Abstract
Organisms adapt their metabolism to meet ever changing environmental conditions. This metabolic adaptation involves at a cellular level the fine-tuning of mitochondrial function, which is mainly under the control of the transcriptional coactivator PGC-1α. Changes in PGC-1α activity coordinate a transcriptional response, that boosts mitochondrial activity in times of energy needs and attenuates it when energy demands are low. Reversible acetylation has emerged as a key way to alter PGC-1α activity. Although it is well-established that PGC-1α is deacetylated and activated by Sirt1 and acetylated and inhibited by GCN5, less is known about how these enzymes themselves are regulated. Recently, it became clear that the energy sensor, AMP-activated kinase (AMPK) translates the effects of energy stress into altered Sirt1 activity by regulating the intracellular level of its co-substrate NAD+. Conversely, the enzyme ATP citrate lyase (ACL), relates energy balance to GCN5, through the control of the nuclear production of acetyl-CoA, the substrate for GCN5’s acetyltransferase activity. We review here how these metabolic signalling pathways, affecting GCN5 and Sirt1 activity, allow the reversible acetylation/deacetylation of PGC-1α and the adaptation of mitochondrial energy homeostasis to energy levels.
Keywords: acetyl transferases, AMPK, ATP citrate lyase, deacetylases, GCN5, mitochondria, oxidative phosphorylation, PGC-1α, SIRT1
Introduction
When energy levels are limiting, such as during fasting and exercise, peripheral tissues switch to use fatty acids as fuel for mitochondrial oxidation in order to preserve blood glucose for cells that strictly rely on glucose as their main energy source, such as neurons and red blood cells. This metabolic flexibility is critically important, as inappropriate adaptation is often the basis for the onset of diseases, such as metabolic disorders and cancer. Metabolic homeostasis is to a large extent controlled by transcriptional regulatory mechanisms, which involve the coordinated action of transcription factors, cofactors and the transcription initiation machinery regulating the expression of genes involved in mitochondrial biogenesis and function (Kelly and Scarpulla 2004). The transcriptional coactivator PGC-1α is the master regulator of mitochondrial biogenesis and function (Puigserver et al 1998, Wu et al 1999). PGC-1α expression and activity is regulated at multiple levels and by diverse signalling events. Within the whole array of regulatory events that converge on PGC-1α, reversible acetylation only recently emerged as the key modifier of PGC-1α activity. The acetylation state of PGC-1α is regulated by the deacetylase Sirt1 and the acetyltransferase GCN5. Interestingly, the activity of both the Sirt1 and GCN5 proteins are modulated by the energy status of the cell. This review will focus on the direct regulators of the PGC1-α acetylation state, Sirt1 and GCN5, as well as on the pathways that provide the substrates for the deacetylation and acetylation reaction, respectively.
PGC-1α, an important regulator of energy homeostasis
As exemplified by its name, peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α) was originally identified as a transcriptional coactivator of the nuclear receptor PPARγ (Puigserver et al 1998). It is now established that PGC-1α interacts with several other transcription factors, including PPARα, glucocorticoid receptor (GR), hepatic nuclear factor 4α (HNF-4α), estrogen receptor related α (ERRα) and FOXO1 (Puigserver et al 2003, Vega et al 2000, Yoon et al 2001). By binding and modulating the activity of these different transcription factors, PGC-1α fine-tunes the expression of a number of genes involved in diverse metabolic pathways, such as fatty acid oxidation, gluconeogenesis, glycolysis and fatty acid synthesis. In brown adipose tissue, PGC-1α controls adaptive thermogenesis by increasing mitochondrial fatty acid oxidation and heat production through the expression of uncoupling protein 1 (UCP1) after cold exposure (Puigserver et al 1998). In liver, fasting increases PGC-1α expression thereby promoting gluconeogenesis and repressing glycolysis, which results in an increased hepatic glucose output (Yoon et al 2001). Fasting and exercise induces PGC-1α-mediated mitochondrial biogenesis and fatty acid oxidation in the muscle (Canto et al 2009, Gerhart-Hines et al 2007, Wu et al 1999). The expression and activity of PGC-1α is regulated by both cell-autonomous factors, such as AMPK and mTOR (Canto et al 2009, Herzig et al 2001, Yoon et al 2001), as well as by systemic factors of hormonal origin, such as insulin, glucagon and glucocorticoids and centrally regulated neural outputs (Puigserver et al 2003). In addition to the regulation of its expression level, PGC-1α activity is controlled by a variety of post-translational modifications, including phosphorylation (Jager et al 2007, Li et al 2007, Puigserver et al 2003), ubiquitination (Olson et al 2008) and acetylation (Lerin et al 2006, Rodgers et al 2005). Of all these post-translational events reversible acetylation has emerged as a key modifier of PGC-1α activity. Tandem mass spectrometry analysis of PGC-1α identified 13 acetylation sites (Rodgers et al 2005) and sofar only two proteins have been unequivocally shown to be involved in the reversible acetylation of PGC-1α, the deacetylase Sirt1 (Nemoto et al 2005, Rodgers et al 2005) and the acetyltransferase GCN5 (Lerin et al 2006).
