Abstract
Lecithin:retinol acyltransferase (LRAT) catalyzes the synthesis of retinyl esters in many tissues and is crucial for the transport and intracellular storage of vitamin A. LRAT expression is highly regulated in the liver. In this study, we have cloned and sequenced the full-length LRAT mRNA from human liver and identified its 5′- and 3′-ends. Full-length LRAT mRNA comprises 5023 nt with a predicted ORF of 230 amino acids, a short 5′ UTR, and a relatively long 3′UTR of 4 kb containing several polyadenylation signals and AU-rich regions. Based on alignment of this mRNA with human genomic DNA in the GenBank database, the human LRAT gene spans about 9.1 kbp and consists of two exons and a relatively long 4-kbp intron. Further analysis of normal liver revealed a minor alternative splicing variant which lacks a 103 nt polynucleotide contained in the 5′UTR of the full-length LRAT transcript. This variant predicts that the LRAT gene is organized into three exons and two introns, as reported for LRAT cloned from retinal pigment epithelium (RPE) cells. These two LRAT mRNA variants are also present in testis, which is known to express LRAT and contain retinyl esters. Major and minor transcription start sites for human liver LRAT mRNA were identified and the sequence of the upstream proximal promoter region was retrieved from the GenBank database and physically analyzed for the presence of putative cis-acting elements essential for basal transcription. This region contains a TATA box, CCAAT box and Sp1 site, which are apparently conserved in mouse and rat LRAT genes. Our results provide evidence that multiple LRAT mRNA transcripts, which are expressed in a tissue-specific manner, may result from several mechanisms including differential splicing of the 5′UTR region and the use of multiple polyadenylation signals in the 3′UTR.
Keywords: Untranslated region, Splicing variant
1. Introduction
Lecithin:retinol acyltransferase (LRAT), a microsomal protein, catalyzes the synthesis of retinyl esters, the principal cellular form of vitamin A, by transferring the sn –1 fatty acid from membrane-associated phosphatidylcholine to retinol which is present in the cytosol bound to members of the cellular retinol-binding protein gene family (Ross, 1993; Ross et al., 2001). LRAT mRNA and enzymatic activity are expressed in several tissues including the small intestine, liver, lung, testes, and retinal pigment epithelium (RPE) (MacDonald and Ong, 1988; Saari and Bredberg, 1988; Yost et al., 1988; Ong et al., 1991; Ross, 1993; Schmitt and Ong, 1993; Ruiz et al., 1999; Zolfaghari and Ross, 2000, 2002). LRAT is crucial for retinyl ester synthesis, as recently shown by the near absence of esterified retinol in the RPE and liver of mice lacking the LRAT gene (Batten et al., 2004).
The LRAT gene is unrelated to any other known mammalian acyltransferase. Genomic analysis has revealed LRAT as a founding member of an ancient gene family (Anantharaman and Aravind, 2003; Jahng et al., 2003). A comparison of the LRAT protein sequence with those of other proteins available in the GenBank database has revealed a significant homology in the putative catalytic region with the sequences of several proteins including tumor suppressors, putative viral proteases, and Egl-26, a developmental regulator in nematodes (Anantharaman and Aravind, 2003; Jahng et al., 2003). All these proteins have been classified in an LRAT-like protein family, which belongs to the N1pC/P60 protein superfamily of cell-wall peptidases represented in archebacteria and various bacterial lineages (Anantharaman and Aravind, 2003).
Similar to many other membrane-bound proteins, LRAT protein has been difficult to purify, but recent successes in the molecular cloning of LRAT cDNA have allowed progress in understanding the LRAT gene and its enzyme product. Initially, LRAT cDNAs of about 2.5–2.7 kb were isolated from human and bovine RPE (Ruiz et al., 1999) and from mouse and rat liver (Zolfaghari and Ross, 2000). Probes based on the cloned LRAT cDNA of ∼2.5 kb were shown by Northern blot analysis to hybridize to two predominantly expressed mRNA species with sizes of 1.5 and 5 kb, respectively, as well as to several minor mRNA species of intermediate sizes (Ruiz et al., 1999; Zolfaghari and Ross, 2000). Subsequently, we reported the cloning of a full-length LRAT cDNA from rat liver (Zolfaghari et al., 2002). Both the larger 5.3 kb cDNA and the shorter 2.5 kb cDNAs contain an identical 696 bp open reading frame (ORF) (Zolfaghari and Ross, 2000). The full-length 5.3 kb LRAT mRNA from rat liver also includes a relatively short (270 bp) 5′-untranslated region (UTR) and a relatively long (4392 bp) 3′UTR (Zolfaghari et al., 2002). Sequence analysis of the 3′UTR of rat liver LRAT cDNA revealed the presence of multiple polyadenylation signals, while Northern blot and RT-PCR analyses showed that the multiplicity of LRAT mRNA species, which are expressed in tissue-specific patterns, is due at least in part to the differential usage of various polyadenylation signals (Zolfaghari et al., 2002).
