Multiple, functionally overlapping regulatory circuits control the sensitivity of the G1/S transition to yeast pheromones. In the absence of the Cdk inhibitor Far1, pheromone-induced G1 arrest depends on the p27 analogue Sic1, transcriptional repression by the Rb analogues Whi5 and Stb1, and induced degradation of the CLN1 transcription factor Tec1.
Abstract
In budding yeast, mating pheromones arrest the cell cycle in G1 phase via a pheromone-activated Cdk-inhibitor (CKI) protein, Far1. Alternate pathways must also exist, however, because deleting the cyclin CLN2 restores pheromone arrest to far1∆ cells. Here we probe whether these alternate pathways require the G1/S transcriptional repressors Whi5 and Stb1 or the CKI protein Sic1, whose metazoan analogues (Rb or p27) antagonize cell cycle entry. Removing Whi5 and Stb1 allows partial escape from G1 arrest in far1∆ cln2∆ cells, along with partial derepression of G1/S genes, which implies a repressor-independent route for inhibiting G1/S transcription. This route likely involves pheromone-induced degradation of Tec1, a transcriptional activator of the cyclin CLN1, because Tec1 stabilization also causes partial G1 escape in far1∆ cln2∆ cells, and this is additive with Whi5/Stb1 removal. Deleting SIC1 alone strongly disrupts Far1-independent G1 arrest, revealing that inhibition of B-type cyclin-Cdk activity can empower weak arrest pathways. Of interest, although far1∆ cln2∆ sic1∆ cells escaped G1 arrest, they lost viability during pheromone exposure, indicating that G1 exit is deleterious if the arrest signal remains active. Overall our findings illustrate how multiple distinct G1/S-braking mechanisms help to prevent premature cell cycle commitment and ensure a robust signal-induced G1 arrest.
INTRODUCTION
Cell cycle progression in all organisms is regulated by both internal and external cues. In eukaryotes, the G1 phase of the cell cycle provides a critical period in which cells monitor whether conditions are appropriate for entry into a new division cycle (Morgan, 2007). Signals that control this decision include positive and negative growth factors, differentiation triggers, nutrient levels, and environmental stresses. These regulatory signals either promote or prevent the transition from a stable G1 state to a new round of DNA synthesis and mitosis. Often, cells become insensitive to these regulatory signals once they initiate the G1/S transition, establishing a cell cycle commitment phenomenon known as Start in yeast and the Restriction Point in animal cells (Hartwell et al., 1974; Pardee, 1974; Cross, 1995; Blagosklonny and Pardee, 2002). Accordingly, signals that influence cell cycle entry generally affect the molecular machinery that controls the G1/S transition. In all eukaryotes cell cycle transitions are promoted by cyclin-dependent kinases (Cdks), as well as by coordinate changes in gene expression, and these promoting factors are often held in check by inhibitory molecules to ensure that cell cycle progression is carefully controlled (Morgan, 2007). The overall architecture of the G1/S regulatory network is strongly conserved throughout eukaryotes, although some of the individual components may have evolved separately in yeasts versus animals (Cross et al., 2011).
In the budding yeast Saccharomyces cerevisiae, a single Cdk, Cdc28, associates with nine different cyclins that help drive distinct cell cycle events (Bloom and Cross, 2007). The transition from G1 to S phase is predominantly controlled by the G1 cyclin Cln3 along with two G1/S cyclins, Cln1 and Cln2, whereas the subsequent events of DNA synthesis and mitosis (in S and M phases) are driven by six B-type cyclins (Clb1–Clb6). The decision of yeast cells to enter a new cell cycle can be profoundly influenced by the presence of an external cue known as mating pheromone, which promotes fusion of two haploid mating partner cells (Hartwell, 1973). During this mating reaction, pheromone activates an intracellular signaling pathway that arrests the cell cycle in G1 phase, before Start (Dohlman and Thorner, 2001; Bardwell, 2005). An important factor in this G1 arrest pathway is the protein Far1, as far1∆ cells do not arrest in response to pheromone (Chang and Herskowitz, 1992). Far1 is believed to be a Cdk inhibitor (CKI) protein that blocks the activity of Cln-Cdc28 complexes and thereby prevents progression through Start (Peter et al., 1993; Peter and Herskowitz, 1994; Tyers and Futcher, 1993; Jeoung et al., 1998), although some findings conflict with this interpretation (Gartner et al., 1998). Of note, however, in some circumstances Far1 is dispensable for G1 arrest. For example, removing the G1/S cyclin Cln2 from far1∆ cells (i.e., far1∆ cln2∆) restores pheromone-induced G1 arrest (Chang and Herskowitz, 1992; Cherkasova et al., 1999). Thus, even in the complete absence of Far1, pheromone signaling still can interfere with the ability of cells to pass Start and enter a new division cycle. Despite early recognition of this fact, the molecular mechanisms responsible for Far1-independent arrest remain obscure.
In this study, we probe the Far1-independent arrest mechanisms more closely in order to better understand how pheromone signaling regulates G1 arrest and provide more general insights into the multiplicity of factors that control cell cycle commitment decisions. We reasoned that pheromone arrest might involve known negative regulators of the G1/S transition that act as “brakes” to antagonize cell cycle entry in many eukaryotes (Morgan, 2007). One such negative regulator is the CKI protein Sic1 (Donovan et al., 1994; Schwob et al., 1994), which is functionally analogous to the mammalian CKI p27(Kip1) (Sherr and Roberts, 1999). During G1, these CKI proteins inhibit Cdks bound to B-type cyclins and thereby prevent premature entry into S phase. This inhibition is eventually released in late G1, when cyclin-Cdk activity reaches levels sufficient to target the CKI for degradation (Schwob et al., 1994; Nash et al., 2001; Cross et al., 2007; Koivomagi et al., 2011). Another mode of negative regulation involves transcriptional repression of genes expressed at the G1/S boundary. In animal cells, transcription of G1/S genes is activated by the E2F family of heterodimeric transcription factors and repressed by members of the retinoblastoma protein (Rb) family (Frolov and Dyson, 2004; van den Heuvel and Dyson, 2008). Yeast cells have an analogous system (Bahler, 2005; Wittenberg and Reed, 2005), in which G1/S transcription is driven by two heterodimeric transcription factors called SBF (Swi4-Swi6) and MBF (Mbp1-Swi6) and is inhibited in early G1 by the repressors Whi5 and Stb1 (Koch et al., 1996; Costanzo et al., 2003, 2004; de Bruin et al., 2004, 2008; Bean et al., 2005). These repressors block the activity of DNA-bound SBF and MBF in part by recruiting histone deacetylases (Huang et al., 2009; Takahata et al., 2009; Wang et al., 2009); in late G1, they are dissociated via Cdk phosphorylation, allowing SBF/MBF-dependent transcription to ensue (Costanzo et al., 2004; de Bruin et al., 2004; Wagner et al., 2009; Doncic et al., 2011). Important targets of SBF and MBF include the G1/S cyclin genes CLN1 and CLN2, which yields increased G1/S Cdk activity, thereby creating a positive feedback loop that helps to ensure a decisive G1/S transition (Cross and Tinkelenberg, 1991; Dirick and Nasmyth, 1991; Skotheim et al., 2008). MBF/SBF-regulated genes are not expressed in pheromone-arrested cells, regardless of whether the arrest is Far1 dependent or independent (Wittenberg et al., 1990; Cherkasova et al., 1999), but it was unclear whether this inhibited transcriptional state is a cause or an effect of G1 arrest.
Here we report that the CKI Sic1 is strongly required for arrest in the absence of Far1, although not when Far1 is present. The transcriptional repressors Whi5 and Stb1 also contribute to Far1-independent arrest, in a manner that is additive with the pheromone-induced degradation of Tec1, a transcriptional activator of CLN1. Our results reveal that multiple, functionally overlapping regulatory circuits control the sensitivity of the G1/S transition to the pheromone arrest signal. Collectively these multiple pathways provide robustness to the G1 arrest response, which may help to ensure that commitment to cell cycle entry occurs decisively rather than tentatively.
RESULTS
Studying Far1-independent G1 arrest using synchronous cultures
To test the contribution of various regulatory factors to pheromone arrest in both the presence and absence of Far1, we used synchronous cultures to measure the duration of G1 phase. The essential cell cycle gene CDC20 was placed under control of a regulated promoter (PGAL1), allowing cells to be arrested in M phase and then released in the presence or absence of pheromone. Cell cycle progression was monitored by measuring DNA content (using flow cytometry) and/or by scoring budding. This approach offered three useful features: 1) by measuring G1 duration, we could detect even relatively subtle and temporary effects on the G1/S transition; 2) by using synchronized cells, we could readily assess the uniformity of phenotypes; and 3) we could distinguish specific effects of pheromone on G1 arrest from effects on other cell cycle stages or cell viability (which will become important later).
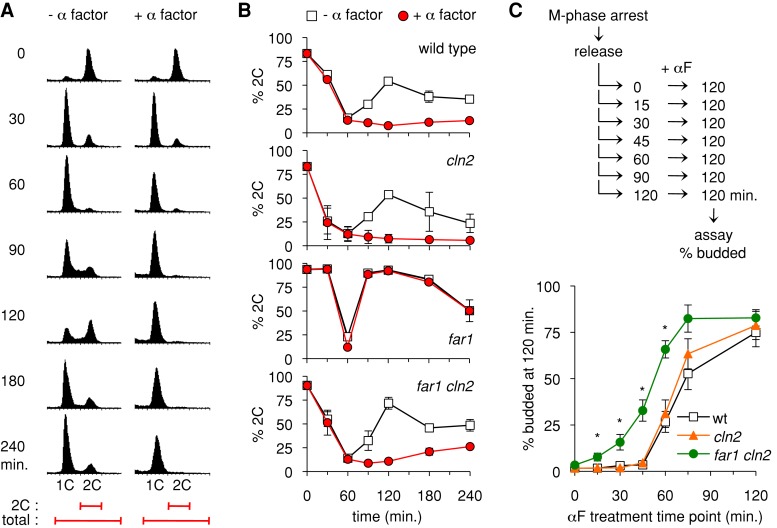
In this experimental context (Figure 1A), most cells finish mitosis and enter G1 (i.e., 1C DNA) by 30–60 min after release from the M-phase block and then begin a new round of DNA synthesis roughly 30 min later (at 90 min). If pheromone is added, wild-type cells complete mitosis and then arrest in G1 (for >3 h). To compare multiple different strains and replicate experiments, we plotted the percentage of cells with 2C DNA content as a function of time (Figure 1B). Generally, M-phase–arrested cultures were 80–90% 2C, and by 60 min after release they had cycled back to G1 and were predominantly 1C (i.e., only 10–15% 2C). As expected, far1∆ cells did not arrest in G1 in the presence of pheromone (Figure 1B). (In addition, with or without pheromone, they showed an accelerated return to the 2C state after completing mitosis, consistent with previous findings that Far1 can alter the timing of the G1/S transition even without pheromone treatment; Alberghina et al., 2004.) In contrast, when far1∆ cln2∆ cells were released in the presence of pheromone, they remained in G1 for an extended period (Figure 1B). This arrest was not as strong as in wild-type or FAR1 cln2∆ strains, as evidenced by the gradual increase in cells with 2C DNA content beginning at 120–150 min after release. Thus G1 arrest in the far1∆ cln2∆ cells is partially leaky, but pheromone clearly imposes a durable G1 delay that affects the majority of cells in the culture.