Deacetylation and activation of PGC-1α by Sirt1
Up to date, the only protein identified to be able to deacetylate PGC-1α is Sirt1 (Gerhart-Hines et al 2007, Nemoto et al 2005, Rodgers and Puigserver 2007). Sirt1 belongs to a family of class III histone/protein deacetylase proteins, compromised of seven members in mammals (i.e. Sirt1-Sirt7), with different cellular localisation. Sirt1 is one of the mammalian homolog of the Silent information regulator 2 (Sir2), which was initially identified as a transacting factor involved in repression of the silent mating type loci in yeast (Shore et al 1984). In addition to silencing, Sir2 activity is linked to lifespan extension in yeast (Kaeberlein et al 1999), worms (Tissenbaum and Guarente 2001) and flies (Rogina and Helfand 2004), indicating that Sir2 has conserved its function throughout evolution. In contrast to mammals, in yeast only five homologues exits; ySir2 and Hst1-4 (homologous to Sir two). Sirt1 is the most studied mammalian orthologue and, as mentioned earlier, responsible for deacetylation of PGC-1α. Unlike histone deacetylation, which condenses chromatin structure and attenuates general transcriptional activity, PGC-1α deacetylation enhances its activity to co-activate transcription factors that in turn will induce transcription of its target genes. Consistent with this fact, overexpression of Sirt1 in livers of mice significantly increases the expression of the gluconeogenic genes that are under the control of PGC-1α, whereas Sirt1 knockdown attenuates this effect (Rodgers and Puigserver 2007). Also in skeletal muscle Sirt1 has been shown to be required for the PGC-1α-mediated induction of the fatty acid oxidation genes in response to fasting and exercise (Canto et al 2009, Gerhart-Hines et al 2007). These findings indicate that PGC-1α is activated by Sirt1-mediated deacetylation in times of energy demand, such as fasting and physical activity.
Since Sirt1 requires the coenzyme Nicotinamide Adenine Dinucleotide (NAD+) as a co-substrate for its function (Imai et al 2000), this suggests that NAD+, NADH or the NAD+ / NADH ratio modulates its activity, thereby linking Sirt1 activity to the energy status of the cell. Intracellular NAD+ levels are regulated by the balance between NAD+ synthesis, either de novo or by recycling via the NAD+ salvage pathway, and NAD+ consumption by NAD+-dependent enzymes (reviewed in (Houtkooper et al 2010)). In addition, AMPK can modulate this balance through altering metabolic pathways, as recently demonstrated (Canto et al 2009, Canto et al 2010, Fulco et al 2008).