Based on the 2.7 kb size of LRAT cDNA cloned from human fetal RPE, Ruiz et al. (2001) reported that human LRAT exists as a single gene with a size of 7.1 kbp which consists of three exons and two introns. However, we believed that neither the 5′UTR nor the 3′UTR were complete, based on finding several EST clones in the GenBank database which align either with the region upstream of the 5′-end or downstream of the 3′-end of the sequence reported for RPE LRAT mRNA. Moreover, in studies of mouse and rat liver LRAT cDNA, we noted evidence for the presence of a single intron. Based on this finding and the physiological evidence that LRAT mRNA is expressed in bands of multiple size and is regulated in tissue-specific patterns (Zolfaghari and Ross, 2000, 2002), we deemed it important to obtain the full-length LRAT cDNA from human liver for analysis of its genomic organization, and for comparison to the LRAT cDNAs cloned from human RPE and rodent liver. In the present studies, we have cloned the full-length sequence of LRAT cDNA from human liver and have characterized the 5′-end as the potential transcription start site. Analysis of the intron–exon boundaries of LRAT cDNAs from several specimens of normal human liver showed the presence of the two exon–one intron structure similar to that we previously deduced for rodent liver, which appears to constitute the major form of LRAT mRNA in human liver. The same specimens, however, also expressed the three exon–two intron structure of the LRAT gene, as described for human RPE, in lower abundance. The presence of differentially processed transcripts suggests an additional mechanism, besides several identified previously (Zolfaghari et al., 2002), by which LRAT may be expressed and regulated in a tissue-specific manner.
2. Materials and methods
2.1. Tissue samples
Human liver samples classified as normal were received in frozen form from the University of Pittsburgh through the Liver Tissue Procurement and Distribution Program (LTPADS). Protocols were approved by the Biosafety Committee of the Pennsylvania State University. The samples were stored at −80 °C until used for RNA extraction.
2.2. RNA extraction and analysis
Total RNA from individual liver samples was extracted using Trizol reagent (Invitrogen Life Technologies, Carlsbad, CA) and then further purified with guanidine hydrochloride and sodium acetate as previously described (Zolfaghari et al., 1993). For preparation of poly[A]+ RNA, the individual total RNA samples were first pooled and then passed through oligo-d[T] cellulose as previously described (Zolfaghari and Ross, 2000). Human testis total RNA was obtained from Clontech (Palo Alto, CA).
For RT-PCR analysis, the first DNA strand was synthesized from DNase-treated total RNA samples with Superscript II RNase H- transcriptase (Invitrogen Life Technologies) and then used for PCR reactions using either Taq Gold DNA polymerase (Applied Biosystems, Foster City, CA) or High Fidelity Platinum Taq DNA polymerase (Invitrogen Life Technologies) following the manufacturer's protocol. The RT-PCR reaction products were analyzed by ethidium bromide-stained agarose gel electrophoresis (Sam-brook and Russell, 2001).
2.3. Cloning of human liver LRAT cDNA
Based on the human RPE LRAT sequence in GenBank database, we designed two primers (see Table 1 for primer positions and sequences), one in the sense direction (primer 1, Table 1) and one in the antisense direction (primer 2, Table 1) to clone the LRAT mRNA from human liver by 3′- and 5′-RACE, respectively (Zolfaghari and Ross, 2000). By 3′-RACE, a clone of 1.4 kb was first amplified, isolated by TA cloning (Promega, Madison, WI) and sequenced. Based on this sequence, three more primers (primers 5, 6, and 7, Table 1) were designed and used to clone the complete region of the 3′-end of the LRAT mRNA by nested-PCR as described previously (Zolfaghari and Ross, 2000; Zolfaghari et al., 2002). This region includes 186 nt of the ORF and all of the 3′UTR of the complete LRAT sequence. For cloning the 5′-end of the LRAT cDNA, primer 2 (Table 1) was used to clone the entire ORF by 5′-RACE.
Table 1. Primers used for human liver LRAT cDNA cloning and analysis.
| No. | Primer name | Region covered in mRNA | Primer sequence (5′→3′) |
|---|---|---|---|
| 1 | HLT22-S | 832–855 | agatatggcaccccgatcagtccc |
| 2 | HLT23-A | 919–942 | agacgccaatcccaagactgctga |
| 3 | LT1-A | 496–515 | atgccatagtgggtcaggtg |
| 4 | LT2-A | 953–972 | agtgtatgataccaagcccg |
| 5 | HLT41-S | 2643–2667 | aaagcaaccactatgagaactaccg |
| 6 | HLT42-S | 2829–2850 | tgaagacccctcattaaaagcc |
| 7 | HLT45-S | 3703–3722 | ttctctgggcttcatccatt |
| 8 | HLT95-S | 104–123 | cttatccgtctcattcccca |
| 9 | HLT96-A | 594–613 | gcttgttggagaccaccttc |
| 10 | HLT105-S | 102–123 | tccttatccgtctcattcccca |
| 11 | HLT107-S | 45–66 | ctccttctccggctgcttgtag |
| 12 | HLT104-A | 4840–4861 | aacaaggacttttgccattgcc |
| 13 | HLT66-A | 446–469 | ctcggtggaaagagctggtttcat |
| 14 | HLT65-A | 505–528 | gtctcctaggtagatgccatagtg |
The putative transcription start site was cloned and identified with GeneRacer kit from Invitrogen using two primers (primers 13 and 14, Table 1). This method is designed to capture only the intact full-length capped 5′-ends of particular transcripts while eliminating any truncated mRNA. For this, pooled poly[A]+ RNA from normal human liver samples was first treated with calf intestinal phosphatase to dephosphorylate the 5′-ends of any truncated mRNA. Following decapping the intact mRNAs with tobacco acid pyrophosphatase, an RNA oligonucleotide was ligated specifically to 5′-end of the decapped mRNAs. After the synthesis of the first strand using the oligo-[dT] primer with Superscript II RNase H− transcriptase, the 5′-end of the LRAT mRNA was amplified by PCR followed by nested PCR according to the procedure described in the GeneRacer kit using primers 13 and 14 (Table 1) as LRAT gene-specific primers, and the 5′primer and nested 5′primer supplied by the kit. The PCR products were isolated, cloned by TA cloning and subjected to sequencing.