FIGURE 1:
Far1-independent arrest and cell cycle commitment in synchronous cultures. (A) Example of synchronous cell cycle progression and G1 arrest. A PGAL1-CDC20 strain was arrested in M phase (by transfer to glucose medium) and then released (by return to galactose medium) in the presence or absence of α factor. At the times indicated, DNA content of cells was assayed by flow cytometry. In each histogram, the horizontal axis represents fluorescence, and the vertical dimension shows the number of cells. Bottom, the range of fluorescence values used to calculate the proportion of cells with replicated DNA (percentage 2C) in subsequent figures. This example uses a cln2∆ strain (YPAP165). (B) The ability of α factor to halt cell cycle progression was analyzed for four strains, using the PGAL1-CDC20 method described in A. Graphs show mean ± range (n = 2) for wild-type and far1∆ or mean ± SD (n = 4) for cln2∆ and far1∆ cln2∆ strains.(C) Cell cycle commitment occurs earlier in the absence of Far1. After releasing PGAL1-CDC20 cultures from the M-phase block, aliquots were removed at 15-min intervals and treated with pheromone. At 120 min, cells were scored for whether they had arrested in G1 (unbudded cells) or entered the cell cycle (budded). Graphs show mean ± SEM (n = 5); asterisks indicate points where the difference between far1∆ cln2∆ and cln2∆ was deemed statistically significant (p < 0.025; two-tailed unpaired t test).
Separately, we tested how Far1 affects the window of time in which cells commit to a new division cycle (Figure 1C). After release of cultures from the M-phase block, aliquots were removed at intervals and treated with pheromone to test whether the cells could still arrest in G1 or were already committed to division. In wild-type and cln2∆ strains, cells transitioned from fully uncommitted (>95% arrest) to substantially committed (<65% arrest) between the 45- and 60-min time points. In the far1∆ cln2∆ strain, two differences were evident (Figure 1C). First, the commitment point occurred roughly 15 min earlier, as judged by the time at which 50% of cells still arrest. Second, the transition was less sharp, as evidenced by a more gradual increase in the fraction of cells that could not arrest. Together these data indicate that Far1 makes the arrest mechanism more potent, as it allows pheromone to arrest cells that have advanced closer to Start, and also more robust, as the arrest is more uniform in FAR1 cln2∆ than far1∆ cln2∆ cells. These findings complement a study in which the commitment point could be delayed by a stabilized form of Far1 (Doncic et al., 2011). Nevertheless, despite these clear effects of Far1, our results show that a commitment point still exists in the absence of Far1, albeit one that is advanced so that the time window in which arrest can be imposed is more limited. In experiments to follow, the Far1-independent arrest phenotype in far1∆ cln2∆ strains serves as sensitized setting in which to test the contribution of other factors that affect the G1/S transition.
Role of G1/S transcriptional repressors in G1 arrest
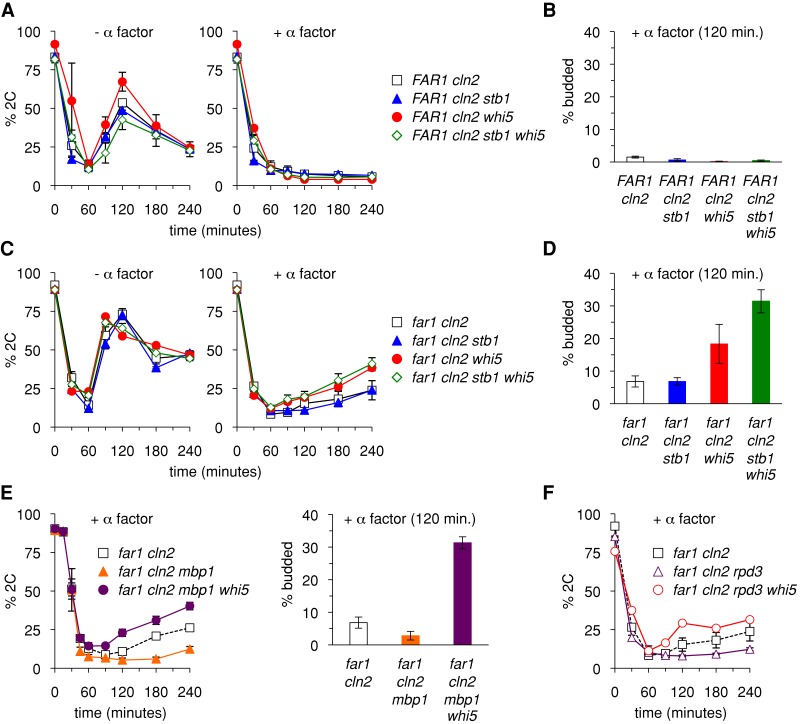
Previous studies found that expression of G1/S transcripts is inhibited in pheromone-arrested cells (Wittenberg et al., 1990), even when the arrest is Far1 independent (Cherkasova et al., 1999). Yet it was unclear whether this inhibition is a cause or a reflection of the G1 arrest. Hence, to determine whether Far1-independent arrest relies on control of G1/S transcription, we probed the role of Whi5 and Stb1, the repressors of the G1/S transcription factors SBF and MBF. Deletion of STB1 alone produced no discernible change in either FAR1 cln2∆ or far1∆ cln2∆ strains (Figure 2, A–D). Deletion of WHI5 had no effect in FAR1 cln2∆ cells (Figure 2A), but in far1∆ cln2∆ cells (Figure 2C) it allowed a greater fraction of cells to escape G1 arrest (e.g., 38 vs. 24% 2C at 240 min). Assays using bud emergence as a marker of cell cycle progression yielded largely similar results, except that the escape phenotype caused by removing Whi5 was even more evident, and there was further enhancement when both Whi5 and Stb1 were removed (Figure 2D). Thus the transcriptional repressors contribute to Far1-independent G1 arrest. Despite these effects, the escape phenotypes were only partial, in that pheromone still imposed a significant G1 delay in the majority of cells.
FIGURE 2:
Partial role for transcriptional repressors in Far1-independent arrest. (A) Removal of Whi5 and/or Stb1 does not affect G1 arrest when Far1 is present. PGAL1-CDC20 strains in the FAR1 cln2∆ background were arrested in M phase and released in the presence or absence of pheromone. Cell cycle progression and G1 arrest were measured by the flow cytometry assay of DNA content. Graphs show mean ± SEM (n = 4–8). (B) The strains in A were tested for G1 arrest by the budding assay. Budding was scored 120 min after release from M-phase arrest in the presence of α factor. Bars, mean ± SEM (n = 3–4). (C) Removal of Whi5 partially compromises Far1-independent arrest. Cell cycle progression and G1 arrest were measured in far1∆ cln2∆ strains by the DNA assay. Graphs show mean ± SEM (n = 4–8). (D) Strains from C were tested for G1 arrest by the budding assay as in B. Bars show mean ± SEM (n = 6). (E) Mbp1 is not required for Far1-independent arrest or for the role of Whi5. G1 arrest was measured by the DNA (left) and budding (right) assays. Data points, mean ± SEM (n = 3). (F) Rpd3 is not required for the role of Whi5. Graphs show mean ± SEM (n = 4) for far1∆ cln2∆ and far1∆ cln2∆ rpd3∆ or mean ± range (n = 2) for far1∆ cln2∆ rpd3∆ whi5∆.
We also tested the role of Mbp1, which is the DNA-binding component of the MBF heterodimer (Bahler, 2005). Although Mbp1 is required for transcriptional activation by MBF, it is also required for full repression of MBF-bound genes (Koch et al., 1993; Bean et al., 2005; de Bruin et al., 2006). We found that removing Mbp1 from far1∆ cln2∆ cells caused a more complete arrest rather than increased escape (Figure 2E), suggesting that either its role as an activator outweighs its role as a repressor in this setting or its removal allows stronger gene repression during pheromone treatment, possibly due to increased binding of SBF to MBF target genes (de Bruin et al., 2006). Deleting WHI5 from these far1∆ cln2∆ mbp1∆ cells allowed increased escape from G1 arrest (Figure 2E), with an especially strong difference (i.e., with vs. without Whi5), although the effect remained partial such that the majority of cells still arrested.
Whi5 and Stb1 repress transcription in part through the recruitment of the histone deacetylase Rpd3 (Huang et al., 2009; Takahata et al., 2009; Wang et al., 2009). To determine whether the reduced arrest proficiency of the far1∆ cln2∆ whi5∆ strain was due to the loss of Rpd3 recruitment, we tested far1∆ cln2∆ rpd3∆ strains (Figure 2F). In fact, deletion of Rpd3 did not increase escape from G1 arrest and instead seemed to make G1 exit even slower, although this may reflect a slightly decreased growth rate of rpd3∆ mutants. Furthermore, Rpd3 is not required for the G1 arrest role of Whi5, because removing Whi5 from the far1∆ cln2∆ rpd3∆ strain still led to increased escape, indicating that the whi5∆ phenotype cannot be attributed solely to a defect in Rpd3 recruitment.
Collectively these results show that Whi5 and Stb1 are not required for the strong G1 arrest seen when Far1 is present, but they contribute to the weaker G1 arrest observed in cells that lack Far1. Even so, their removal from far1∆ cln2∆ cells causes only a partial defect in G1 arrest, indicating that pheromone signaling can still inhibit the G1/S transition in the absence of both Far1 and these transcriptional repressors.
Loss of repressors only partially derepresses transcription
We considered two possible explanations for why removing the transcriptional repressors did not fully eliminate G1 arrest in far1∆ cln2∆ cells: 1) pheromone signaling might still be able to inhibit G1/S transcription even without the repressors; and 2) the G1/S transcripts could be fully derepressed, but pheromone signaling might exert nontranscriptional effects that inhibit exit from G1. To test these possibilities, we analyzed G1/S transcript levels via real-time quantitative PCR (RT-qPCR). We chose five representative genes (CLN1, YOX1, POL1, RNR1, and CDC21) that are induced by SBF and/or MBF at the G1/S transition (Bean et al., 2005; de Bruin et al., 2006; Eser et al., 2011). For comparison, we also monitored a gene expressed at the earlier M/G1 boundary (SIC1). We first conducted single-time-course experiments for eight different strains, in which we measured transcript levels at numerous time points in synchronous cultures (Figure 3). Then we analyzed the most informative time points in multiple independent trials (Figure 4).
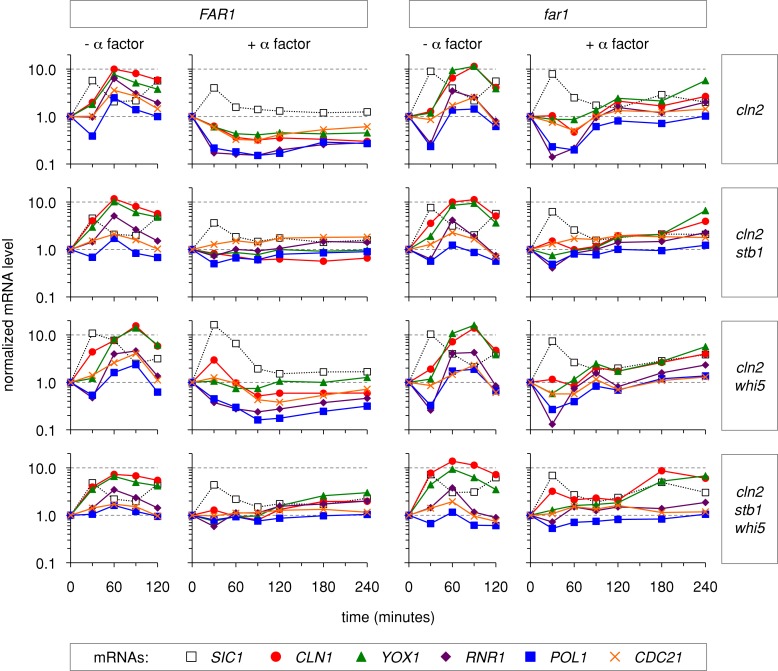
FIGURE 3:
Effects of Far1, Whi5, and Stb1 on G1/S mRNA levels. The effects of Whi5 and Stb1 on G1/S transcript levels were measured in FAR1 cln2∆ (left) and far1∆ cln2∆ (right) backgrounds. PGAL1-CDC20 strains were arrested in M phase and released with or without α factor. At 30-min intervals, mRNA levels were measured by RT-qPCR (see Materials and Methods). Five G1/S transcripts (CLN1, YOX1, RNR1, POL1, and CDC21) and one M/G1 transcript (SIC1) were monitored. mRNA levels at each time point were plotted relative to the levels present in the M-phase–arrested cultures (t = 0). The drop in G1/S transcript levels from M phase (t = 0) to G1 (t = 30 min) was unexpected because these genes are not believed to be active during mitosis; this behavior might reflect imperfect synchronization in M phase, or it might indicate that maximal repression of these genes requires nuclear localization of Swi6 and DNA binding by SBF/MBF, which are inhibited by high Cdk activity in M phase (Sidorova et al., 1995; Koch et al., 1996; Queralt and Igual, 2003; Geymonat et al., 2004). See Figure 4 for further analyses.
