metastatic competence in experimental models.
In prostate cancer, reports have shown roles for Ral in model phenotypes of progression (7), including metastasis to bone (38) and induction of VEGF under androgen ablation (12). Here we found significant association of the Ral signature with emergence of androgen independence in vitro and in vivo in experimental models and in two patient cohorts. These findings implicate Ral in the very center of perhaps the most clinically important aspect of prostate cancer management, namely recurrence under androgen ablation therapy, a key driver of mortality in this disease. Targeting the Ral pathway in simultaneous combination with androgen deprivation might reduce the emergence of the hormone refractory state and deserves investigation as a therapeutic strategy. Importantly, excepting in the Swedish Watchful Waiting cohort of incidentally diagnosed (i.e., not PSA screened, as modern cohorts are) low stage disease, we found no correlation between the Ral signature score and Gleason grade. While this exception identifies another molecular difference between the Swedish cohort and modern PSA-screened biopsy populations (such as have been seen before regarding prevalence of ERG rearrangements), overall the Ral signature was uncorrelated to Gleason grade, suggesting that it could be adapted to provide independent or complementary prognostic data.
In summary, these findings provide the first conclusive evidence from human tumors that Ral GTPase status is clinically important. Furthermore, they provide a new tool to the scientific community, the Ral transcriptional signature score, which can be evaluated and compared to other prognostic tools in evaluating patients with cancers where Ral has been shown to have a driving role in model systems. In particular, these scores require validation in prospective cohorts and comparison to Ral activation in parallel aliquots of tumor, which despite the difficulty of biochemical activation assays (8) may become feasible through ELISA or activation state specific probes. Most importantly, by demonstrating the clinical relevance of Ral in human tumors, our work makes a strong case for investigation of strategies to interrupt Ral function. Irrespective of which therapeutic strategies should succeed, all would benefit from rational cohort selection for clinical trials based on the Ral signature score described herein.
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge Dr. Christopher Moskaluk, Dr. Henry Frierson, Ms. Sharon Birdsall and the University of Virginia Tissue Repository for their expertise in development and performance of immunohistochemistry. The authors would like to thank numerous members of the Theodorescu Lab and Dr. Scott Arthur Tomlins of the University of Michigan Department of Pathology for helpful discussions.
GRANT SUPPORT
National Institutes of Health grant CA075115 to DT.
Abbreviations
- UC
urothelial carcinoma
- NMIBC
non-muscle invasive bladder cancer
- MIBC
muscle invasive bladder cancer
- SCC
squamous cell carcinoma
- WNN
weighted K-nearest neighbor
Footnotes
Conflict of Interest: The Authors declare there are none.
REFERENCES
- 1.Bodemann BO, White MA. Ral GTPases and cancer: linchpin support of the tumorigenic platform. Nat Rev Cancer. 2008;8:133–40. doi: 10.1038/nrc2296. [DOI] [PubMed] [Google Scholar]
- 2.Feig LA. Ral-GTPases: approaching their 15 minutes of fame. Trends Cell Biol. 2003;13:419–25. doi: 10.1016/s0962-8924(03)00152-1. [DOI] [PubMed] [Google Scholar]