3. Results
3.1. Cloning of full-length LRAT cDNA and identification of its 3′- and 5′-ends
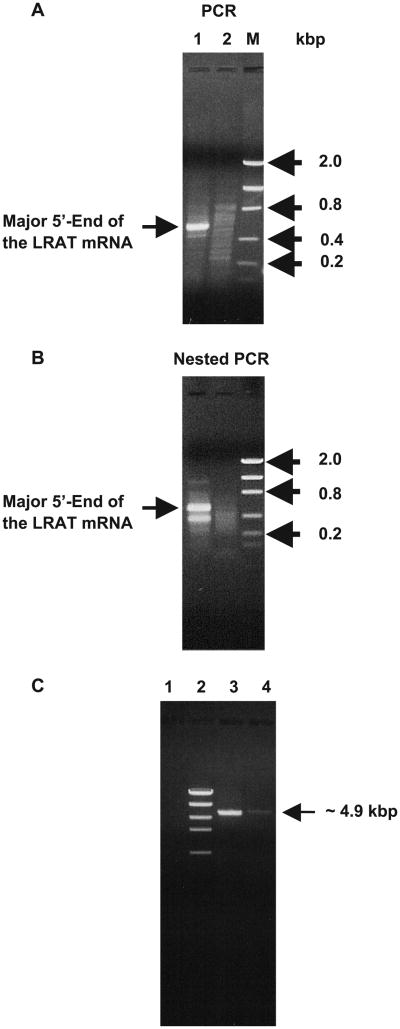
Based on the sequences available in the GenBank database, two primers were initially designed to clone the entire 3′- and 5′-ends of the LRAT cDNA from human liver (see Materials and methods and Table 1 for primer sequences). The transcription start site was cloned and identified from human liver RNA with GeneRacer kit using two primers (primers 13 and 14, Table 1) as described in the Materials and methods. Following the synthesis of the first-strand cDNA from human liver RNA, the 5′-end of the LRAT mRNA was amplified by PCR according to the procedure described in Materials and methods using primer 14 (Table 1) as an LRAT gene-specific primer, and the 5′ primer supplied in the GeneRacer kit. The PCR product (Fig. 1A) comprised one major band with a size of 572 bp when the primer pair of the LRAT gene-specific primer with the 5′-GeneRacer primer was used (lane 1). No such a band was observed when only the GeneRacer 5′-primer (lane 2) or the gene-specific primer alone (not shown) was used. The DNA from the major band was extracted from the gel and cloned (Zolfaghari and Ross, 2000; Zolfaghari et al., 2002). The major 5′-end of the human liver LRAT mRNA was identified after sequencing the cloned DNA piece. Another LRAT mRNA 5′-end was also identified when nested PCR was run on the PCR product using primer 13 (Table 1) as another gene-specific primer, with the GeneRacer 5′-nested primer (Fig. 1B, lane 1 on Nested PCR gel). This minor 5′-end starts 115 bp downstream of the major 5′-end of the LRAT mRNA.
Fig. 1.
Cloning of full-length LRAT mRNA from human liver. (A, B) Identification of the 5′-end of the LRAT mRNA. Ethidium bromide-stained agarose gel after electrophoresis of the PCR products (A) and nested PCR products (B) of the 5′-end of LRAT mRNA as cloned by the GeneRacer kit (see text for details). M, low-mass DNA marker. (C) Amplification of full-length LRAT mRNA from human liver. Ethidium bromide-stained agarose gel of the LRAT RT-PCR product amplified from human liver poly[A]+ RNA. Either of two sense primers (primers 10 and 11) with an antisense primer (primer 12) was designed from the 5′- and 3′-ends of the LRAT mRNA (lane 3, primer pair 10 and 12; lane 4, primer pair 11 and 12, see Table 1 for primer identification), and used for RT-PCR as described in Materials and methods. The LRAT amplicons from human liver had the expected size of about 4.8–4.9 kb. Lane 1, negative control; lane 2, high-mass DNA marker.
