Abstract
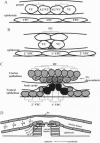
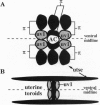
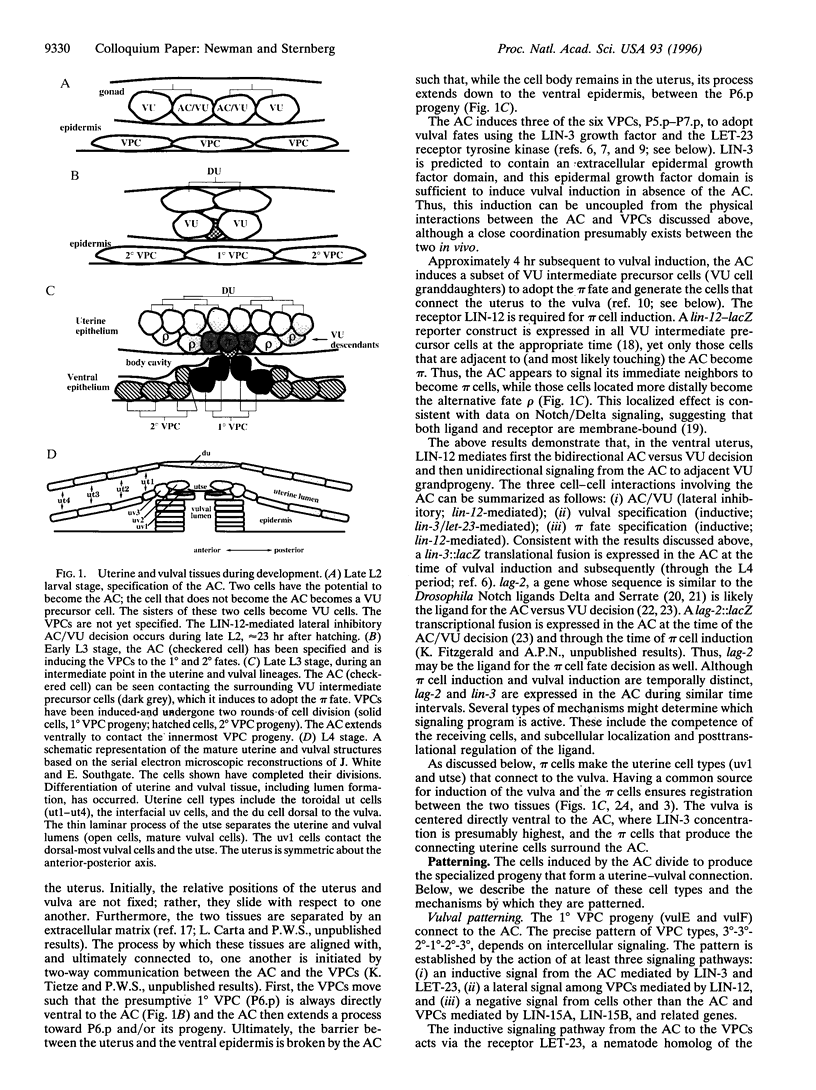
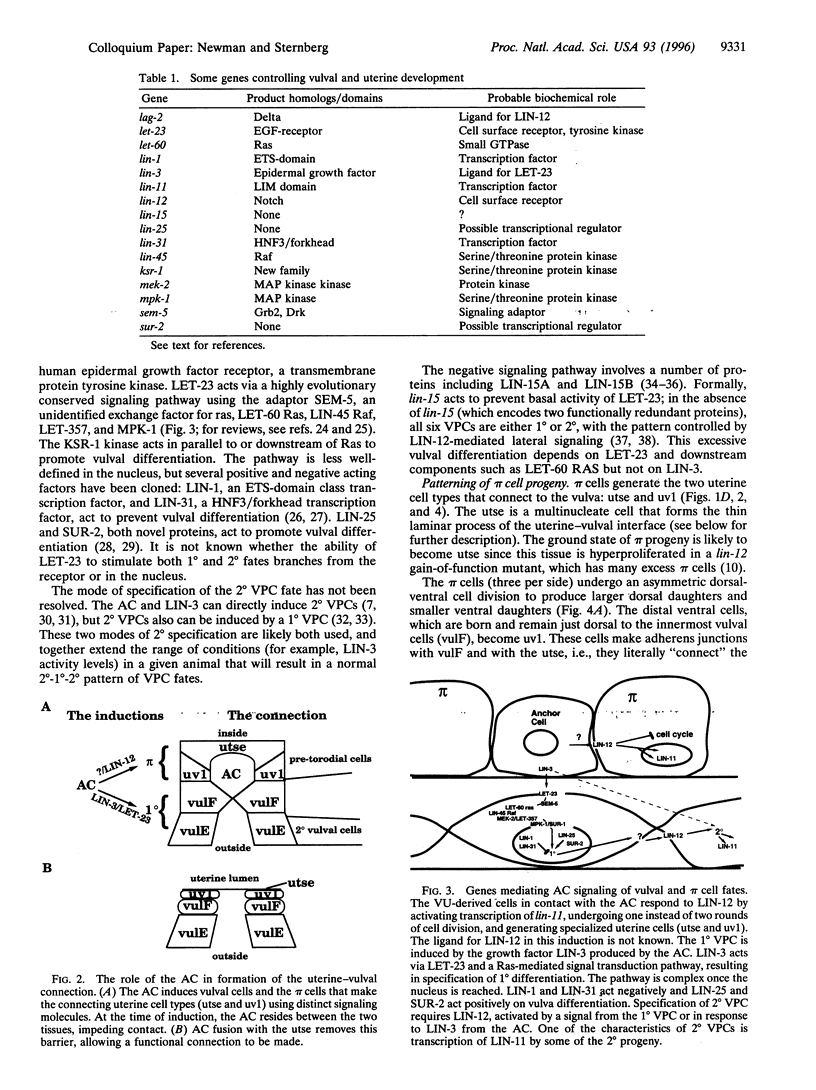
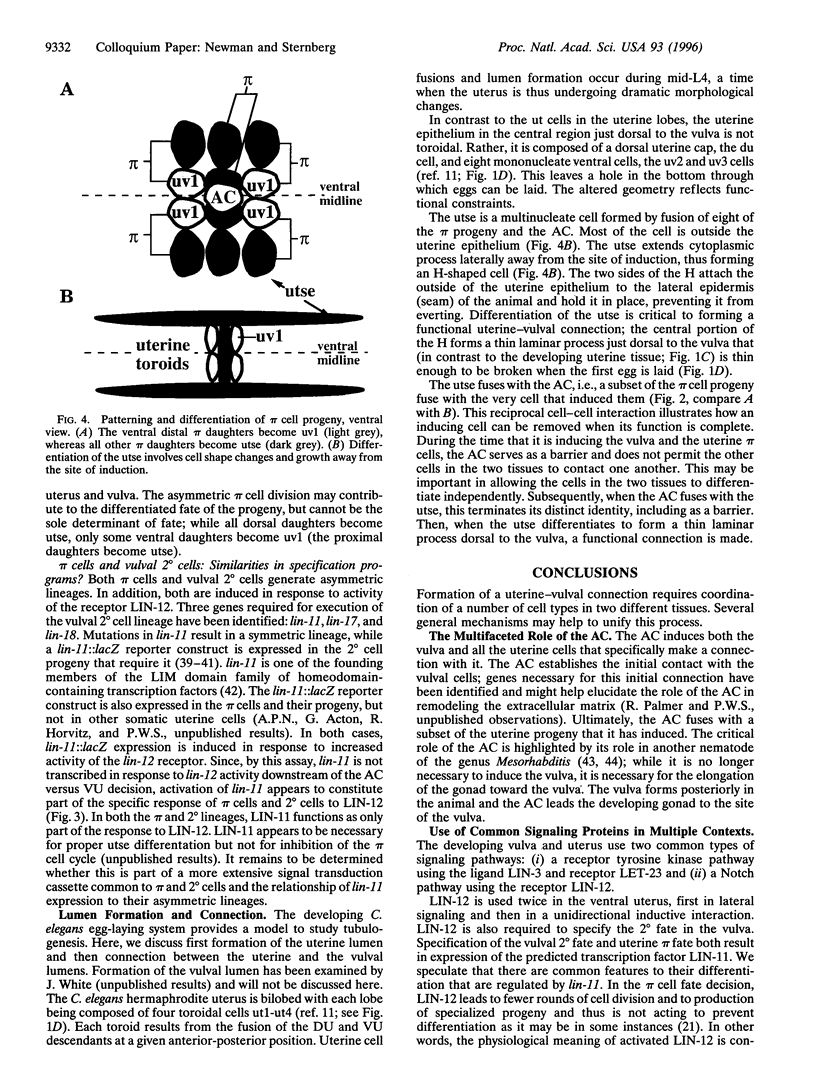
Development of the nematode egg-laying system requires the formation of a connection between the uterine lumen and the developing vulval lumen, thus allowing a passage for eggs and sperm. This relatively simple process serves as a model for certain aspects of organogenesis. Such a connection demands that cells in both tissues become specialized to participate in the connection, and that the specialized cells are brought in register. A single cell, the anchor cell, acts to induce and to organize specialization of the epidermal and uterine epithelia, and registrates these tissues. The inductions act via evolutionarily conserved intercellular signaling pathways. The anchor cell induces the vulva from ventral epithelial cells via the LIN-3 growth factor and LET-23 transmembrane tyrosine kinase. It then induces surrounding uterine intermediate precursors via the receptor LIN-12, a founding member of the Notch family of receptors. Both signaling pathways are used multiple times during development of Caenorhabditis elegans. The outcome of the signaling is context-dependent. Both inductions are reciprocated. After the anchor cell has induced the vulva, it stretches toward the induced vulval cells. After the anchor cell has induced specialized uterine intermediate precursor cells, it fuses with a subset of their progeny.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aroian R. V., Koga M., Mendel J. E., Ohshima Y., Sternberg P. W. The let-23 gene necessary for Caenorhabditis elegans vulval induction encodes a tyrosine kinase of the EGF receptor subfamily. Nature. 1990 Dec 20;348(6303):693–699. doi: 10.1038/348693a0. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S., Matsuno K., Fortini M. E. Notch signaling. Science. 1995 Apr 14;268(5208):225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- Beitel G. J., Tuck S., Greenwald I., Horvitz H. R. The Caenorhabditis elegans gene lin-1 encodes an ETS-domain protein and defines a branch of the vulval induction pathway. Genes Dev. 1995 Dec 15;9(24):3149–3162. doi: 10.1101/gad.9.24.3149. [DOI] [PubMed] [Google Scholar]