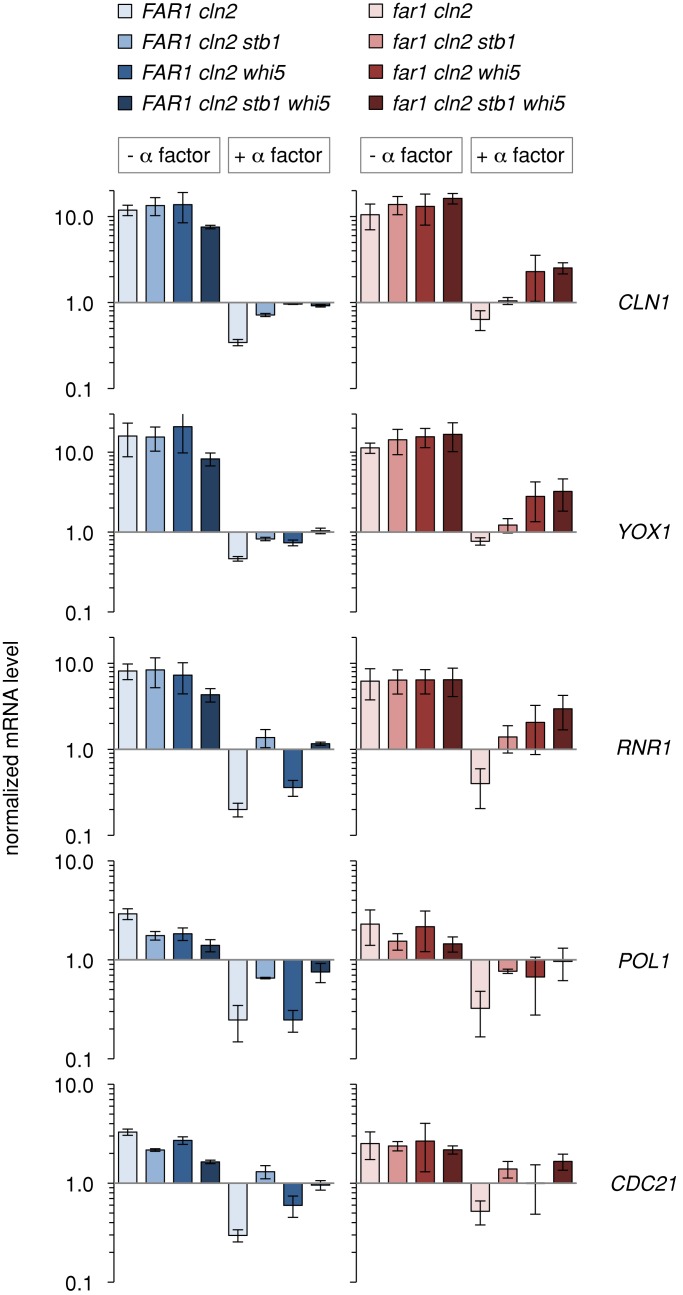
FIGURE 4:
Loss of repressors only partially derepresses transcription. G1/S transcripts were assayed at a fixed time corresponding to the transition from G1 to S phase. The PGAL1-CDC20 arrest/release experiments shown in Figure 3 were repeated three times, and mRNA levels were measured before and 60 min after release in either the presence or absence of α factor. Bars, mRNA levels (mean ± SD; n = 3) at the 60-min time points, expressed relative to the levels in the M-phase–arrested cells. The effects of Whi5 and Stb1 were compared in FAR1 cln2∆ (left) and far1∆ cln2∆ (right) backgrounds. Statistical analysis of all 120 pairwise comparisons, using the Benjamini–Hochberg false discovery rate method, is provided in Supplemental Table S1. The most pertinent points are 1) the inhibition by α factor is statistically significant (q < 0.05) for each of the eight individual genotypes and 2) the derepression due to removal of Whi5/Stb1 is significant in most cases except for comparisons involving the far1∆ cln2∆ whi5∆ strain, for which greater variability prevents a firm conclusion, but the effect of Whi5 is separately supported by comparing far1∆ cln2∆ stb1∆ with far1∆ cln2∆ stb1∆ whi5∆ (e.g., q = 0.007 for CLN1 mRNA).
In cells released from mitosis without pheromone, the M/G1 and G1/S transcripts peaked at 30 and 60 min, respectively (Figure 3). This agrees with the timing of DNA synthesis and budding (which begin at 60–90 min). Adding pheromone prevented the G1/S peak in FAR1 cln2∆ cells, and instead these transcripts declined to a minimum at 60 min and remained low for up to 4 h (Figure 3). In the absence of Far1 (far1∆ cln2∆ cells), pheromone still inhibited the G1/S peak, but after 60 min these transcripts gradually increased, reaching levels that were higher than the corresponding FAR1 cln2∆ cells but below the peak levels in untreated cells. This gradual increase is consistent with the leaky arrest phenotype of far1∆ cln2∆ cells. As expected for transcriptional repressors, removal of Whi5 and/or Stb1 allowed G1/S transcripts to start increasing earlier and/or to reach elevated levels in pheromone-treated cells. More specifically, the stb1∆ mutation seemed to reduce the pheromone-mediated decrease in all transcripts upon release from the mitotic block, whereas the whi5∆ mutation was more selective for specific transcripts. Nevertheless, in the absence of either repressor, or both simultaneously, pheromone still interfered with peak gene expression. These patterns of partial derepression were seen in both FAR1 cln2∆ and far1∆ cln2∆ backgrounds, although inhibition by pheromone was generally more potent and durable when Far1 was present. Clearly, pheromone signaling can prevent peak G1/S transcription even without Whi5 and Stb1.
For further analysis we performed multiple repetitions of each time course experiment and then measured mRNA levels at 60 min after release from M phase (Figure 4), as this was when expression peaked in the absence of pheromone. Without pheromone, G1/S transcripts reached roughly the same peak level in all strains (Figure 4), consistent with previous findings that Whi5 and Stb1 only slightly affect peak expression (Costanzo et al., 2003, 2004; de Bruin et al., 2004, 2008; Takahata et al., 2009). However, derepression was clearly evident in the pheromone-treated samples, as G1/S transcripts were no longer fully repressed when Whi5 and/or Stb1 were absent. Of note, however, even in the absence of both Whi5 and Stb1, G1/S transcript levels in pheromone-treated cells did not reach the maximum seen in untreated samples. Therefore inactivation of these repressors is not sufficient for full expression of G1/S transcripts, as pheromone signaling can still prevent their full expression. For several mRNAs the highest levels were seen in the far1∆ cln2∆ whi5∆ and far1∆ cln2∆ stb1∆ whi5∆ strains, which agrees with the finding that these strains have the strongest G1 escape phenotype, yet the ability of pheromone to prevent peak expression in these strains also agrees with the finding that even the strongest G1 escape phenotypes were partial.
The mRNA analyses revealed striking synergy between Far1 and Whi5 (Figure 4). That is, for several genes there was an additive effect of removing both proteins (i.e., far1∆ cln2∆ whi5∆), whereas pheromone treatment could still exert a strong repressive effect if either one was present (i.e., FAR1 cln2∆ whi5∆ or far1∆ cln2∆ WHI5). Again, this correlates with the arrest behavior, in that removing Whi5 caused a notable escape phenotype only when Far1 was absent. By contrast, the derepression caused by removing Stb1 was not further enhanced when Far1 was also removed (compare FAR1 cln2∆ stb1∆ with far1∆ cln2∆ stb1∆), and the additive relationship between Far1 and Whi5 did not require Stb1 (compare FAR1 cln2∆ stb1∆ whi5∆ with far1∆ cln2∆ stb1∆ whi5∆). Together these results suggest that Far1 and Whi5 contribute additively to transcriptional repression, with a corresponding additive effect on G1 arrest. Because Far1 is believed to inhibit Cln-Cdk activity (Peter et al., 1993; Peter and Herskowitz, 1994; Tyers and Futcher, 1993; Jeoung et al., 1998), the finding that it contributes to transcriptional repression even in the absence of Whi5 and Stb1 suggests that Cdk activity promotes G1/S transcription by additional mechanisms distinct from repressor displacement (see Discussion).
The CLN1 transcription factor Tec1 antagonizes G1 arrest
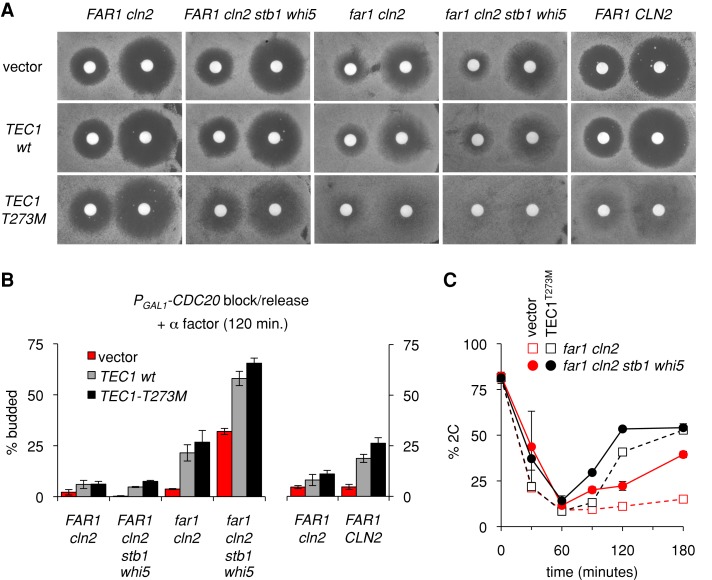
One route by which pheromone might inhibit G1/S transcription could involve the ability of the pheromone-activated mitogen-activated protein kinase (MAPK) Fus3 to trigger degradation of the transcription factor Tec1 (Bao et al., 2004; Bruckner et al., 2004; Chou et al., 2004). Because Tec1 positively regulates CLN1 expression (Madhani et al., 1999; Bruckner et al., 2004), its pheromone-induced degradation could indirectly dampen G1/S transcripts that are targets of Cln1-Cdc28 (Eser et al., 2011), which might be especially consequential in cln2∆ cells. To explore these notions, we first used halo assays to monitor how Tec1 affects growth arrest in both the presence and absence of Far1 (Figure 5A). We compared plasmids containing an extra copy of wild-type Tec1 or a mutant form, Tec1-T273M, which is resistant to pheromone-induced degradation (Bao et al., 2004; Bruckner et al., 2004; Chou et al., 2004) and disrupts the ability of pheromone to inhibit CLN1 expression (Bruckner et al., 2004). We observed a gradual reduction in arrest proficiency as more regulatory pathways were inactivated. Specifically, the Tec1-T273M mutant caused only a mild arrest defect in FAR1 cln2∆ cells (visible as a reduction in halo clarity and sharpness), but it caused a strong arrest defect in far1∆ cln2∆ cells, which was even stronger when Whi5/Stb1 were also absent (Figure 5A). By comparison, the extra copy of wild-type Tec1 had a milder effect in these assays. Overall the results suggest that pheromone inhibition of Tec1 activity becomes especially important when the Far1-dependent pathway is disrupted.