From the sequences and assembly of the cloned cDNA pieces resulting from 5′- and 3′-RACE (for detail, see Materials and methods), the LRAT mRNA from human liver was constructed. This cDNA clone, which is 5023 nt in size1 includes a small ORF of 693 nt, as previously reported (Ruiz et al., 1999) and a long 3′UTR of 4 kb nt. The first ATG and sequence of the ORF of the human liver LRAT exactly matches with that of human RPE LRAT except for the predicted lysine (aag) at residue number 32, which is substituted with glutamic acid (gaa) in the LRAT ORF of the human liver samples we analyzed. The 3′UTR of the human liver LRAT cDNA extends more than 2000 nt beyond the 3′-end of the RPE LRAT cDNA. As was the case for rat LRAT mRNA which we characterized previously (Zolfaghari and Ross, 2000), the 3′UTR region of human liver LRAT mRNA contains 3 polyadenylation signals as well as 20 AU-rich motif regions which, in other genes, have been shown to play an important role in mRNA stability (Chen and Shyu, 1995; Conne et al., 2000). The 5′UTR of LRAT mRNA is relatively short and ends at two sites, which yield either a 327 nt 5′-end (the major transcript) or a 212 nt 5′-end (the minor form). Both of these sites are located further upstream than the 5′-end of the human RPE LRAT cDNA previously described (Ruiz et al., 1999).
In addition, the human liver LRAT cDNA clone contains a polynucleotide of 103 bases starting from nt 224 to nt 326, which is missing in the human RPE mRNA sequence (Ruiz et al., 1999). Based on the sequence of the human RPE LRAT, this polynucleotide was reported as the first intron for the human LRAT gene (Ruiz et al., 2001). To determine whether such an mRNA species is present in intact form in human liver, either of the two sense primers (primers 10 and 11, Table 1) with an antisense primer (primer 12, Table 1) were designed from the extreme regions of this clone and used to amplify this large fragment by RT-PCR. As shown in Fig. 1C, presence of fragment of 4.8–4.9 kb are revealed when the two primer sets are used.
3.2. Genomic organization of human LRAT
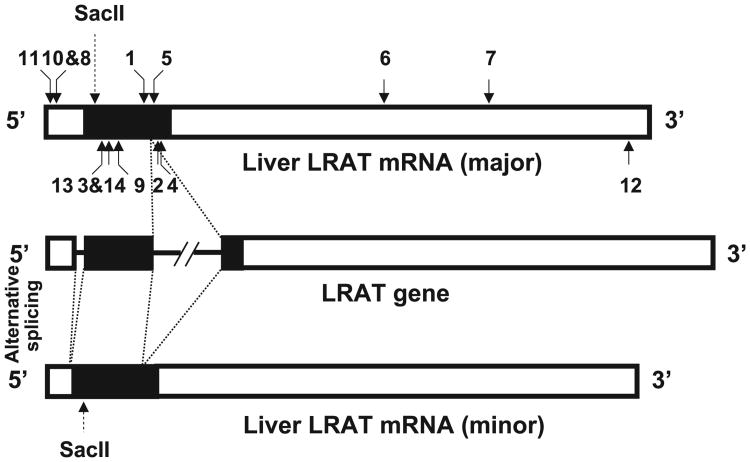
In order to establish the genomic organization of human LRAT based on the human liver LRAT mRNA, we aligned the sequence of the liver LRAT cDNA to the human genomic sequence available in the GenBank database of the National Center for Biotechnology Information (NCBI) using the Basic Local Alignment Search Tool (BLAST) program. Based on the sequence of liver LRAT mRNA, the human LRAT gene consists of two exons and a long intron (about 4.1 kbp), and spans 9.1 kbp (Fig. 2). Based on the sequence of RPE LRAT mRNA, the human LRAT gene consists of three exons and two introns. The first intron which is small (103 nt) and located in the 5′UTR of human RPE LRAT mRNA is apparently absent from the liver LRAT mRNA (Fig. 2). This intron apparently is formed through an alternative splicing process.
Fig. 2.
Organization of human LRAT gene. Schematic organization of LRAT gene relative to the full-length and spliced forms of the LRAT mRNA transcripts. The empty box represents the UTR and the filled box indicates the coding region. The box and the thick line in the LRAT gene represent exon and intron, respectively. The solid arrows indicate the position of the primers used to clone and analyze the LRAT cDNA and the numbers on the top and bottom of the LRAT transcript represent the sense and antisense primers (see Table 1), respectively.
