Abstract
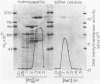
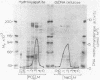
Dye-ligand chromatography on Cibacron blue F3GA-agarose has been used to resolve two species of DNA (cytosine-5-)-methyltransferase from nuclear extracts of uninduced Friend murine erythroleukemia cells. Each species has been highly purified; the activities in the first and second peaks were associated with polypeptides of Mr 150,000 and 175,000, respectively. Analysis of substrate specificity with synthetic DNAs and restriction fragments of phi X174 replicative form DNA and pBR322 DNA showed that neither enzyme had dependence on the sequence context of CpG dinucleotides; poly(dG-dC) had the greatest methyl-accepting activity of any unmethylated DNA substrate tested. De novo methylation by both enzymes was inefficient relative to methylation of hemimethylated sites. Methyl-accepting activity was strongly dependent on DNA chain length. This observation suggests that binding to DNA, followed by one-dimensional diffusion of enzyme along the DNA molecule, is important in the mechanism by which DNA methyltransferase locates its recognition sites.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams R. L., McKay E. L., Craig L. M., Burdon R. H. Mouse DNA methylase: methylation of native DNA. Biochim Biophys Acta. 1979 Feb 27;561(2):345–357. doi: 10.1016/0005-2787(79)90143-6. [DOI] [PubMed] [Google Scholar]
- Berg O. G., Winter R. B., von Hippel P. H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981 Nov 24;20(24):6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- Bird A. P., Southern E. M. Use of restriction enzymes to study eukaryotic DNA methylation: I. The methylation pattern in ribosomal DNA from Xenopus laevis. J Mol Biol. 1978 Jan 5;118(1):27–47. doi: 10.1016/0022-2836(78)90242-5. [DOI] [PubMed] [Google Scholar]
- Bonner W. M., Stedman J. D. Efficient fluorography of 3H and 14C on thin layers. Anal Biochem. 1978 Aug 15;89(1):247–256. doi: 10.1016/0003-2697(78)90747-9. [DOI] [PubMed] [Google Scholar]
- Christman J. K., Weich N., Schoenbrun B., Schneiderman N., Acs G. Hypomethylation of DNA during differentiation of Friend erythroleukemia cells. J Cell Biol. 1980 Aug;86(2):366–370. doi: 10.1083/jcb.86.2.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B. Nature of Col E 1 plasmid replication in Escherichia coli in the presence of the chloramphenicol. J Bacteriol. 1972 May;110(2):667–676. doi: 10.1128/jb.110.2.667-676.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drahovský D., Morris N. R. Mechanism of action of rat liver DNA methylase. II. Interaction with single-stranded methyl-acceptor DNA. J Mol Biol. 1971 Oct 28;61(2):343–356. doi: 10.1016/0022-2836(71)90384-6. [DOI] [PubMed] [Google Scholar]
- Groudine M., Eisenman R., Weintraub H. Chromatin structure of endogenous retroviral genes and activation by an inhibitor of DNA methylation. Nature. 1981 Jul 23;292(5821):311–317. doi: 10.1038/292311a0. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y., Cedar H., Razin A. Substrate and sequence specificity of a eukaryotic DNA methylase. Nature. 1982 Feb 18;295(5850):620–622. doi: 10.1038/295620a0. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y., Stein R., Cedar H., Razin A. Methylation of CpG sequences in eukaryotic DNA. FEBS Lett. 1981 Feb 9;124(1):67–71. doi: 10.1016/0014-5793(81)80055-5. [DOI] [PubMed] [Google Scholar]
- Haigh L. S., Owens B. B., Hellewell O. S., Ingram V. M. DNA methylation in chicken alpha-globin gene expression. Proc Natl Acad Sci U S A. 1982 Sep;79(17):5332–5336. doi: 10.1073/pnas.79.17.5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack W. E., Terry B. J., Modrich P. Involvement of outside DNA sequences in the major kinetic path by which EcoRI endonuclease locates and leaves its recognition sequence. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4010–4014. doi: 10.1073/pnas.79.13.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalousek F., Morris N. R. Deoxyribonucleic acid methylase activity in rat spleen. J Biol Chem. 1968 May 10;243(9):2440–2442. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mandel J. L., Chambon P. DNA methylation: organ specific variations in the methylation pattern within and around ovalbumin and other chicken genes. Nucleic Acids Res. 1979 Dec 20;7(8):2081–2103. doi: 10.1093/nar/7.8.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks P. A., Rifkind R. A. Erythroleukemic differentiation. Annu Rev Biochem. 1978;47:419–448. doi: 10.1146/annurev.bi.47.070178.002223. [DOI] [PubMed] [Google Scholar]