FIGURE 5:
The CLN1 transcription factor Tec1 antagonizes Far1-independent G1 arrest. (A) Empty vector or TEC1 plasmids (pPP681, pPP4042, pPP4043) were introduced into the indicated strains (PPY1716, PPY1789, PPY1867, YPAP157, YPAP161). Cells were spread on selective media (SC –Ura), overlaid with filter disks containing 20 μl of α factor (20 or 100 μM), and then incubated at 30°C for 2 d. (B) Empty vector or TEC1 plasmids (pPP680, pPP4050, pPP4051) were introduced into the indicated PGAL1-CDC20 strains (PPY2014, PPY2063, YPAP165, YPAP167, YPAP171). Cells were released from the mitotic block into medium containing α factor, and the percentage of cells that escaped G1 arrest was measured by scoring budding after 120 min. Bars, mean ± range; the two strains at right (n = 3) were assayed together in a set of experiments separate from the four strains at left (n = 2–5). In each strain, differences between vector and TEC1 (wild type and T273M) sets were ranked significant by a two-tailed unpaired t test (from left to right, p = 0.008, 0.009, 0.014, 0.002, 0.005, 0.0002). (C) PGAL1-CDC20 strains (PPY2014, YPAP171) contained empty vector or a TEC1-T273M plasmid (pPP680, pPP4051). Cells were released from the mitotic block into medium containing α factor, and DNA content was monitored at the indicated times. Graphs show mean ± range of two separate experiments (some error bars are smaller than symbols).
Related phenotypes were observed when cell cycle entry was analyzed in synchronous PGAL1-CDC20 cultures. Namely, extra Tec1 or stabilized Tec1-T273M caused only a mild escape from G1 arrest in FAR1 cln2∆ cells but caused a substantial escape in far1∆ cln2∆ cells and had an additive effect with Whi5/Stb1 removal, such that the majority of far1∆ cln2∆ stb1∆ whi5∆ cells now escaped G1 arrest (Figure 5B). This additive effect was also clearly evident when the G1/S transition was monitored using DNA content (Figure 5C), for which we found that the degree of G1 escape was maximal upon the simultaneous removal of Whi5/Stb1 and stabilization of Tec1. It is not known why an extra copy of TEC1 mimics TEC1-T273M more closely in synchronous assays (Figure 5B) than in halo assays (Figure 5A), but it is possible that the synchronization protocol imparts greater sensitivity to TEC1 dose because of the transfer to glucose-free media, which increases expression of TEC1 and Tec1-dependent genes (Cullen and Sprague, 2012). Nonetheless, the similar effects of both extra and stabilized Tec1 agree with the notion that pheromone arrest depends in part on blocking Tec1 accumulation. Taken together, our results indicate that Far1-independent arrest is promoted by two parallel pathways that act additively to reduce G1/S gene expression: Whi5/Stb1-mediated repression of SBF/MBF genes, and pheromone-triggered degradation of the CLN1 transcription factor Tec1.
Finally, to assess how strongly Far1-independent pathways can contribute to arrest in an otherwise wild-type cell, we tested the same TEC1 plasmids in a strain with intact copies of both FAR1 and CLN2. Remarkably, the Tec1-T273M mutant caused much stronger pheromone resistance in FAR1 CLN2 cells than in FAR1 cln2∆ cells (Figure 5, A, leftmost vs. rightmost columns, and B, right). Thus the same pathway can promote arrest in both far1∆ cln2∆ cells and wild-type cells. In addition, these results reveal an interesting parallel between Far1-dependent and Far1-independent pathways, as the pheromone resistance phenotype of either far1∆ or TEC1-T273M is suppressed by removing Cln2.
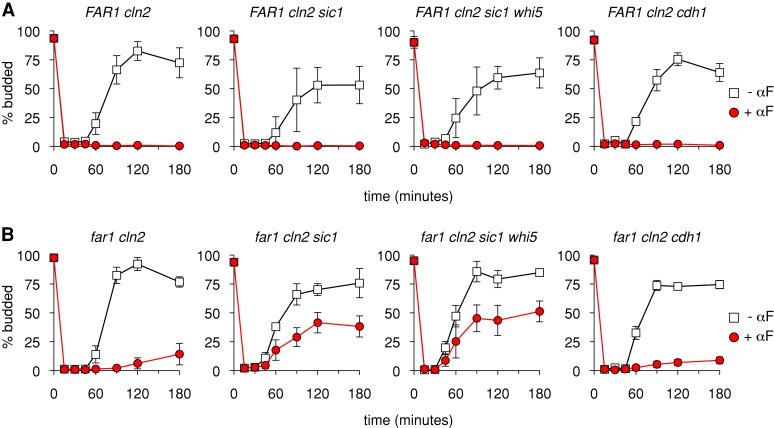
Sic1 plays a strong role in Far1-independent arrest
Another regulator of the G1/S transition is the protein Sic1, a CKI that inhibits S-phase cyclin-Cdks during early G1 and is inactivated in late G1 via phosphorylation by G1/S cyclin-Cdks. Because of its role as an antagonist of the G1/S transition, we tested whether Sic1 is involved in the Far1-independent arrest by pheromone (Figure 6). (These experiments exclusively used the budding assay because the sic1∆ mutation caused extremely broad DNA flow cytometry profiles in PGAL1-CDC20 strains, which obscured the analysis.) Indeed, although deleting SIC1 had no effect on G1 arrest when Far1 was present (Figure 6A), it had a substantial effect on Far1-independent arrest (Figure 6B). That is, in far1∆ cln2∆ sic1∆ cells the ability of pheromone to impose a G1 arrest was strongly disrupted, although it was not eliminated. The residual arrest was not eliminated by further deletion of WHI5 (Figure 6B), and hence we saw no evidence for synergy between Sic1 and Whi5. Furthermore, removing both Sic1 and Whi5 in a FAR1 background had no phenotype (Figure 6A). Thus removing any two antagonists of the G1/S transition is not sufficient to cause a G1 escape phenotype. Instead, this phenotype is only seen when combining far1∆ with either whi5∆ or sic1∆ mutations. We conclude that the G1/S-braking mechanism provided by Sic1 allows pheromone to activate a weakened G1 arrest response when Far1 is absent.
FIGURE 6:
Sic1 plays a strong role in Far1-independent arrest. PGAL1-CDC20 strains of the indicated genotypes were synchronized and then released into the presence or absence of α factor (αF). Cell cycle progression and G1 arrest was assayed by budding. (A, B) Results in the FAR1 cln2∆ and far1∆ cln2∆ backgrounds, respectively. All graphs show the mean ± SD (n = 4–6). Note that far1∆ cln2∆ cdh1∆ strains showed phenotypic heterogeneity that was isolate dependent. Specifically, we tested 12 isolates: six PGAL1-CDC20 derivatives from each of two independent far1∆ cln2∆ cdh1∆ strains. The results shown are an average of three strains (YPAP242, 244, 245) that displayed the majority phenotype seen in 10 of 12 isolates. In two of 12 isolates, both derived from the same initial far1∆ cln2∆ cdh1∆ parent strain, we observed a notable escape phenotype (e.g., for YPAP243, ∼40% budded cells after 120–180 min in α factor). The reason for this heterogeneity is unknown, but the observation of the escape phenotype in only a minority of derivatives (2/6) of one parent strain and in no derivatives (0/6) of the other suggests that a rare enhancer mutation may be responsible.
For comparison to the findings with Sic1, we tested another antagonist of S- and M-phase cyclins, namely, the APC component Cdh1, which inhibits accumulation of B-type cyclins during G1 (Visintin et al., 1997; Morgan, 2007). We found that removal of Cdh1 in the far1∆ cln2∆ background caused a negligible change in G1 escape (Figure 6B), although for unknown reasons we did observe an increased escape phenotype in rare isolates (legend to Figure 6). Therefore these findings suggest that Far1-independent arrest depends more strongly on the inhibitor of Clb-Cdk activity (Sic1) than on the inhibitor of Clb protein accumulation (Cdh1). Note, however, that we cannot rule out the possibility that the absence of Cdh1 was suppressed in our experiments by ectopic expression of its functional relative, Cdc20, which enabled the cell synchronization protocol; for example, PGAL1-CDC20 expression may limit accumulation of the B-type cyclin Clb2, which was shown previously to allow premature escape from pheromone arrest in a fraction of cdh1 (hct1-3) mutant cells (Schwab et al., 1997).
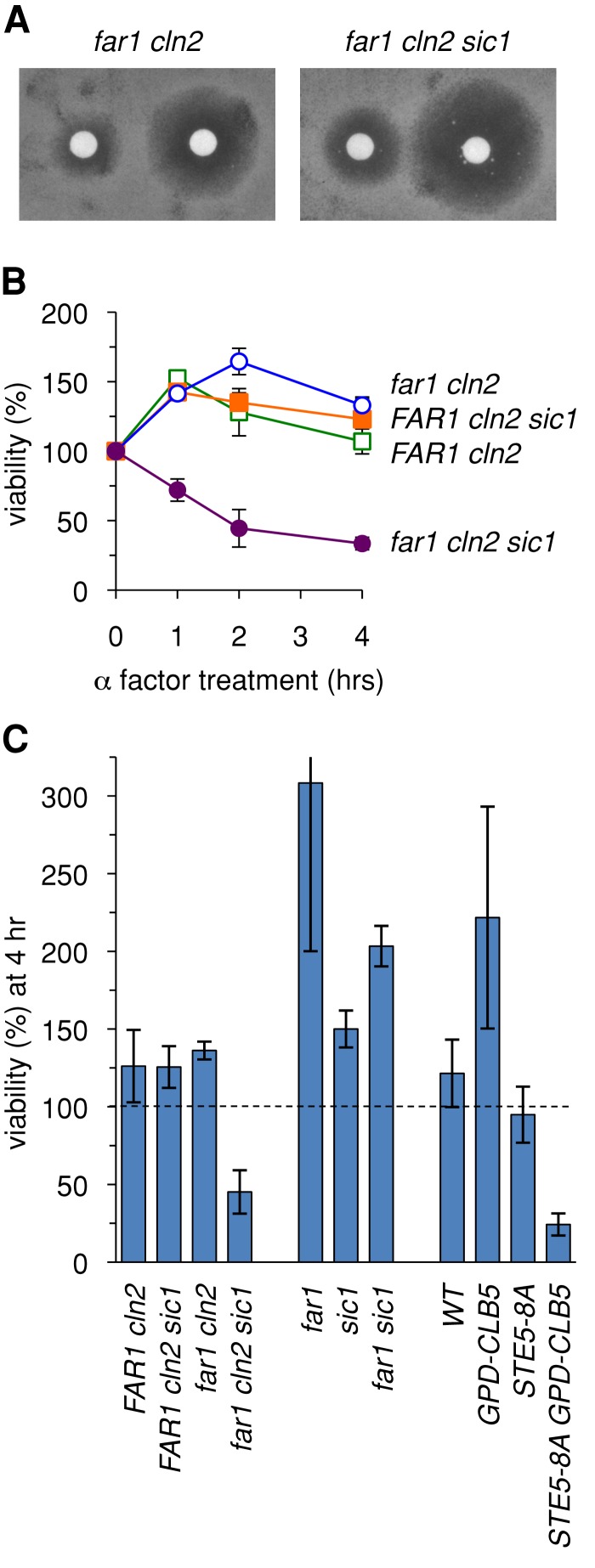
G1 arrest failure can compromise cell viability
Although removing Sic1 allowed far1∆ cln2∆ cells to escape pheromone-induced G1 arrest more readily, it appeared to cause enhanced sensitivity to pheromone when growth arrest was measured by a long-term halo assay (Figure 7A). This paradox led us to consider the possibility that the failure of these cells to arrest in G1 causes reduced viability during continuous incubation with pheromone. Therefore we assayed cell viability in asynchronous cultures treated with pheromone for various times (Figure 7B). Wild-type strains maintained viability for several hours, but the far1∆ cln2∆ sic1∆ strain showed a clear loss of viability after only a short period (2–4 h). This reduced viability was not seen in the absence of pheromone (legend to Figure 7) or when either Far1 or Sic1 was present (i.e., FAR1 cln2∆ sic1∆ or far1∆ cln2∆ SIC1 strains), suggesting that it is not the presence or absence of either protein per se but rather the rapid escape from G1 that ultimately causes reduced viability in the far1∆ cln2∆ sic1∆ strain. Furthermore, simultaneous absence of both proteins was tolerated if strains retained CLN2 (i.e., far1∆ sic1∆ strains; Figure 7C). Because Cln2 plays a prominent role in blocking pheromone response as cells pass Start (Oehlen and Cross, 1994; Strickfaden et al., 2007), the combined results suggest that the observed inviability is due to cells exiting G1 without down-regulating pheromone signaling.