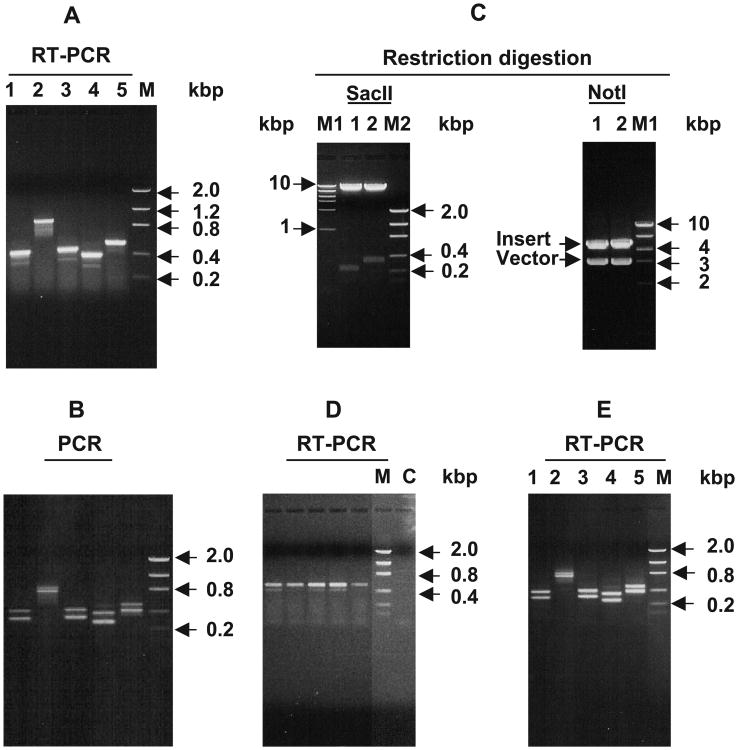
In order to further confirm and characterize the splicing pattern of the LRAT gene in human liver LRAT we performed 3 different experiments. First, RT-PCR reactions were designed using one sense primer (primer 8, Table 1) coupled with any one of 5 different antisense primers (primers 3, 4, 13, 14, and 9, Table 1) designed to flank the regions in the first intron. As shown in Fig. 3A, each RT-PCR reaction resulted in amplicons of two different sizes, a large band of major intensity and a smaller band of lesser intensity. The individual bands were cut, cloned and then sequenced. The clone corresponding to the major, larger-sized, band in each reaction contains the region including the polynucleotide corresponding to the small intron described above, whereas the clone containing the minor smaller band consists of the same region but lacks the oligonucleotide corresponding to the first intron of LRAT mRNA from RPE. These results show that, as in RPE, alternative splicing may occur in the liver but apparently at a comparatively minor level.
Fig. 3.
Identification of the full-length and spliced forms of the LRAT mRNA transcripts in human liver and testis. (A, B) The presence and absence of the polynucleotide corresponding to the first intron of the LRAT gene in the LRAT mRNA transcripts. Ethidium bromide-stained agarose gel electrophoresis of RT-PCR products of human liver poly[A]+ RNA (A) and of PCR products from the DNA template extracted from the gel shown in Fig. 1C (B). Primer pairs used are as follows: lane 1, primers 8 and 3; lane 2, primers 8 and 4; lane 3; primers 8 and 14; lane 4, primers 8 and 13; lane 5, primers 8 and 9 (see Table 1 for position of the primers). M, low-mass DNA marker. (C) Cloning and analysis of the full-length and spliced form of the human liver LRAT mRNA transcripts. The DNA from the gel shown in lane 3 of Fig. 1C was cut, extracted and cloned by TA cloning as described in Materials and methods. Two clones were picked up for digestion with SacII and NotI enzymes. Lane 1, spliced form clone; lane 2, the intron inclusion form clone. M1 and M2 are high- and low-mass DNA markers, respectively. (D) Expression of LRAT mRNA in individual human liver samples. Ethidium bromide-stained agarose gel electrophoresis of RT-PCR products of total RNA from liver samples, classified as normal, of five individuals. Total RNA from individual samples was extracted and treated with DNase to remove any genomic DNA followed by RT-PCR analysis as described in detail in Materials and methods using primers number 8 and 9 (see Table 1 for sequence and position of the primers). M, low-mass DNA marker; C, negative control. (E) Expression of LRAT spliced forms in human testis. Ethidium bromide-stained agarose gel electrophoresis of RT-PCR products of human testis total RNA using the same sets of primers used in A and B.
Second, since the sense primer (primer 8, Table 1) that we used to amplify the large LRAT transcript (Fig. 1C) is located upstream of the small intron, we reasoned that the 4.9 kbp nt DNA band should have contained two fragments if there is also alternative splicing is also occurring in human liver. To test this, the DNA from the band shown in Fig. 1C was extracted from the gel and used as template to perform PCR using the same sets of primers used for the RT-PCR analysis as shown in Fig. 3A. As shown in Fig. 3B, similar to the RT-PCR reaction, each PCR reaction resulted in two bands of similar size to those from the RT-PCR reaction.
Third, the large DNA band from Fig. 1C was cut from the gel, cloned, and two clones were subjected to either NotI or SacII digestion. As shown in Fig. 3C, both clones contain the DNA insert with a size of about 4.8–4.9 kb, similar to the band size in Fig. 1C. However, upon digestion with SacII, clone 1 resulted in a band with a size of about 245 nt (an alternatively spliced clone) and clone 2 in a band of about 348 nt, consistent with it containing the region corresponding to the first intron of RPE LRAT. By sequencing from both ends, clone 1 was confirmed to lack the nucleotide region corresponding to the first intron region of RPE LRAT, whereas clone 2 contained that region.
To evaluate whether alternative splicing is present in each of the human samples used to prepare our liver poly[A]+ RNA pool, individual total RNA samples from 5 individual livers were subjected to RT-PCR using primers 8 and 9 (Table 1). The alternative splicing (three exon–two intron structure) was apparent in each of the liver samples, but at minor level in all of them (Fig. 3D). To examine whether such splicing occurs in other organs, RT-PCR was performed in total RNA from human testis using the same sets of primers used in the liver shown in Fig. 3A. As shown in Fig. 3E, both splicing patterns seem to occur at relatively same levels in testis as compared to those in liver (Fig. 3A).