- McGeady M. L., Jhappan C., Ascione R., Vande Woude G. F. In vitro methylation of specific regions of the cloned Moloney sarcoma virus genome inhibits its transforming activity. Mol Cell Biol. 1983 Mar;3(3):305–314. doi: 10.1128/mcb.3.3.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi H., Prem veer Reddy G., Pardee A. B. Rapid incorporation of label from ribonucleoside disphosphates into DNA by a cell-free high molecular weight fraction from animal cell nuclei. Cell. 1983 Feb;32(2):443–451. doi: 10.1016/0092-8674(83)90464-6. [DOI] [PubMed] [Google Scholar]
- Park C. S., Hillel Z., Wu C. W. Molecular mechanism of promoter selection in gene transcription. I. Development of a rapid mixing-photocrosslinking technique to study the kinetics of Escherichia coli RNA polymerase binding to T7 DNA. J Biol Chem. 1982 Jun 25;257(12):6944–6949. [PubMed] [Google Scholar]
- Peck L. J., Nordheim A., Rich A., Wang J. C. Flipping of cloned d(pCpG)n.d(pCpG)n DNA sequences from right- to left-handed helical structure by salt, Co(III), or negative supercoiling. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4560–4564. doi: 10.1073/pnas.79.15.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy P. H., Weissbach A. DNA methylase from HeLa cell nuclei. Nucleic Acids Res. 1975 Oct;2(10):1669–1684. doi: 10.1093/nar/2.10.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Air G. M., Barrell B. G., Brown N. L., Coulson A. R., Fiddes C. A., Hutchison C. A., Slocombe P. M., Smith M. Nucleotide sequence of bacteriophage phi X174 DNA. Nature. 1977 Feb 24;265(5596):687–695. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- Sano H., Sager R. Tissue specificity and clustering of methylated cystosines in bovine satellite I DNA. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3584–3588. doi: 10.1073/pnas.79.11.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffery M., Rifkind R. A., Marks P. A. Murine erythroleukemia cell differentiation: DNase I hypersensitivity and DNA methylation near the globin genes. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1180–1184. doi: 10.1073/pnas.79.4.1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon D., Grunert F., von Acken U., Döring H. P., Kröger H. DNA-methylase from regenerating rat liver: purification and characterisation. Nucleic Acids Res. 1978 Jun;5(6):2153–2167. doi: 10.1093/nar/5.6.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneider T. W., Teague W. M., Rogachevsky L. M. S-adenosylmethionine: DNA-cytosine 5-methyltransferase from a Novikoff rat hepatoma cell line. Nucleic Acids Res. 1975 Oct;2(10):1685–1700. doi: 10.1093/nar/2.10.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein R., Razin A., Cedar H. In vitro methylation of the hamster adenine phosphoribosyltransferase gene inhibits its expression in mouse L cells. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3418–3422. doi: 10.1073/pnas.79.11.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Taylor S. M., Jones P. A. Mechanism of action of eukaryotic DNA methyltransferase. Use of 5-azacytosine-containing DNA. J Mol Biol. 1982 Dec 15;162(3):679–692. doi: 10.1016/0022-2836(82)90395-3. [DOI] [PubMed] [Google Scholar]
- Wigler M. H. The inheritance of methylation patterns in vertebrates. Cell. 1981 May;24(2):285–286. doi: 10.1016/0092-8674(81)90317-2. [DOI] [PubMed] [Google Scholar]
- Wigler M., Levy D., Perucho M. The somatic replication of DNA methylation. Cell. 1981 Apr;24(1):33–40. doi: 10.1016/0092-8674(81)90498-0. [DOI] [PubMed] [Google Scholar]
- van der Ploeg L. H., Flavell R. A. DNA methylation in the human gamma delta beta-globin locus in erythroid and nonerythroid tissues. Cell. 1980 Apr;19(4):947–958. doi: 10.1016/0092-8674(80)90086-0. [DOI] [PubMed] [Google Scholar]