NAD+ synthesis and NAD+ consumption
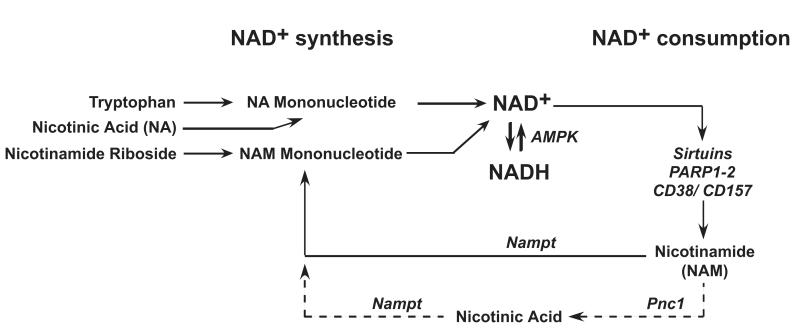
Although NAD+ can be synthesized de novo from the amino acid tryptophan derived from the diet, it is assumed that the main source of NAD+ is produced via the so-called NAD+ salvage pathways. This requires the dietary uptake of NAD+ precursors, such as the niacin derived Nicotinic Acid (NA), Nicotinamide (NAM) and Nicotinamide Riboside, which in mammals are converted into NAD+ through the salvage pathway (see figure 1). In this pathway, Nicotinamide is thought to be the most important contributor to NAD+ synthesis. The conversion of Nicotinamide to NAD+ is different between yeast and mammals. In yeast, Nicotinamide, the end product of reactions catalyzed by NAD+ consuming enzymes, is converted to Nicotinic Acid by the enzyme pyrazinamidase/nicotinamidase 1 (Pnc1), followed by the conversion to NAM mononucleotide. In contrast, in mammals Nicotinamide is directly converted to NAM Mononucleotide by one of the NAM phosphoribosyltransferase (Nampt) enzymes. In both yeast and mammals, NAM Mononucleotide is subsequently converted to NAD+. Interestingly, mutating either Sir2 or Pnc1 abolishes lifespan expansion after caloric restriction in yeast (Lin et al 2000). In addition, in skeletal muscle the level of Nampt increases upon exercise (Canto et al 2010, Costford et al 2009) as well as in muscle of mice upon fasting (Canto et al 2010), all conditions in which Sirt1 is active. This might suggest that an increase in the Nampt-dependent NAD+ salvage pathway contributes to the increased Sirt1 activity under these conditions. This could occur, either by an increase of NAD+ levels, which activates Sirt1, or by a decrease in the levels of Nicotinamide, which act as a potent inhibitor of Sirt1 (Anderson et al 2003, Bitterman et al 2002). In addition to Nicotinamide, Nicotinic Acid and Nicotinamide Riboside can also function as precursors in the salvage pathway. For Nicotinic Acid, the pathway starts with the conversion of Nicotinic Acid to NA Mononucleotide and then converges with the pathway of de novo NAD+ synthesis from tryptophan. When Nicotinamide Riboside is used, NAM Mononucleotide formation from Nicotinamide Riboside is first required, before entering the Nicotinamide-dependent salvage pathway (figure 1).
Figure 1. Regulation of intracellular NAD+ levels.
Intracellular NAD+ levels are regulated by the balance between NAD+ synthesis and consumption. NAD+ synthesis can occur either de novo from tryptophan, or via the “salvage pathways” that use Nicotinic Acid (NA), Nicotinamide Riboside, or Nicotinamide (NAM), which is produced by enzymes that produce Nicotinamide. NAD+ is consumed by enzymes, such as the sirtuins, CD38, CD157, PARP1 and PARP2, that use it as a substrate for their catalytic reaction and convert it into Nicotinamide. Furthermore the ratio of NAD+ to NADH can be modulated by metabolic pathways, such as those activated by AMPK.
NAD+ is not only synthesized, it is also consumed by several enzymes. In addition to the sirtuins, poly(ADP-ribose) polymerases (PARPs) and two cADP-ribose synthetases, CD38 and CD157, use NAD+ as a substrate. The PARPs, of which PARP1 and PARP2 are most widely studied, catalyze a reaction in which the ADP-ribose moiety of NAD+ is transferred to a substrate protein. In addition, the multifunctional enzymes, CD38 and CD157 use NAD+ as a substrate to generate second messengers, like cADP-ribose, which contributes to calcium mobilization. The functions of these enzymes, which also generate Nicotinamide as a by-product, will not be discussed here as they have been reviewed elsewhere (Malavasi et al 2008, Ortolan et al 2002, Schreiber et al 2006). By using NAD+ as a substrate, these enzymes seem under certain conditions to be able to modulate intracellular NAD+ and Nicotinamide levels, as recently reviewed (Houtkooper et al 2010).