3.3. Physical analysis of sequence of human LRAT putative proximal promoter
Since we identified the major transcription start site of the human liver LRAT, the sequence located upstream of that site was retrieved from the GenBank database and analyzed for the presence of DNA response elements for any basal transcription factor using Transcription Element Search Software (http://www.cbil.upenn.edu/tess). Analysis of about 100 nt upstream of the major transcription start site identified a TATA box (from −33 to −24 from transcription start site), SP1 site (−52 to −41), and CCAAT box (−87 to −80).
4. Discussion
Many examples are known of gene families in which family members display tissue-specific patterns of expression which, often times, are important for the functions of the gene family. LRAT, however, appears to be different. In the case of LRAT, only a single gene is known, yet several different patterns of expression are seen in LRAT-expressing tissues. In the liver, LRAT mRNA and enzyme activity are strongly regulated by nutritional and hormonal factors, including by metabolites produced from retinol (Ross, 2003; Ross and Zolfaghari, 2004). Further variations in the level and/or patterns of expression have been reported between normal cells and tumor cell lines (Andreola et al., 2000; Guo et al., 2000, 2001, 2002; Simmons et al., 2002; Zhan et al., 2003). Thus, understanding the processing of the single LRAT gene is important for understanding its overall mode of regulation by dietary, hormonal and other physiological factors, and by tumorigenic transformation. The recent observation that LRAT knockout mice are severely deficient in retinyl esters in the retina and liver suggests a lack of redundancy for the retinol esterifying functions of LRAT in these tissues (Batten et al., 2004).
In this report, we cloned and sequenced the full-length LRAT cDNA from human liver and aligned its sequence with that available in GenBank database to lay out the genomic organization of LRAT in human. The sequence information of full-length LRAT mRNA in liver has been reported for rat (Zolfaghari et al., 2002) but is not available for other species including human. The complete sequence of LRAT mRNA in the liver of human shows that the full-length liver LRAT cDNA is 5023 nt for human, as compared to 5358 nt for rat liver LRAT (Zolfaghari et al., 2002). The predicted ORF for human liver LRAT contains 230 amino acids as compared to 231 in mouse and rat liver LRAT. There is about 80% identity and 86% similarity in predicted sequence of LRAT protein between human and rodent liver and more than 90% identity and about 94% similarity in amino acid residues between mouse and rat liver LRAT. Similar to the 5′UTR of rat and mouse LRAT mRNA, the 5′UTR of human liver LRAT mRNA is relatively short. However, there is no significant similarity between the sequence of the 5′UTR of human liver LRAT mRNA as compared with those of either mouse or rat. The 3′UTR region is relatively long (about 4 kb for human and 4.4 kb for rat LRAT) and contains several polyadenylation signals and numerous AU-rich regions, which may play important role in destabilization and turnover of LRAT mRNA. Similar to the 5′UTR, there was no significant similarity in the sequence of the 3′UTR of human LRAT mRNA as compared to that of rat. Thus, the ORF regions have remained well conserved, while the 5′ and 3′UTR appear not to have been conserved during speciation.
During cloning of the human liver LRAT mRNA, we made comparisons to the sequence of LRAT cloned from human fetal RPE (Ruiz et al., 1999) This resulted into several new findings and interesting comparisons. First, we believe that we have identified at least two 5′-ends of human LRAT mRNA as transcription start sites; one is the major site based on its higher abundance and the other, with minor abundance, begins 115 bp downstream of the major 5′ -end. The major site is located 327 nt upstream of ATG codon. This site is 131 nt upstream of the 5′-end of the RPE LRAT cDNA sequence (Ruiz et al., 1999), and 11 nt upstream of the 5′-end of an human EST (accession no. AA243120) reported for human brain. Second, although the majority of the human liver LRAT mRNA transcript contains a polynucleotide segment of 103 nt which is located between nt 224 to nt 326 (ending just −2 from ATG initiation codon), there is also a detectable LRAT mRNA transcript which lacks this polynucleotide. Such a polynucleotide is also absent in the human RPE LRAT mRNA and the human brain EST sequence which is aligned to the 5′-end of the human LRAT mRNA. This polynucleotide piece has been reported as the first intron for the human LRAT gene when the RPE LRAT cDNA was aligned with the sequences available in GenBank (Ruiz et al., 2001). This intron may be the result of alternative splicing in the 5′UTR of LRAT mRNA in the RPE or brain. Similar alternative splicing does apparently occur in human liver as well as in human testis but at a lower rate in the liver as compared with that in testis as inferred by the lower intensity of this transcript in each of several normal human liver samples (compare Fig. 3A and D with E). Based on this result, we checked the 5′UTR of rat LRAT mRNA in several tissues including adrenal gland, eye, small intestine, liver, lung, mammary tissue, and testis, but no differences were found in this regard (data not shown). Whether such an alternative splicing process is specific for human tissues remains to be determined. Third, a septa-nucleotide cctgcag located from −9 to −2 from ATG initiation codon is present in both of the LRAT transcript variants. This sequence may play an important role as a Kozak sequence (Kozak, 1987) in the translation of both LRAT transcript variants. Fourth, the human liver ORF uses the same ATG initiation codon as that in human RPE LRAT (Ruiz et al., 1999) and the ORF sequence of human liver matches exactly with that of human RPE except for the prediction of lysine residue 32 which is substituted with glutamic acid in human liver LRAT. This result is based on the sequencing results from several different clones of human liver samples and matches with that in human genome sequence present in the GenBank database.