- Birchmeier C., Birchmeier W. Molecular aspects of mesenchymal-epithelial interactions. Annu Rev Cell Biol. 1993;9:511–540. doi: 10.1146/annurev.cb.09.110193.002455. [DOI] [PubMed] [Google Scholar]
- Chamberlin H. M., Sternberg P. W. The lin-3/let-23 pathway mediates inductive signalling during male spicule development in Caenorhabditis elegans. Development. 1994 Oct;120(10):2713–2721. doi: 10.1242/dev.120.10.2713. [DOI] [PubMed] [Google Scholar]
- Clark S. G., Lu X., Horvitz H. R. The Caenorhabditis elegans locus lin-15, a negative regulator of a tyrosine kinase signaling pathway, encodes two different proteins. Genetics. 1994 Aug;137(4):987–997. doi: 10.1093/genetics/137.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downward J. KSR: a novel player in the RAS pathway. Cell. 1995 Dec 15;83(6):831–834. doi: 10.1016/0092-8674(95)90198-1. [DOI] [PubMed] [Google Scholar]
- Fehon R. G., Kooh P. J., Rebay I., Regan C. L., Xu T., Muskavitch M. A., Artavanis-Tsakonas S. Molecular interactions between the protein products of the neurogenic loci Notch and Delta, two EGF-homologous genes in Drosophila. Cell. 1990 May 4;61(3):523–534. doi: 10.1016/0092-8674(90)90534-l. [DOI] [PubMed] [Google Scholar]
- Ferguson E. L., Horvitz H. R. Identification and characterization of 22 genes that affect the vulval cell lineages of the nematode Caenorhabditis elegans. Genetics. 1985 May;110(1):17–72. doi: 10.1093/genetics/110.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson E. L., Horvitz H. R. The multivulva phenotype of certain Caenorhabditis elegans mutants results from defects in two functionally redundant pathways. Genetics. 1989 Sep;123(1):109–121. doi: 10.1093/genetics/123.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson E. L., Sternberg P. W., Horvitz H. R. A genetic pathway for the specification of the vulval cell lineages of Caenorhabditis elegans. Nature. 1987 Mar 19;326(6110):259–267. doi: 10.1038/326259a0. [DOI] [PubMed] [Google Scholar]
- Freyd G., Kim S. K., Horvitz H. R. Novel cysteine-rich motif and homeodomain in the product of the Caenorhabditis elegans cell lineage gene lin-11. Nature. 1990 Apr 26;344(6269):876–879. doi: 10.1038/344876a0. [DOI] [PubMed] [Google Scholar]