FIGURE 7:
Failure to arrest in G1 causes loss of viability during pheromone exposure. (A) Removing Sic1 from far1∆ cln2∆ cells causes enhanced pheromone sensitivity when measured by a chronic growth arrest (halo) assay. Cells were spread on solid growth medium, overlaid with filter disks containing 20 μl of α factor (20 or 100 μM), and then incubated at 30°C for 2 d. (B) Pheromone treatment causes loss of viability in far1∆ cln2∆ sic1∆ cells. Asynchronous liquid cultures were incubated with pheromone for 1–4 h, and then cell viability was measured by plating on medium lacking pheromone and counting colony formation (see Materials and Methods). Viable cells at each time point were expressed relative to the number present before treatment (t = 0). Graphs show mean ± range (n = 2). In parallel cultures incubated without pheromone, no differences in viability were observed among these strains (unpublished data ). (C) Loss of cell viability is a consequence of escaping G1 arrest without inhibiting pheromone signaling. Asynchronous cultures were incubated with pheromone for 4 h, and viable cells were measured and expressed relative to the pretreated cultures (t = 0) as in B. Bars, mean ± SD (n = 4). Strains that continue dividing in the presence of pheromone (e.g., far1∆) show an increased number of viable cells at 4 h compared with pretreated culture. See the text for further explanation.
These implications were further corroborated by other experiments that did not involve Far1, Sic1, or Cln2. In particular, overproduction of the S-phase cyclin Clb5 can override G1 arrest by pheromone (Oehlen et al., 1998), and a Cdk-resistant form of the signaling protein Ste5 (Ste5-8A) prevents pheromone response from being shut down in post-Start cells (Strickfaden et al., 2007). The presence of both features together (i.e., STE5–8A PGPD1-CLB5 double mutants) caused cells to lose viability during pheromone exposure, whereas the single mutants did not (Figure 7C), arguing that inviability is a specific consequence of allowing pheromone signaling to continue in cells that escape the G1 arrest. Taken together, these findings illustrate the physiological importance of maintaining a robust G1 arrest in cells that are still transmitting the arrest signal. They also reveal that, under some circumstances, mutations that compromise G1 arrest would not be identified using only a growth arrest assay (see Discussion).
DISCUSSION
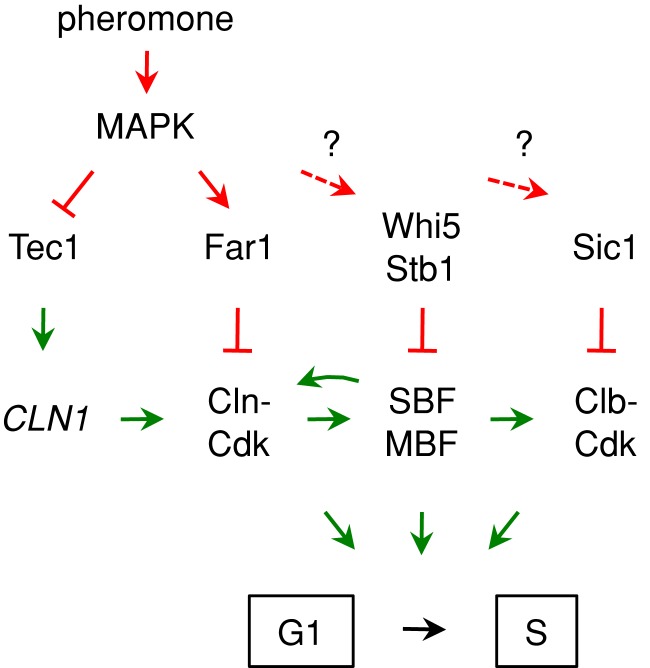
In this study we sought to increase our understanding of the mechanisms that yeast cells use to activate G1 arrest in response to extracellular mating pheromones. Previous findings indicate that this arrest does not absolutely require the pheromone-activated CKI Far1, because removal of Cln2 restores pheromone arrest to far1∆ cells. Thus here we compared Far1-dependent and Far1-independent G1 arrest in terms of their molecular phenotypes and their dependence on other inhibitory factors. We found that Far1 is not absolutely required for establishing a commitment point at Start, but it lengthens the time window in which pheromone can block this commitment step. Far1 also makes the G1 arrest process less dependent on other antagonists of the G1/S transition, such as repressors of G1/S transcription and inhibitors of S- and M-phase Cdk activity. Conversely, these repressors and inhibitors are not absolutely required for G1 arrest, but they reduce the dependence on Far1. Thus G1 arrest is made robust by functional overlap among multiple distinct G1/S inhibitory pathways (Figure 8).
FIGURE 8:
Simple illustration of multiple pathways contributing to pheromone arrest. Regulatory effects that inhibit or promote the G1/S transition are indicated in red or green, respectively. Dashed arrows with question marks emphasize that, although we found roles for Whi5/Stb1 and Sic1 in Far1-independent arrest, it is not known whether pheromone signaling enhances their inhibitory activity or simply depends on their constitutive effects. This simplified view is elaborated in greater detail in Supplemental Figure S1, which also includes a comparison of expected differences in wild-type, far1∆, and far1∆ cln2∆ cells. See the text for further discussion.
In many eukaryotes it is believed that inhibition of G1/S transcription by Rb-like repressors and inhibition of S- and M-phase Cdk activity by Cip/Kip-like CKIs are key factors that restrain cell cycle entry and thereby impose a G1 waiting period (Morgan, 2007). In this study we found that pheromone-induced G1 arrest in budding yeast can still be imposed in the absence of the transcriptional repressors Whi5/Stb1 or the CKI Sic1, or even both Whi5 and Sic1 simultaneously, but these negative factors become important for pheromone arrest in far1∆ cells. The existence of multiple braking mechanisms may help to prevent premature cell cycle entry when antiproliferative signals are present and expand the commitment decision period, perhaps by imposing a requirement that G1/S Cdk activity exceeds a sufficiently high threshold to counteract multiple antagonists. Such functional overlap, while in principle not required for a basic cell cycle, may increase the opportunities for regulatory control. Indeed, in animals there is evidence that Rb and CKIs can have additive effects in restraining proliferation of undifferentiated cells or redundant effects in terminally differentiated cells (Brugarolas et al., 1998; Buttitta et al., 2010; Wirt et al., 2010).
It is noteworthy that removal of Whi5 and Stb1 was not sufficient for full expression of G1/S transcripts, despite evident derepression. Instead, we found that pheromone signaling inhibits expression of G1/S genes even in the absence of both repressors, similar to previous results in whi5∆ or stb1∆ single mutants (Costanzo et al., 2004; de Bruin et al., 2008). Because this inhibition was maximal when Far1 was present, it suggests that Cdk activity promotes full G1/S transcription by an additional route separate from its role in releasing repression by Whi5/Stb1, perhaps via direct phosphorylation of SBF and/or MBF. Indeed, Cdk sites have been identified in both Swi4 and Swi6, but direct evidence that Cdk phosphorylation of SBF/MBF enhances their activity is lacking (Sidorova et al., 1995; Wijnen et al., 2002); instead, mutational analyses suggest that Cdk phosphorylation of Swi6 or Whi5 acts redundantly to block repression (Costanzo et al., 2004; de Bruin et al., 2004; Wagner et al., 2009). A further Cdk-regulated mechanism could affect the activity of SBF/MBF itself, an associated factor such as Msa1 (Ashe et al., 2008), or a separate factor that cooperates with SBF/MBF such as Bck2 or Spt10 (Wijnen and Futcher, 1999; Eriksson et al., 2011). The existence of such a mechanism also fits with the finding that stabilization of Tec1, which promotes CLN1 expression and hence Cln1-Cdk activity, was able to increase escape from G1 arrest even when Whi5/Stb1 were already absent.
Although Whi5/Stb1 and Sic1 contribute to pheromone-induced arrest when Far1 is absent, it is not clear whether they are regulated by pheromone or function instead as constitutive inhibitors (see question marks in Figure 8). Because Sic1 can be phosphorylated and stabilized by another MAPK, Hog1 (Escote et al., 2004), a similar effect could be exerted by the pheromone-regulated MAPK, Fus3. It is unlikely that the identical mechanism is used because the Hog1-phosphorylated form of Sic1 was absent in pheromone-arrested cells (Escote et al., 2004), but another study did observe partial phosphorylation of the Sic1 N-terminus in pheromone-arrested cells (Koivomagi et al., 2011), although the sites and responsible kinases are unknown. The effects of pheromone on G1/S transcription may be caused primarily by Far1-mediated inhibition of Cdk activity and MAPK-mediated degradation of Tec1, although it is conceivable that MAPK phosphorylation of SBF/MBF or Whi5/Stb1 provides another route for inhibition. Other pertinent mechanisms may include pheromone-induced reduction in Cln protein levels (Valdivieso et al., 1993) or reductions in total protein synthesis rates (Goranov et al., 2009; see question marks in Supplemental Figure S1), which may cause a rapid drop in the levels of short-lived cyclins and hence reduce cyclin-Cdk activity.
Which molecular targets can explain the escape phenotypes we obtained by inactivating inhibitory circuits in far1∆ cln2∆ cells? Ultimately, the key factor dictating whether cells can pass the G1/S transition despite the presence of pheromone may be acquisition of B-type Cdk activity, as pheromone cannot arrest cells in G1 if the S-phase cyclin Clb5 is expressed from a strong foreign promoter (Oehlen et al., 1998; Strickfaden et al., 2007). This view can explain the effects of removing Sic1, which directly inhibits Clb5-Cdk activity (Schwob et al., 1994). Furthermore, CLB5 is an SBF-regulated gene, as are CLN1 and CLN2, which engage in a positive feedback loop that enhances their own expression plus other SBF/MBF-dependent genes (Cross and Tinkelenberg, 1991; Dirick and Nasmyth, 1991; Skotheim et al., 2008). Thus, in far1∆ cln2∆ cells, levels of CLN1 and CLB5 expression may determine whether sufficient Clb5-Cdk activity accumulates to pass Start, and hence their increased expression upon removal of Whi5/Stb1 and/or addition of stabilized Tec1 may cause the escape phenotype. In far1∆ single mutants, the presence of Cln2 may tip the balance in favor of Start by a mix of several effects: 1) it might simply increase the total cyclin dosage, equal to an extra copy of CLN1; 2) Cln2 might be a more effective promoter of Start than Cln1 (Tyers and Futcher, 1993), perhaps by providing greater Cdk activity toward key substrates like Whi5 and Sic1; and 3) Cln2 counteracts pheromone arrest by directly inhibiting pheromone signaling, and it does so more potently than does Cln1 (Oehlen and Cross, 1994; Strickfaden et al., 2007). Thus, when Far1 is absent, the weaker Far1-independent arrest mechanisms can be overridden by Cln2 or genetic changes that increase Clb5-Cdk activity. When Far1 is present, the stronger inhibition of G1/S transcription may dampen Clb5 accumulation to a degree such that Sic1 is not needed for pheromone arrest. A comparison of these scenarios is illustrated in Supplemental Figure S1.
Finally, our findings reveal that robust G1 arrest is physiologically important, because if cell cycle entry occurs when cells are responding to pheromone, it can lead to irreversible, lethal effects. In particular, this was observed when far1∆ cln2∆ sic1∆ cells were exposed to pheromone; they were unable to arrest in G1, but they were nevertheless unable to divide and grow. Similar inviability was observed during pheromone treatment of STE5–8A PGPD1-CLB5 cells, in which G1 arrest is bypassed but signaling cannot be shut down. The cause of this inviability is not known, but ongoing studies suggest that pheromone signaling disrupts the function of the microtubule cytoskeleton during nuclear division (unpublished observations). Our findings clarify earlier results that implied that Sic1 was not required for Far1-independent arrest: namely, pheromone was able to arrest growth of cln1∆ cln2∆ cln3∆ far1∆ sic1∆ cells (Tyers, 1996). In retrospect, those cells may not have arrested in G1 but instead became inviable due to failed G1 arrest. These notions also illuminate an important consideration when analyzing other mutants for defects in pheromone-mediated arrest: if they allow slippage past the G1/S transition but do not prevent pheromone signaling, then the G1 arrest defect will not be noticeable by growth arrest assays. Hence it is conceivable that a distinct class of pheromone arrest mutants exists that would have gone undetected in prior genetic screens.