AMPK modulates Sirt1 and NAD+ levels
Very recently, activation of AMPK has been shown to increase intracellular NAD+ levels in C2C12 myotubes and mouse skeletal muscle (Canto et al 2009, Canto et al 2010, Fulco et al 2008). In eukaryotic cells, AMPK plays an essential role in the maintenance of cellular energy stores. Upon activation, AMPK switches on catabolic pathways that produce ATP, such as oxidative phosphorylation, while switching off anabolic pathways that consume ATP (Hardie 2007). In line with these actions, AMPK activation increases PGC-1α expression (Suwa et al 2003), and activates PGC-1α by direct phosphorylation (Jager et al 2007). In addition, AMPK activation also induces Sirt1-mediated PGC-1α deacetylation (Canto et al 2009). In keeping with the dependence of Sirt1 on cellular NAD+, the NAD+ levels in C2C12 myotubes and skeletal muscle were increased upon AMPK activation (Canto et al 2009, Canto et al 2010). Interestingly, the AMPK-mediated phosphorylation of PGC-1α serves as a priming signal for the subsequent PGC-1α deacetylation that AMPK induces through boosting intracellular NAD+ levels (Canto et al 2009). The question remains how AMPK activation leads to an increase in intracellular NAD+ levels. An increase in NAD+ might be achieved by an increase in the Nicotinamide-dependent NAD+ salvage pathway. In line with this, Nampt expression increases after AICAR-mediated AMPK activation (Fulco et al 2008) as well as after exercise in muscle of mice after a 20-hour fast (Canto et al 2010). Consistent with the importance of Nampt in generating NAD+, shRNA-mediated knockdown of Nampt blocked the increase in NAD+/NADH ratio in 24-hours glucose restricted cells in which AMPK is activated (Fulco et al 2008). In apparent contradiction to this last observation, acute pharmacological inhibition of Nampt activity did not seem to affect AICAR’s capacity to modulate PGC1α activity or NAD+/NADH ratio (Canto et al 2009). Rather inhibition of mitochondrial fatty acid uptake blocked the acute AICAR-mediated increase in NAD+/NADH ratio (Canto et al 2009), suggesting that under these conditions an increase in mitochondrial oxidative phosphorylation is required for AMPK to increase the NAD+/NADH ratio. Combining the above evidence, it is likely that the AMPK-mediated increase in NAD+ levels is boosted through a “faster” metabolic adaptation (Canto et al 2009), and sustained by the “slower” induction of Nampt transcription, which maintains NAD+ levels for a longer period (Canto et al 2010, Fulco et al 2008).
Acetylation and inhibition of PGC-1α by GCN5
p300, SRC-1, GCN5 and SRC-3 are all acetyltransferases that have been shown to interact with PGC1α (Coste et al 2008, Lerin et al 2006, Puigserver et al 1999), but only GCN5 was shown to acetylate and inhibit PGC-1α (Lerin et al 2006). GCN5 was discovered as a histone acetyltransferase (HAT) in Tetrahymena and shown to be the homolog of yeast Gcn5, thereby being the first enzyme linking transcriptional activation to histone acetylation (Brownell et al 1996). Histone acetylation results in unwinding the chromatin making it accessible for the transcriptional machinery to initiate transcription (Kouzarides 2000). Besides histone acetylation, GCN5 has been shown to acetylate a large array of transcription factors (reviewed in (Nagy and Tora 2007)) including the transcriptional cofactors, PGC-1α (Lerin et al 2006) and PGC-1β (Kelly et al 2009). GCN5-mediated PGC-1α acetylation suppresses its transcriptional activity in vitro and in vivo resulting in reduced gluconeogenesis and hepatic glucose output (Lerin et al 2006). Although, it is clear that GCN5 acetylates PGC-1α, it cannot be excluded that other acetyltransferases can acetylate PGC-1α as well under certain conditions.
In addition to the expression of GCN5, which is regulated by nutritional/metabolic factors (Coste et al 2008), another link between GCN5 and metabolism could come from the fact that, like all acetyltransferases, it requires acetyl-CoA as a substrate for the acetylation reaction. Remarkably, the regulation of the acetylation reaction and its dependence on acetyl-CoA as a substrate is still not well understood.