- 3.Hamad NM, Elconin JH, Karnoub AE, et al. Distinct requirements for Ras oncogenesis in human versus mouse cells. Genes Dev. 2002;16:2045–57. doi: 10.1101/gad.993902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lim KH, Baines AT, Fiordalisi JJ, et al. Activation of RalA is critical for Ras-induced tumorigenesis of human cells. Cancer Cell. 2005;7:533–45. doi: 10.1016/j.ccr.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 5.Wang H, Owens C, Chandra N, Conaway MR, Brautigan DL, Theodorescu D. Phosphorylation of RalB is important for bladder cancer cell growth and metastasis. Cancer Res. 2010;70:8760–9. doi: 10.1158/0008-5472.CAN-10-0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oxford G, Owens CR, Titus BJ, et al. RalA and RalB: antagonistic relatives in cancer cell migration. Cancer Res. 2005;65:7111–20. doi: 10.1158/0008-5472.CAN-04-1957. [DOI] [PubMed] [Google Scholar]
- 7.Wu Z, Owens C, Chandra N, Popovic K, Conaway M, Theodorescu D. RalBP1 is necessary for metastasis of human cancer cell lines. Neoplasia. 2010;12:1003–12. doi: 10.1593/neo.101080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith SC, Oxford G, Baras AS, et al. Expression of ral GTPases, their effectors, and activators in human bladder cancer. Clin Cancer Res. 2007;13:3803–13. doi: 10.1158/1078-0432.CCR-06-2419. [DOI] [PubMed] [Google Scholar]
- 9.Varambally S, Yu J, Laxman B, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 10.Wang H, Owens C, Chandra N, Conaway MR, Brautigan DL, Theodorescu D. Phosphorylation of RalB is important for bladder cancer cell growth and metastasis. Cancer Res. 2010;70:8760–9. doi: 10.1158/0008-5472.CAN-10-0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henry DO, Moskalenko SA, Kaur KJ, et al. Ral GTPases contribute to regulation of cyclin D1 through activation of NF-kappaB. Mol Cell Biol. 2000;20:8084–92. doi: 10.1128/mcb.20.21.8084-8092.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rinaldo F, Li J, Wang E, Muders M, Datta K. RalA regulates vascular endothelial growth factor-C (VEGF-C) synthesis in prostate cancer cells during androgen ablation. Oncogene. 2006 doi: 10.1038/sj.onc.1209971. [DOI] [PubMed] [Google Scholar]
- 13.Smith SC, Oxford G, Wu Z, et al. The metastasis-associated gene CD24 is regulated by Ral GTPase and is a mediator of cell proliferation and survival in human cancer. Cancer Res. 2006;66:1917–22. doi: 10.1158/0008-5472.CAN-05-3855. [DOI] [PubMed] [Google Scholar]
- 14.Lee JK, Havaleshko DM, Cho H, et al. A strategy for predicting the chemosensitivity of human cancers and its application to drug discovery. Proc Natl Acad Sci U S A. 2007;104:13086–91. doi: 10.1073/pnas.0610292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dancik GM, Ru Y, Owens CR, Theodorescu D. A framework to select clinically relevant cancer cell lines for investigation by establishing their molecular similarity with primary human cancers. Cancer Res. 2011;71:7398–409. doi: 10.1158/0008-5472.CAN-11-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith SC, Nicholson B, Nitz M, et al. Profiling Bladder Cancer Organ Site-Specific Metastasis Identifies LAMC2 as a Novel Biomarker of Hematogenous Dissemination. Am J Pathol. 2009;174:371–79. doi: 10.2353/ajpath.2009.080538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chien Y, White MA. RAL GTPases are linchpin modulators of human tumour-cell proliferation and survival. EMBO Rep. 2003;4:800–6. doi: 10.1038/sj.embor.embor899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oxford G, Smith SC, Hampton G, Theodorescu D. Expression profiling of Ral-depleted bladder cancer cells identifies RREB-1 as a novel transcriptional Ral effector. Oncogene. 2007;26:7143–52. doi: 10.1038/sj.onc.1210521. [DOI] [PubMed] [Google Scholar]