MATERIALS AND METHODS
Yeast strains and plasmids
Standard procedures were used for growth and genetic manipulation of yeast (Rothstein, 1991; Sherman, 2002). Yeast cultures were grown at 30°C. Strains and plasmids are listed in Tables 1 and 2, respectively; all yeast strains were derived from the W303 background (Thomas and Rothstein, 1989) and harbored the bar1∆ mutation to block α factor degradation. PCR-mediated gene targeting used methods described previously (Longtine et al., 1998); selectable markers included antibiotic resistance genes (kanMX6, natMX6) and orthologues of biosynthesis genes from other yeasts (Saccharomyces kluyveri HIS3, Candida glabrata TRP1, Kluyveromyces lactis URA3). To ensure that genetic effects were reproducible, independently derived strains of identical genotype were tested in parallel, and the combined results were averaged. For cell synchronization, the promoter of the essential cell cycle gene CDC20 was replaced with a regulated promoter (PGAL1) using a PCR-generated cassette marked with the K. lactis URA3 gene (URA3Kl).
TABLE 1:
Yeast strains used in this study.
| Name | Relevant genotypea | Source |
|---|---|---|
| PPY1716 | MATa bar1 | Zimmerman and Kellogg (2001) |
| PPY1748 | MATa bar1 STE5-8A | Strickfaden et al. (2007) |
| PPY1777 | MATa bar1 far1∆::ADE2 | Strickfaden et al. (2007) |
| PPY1789 | MATa bar1 far1∆::ADE2 cln2∆::kanMX6 | Strickfaden et al. (2007) |
| PPY1867 | MATa bar1 cln2∆::kanMX6 | This study |
| PPY1913 | MATa bar1 HIS3::PGPD1-CLB5 | Strickfaden et al. (2007) |
| PPY1918 | MATa bar1 STE5-8A HIS3::PGPD1-CLB5 | Strickfaden et al. (2007) |
| PPY2013 | MATa bar1 far1∆::ADE2 cln2∆::kanMX6 PGAL1-CDC20::URA3Kl | This study |
| PPY2014 | MATa bar1 far1∆::ADE2 cln2∆::kanMX6 PGAL1-CDC20::URA3Kl | This study |
| PPY2019 | MATa bar1 far1∆::ADE2 cln2∆::kanMX6 mbp1∆::TRP1Cg PGAL1-CDC20::URA3Kl | This study |
| PPY2020 | MATa bar1 far1∆::ADE2 cln2∆::kanMX6 mbp1∆::TRP1Cg PGAL1-CDC20::URA3Kl | This study |
| PPY2043 | MATa bar1 far1∆::ADE2 cln2∆::kanMX6 sic1∆::TRP1Cg | This study |
| PPY2063 | MATa bar1 PGAL1-CDC20::URA3Kl | This study |
| PPY2064 | MATa bar1 PGAL1-CDC20::URA3Kl | This study |
| PPY2068 | MATa bar1 far1∆::ADE2 cln2∆::kanMX6 sic1∆::TRP1Cg PGAL1-CDC20::URA3Kl | This study |
| PPY2069 | MATa bar1 far1∆::ADE2 cln2∆::kanMX6 sic1∆::TRP1Cg PGAL1-CDC20::URA3Kl | This study |
| PPY2082 | MATa bar1 far1∆::ADE2 PGAL1-CDC20::URA3Kl | This study |
| PPY2083 | MATa bar1 far1∆::ADE2 PGAL1-CDC20::URA3Kl | This study |
| PPY2085 | MATa bar1 sic1∆::TRP1Cg | This study |
| PPY2087 | MATa bar1 far1∆::ADE2 sic1∆::TRP1Cg | This study |
| PPY2090 | MATa bar1 far1∆::ADE2 cln2∆::kanMX6 mbp1∆::TRP1Cg whi5∆::HIS3Sk PGAL1-CDC20::URA3Kl | This study |
| PPY2091 | MATa bar1 far1∆::ADE2 cln2∆::kanMX6 mbp1∆::TRP1Cg whi5∆::HIS3Sk PGAL1-CDC20::URA3Kl | This study |
| PPY2128 | MATa bar1 far1∆::ADE2 cln2∆::kanMX6 mbp1∆::TRP1Cg whi5∆::HIS3Sk PGAL1-CDC20::URA3Kl | This study |
| YPAP137 | MATa bar1 cln2∆::kanMX6 stb1∆::natMX6 PGAL1-CDC20::URA3Kl | This study |
| YPAP138 | MATa bar1 cln2∆::kanMX6 stb1∆::natMX6 PGAL1-CDC20::URA3Kl | This study |
| YPAP141 | MATa bar1 far1∆::ADE2 cln2∆::kanMX6 stb1∆::natMX6 PGAL1-CDC20::URA3Kl | This study |
| YPAP142 | MATa bar1 far1∆::ADE2 cln2∆::kanMX6 stb1∆::natMX6 PGAL1-CDC20::URA3Kl | This study |
| YPAP143 | MATa bar1 far1∆::ADE2 cln2∆::kanMX6 rpd3∆::HIS3Sk PGAL1-CDC20::URA3Kl | This study |
| YPAP144 | MATa bar1 far1∆::ADE2 cln2∆::kanMX6 rpd3∆::HIS3Sk PGAL1-CDC20::URA3Kl | This study |
| YPAP151 | MATa bar1 far1∆::ADE2 cln2∆::kanMX6 whi5∆::HIS3Sk PGAL1-CDC20::URA3Kl | This study |
| YPAP152 | MATa bar1 far1∆::ADE2 cln2∆::kanMX6 whi5∆::HIS3Sk PGAL1-CDC20::URA3Kl | This study |
| YPAP153 | MATa bar1 cln2∆::kanMX6 whi5∆::HIS3Sk PGAL1-CDC20::URA3Kl | This study |
| YPAP156 | MATa bar1 cln2∆::kanMX6 whi5∆::HIS3Sk PGAL1-CDC20::URA3Kl | This study |
| YPAP157 | MATa bar1 far1∆::ADE2 cln2∆::kanMX6 stb1∆::natMX6 whi5∆::HIS3Sk | This study |
| YPAP161 | MATa bar1 cln2∆::kanMX6 stb1∆::natMX6 whi5∆::HIS3Sk | This study |
| YPAP165 | MATa bar1 cln2∆::kanMX6 PGAL1-CDC20::URA3Kl | This study |
| YPAP166 | MATa bar1 cln2∆::kanMX6 PGAL1-CDC20::URA3Kl | This study |
| YPAP167 | MATa bar1 cln2∆::kanMX6 stb1∆::natMX6 whi5∆::HIS3Sk PGAL1-CDC20::URA3Kl | This study |
| YPAP168 | MATa bar1 cln2∆::kanMX6 stb1∆::natMX6 whi5∆::HIS3Sk PGAL1-CDC20::URA3Kl | This study |
| YPAP171 | MATa bar1 far1∆::ADE2 cln2∆::kanMX6 stb1∆::natMX6 whi5∆::HIS3Sk PGAL1-CDC20::URA3Kl | This study |
| YPAP172 | MATa bar1 far1∆::ADE2 cln2∆::kanMX6 stb1∆::natMX6 whi5∆::HIS3Sk PGAL1-CDC20::URA3Kl | This study |
| YPAP203 | MATa bar1 far1∆::ADE2 cln2∆::kanMX6 rpd3∆::HIS3Sk whi5∆::TRP1Cg PGAL1-CDC20::URA3Kl | This study |
| YPAP204 | MATa bar1 far1∆::ADE2 cln2∆::kanMX6 rpd3∆::HIS3Sk whi5∆::TRP1Cg PGAL1-CDC20::URA3Kl | This study |
| YPAP208 | MATa bar1 cln2∆::natMX6 sic1∆::TRP1Cg | This study |
| YPAP209 | MATa bar1 far1∆::ADE2 cln2∆::kanMX6 sic1∆::TRP1Cg whi5∆::HIS3Sk PGAL1-CDC20::URA3Kl | This study |
| YPAP210 | MATa bar1 far1∆::ADE2 cln2∆::kanMX6 sic1∆::TRP1Cg whi5∆::HIS3Sk PGAL1-CDC20::URA3Kl | This study |
| YPAP236 | MATa bar1 cln2∆::natMX6 sic1∆::TRP1Cg PGAL1-CDC20::URA3Kl | This study |
| YPAP237 | MATa bar1 cln2∆::natMX6 sic1∆::TRP1Cg PGAL1-CDC20::URA3Kl | This study |
| YPAP238 | MATa bar1 cln2∆::natMX6 sic1∆::TRP1Cg whi5∆::HIS3Sk PGAL1-CDC20::URA3Kl | This study |
| YPAP239 | MATa bar1 cln2∆::natMX6 sic1∆::TRP1Cg whi5∆::HIS3Sk PGAL1-CDC20::URA3Kl | This study |
| YPAP240 | MATa bar1 cln2∆::kanMX6 cdh1∆::HIS3Sk PGAL1-CDC20::URA3Kl | This study |
| YPAP241 | MATa bar1 cln2∆::kanMX6 cdh1∆::HIS3Sk PGAL1-CDC20::URA3Kl | This study |
| YPAP242 | MATa bar1 far1∆::ADE2 cln2∆::kanMX6 cdh1∆::HIS3Sk PGAL1-CDC20::URA3Kl | This study |
| YPAP243 | MATa bar1 far1∆::ADE2 cln2∆::kanMX6 cdh1∆::HIS3Sk PGAL1-CDC20::URA3Kl | This study |
| YPAP244 | MATa bar1 far1∆::ADE2 cln2∆::kanMX6 cdh1∆::HIS3Sk PGAL1-CDC20::URA3Kl | This study |
| YPAP245 | MATa bar1 far1∆::ADE2 cln2∆::kanMX6 cdh1∆::HIS3Sk PGAL1-CDC20::URA3Kl | This study |
aAll strains are in the W303 background (ade2-1 his3-11,15 leu2-3112 trp1-1 ura3-1 can1). In the PGAL1-CDC20 strains, a cassette containing the URA3Kl marker and GAL1 promoter is inserted in place of the CDC20 promoter at the native CDC20 locus.
TABLE 2:
Plasmids used in this study.
| Name | Alias | Description | Source |
|---|---|---|---|
| pPP680 | pRS315 | CEN/ARS LEU2 vector | Sikorski and Hieter (1989) |
| pPP681 | pRS316 | CEN/ARS URA3 vector | Sikorski and Hieter (1989) |
| pPP3025 | pFA6a-URA3-PGAL1 | PCR template for URA3Kl-PGAL1 promoter insertion | This study |
| pPP4042 | YCplac33-TEC1 | CEN/ARS URA3 TEC1 | Bao et al. (2004) |
| pPP4043 | YCplac33-tec1-T273M | CEN/ARS URA3 TEC1-T273M | Bao et al. (2004) |
| pPP4050 | pL-TEC1-WT | CEN/ARS LEU2 TEC1 | This study |
| pPP4051 | pL-TEC1-T273M | CEN/ARS LEU2 TEC1-T273M | This study |
Synchronous culture assays
To synchronize cell cultures, we placed the CDC20 gene under control of the GAL1 promoter (Cosma et al., 2001; Bean et al., 2005; Takahata et al., 2009). These PGAL1-CDC20 strains were grown asynchronously in liquid YPGal medium (containing 2% galactose), and then cultures were arrested in M phase by pelleting and resuspending in YPD medium (2% glucose), followed by incubation for 3 h. Cultures were released from the M-phase block by two rounds of pelleting and washing in YPGal, resuspension in YPGal either with or without α factor (0.2 μM), and incubation (with shaking) for 0–240 min. In experiments using TEC1 plasmids, before the arrest in YPD and release in YPGal, cultures were grown overnight in synthetic media (SC –Leu, with 2% raffinose and 2% galactose) to maintain selection for LEU2 plasmids.