Although LRAT mRNA is relatively long compared to an average size of mRNA, the LRAT gene seems to be relatively short. The human LRAT gene is about 9.1 kbp, which is similar to that of rat gene (data not shown). Experimentally, the human LRAT has shown to arise from a single gene when RPE LRAT cDNA was used as a probe in Southern blot analysis (Ruiz et al., 2001). As a result of alignment the gene has been located on human chromosome 4. Based on the liver LRAT cDNA sequence and the GenBank sequences for the human genome, the LRAT gene consists of two exons and a long intron. However, we have shown that an alternative splicing process results in an extra small intron, as the first intron, in the 5′UTR of the LRAT transcript. Whether such an alternative processing is important in regulation of LRAT transcription remains to be determined. This is the first alternative splicing process reported for the LRAT gene. Since the GenBank database contains two ESTs from human testis (accession nos. BU568311 and AA393342) which matches the nucleotide area of the long intron of the LRAT gene, we speculate that other alternative splicing processes may occur in the LRAT gene. Studies are now under way in our laboratory to find whether a further splicing process occurs in the human LRAT gene, which might also lend diversity to the LRAT mRNA sequence and, possibly, protein.
The major transcription start site of the LRAT gene had not previously been identified. The present study identified both a major and a minor 5′-end based for the human liver LRAT mRNA. The physical analysis of the proximal region upstream of the putative start site revealed the presence of cis-acting elements necessary for the binding of basal transcription factors including a TATA box, CCAAT box, and an SP1 site. Comparison of the DNA upstream of the human LRAT gene with those of mouse and rat gene showed that those elements are conserved in all 3 species (not shown). In addition to the major transcription start site, another transcription start site, apparently used less often, is located 115 nt downstream of the major site. Future studies will address whether those cis acting elements have any physiological function in the regulation of the expression of LRAT mRNA under various physiological and pathological conditions.
In summary, based on the complete sequence of human liver LRAT mRNA, the LRAT gene consists of two exons and one intron. However, based on the RPE LRAT mRNA, the human LRAT gene consists of three exons and two introns, with the extra small intron located in the 5′UTR of the LRAT mRNA. This may be the result of a specific alternative splicing which occurs in RPE as well as in the human brain. We also found that such an alternative splicing does occur in human liver although at much lower level and in the testis. This splicing process may play an important role in the regulation of LRAT expression.
Acknowledgments
We thank the LTPADS Program for providing human liver specimens, and Dr. Deborah Grove for sequencing the LRAT cDNA clones in the Nucleic Acid Facility at the Pennsylvania State University. Supported by grant NIH R01 CA-90214.
Abbreviations
- LRAT
lecithin:retinol acyltransferase
- RPE
retinal pigment epithelium
- UTR
untranslated region
- PCR
polymerase chain reaction
- RACE
rapid amplification of cDNA ends
Footnotes
Contributor Information
Reza Zolfaghari, Email: rxz7@psu.edu.
A. Catharine Ross, Email: acr6@psu.edu.
References
- Anantharaman V, Aravind L. Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol. 2003;4:R11. doi: 10.1186/gb-2003-4-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreola F, Giandomenico V, Spero R, De Luca LM. Expression of a smaller lecithin:retinol acyl transferase transcript and reduced retinol esterification in MCF-7 cells. Biochem Biophys Res Commun. 2000;279:920–924. doi: 10.1006/bbrc.2000.3995. [DOI] [PubMed] [Google Scholar]
- Batten ML, Imanishi Y, Maeda T, Tu DC, Moise AR, Bronson D, Possin D, Van Gelder RN, Baehr W, Palczewski K. Lecithin–retinol acyltransferase is essential for accumulation of all-trans-retinyl esters in the eye and in the liver. J Biol Chem. 2004;279:10422–10432. doi: 10.1074/jbc.M312410200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- Conne B, Stutz A, Vassalli JD. The 3′ untranslated region of messenger RNA: a molecular dhotspotT for pathology? Nat Med. 2000;6:637–641. doi: 10.1038/76211. [DOI] [PubMed] [Google Scholar]
- Guo X, Ruiz A, Rando RR, Bok D, Gudas LJ. Esterification of all-trans-retinol in normal human epithelial cell strains and carcinoma lines from oral cavity, skin and breast: reduced expression of lecithin:retinol acyltransferase in carcinoma lines. Carcinogenesis. 2000;21:1925–1933. doi: 10.1093/carcin/21.11.1925. [DOI] [PubMed] [Google Scholar]