Acetyl-CoA synthetase regulates nucleocytosolic acetyl-CoA levels in yeast
In yeast acetyl-CoA can be produced by at least three major pathways. Using yeast strains in which the critical enzymes of either of these pathways were deleted, acetyl-CoA synthetase 2 (Acs2p) was shown to be essential for histone acetylation by histone acetyl transferases (HATs) (Takahashi et al 2006). Two different acetyl-CoA synthetases, which catalyze the ligation of acetate and CoA to acetyl-CoA, exist in yeast, i.e. Acs1p and Acs2p. Consistent with the presence of a putative mitochondrial localization signal, Acs1p localizes to mitochondria (Kumar et al 2002), whereas Acs2p is present in the nucleus (Kals et al 2005, Takahashi et al 2006). Although glucose represses the mitochondrial Acs1p, Acs2p is active in the presence of glucose (van den Berg et al 1996), suggesting that high nutrient availability drives acetyl-CoA production and concomitant histone acetylation. Indeed, glucose induces overall histone acetylation mainly by the stimulation of the two HATs, Gcn5 (GCN5) and Esa1 (TIP60), acting in 2 different complexes (Friis et al 2009). Interestingly, this required glycolysis, indicating that glucose-induced overall histone acetylation is due to direct metabolic induction of HATs. In line with this, PKA and TORC signal transduction pathways had little effect on the glucose-induced overall histone acetylation, whereas the deletion of either Acs2p or Gcn5 in yeast abolished the glucose-induced H3 acetylation (Friis et al 2009). All together this indicates that H3 acetylation depends on the enzyme that require acetyl-CoA (Gcn5) and the enzyme that generate the nuclear acetyl-CoA (Acs2p).
In mammals ACL regulates nuclear acetyl-CoA levels
In mammals also two acetyl-CoA synthetases exist, AceCS1 and AceCS2, which are the homologs of yeast Acs2p and Acs1p, respectively. AceCS2 is mainly expressed in brown adipose tissue, skeletal muscle and heart and localizes to the mitochondria, where it generates acetyl-CoA for oxidation in the tricarboxylic acid (TCA) cycle to generate ATP and CO2 (Fujino et al 2001). In contrast, AceCS1 is highly expressed in the liver (Luong et al 2000) and present in the cytoplasm, where it produces acetyl-CoA for fatty acid synthesis. Although, a homolog of yeast AceCS2 is thus present in higher eukaryotes, the cellular concentrations of acetate are relatively low in mammals, suggesting that the contribution of AceCS1 to the generation of the nuclear acetyl-CoA pool is relatively low. Interestingly, both AceCS2 and AceCS1 are acetylated proteins themselves and are deacetylated by SIRT3 and SIRT1, respectively (Hallows et al 2006).
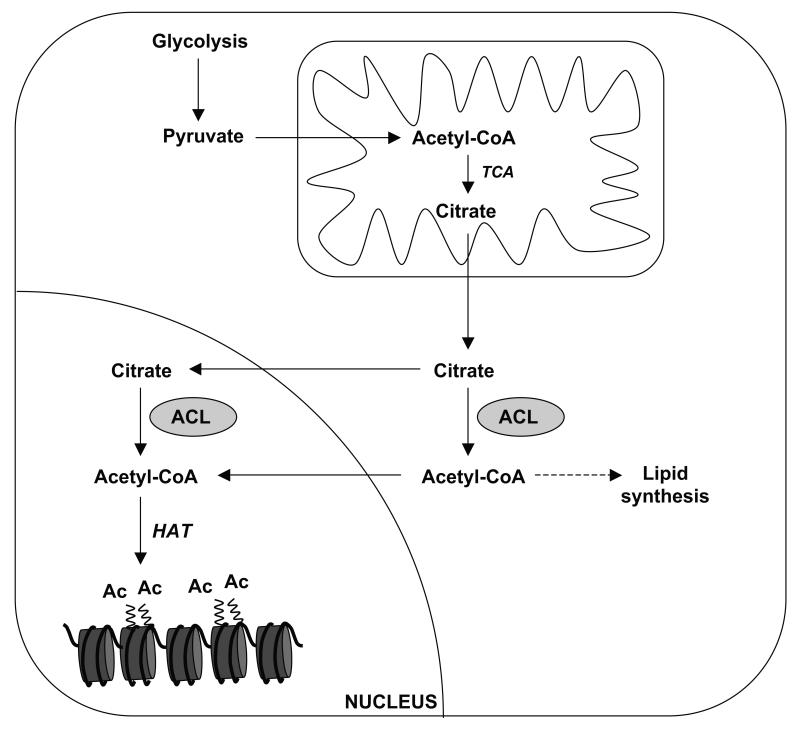