- Félix M. A., Sternberg P. W. Symmetry breakage in the development of one-armed gonads in nematodes. Development. 1996 Jul;122(7):2129–2142. doi: 10.1242/dev.122.7.2129. [DOI] [PubMed] [Google Scholar]
- Greenwald I. S., Sternberg P. W., Horvitz H. R. The lin-12 locus specifies cell fates in Caenorhabditis elegans. Cell. 1983 Sep;34(2):435–444. doi: 10.1016/0092-8674(83)90377-x. [DOI] [PubMed] [Google Scholar]
- Hill R. J., Sternberg P. W. The gene lin-3 encodes an inductive signal for vulval development in C. elegans. Nature. 1992 Aug 6;358(6386):470–476. doi: 10.1038/358470a0. [DOI] [PubMed] [Google Scholar]
- Horvitz H. R., Sulston J. E. Isolation and genetic characterization of cell-lineage mutants of the nematode Caenorhabditis elegans. Genetics. 1980 Oct;96(2):435–454. doi: 10.1093/genetics/96.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L. S., Tzou P., Sternberg P. W. The lin-15 locus encodes two negative regulators of Caenorhabditis elegans vulval development. Mol Biol Cell. 1994 Apr;5(4):395–411. doi: 10.1091/mbc.5.4.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessell T. M., Dodd J. Floor plate-derived signals and the control of neural cell pattern in vertebrates. Harvey Lect. 1990;86:87–128. [PubMed] [Google Scholar]
- Jongeward G. D., Clandinin T. R., Sternberg P. W. sli-1, a negative regulator of let-23-mediated signaling in C. elegans. Genetics. 1995 Apr;139(4):1553–1566. doi: 10.1093/genetics/139.4.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz W. S., Hill R. J., Clandinin T. R., Sternberg P. W. Different levels of the C. elegans growth factor LIN-3 promote distinct vulval precursor fates. Cell. 1995 Jul 28;82(2):297–307. doi: 10.1016/0092-8674(95)90317-8. [DOI] [PubMed] [Google Scholar]
- Katz W. S., Lesa G. M., Yannoukakos D., Clandinin T. R., Schlessinger J., Sternberg P. W. A point mutation in the extracellular domain activates LET-23, the Caenorhabditis elegans epidermal growth factor receptor homolog. Mol Cell Biol. 1996 Feb;16(2):529–537. doi: 10.1128/mcb.16.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayne P. S., Sternberg P. W. Ras pathways in Caenorhabditis elegans. Curr Opin Genet Dev. 1995 Feb;5(1):38–43. doi: 10.1016/s0959-437x(95)90051-9. [DOI] [PubMed] [Google Scholar]