- 19.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith SC, Baras AS, Lee JK, Theodorescu D. The COXEN principle: translating signatures of in vitro chemosensitivity into tools for clinical outcome prediction and drug discovery in cancer. Cancer Res. 2010;70:1753–8. doi: 10.1158/0008-5472.CAN-09-3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith SC, Baras AS, Dancik G, et al. A 20-gene model for molecular nodal staging of bladder cancer: development and prospective assessment. Lancet Oncol. 2011;12:137–43. doi: 10.1016/S1470-2045(10)70296-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanchez-Carbayo M, Socci ND, Lozano J, Saint F, Cordon-Cardo C. Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. J Clin Oncol. 2006;24:778–89. doi: 10.1200/JCO.2005.03.2375. [DOI] [PubMed] [Google Scholar]
- 23.Neel NF, Martin TD, Stratford JK, Zand TP, Reiner DJ, Der CJ. The RalGEF-Ral Effector Signaling Network: The Road Less Traveled for Anti-Ras Drug Discovery. Genes Cancer. 2011;2:275–87. doi: 10.1177/1947601911407329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nitz MD, Harding MA, Smith SC, Thomas S, Theodorescu D. RREB1 Transcription Factor Splice Variants in Urologic Cancer. Am J Pathol. 2011;179:477–86. doi: 10.1016/j.ajpath.2011.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams PD, Cheon S, Havaleshko DM, et al. Concordant gene expression signatures predict clinical outcomes of cancer patients undergoing systemic therapy. Cancer Res. 2009;69:8302–9. doi: 10.1158/0008-5472.CAN-09-0798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang JT, Carvalho C, Mori S, et al. A genomic strategy to elucidate modules of oncogenic pathway signaling networks. Mol Cell. 2009;34:104–14. doi: 10.1016/j.molcel.2009.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dyrskjot L, Thykjaer T, Kruhoffer M, et al. Identifying distinct classes of bladder carcinoma using microarrays. Nat Genet. 2003;33:90–6. doi: 10.1038/ng1061. [DOI] [PubMed] [Google Scholar]
- 28.Stransky N, Vallot C, Reyal F, et al. Regional copy number-independent deregulation of transcription in cancer. Nat Genet. 2006;38:1386–96. doi: 10.1038/ng1923. [DOI] [PubMed] [Google Scholar]
- 29.Lindgren D, Frigyesi A, Gudjonsson S, et al. Combined gene expression and genomic profiling define two intrinsic molecular subtypes of urothelial carcinoma and gene signatures for molecular grading and outcome. Cancer Res. 2010;70:3463–72. doi: 10.1158/0008-5472.CAN-09-4213. [DOI] [PubMed] [Google Scholar]
- 30.Kim WJ, Kim EJ, Kim SK, et al. Predictive value of progression-related gene classifier in primary non-muscle invasive bladder cancer. Mol Cancer. 2010;9:3. doi: 10.1186/1476-4598-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riester M, Taylor JM, Feifer A, et al. Combination of a novel gene expression signature with a clinical nomogram improves the prediction of survival in high-risk bladder cancer. Clin Cancer Res. 2012;18:1323–33. doi: 10.1158/1078-0432.CCR-11-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Overdevest JB, Thomas S, Kristiansen G, Hansel DE, Smith SC, Theodorescu D. CD24 offers a therapeutic target for control of bladder cancer metastasis based on a requirement for lung colonization. Cancer Res. 2011;71:3802–11. doi: 10.1158/0008-5472.CAN-11-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dyrskjot L, Zieger K, Kruhoffer M, et al. A molecular signature in superficial bladder carcinoma predicts clinical outcome. Clin Cancer Res. 2005;11:4029–36. doi: 10.1158/1078-0432.CCR-04-2095. [DOI] [PubMed] [Google Scholar]