Unlike in yeast, mammals can produce acetyl-CoA by the enzyme ATP-citrate lyase (ACL), which generates acetyl-CoA from TCA-derived citrate in the cytoplasm. Interestingly, ACL and AceCS1 were recently also reported to be present in the nucleus (Wellen et al 2009). Since both acetate and citrate are small molecules able to diffuse freely through the nucleopore complex (Paine et al 1975), this suggests that acetyl-CoA production may occur in both the cytoplasm and nucleus (see figure 2). Given that siRNA-mediated knockdown of ACL decreased histone acetylation, it seems that ACL is critical for nuclear acetyl-CoA production and that AceCS1 cannot compensate under basal conditions (Wellen et al 2009). ACL knockdown in 3T3-L1 cells moreover blocks adipocyte differentation (Wellen et al 2009), a process in which histone acetylation normally increases (Yoo et al 2006), whereas it induces differentiation of C2C12, which is normally associated with reduced histone acetylation (Bracha et al 2010). Furthermore, under glucose-deprived conditions histone acetylation decreases in MEF cells (Wellen et al 2009). Under these conditions cells can use fatty acid oxidation as a source of energy. However, fatty acid oxidation results in the production of mitochondrial, but not nucleocytosolic acetyl-CoA and consistent with this notion, supplementation of fatty acids to glucose-starved cells could not rescue histone acetylation (Wellen et al 2009). This suggests that, like in yeast, a mitochondrial and a nuclear/cytosolic pool of acetyl-CoA exist regulating acetylation reactions in different cellular compartments. Interestingly, ACL seems to provide acetyl-CoA only for certain acetyl transferases, as acetylation of p53, which is regulated by the p300, was not affected by ACL knockdown. Since the combined knockdown of ACL and GCN5 did not result in an additive inhibition of histone acetylation, GCN5 and ACL function in the same pathway (Wellen et al 2009). As mentioned earlier, GCN5 can also acetylate non-histone proteins, such as PGC-1α. Conspicuously, ACL mediates histone acetylation when glucose (energy) is present, conditions in which PGC-1α is also acetylated. This suggests that ACL may not only provide acetyl-CoA for GCN5-mediated histone acetylation, but also for the GCN5-mediated acetylation of non-histone proteins such as PGC-1α and that this occurs in a nutrient-dependent manner.
Figure 2. ACL generates a nuclear/cytosolic pool of acetyl-CoA in mammals.
In times when glucose is around, TCA-derived citrate can be converted into aceteyl-CoA by ACL, both in the nucleus and cytosol, thereby providing a nuclear/cytosolic pool of acetyl-CoA. This pool of acetyl-CoA, which seems to be independent of the mitochondrial acetyl-CoA pool, is used for histone acetylation by histone acetyltransferases, such as GCN5.