Flow cytometry and budding assays
DNA content was measured by flow cytometry using previous methods (Haase and Reed, 2002; Strickfaden et al., 2007). Briefly, cell aliquots (0.5 ml) were harvested by centrifugation, resuspended in 0.3 ml water, fixed by addition of 0.7 ml of 100% ethanol, mixed by inversion, and incubated overnight at 4°C. Fixed cells were pelleted, washed once with water, resuspended in 0.5 ml of freshly prepared RNase solution (2 mg/ml RNase A in 50 mM Tris-HCl, pH 8.0, 15 mM NaCl), and incubated for 2 h at 36°C. They were then pelleted and resuspended in 0.2 ml of fresh proteinase solution (1 mg/ml proteinase K in 50 mM Tris-HCl, pH 8.0), incubated for 1 h at 36°C, then pelleted and resuspended in 0.5 ml of buffer (50 mM Tris-HCl, pH 7.5), and stored at 4°C. Before analysis, suspensions were sonicated (10 pulses with a microtip probe), and then 50 μl was mixed with 1 ml of fresh Sytox Green solution (1 μM in 50 mM Tris-HCl, pH 7.5), gently vortexed, and analyzed with a FACScan flow cytometer (BD, Franklin Lakes, NJ). For experiments that were directly compared with each other, a uniform range of fluorescence values was defined for both the 2C peak (generally 100–150 U) and the total (including 1C, 2C, and intermediate; generally 40–200 U), and then percentage 2C was calculated as 100% × 2C/total.
To analyze cell cycle position by budding, cells aliquots (0.5 ml) were fixed by the addition of formaldehyde to 3.7% final concentration, incubated on a nutator (room temperature, 10 min), washed three times with phosphate-buffered saline (PBS), and resuspended in 500 μl of PBS. Fixed cells were spotted onto glass slides and viewed microscopically to score budded and unbudded cells; for each experimental condition, 200 cells were counted.
mRNA preparation and RT-qPCR analysis
RNA was prepared as described previously (de Bruin et al., 2008) using an RNeasy Plus Mini Kit (74134; Qiagen, Valencia, CA). Cells (∼5 × 107) were harvested by centrifugation and frozen in liquid nitrogen. Cell pellets were resuspended in 400 μl of Qiagen RLT-plus buffer freshly supplemented with β-mercaptoethanol (10 μl/ml of buffer) and transferred to a microcentrifuge tube. Approximately 400 μl of acid-washed glass beads was added, and cells were lysed by vortexing (four cycles of 1 min, interspersed with rest periods of 3 min on ice). The tube was punctured at the bottom with a needle, placed in another tube, and centrifuged briefly (10 s) to transfer the lysate to the fresh tube. Then cell debris was removed by centrifugation (2 min, full speed). The supernatant was loaded onto Qiagen gDNA Eliminator columns, and then mRNA was prepared according to the manufacturer's instructions, diluted to equal concentrations (0.5 μg/μl), and stored at −70°C. cDNA was synthesized from the RNA samples using a SuperScript VILO cDNA synthesis kit (11754; Invitrogen, Carlsbad, CA) as per the manufacturer's instructions, using ∼2 μg of RNA per reaction. Products were diluted to 2.5 ng/μl.
Quantitative real-time PCRs were performed using Power SYBR Green PCR Master Mix (4367659; Applied Biosystems, Foster City, CA). Reaction mixtures (15 μl) contained 7.5 μl of SYBR Green reaction mix, 2 μl of primer mix (3 μM each primer), 1 μl of cDNA (2.5 ng), and 4.5 μl of water. Reactions were performed in 96-well plates, in duplicate, using an Applied Biosystems StepOnePlus instrument. Preliminary trials using multiple primers for each gene were performed to identify primer sets with optimal properties (linearity of amplification) for use in all subsequent experiments. Primers are listed in Table 3. The ∆∆CT method was used to convert real-time amplification kinetics into relative mRNA levels; ACT1 mRNA served as the internal control.
TABLE 3:
Oligonucleotide primers used for RT-qPCR analysis.
| Primer name | Sequence (5′ to 3′) |
|---|---|
| CLN1-fw1 | CTTTGGTTAGCGGCCAAAAC |
| CLN1-rev1 | AGAAAGGCGTGGAATACGAG |
| YOX1-up1 | AAATAGGCGCTCATCCACAC |
| YOX1-dn1 | ACGTTTTCACGGGAGTCAAC |
| RNR1-up1 | TCGAGGCTGCTTTAGAAACG |
| RNR1-dn1 | GGCAACCAAGAAACAAGAGG |
| POL1-fw1 | TGACATTTGCTCTGGTAGGC |
| POL1-rev1 | CGGCTTATGCTCCTTTTCAC |
| CDC21-fw1 | GGAACCCAGCTGATTTTGAC |
| CDC21-rev1 | CGGATCCTTCTCCTTCTTTG |
| SIC1-fw1 | CCAAAAGCCTTCACAGAACC |
| SIC1-rev1 | GAGAGGTCATACCCATGTTCG |
| ACT1-fw1 | TTCCAGCCTTCTACGTTTCC |
| ACT1-rev1 | CCAGCGTAAATTGGAACGAC |
Cell viability assays
Asynchronous cultures were treated with pheromone (0.2 μM) for 1–4 h. Aliquots were collected before and after treatment, sonicated (seven pulses with a microtip probe), diluted in sterile PBS, spread on solid synthetic medium, and incubated for 2 d. Viable colonies were counted and expressed as a percentage of the number obtained from equivalent aliquots of the initial, untreated cultures.
Supplementary Material
Acknowledgments
We thank Hiten Madhani for TEC1 plasmids, Nick Rhind for use of his flow cytometer, and Curt Wittenberg for mRNA/RT-qPCR protocols. This work was supported by a grant from the National Institutes of Health (GM57769) to P.M.P.
Abbreviations used:
- Cdk
cyclin-dependent kinase
- CKI
Cdk inhibitor
- MAPK
mitogen-activated protein kinase
- MBF
MluI-box binding factor
- RT-qPCR
real-time quantitative PCR
- SBF
Swi4/6-box binding factor
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-07-0373) on October 2, 2013.
REFERENCES
- Alberghina L, Rossi RL, Querin L, Wanke V, Vanoni M. A cell sizer network involving Cln3 and Far1 controls entrance into S phase in the mitotic cycle of budding yeast. J Cell Biol. 2004;167:433–443. doi: 10.1083/jcb.200405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe M, de Bruin RA, Kalashnikova T, McDonald WH, Yates JR, 3rd, Wittenberg C. The SBF- and MBF-associated protein Msa1 is required for proper timing of G1-specific transcription in Saccharomyces cerevisiae. J Biol Chem. 2008;283:6040–6049. doi: 10.1074/jbc.M708248200. [DOI] [PubMed] [Google Scholar]
- Bahler J. Cell-cycle control of gene expression in budding and fission yeast. Annu Rev Genet. 2005;39:69–94. doi: 10.1146/annurev.genet.39.110304.095808. [DOI] [PubMed] [Google Scholar]
- Bao MZ, Schwartz MA, Cantin GT, Yates JR, 3rd, Madhani HD. Pheromone-dependent destruction of the Tec1 transcription factor is required for MAP kinase signaling specificity in yeast. Cell. 2004;119:991–1000. doi: 10.1016/j.cell.2004.11.052. [DOI] [PubMed] [Google Scholar]
- Bardwell L. A walk-through of the yeast mating pheromone response pathway. Peptides. 2005;26:339–350. doi: 10.1016/j.peptides.2004.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean JM, Siggia ED, Cross FR. High functional overlap between MluI cell-cycle box binding factor and Swi4/6 cell-cycle box binding factor in the G1/S transcriptional program in Saccharomyces cerevisiae. Genetics. 2005;171:49–61. doi: 10.1534/genetics.105.044560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagosklonny MV, Pardee AB. The restriction point of the cell cycle. Cell Cycle. 2002;1:103–110. [PubMed] [Google Scholar]
- Bloom J, Cross FR. Multiple levels of cyclin specificity in cell-cycle control. Nat Rev Mol Cell Biol. 2007;8:149–160. doi: 10.1038/nrm2105. [DOI] [PubMed] [Google Scholar]
- Bruckner S, Kohler T, Braus GH, Heise B, Bolte M, Mosch HU. Differential regulation of Tec1 by Fus3 and Kss1 confers signaling specificity in yeast development. Curr Genet. 2004;46:331–342. doi: 10.1007/s00294-004-0545-1. [DOI] [PubMed] [Google Scholar]
- Brugarolas J, Bronson RT, Jacks T. p21 is a critical CDK2 regulator essential for proliferation control in Rb-deficient cells. J Cell Biol. 1998;141:503–514. doi: 10.1083/jcb.141.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttitta LA, Katzaroff AJ, Edgar BA. A robust cell cycle control mechanism limits E2F-induced proliferation of terminally differentiated cells in vivo. J Cell Biol. 2010;189:981–996. doi: 10.1083/jcb.200910006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F, Herskowitz I. Phosphorylation of FAR1 in response to alpha-factor: a possible requirement for cell-cycle arrest. Mol Biol Cell. 1992;3:445–450. doi: 10.1091/mbc.3.4.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkasova V, Lyons DM, Elion EA. Fus3p and Kss1p control G1 arrest in Saccharomyces cerevisiae through a balance of distinct arrest and proliferative functions that operate in parallel with Far1p. Genetics. 1999;151:989–1004. doi: 10.1093/genetics/151.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S, Huang L, Liu H. Fus3-regulated Tec1 degradation through SCFCdc4 determines MAPK signaling specificity during mating in yeast. Cell. 2004;119:981–990. doi: 10.1016/j.cell.2004.11.053. [DOI] [PubMed] [Google Scholar]
- Cosma MP, Panizza S, Nasmyth K. Cdk1 triggers association of RNA polymerase to cell cycle promoters only after recruitment of the mediator by SBF. Mol Cell. 2001;7:1213–1220. doi: 10.1016/s1097-2765(01)00266-0. [DOI] [PubMed] [Google Scholar]
- Costanzo M, Nishikawa JL, Tang X, Millman JS, Schub O, Breitkreuz K, Dewar D, Rupes I, Andrews B, Tyers M. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell. 2004;117:899–913. doi: 10.1016/j.cell.2004.05.024. [DOI] [PubMed] [Google Scholar]
- Costanzo M, Schub O, Andrews B. G1 transcription factors are differentially regulated in Saccharomyces cerevisiae by the Swi6-binding protein Stb1. Mol Cell Biol. 2003;23:5064–5077. doi: 10.1128/MCB.23.14.5064-5077.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross FR. Starting the cell cycle: what's the point. Curr Opin Cell Biol. 1995;7:790–797. doi: 10.1016/0955-0674(95)80062-x. [DOI] [PubMed] [Google Scholar]
- Cross FR, Buchler NE, Skotheim JM. Evolution of networks and sequences in eukaryotic cell cycle control. Philos Trans R Soc Lond B Biol Sci. 2011;366:3532–3544. doi: 10.1098/rstb.2011.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross FR, Schroeder L, Bean JM. Phosphorylation of the Sic1 inhibitor of B-type cyclins in Saccharomyces cerevisiae is not essential but contributes to cell cycle robustness. Genetics. 2007;176:1541–1555. doi: 10.1534/genetics.107.073494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross FR, Tinkelenberg AH. A potential positive feedback loop controlling CLN1 and CLN2 gene expression at the start of the yeast cell cycle. Cell. 1991;65:875–883. doi: 10.1016/0092-8674(91)90394-e. [DOI] [PubMed] [Google Scholar]
- Cullen PJ, Sprague GF., Jr The regulation of filamentous growth in yeast. Genetics. 2012;190:23–49. doi: 10.1534/genetics.111.127456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin RA, Kalashnikova TI, Chahwan C, McDonald WH, Wohlschlegel J, Yates J, 3rd, Russell P, Wittenberg C. Constraining G1-specific transcription to late G1 phase: the MBF-associated corepressor Nrm1 acts via negative feedback. Mol Cell. 2006;23:483–496. doi: 10.1016/j.molcel.2006.06.025. [DOI] [PubMed] [Google Scholar]