- Kimble J. Alterations in cell lineage following laser ablation of cells in the somatic gonad of Caenorhabditis elegans. Dev Biol. 1981 Oct 30;87(2):286–300. doi: 10.1016/0012-1606(81)90152-4. [DOI] [PubMed] [Google Scholar]
- Kimble J., Hirsh D. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev Biol. 1979 Jun;70(2):396–417. doi: 10.1016/0012-1606(79)90035-6. [DOI] [PubMed] [Google Scholar]
- Koga M., Ohshima Y. Mosaic analysis of the let-23 gene function in vulval induction of Caenorhabditis elegans. Development. 1995 Aug;121(8):2655–2666. doi: 10.1242/dev.121.8.2655. [DOI] [PubMed] [Google Scholar]
- Lambie E. J., Kimble J. Two homologous regulatory genes, lin-12 and glp-1, have overlapping functions. Development. 1991 May;112(1):231–240. doi: 10.1242/dev.112.1.231. [DOI] [PubMed] [Google Scholar]
- Miller L. M., Gallegos M. E., Morisseau B. A., Kim S. K. lin-31, a Caenorhabditis elegans HNF-3/fork head transcription factor homolog, specifies three alternative cell fates in vulval development. Genes Dev. 1993 Jun;7(6):933–947. doi: 10.1101/gad.7.6.933. [DOI] [PubMed] [Google Scholar]
- Newman A. P., White J. G., Sternberg P. W. The Caenorhabditis elegans lin-12 gene mediates induction of ventral uterine specialization by the anchor cell. Development. 1995 Feb;121(2):263–271. doi: 10.1242/dev.121.2.263. [DOI] [PubMed] [Google Scholar]
- Seydoux G., Greenwald I. Cell autonomy of lin-12 function in a cell fate decision in C. elegans. Cell. 1989 Jun 30;57(7):1237–1245. doi: 10.1016/0092-8674(89)90060-3. [DOI] [PubMed] [Google Scholar]
- Simske J. S., Kim S. K. Sequential signalling during Caenorhabditis elegans vulval induction. Nature. 1995 May 11;375(6527):142–146. doi: 10.1038/375142a0. [DOI] [PubMed] [Google Scholar]
- Singh N., Han M. sur-2, a novel gene, functions late in the let-60 ras-mediated signaling pathway during Caenorhabditis elegans vulval induction. Genes Dev. 1995 Sep 15;9(18):2251–2265. doi: 10.1101/gad.9.18.2251. [DOI] [PubMed] [Google Scholar]
- Sommer R. J., Sternberg P. W. Changes of induction and competence during the evolution of vulva development in nematodes. Science. 1994 Jul 1;265(5168):114–118. doi: 10.1126/science.8016644. [DOI] [PubMed] [Google Scholar]
- Stern M. J., DeVore D. L. Extending and connecting signaling pathways in C. elegans. Dev Biol. 1994 Dec;166(2):443–459. doi: 10.1006/dbio.1994.1328. [DOI] [PubMed] [Google Scholar]