- 34.Sowalsky AG, Alt-Holland A, Shamis Y, Garlick JA, Feig LA. RalA suppresses early stages of Ras-induced squamous cell carcinoma progression. Oncogene. 2010;29:45–55. doi: 10.1038/onc.2009.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su H, Hu N, Yang HH, et al. Global Gene Expression Profiling and Validation in Esophageal Squamous Cell Carcinoma and its Association with Clinical Phenotypes. Clin Cancer Res. 2011 doi: 10.1158/1078-0432.CCR-10-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ye H, Yu T, Temam S, et al. Transcriptomic dissection of tongue squamous cell carcinoma. BMC Genomics. 2008;9:69. doi: 10.1186/1471-2164-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhai Y, Kuick R, Nan B, et al. Gene expression analysis of preinvasive and invasive cervical squamous cell carcinomas identifies HOXC10 as a key mediator of invasion. Cancer Res. 2007;67:10163–72. doi: 10.1158/0008-5472.CAN-07-2056. [DOI] [PubMed] [Google Scholar]
- 38.Yin J, Pollock C, Tracy K, et al. Activation of the RalGEF/Ral pathway promotes prostate cancer metastasis to bone. Mol Cell Biol. 2007;27:7538–50. doi: 10.1128/MCB.00955-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sboner A, Demichelis F, Calza S, et al. Molecular sampling of prostate cancer: a dilemma for predicting disease progression. BMC Med Genomics. 2010;3:8. doi: 10.1186/1755-8794-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor BS, Schultz N, Hieronymus H, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 42.Wallace TA, Prueitt RL, Yi M, et al. Tumor immunobiological differences in prostate cancer between African-American and European-American men. Cancer Res. 2008;68:927–36. doi: 10.1158/0008-5472.CAN-07-2608. [DOI] [PubMed] [Google Scholar]
- 43.D’Antonio JM, Ma C, Monzon FA, Pflug BR. Longitudinal analysis of androgen deprivation of prostate cancer cells identifies pathways to androgen independence. Prostate. 2008;68:698–714. doi: 10.1002/pros.20677. [DOI] [PubMed] [Google Scholar]
- 44.Terada N, Shimizu Y, Kamba T, et al. Identification of EP4 as a potential target for the treatment of castration-resistant prostate cancer using a novel xenograft model. Cancer Res. 2010;70:1606–15. doi: 10.1158/0008-5472.CAN-09-2984. [DOI] [PubMed] [Google Scholar]
- 45.Best CJ, Gillespie JW, Yi Y, et al. Molecular alterations in primary prostate cancer after androgen ablation therapy. Clin Cancer Res. 2005;11:6823–34. doi: 10.1158/1078-0432.CCR-05-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei Q, Li M, Fu X, et al. Global analysis of differentially expressed genes in androgen-independent prostate cancer. Prostate Cancer Prostatic Dis. 2007;10:167–74. doi: 10.1038/sj.pcan.4500933. [DOI] [PubMed] [Google Scholar]
- 47.Stanbrough M, Bubley GJ, Ross K, et al. Increased expression of genes converting adrenal androgens to testosterone in androgen-independent prostate cancer. Cancer Res. 2006;66:2815–25. doi: 10.1158/0008-5472.CAN-05-4000. [DOI] [PubMed] [Google Scholar]
- 48.Dayyani F, Gallick GE, Logothetis CJ, Corn PG. Novel therapies for metastatic castrate-resistant prostate cancer. J Natl Cancer Inst. 2011;103:1665–75. doi: 10.1093/jnci/djr362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Levidou G, Gakiopoulou H, Kavantzas N, et al. Prognostic significance of replication protein A (RPA) expression levels in bladder urothelial carcinoma. BJU Int. 2011;108:E59–65. doi: 10.1111/j.1464-410X.2010.09828.x. [DOI] [PubMed] [Google Scholar]
- 50.Raveh S, Gavert N, Ben-Ze’ev A. L1 cell adhesion molecule (L1CAM) in invasive tumors. Cancer Lett. 2009;282:137–45. doi: 10.1016/j.canlet.2008.12.021. [DOI] [PubMed] [Google Scholar]
- 51.He X, Marchionni L, Hansel DE, et al. Differentiation of a highly tumorigenic basal cell compartment in urothelial carcinoma. Stem Cells. 2009;27:1487–95. doi: 10.1002/stem.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shankar S, Nall D, Tang SN, et al. Resveratrol inhibits pancreatic cancer stem cell characteristics in human and KrasG12D transgenic mice by inhibiting pluripotency maintaining factors and epithelial-mesenchymal transition. PLoS One. 2011;6:e16530. doi: 10.1371/journal.pone.0016530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.