- de Bruin RA, Kalashnikova TI, Wittenberg C. Stb1 collaborates with other regulators to modulate the G1-specific transcriptional circuit. Mol Cell Biol. 2008;28:6919–6928. doi: 10.1128/MCB.00211-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin RA, McDonald WH, Kalashnikova TI, Yates J, 3rd, Wittenberg C. Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell. 2004;117:887–898. doi: 10.1016/j.cell.2004.05.025. [DOI] [PubMed] [Google Scholar]
- Dirick L, Nasmyth K. Positive feedback in the activation of G1 cyclins in yeast. Nature. 1991;351:754–757. doi: 10.1038/351754a0. [DOI] [PubMed] [Google Scholar]
- Dohlman HG, Thorner JW. Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu Rev Biochem. 2001;70:703–754. doi: 10.1146/annurev.biochem.70.1.703. [DOI] [PubMed] [Google Scholar]
- Doncic A, Falleur-Fettig M, Skotheim JM. Distinct interactions select and maintain a specific cell fate. Mol Cell. 2011;43:528–539. doi: 10.1016/j.molcel.2011.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan JD, Toyn JH, Johnson AL, Johnston LH. P40SDB25, a putative CDK inhibitor, has a role in the M/G1 transition in Saccharomyces cerevisiae. Genes Dev. 1994;8:1640–1653. doi: 10.1101/gad.8.14.1640. [DOI] [PubMed] [Google Scholar]
- Eriksson PR, Ganguli D, Clark DJ. Spt10 and Swi4 control the timing of histone H2A/H2B gene activation in budding yeast. Mol Cell Biol. 2011;31:557–572. doi: 10.1128/MCB.00909-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escote X, Zapater M, Clotet J, Posas F. Hog1 mediates cell-cycle arrest in G1 phase by the dual targeting of Sic1. Nat Cell Biol. 2004;6:997–1002. doi: 10.1038/ncb1174. [DOI] [PubMed] [Google Scholar]
- Eser U, Falleur-Fettig M, Johnson A, Skotheim JM. Commitment to a cellular transition precedes genome-wide transcriptional change. Mol Cell. 2011;43:515–527. doi: 10.1016/j.molcel.2011.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov MV, Dyson NJ. Molecular mechanisms of E2F-dependent activation and pRB-mediated repression. J Cell Sci. 2004;117:2173–2181. doi: 10.1242/jcs.01227. [DOI] [PubMed] [Google Scholar]
- Gartner A, Jovanovic A, Jeoung DI, Bourlat S, Cross FR, Ammerer G. Pheromone-dependent G1 cell cycle arrest requires Far1 phosphorylation, but may not involve inhibition of Cdc28-Cln2 kinase, in vivo. Mol Cell Biol. 1998;18:3681–3691. doi: 10.1128/mcb.18.7.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geymonat M, Spanos A, Wells GP, Smerdon SJ, Sedgwick SG. Clb6/Cdc28 and Cdc14 regulate phosphorylation status and cellular localization of Swi6. Mol Cell Biol. 2004;24:2277–2285. doi: 10.1128/MCB.24.6.2277-2285.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goranov AI, Cook M, Ricicova M, Ben-Ari G, Gonzalez C, Hansen C, Tyers M, Amon A. The rate of cell growth is governed by cell cycle stage. Genes Dev. 2009;23:1408–1422. doi: 10.1101/gad.1777309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase SB, Reed SI. Improved flow cytometric analysis of the budding yeast cell cycle. Cell Cycle. 2002;1:132–136. [PubMed] [Google Scholar]
- Hartwell LH. Synchronization of haploid yeast cell cycles, a prelude to conjugation. Exp Cell Res. 1973;76:111–117. doi: 10.1016/0014-4827(73)90425-4. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Culotti J, Pringle JR, Reid BJ. Genetic control of the cell division cycle in yeast. Science. 1974;183:46–51. doi: 10.1126/science.183.4120.46. [DOI] [PubMed] [Google Scholar]
- Huang D, et al. Dual regulation by pairs of cyclin-dependent protein kinases and histone deacetylases controls G1 transcription in budding yeast. PLoS Biol. 2009;7:e1000188. doi: 10.1371/journal.pbio.1000188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeoung DI, Oehlen LJ, Cross FR. Cln3-associated kinase activity in Saccharomyces cerevisiae is regulated by the mating factor pathway. Mol Cell Biol. 1998;18:433–441. doi: 10.1128/mcb.18.1.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C, Moll T, Neuberg M, Ahorn H, Nasmyth K. A role for the transcription factors Mbp1 and Swi4 in progression from G1 to S phase. Science. 1993;261:1551–1557. doi: 10.1126/science.8372350. [DOI] [PubMed] [Google Scholar]
- Koch C, Schleiffer A, Ammerer G, Nasmyth K. Switching transcription on and off during the yeast cell cycle: Cln/Cdc28 kinases activate bound transcription factor SBF (Swi4/Swi6) at start, whereas Clb/Cdc28 kinases displace it from the promoter in G2. Genes Dev. 1996;10:129–141. doi: 10.1101/gad.10.2.129. [DOI] [PubMed] [Google Scholar]
- Koivomagi M, Valk E, Venta R, Iofik A, Lepiku M, Balog ER, Rubin SM, Morgan DO, Loog M. Cascades of multisite phosphorylation control Sic1 destruction at the onset of S phase. Nature. 2011;480:128–131. doi: 10.1038/nature10560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, 3rd, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Madhani HD, Galitski T, Lander ES, Fink GR. Effectors of a developmental mitogen-activated protein kinase cascade revealed by expression signatures of signaling mutants. Proc Natl Acad Sci USA. 1999;96:12530–12535. doi: 10.1073/pnas.96.22.12530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan DO. The Cell Cycle: Principles of Control. London: New Science Press; 2007. [Google Scholar]
- Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- Oehlen LJ, Cross FR. G1 cyclins CLN1 and CLN2 repress the mating factor response pathway at Start in the yeast cell cycle. Genes Dev. 1994;8:1058–1070. doi: 10.1101/gad.8.9.1058. [DOI] [PubMed] [Google Scholar]
- Oehlen LJ, Jeoung DI, Cross FR. Cyclin-specific START events and the G1-phase specificity of arrest by mating factor in budding yeast. Mol Gen Genet. 1998;258:183–198. doi: 10.1007/s004380050722. [DOI] [PubMed] [Google Scholar]
- Pardee AB. A restriction point for control of normal animal cell proliferation. Proc Natl Acad Sci USA. 1974;71:1286–1290. doi: 10.1073/pnas.71.4.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter M, Gartner A, Horecka J, Ammerer G, Herskowitz I. FAR1 links the signal transduction pathway to the cell cycle machinery in yeast. Cell. 1993;73:747–760. doi: 10.1016/0092-8674(93)90254-n. [DOI] [PubMed] [Google Scholar]
- Peter M, Herskowitz I. Direct inhibition of the yeast cyclin-dependent kinase Cdc28-Cln by Far1. Science. 1994;265:1228–1231. doi: 10.1126/science.8066461. [DOI] [PubMed] [Google Scholar]
- Queralt E, Igual JC. Cell cycle activation of the Swi6p transcription factor is linked to nucleocytoplasmic shuttling. Mol Cell Biol. 2003;23:3126–3140. doi: 10.1128/MCB.23.9.3126-3140.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Schwab M, Lutum AS, Seufert W. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- Schwob E, Bohm T, Mendenhall MD, Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 2002;350:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- Sidorova JM, Mikesell GE, Breeden LL. Cell cycle-regulated phosphorylation of Swi6 controls its nuclear localization. Mol Biol Cell. 1995;6:1641–1658. doi: 10.1091/mbc.6.12.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skotheim JM, Di Talia S, Siggia ED, Cross FR. Positive feedback of G1 cyclins ensures coherent cell cycle entry. Nature. 2008;454:291–296. doi: 10.1038/nature07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickfaden SC, Winters MJ, Ben-Ari G, Lamson RE, Tyers M, Pryciak PM. A mechanism for cell-cycle regulation of MAP kinase signaling in a yeast differentiation pathway. Cell. 2007;128:519–531. doi: 10.1016/j.cell.2006.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata S, Yu Y, Stillman DJ. The E2F functional analogue SBF recruits the Rpd3(L) HDAC, via Whi5 and Stb1, and the FACT chromatin reorganizer, to yeast G1 cyclin promoters. EMBO J. 2009;28:3378–3389. doi: 10.1038/emboj.2009.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Tyers M. The cyclin-dependent kinase inhibitor p40SIC1 imposes the requirement for Cln G1 cyclin function at Start. Proc Natl Acad Sci USA. 1996;93:7772–7776. doi: 10.1073/pnas.93.15.7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M, Futcher B. Far1 and Fus3 link the mating pheromone signal transduction pathway to three G1-phase Cdc28 kinase complexes. Mol Cell Biol. 1993;13:5659–5669. doi: 10.1128/mcb.13.9.5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivieso MH, Sugimoto K, Jahng KY, Fernandes PM, Wittenberg C. FAR1 is required for posttranscriptional regulation of CLN2 gene expression in response to mating pheromone. Mol Cell Biol. 1993;13:1013–1022. doi: 10.1128/mcb.13.2.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel S, Dyson NJ. Conserved functions of the pRB and E2F families. Nat Rev Mol Cell Biol. 2008;9:713–724. doi: 10.1038/nrm2469. [DOI] [PubMed] [Google Scholar]
- Visintin R, Prinz S, Amon A. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- Wagner MV, Smolka MB, de Bruin RA, Zhou H, Wittenberg C, Dowdy SF. Whi5 regulation by site specific CDK-phosphorylation in Saccharomyces cerevisiae. PLoS One. 2009;4:e4300. doi: 10.1371/journal.pone.0004300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Carey LB, Cai Y, Wijnen H, Futcher B. Recruitment of Cln3 cyclin to promoters controls cell cycle entry via histone deacetylase and other targets. PLoS Biol. 2009;7:e1000189. doi: 10.1371/journal.pbio.1000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnen H, Futcher B. Genetic analysis of the shared role of CLN3 and BCK2 at the G(1)-S transition in Saccharomyces cerevisiae. Genetics. 1999;153:1131–1143. doi: 10.1093/genetics/153.3.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnen H, Landman A, Futcher B. The G(1) cyclin Cln3 promotes cell cycle entry via the transcription factor Swi6. Mol Cell Biol. 2002;22:4402–4418. doi: 10.1128/MCB.22.12.4402-4418.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirt SE, et al. G1 arrest and differentiation can occur independently of Rb family function. J Cell Biol. 2010;191:809–825. doi: 10.1083/jcb.201003048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenberg C, Reed SI. Cell cycle-dependent transcription in yeast: promoters, transcription factors, and transcriptomes. Oncogene. 2005;24:2746–2755. doi: 10.1038/sj.onc.1208606. [DOI] [PubMed] [Google Scholar]
- Wittenberg C, Sugimoto K, Reed SI. G1-specific cyclins of S. cerevisiae: cell cycle periodicity, regulation by mating pheromone, and association with the p34CDC28 protein kinase. Cell. 1990;62:225–237. doi: 10.1016/0092-8674(90)90361-h. [DOI] [PubMed] [Google Scholar]
- Zimmerman ZA, Kellogg DR. The Sda1 protein is required for passage through start. Mol Biol Cell. 2001;12:201–219. doi: 10.1091/mbc.12.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.