- Guo X, Nanus DM, Ruiz A, Rando RR, Bok D, Gudas LJ. Reduced levels of retinyl esters and vitamin A in human renal cancers. Cancer Res. 2001;61:2774–2781. [PubMed] [Google Scholar]
- Guo X, Knudsen BS, Peehl DM, Ruiz A, Bok D, Rando RR, Rhim JS, Nanus DM, Gudas LJ. Retinol metabolism and lecithin:retinol acyltransferase levels are reduced in cultured human prostate cancer cells and tissue specimens. Cancer Res. 2002;62:1654–1661. [PubMed] [Google Scholar]
- Jahng WJ, Xue L, Rando RR. Lecithin retinol acyltransferase is a founder member of a novel family of enzymes. Biochemistry. 2003;42:12805–12812. doi: 10.1021/bi035370p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald PN, Ong DE. A lecithin:retinol acyltransferase activity in human and rat liver. Biochem Biophys Res Commun. 1988;156:157–163. doi: 10.1016/s0006-291x(88)80818-0. [DOI] [PubMed] [Google Scholar]
- Ong DE, Lucas PC, Kakkad B, Quick TC. Ontogeny of two vitamin A-metabolizing enzymes and two retinol-binding proteins present in the small intestine of the rat. J Lipid Res. 1991;32:1521–1527. [PubMed] [Google Scholar]
- Ross AC. Cellular metabolism and activation of retinoids: roles of cellular retinoid-binding proteins. FASEB J. 1993;7:317–327. doi: 10.1096/fasebj.7.2.8440409. [DOI] [PubMed] [Google Scholar]
- Ross AC. Retinoid production and catabolism: role of diet in regulating retinol esterification and retinoic acid oxidation. J Nutr. 2003;133:291S–296S. doi: 10.1093/jn/133.1.291S. [DOI] [PubMed] [Google Scholar]
- Ross AC, Zolfaghari R. Regulation of hepatic retinol metabolism: perspectives from studies on vitamin A status. J Nutr. 2004;134:269S–275S. doi: 10.1093/jn/134.1.269S. [DOI] [PubMed] [Google Scholar]
- Ross AC, Zolfaghari R, Weisz J. Vitamin A: recent advances in the biotransformation, transport, and metabolism of retinoids. Curr Opin Gastroenterol. 2001;17:184–192. doi: 10.1097/00001574-200103000-00015. [DOI] [PubMed] [Google Scholar]
- Ruiz A, Winston A, Lim YH, Gilbert BA, Rando RR, Bok D. Molecular and biochemical characterization of lecithin retinol acyltransferase. J Biol Chem. 1999;274:3834–3841. doi: 10.1074/jbc.274.6.3834. [DOI] [PubMed] [Google Scholar]
- Ruiz A, Kuehn MH, Andorf JL, Stone E, Hageman GS, Bok D. Genomic organization and mutation analysis of the gene encoding lecithin retinol acyltransferase in human retinal pigment epithelium. Invest Ophthalmol Visual Sci. 2001;42:31–37. [PubMed] [Google Scholar]
- Saari JC, Bredberg DL. CoA- and non-CoA-dependent retinol esterification in retinal pigment epithelium. J Biol Chem. 1988;263:8084–8090. [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- Schmitt MC, Ong DE. Expression of cellular retinol-binding protein and lecithin–retinol acyltransferase in developing rat testis. Biol Reprod. 1993;49:972–979. doi: 10.1095/biolreprod49.5.972. [DOI] [PubMed] [Google Scholar]
- Simmons DP, Andreola F, De Luca LM. Human melanomas of fibroblast and epithelial morphology differ widely in their ability to synthesize retinyl esters. Carcinogenesis. 2002;23:1821–1830. doi: 10.1093/carcin/23.11.1821. [DOI] [PubMed] [Google Scholar]
- Yost RW, Harrison EH, Ross AC. Esterification by rat liver microsomes of retinol bound to cellular retinol-binding protein. J Biol Chem. 1988;263:18693–18701. [PubMed] [Google Scholar]
- Zhan HC, Gudas LJ, Bok D, Rando R, Nanus DM, Tickoo SK. Differential expression of the enzyme that esterifies retinol, lecithin:retinol acyltransferase, in subtypes of human renal cancer and normal kidney. Clin Cancer Res. 2003;9:4897–4905. [PubMed] [Google Scholar]
- Zolfaghari R, Ross AC. Lecithin:retinol acyltransferase from mouse and rat liver. CDNA cloning and liver-specific regulation by dietary vitamin a and retinoic acid. J Lipid Res. 2000;41:2024–2034. [PubMed] [Google Scholar]
- Zolfaghari R, Ross AC. Lecithin:retinol acyltransferase expression is regulated by dietary vitamin A and exogenous retinoic acid in the lung of adult rats. J Nutr. 2002;132:1160–1164. doi: 10.1093/jn/132.6.1160. [DOI] [PubMed] [Google Scholar]
- Zolfaghari R, Chen X, Fisher EA. Simple method for extracting RNA from cultured cells and tissue with guanidine salts. Clin Chem. 1993;39:1408–1411. [PubMed] [Google Scholar]
- Zolfaghari R, Wang Y, Chen Q, Sancher A, Ross AC. Cloning and molecular expression analysis of large and small lecithin:retinol acyltransferase mRNAs in the liver and other tissues of adult rats. Biochem J. 2002;368:621–631. doi: 10.1042/BJ20020918. [DOI] [PMC free article] [PubMed] [Google Scholar]