- Sternberg P. W. Control of cell fates within equivalence groups in C. elegans. Trends Neurosci. 1988 Jun;11(6):259–264. doi: 10.1016/0166-2236(88)90106-3. [DOI] [PubMed] [Google Scholar]
- Sternberg P. W., Horvitz H. R. Pattern formation during vulval development in C. elegans. Cell. 1986 Mar 14;44(5):761–772. doi: 10.1016/0092-8674(86)90842-1. [DOI] [PubMed] [Google Scholar]
- Sternberg P. W., Horvitz H. R. The combined action of two intercellular signaling pathways specifies three cell fates during vulval induction in C. elegans. Cell. 1989 Aug 25;58(4):679–693. doi: 10.1016/0092-8674(89)90103-7. [DOI] [PubMed] [Google Scholar]
- Struhl G., Fitzgerald K., Greenwald I. Intrinsic activity of the Lin-12 and Notch intracellular domains in vivo. Cell. 1993 Jul 30;74(2):331–345. doi: 10.1016/0092-8674(93)90424-o. [DOI] [PubMed] [Google Scholar]
- Tax F. E., Yeargers J. J., Thomas J. H. Sequence of C. elegans lag-2 reveals a cell-signalling domain shared with Delta and Serrate of Drosophila. Nature. 1994 Mar 10;368(6467):150–154. doi: 10.1038/368150a0. [DOI] [PubMed] [Google Scholar]
- Tuck S., Greenwald I. lin-25, a gene required for vulval induction in Caenorhabditis elegans. Genes Dev. 1995 Feb 1;9(3):341–357. doi: 10.1101/gad.9.3.341. [DOI] [PubMed] [Google Scholar]
- White J. G., Southgate E., Thomson J. N., Brenner S. The structure of the ventral nerve cord of Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1976 Aug 10;275(938):327–348. doi: 10.1098/rstb.1976.0086. [DOI] [PubMed] [Google Scholar]
- Wilkinson H. A., Fitzgerald K., Greenwald I. Reciprocal changes in expression of the receptor lin-12 and its ligand lag-2 prior to commitment in a C. elegans cell fate decision. Cell. 1994 Dec 30;79(7):1187–1198. doi: 10.1016/0092-8674(94)90010-8. [DOI] [PubMed] [Google Scholar]
- Wilkinson H. A., Greenwald I. Spatial and temporal patterns of lin-12 expression during C. elegans hermaphrodite development. Genetics. 1995 Oct;141(2):513–526. doi: 10.1093/genetics/141.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yochem J., Weston K., Greenwald I. The Caenorhabditis elegans lin-12 gene encodes a transmembrane protein with overall similarity to Drosophila Notch. Nature. 1988 Oct 6;335(6190):547–550. doi: 10.1038/335547a0. [DOI] [PubMed] [Google Scholar]