Other regulators of GCN5 activity
Steroid Receptor Coactivators (SRC1-3) are transcriptional coactivators that bind to nuclear receptors and other transcription factors thereby enhancing their transcriptional activity (Spiegelman and Heinrich 2004, Xu et al 2009). Although SRCs have been reported to possess HAT activity (Spencer, 194; Chen 569), this activity is much weaker than that of other HATs. Consistent with this, SRCs do not seem to contain a prototypical acetyl-CoA binding site as other HATs, such as GCN5 (Trievel et al 1999). These SRC proteins therefore rather seem to act as platform proteins that bind to transcription factors and recruit other proteins with HAT activity, thereby enhancing transcription. Interestingly, SRC-3 has been reported to positively influence GCN5 activity in mammalian cells (Coste et al 2008). GCN5 expression was decreased in muscle and brown adipose tissue of SRC-3−/− mice. In line with the reduction in GCN5, mitochondrial function and energy expenditure was increased in SRC-3−/− mice, which nicely correlated with a decrease in PGC-1α acetylation levels (Coste et al 2008). Whereas overexpression of SRC-3 by itself did not increase PGC-1α acetylation in myotubes, it enhanced the GCN5-mediated PGC-1α acetylation. Together these data indicate that SRC-3 acts as a modulator of GCN5 activity. Supportive of a physiological role of SRC-3/GCN5 in the control of PGC-1α activity, the expression of both SRC-3 and GCN5 was shown to increase concomitant with the induction of PGC-1α acetylation levels in muscle of mice on a high fat diet. The converse, a reduction in the expression of GCN5 and SRC-3, associated with PGC-1α deacetylation and activation, was seen in muscle of fasted animals (Coste et al 2008). These observations indicate that SRC-3 and GCN5 expression could in fact contribute to modulate PGC-1α acetylation state and mitochondrial activity pending on energy levels, in a synergistic fashion with Sirt1, ultimately resulting in the inhibition of mitochondrial function in situations with high energy supplies favouring fat accumulation (Coste et al 2008). Although, SRC-1 has been shown to interact with PGC-1α (Puigserver et al 1999), and SRC-2−/− mice have increased energy expenditure, a phenotype that was reminiscent of Sirt1 activation (Picard et al 2002), it remains to be investigated, whether these SRC proteins also impact on GCN5 activity in a manner analogous to SRC3.
A role for PGC-1α and its partners in cancer
A common feature of human cancer is a metabolic shift from the more efficient oxidative metabolism to the faster glycolytic metabolism to meet the high energy demand of tumour cells. Since the transcriptional coactivator PGC-1α triggers a shift from glycolytic to oxidative metabolism through the increase in mitochondrial biogenesis and activity, this might suggest that PGC-1α expression levels are down regulated in cancer. This hypothesis is supported by the reduced PGC-1α expression levels in a variety of cancers, including breast (Jiang et al 2003, Watkins et al 2004), colon (Feilchenfeldt et al 2004), ovary (Zhang et al 2007) and liver cancer (Yin et al 2004). In addition, PGC-1α overexpression suppressed cancer cell growth (Zhang et al 2007) further supporting a role for PGC-1α in cancer. As described earlier, the activity of PGC-1α is modulated by reversible acetylation/deacetylation. The deacetylase Sirt1 activates PGC-1α in times of energy demand thereby switching on oxidative phosphorylation, whereas the acetyltransferase GCN5 represses PGC-1α in excess of energy. In turn, the activity of Sirt1 and GCN5 seem to depend on the activity of AMPK and ACL, the metabolic enzymes that provide the substrates for the deacetylation and acetylation reaction, respectively. This implies that modulation of the activity of Sirt1, GCN5, AMPK and/or ACL might potentially prevent and/or suppress tumour formation or growth. In line with this, overexpression of Sirt1 suppresses intestinal tumour formation and growth (Firestein et al 2008) and the AMPK activator Metformin inhibits the growth of human breast cancer cells (Zakikhani et al 2006). In addition, the use of metformin in the treatment of type II diabetes and physical activity, which are both known to activate AMPK, have been associated with a reduced risk in the development of cancer (Brown et al 2007, Evans et al 2005, Libby et al 2009). Interestingly, the tumour suppressor LKB-1 that is mutated in the human Peutz-Jeghers cancer syndrome (Hawley et al 2003), has been identified as the upstream activating kinase of AMPK, supporting the idea that modulation of AMPK might be anti-cancerous. Next to activation of LKB-1, AMPK and/or Sirt1, inhibition of ACL and/or GCN5 will result in PGC-1α activation and potentially prevent carcinogenesis. At present, it is unknown of GCN5 inhibition can prevent tumorigenesis as specific GCN5 inhibitors are lacking. However, pharmacological inhibition of ACL has been shown to suppress tumour cell growth (Hatzivassiliou et al 2005). Moreover, ACL expression is increased in breast and bladder carcinomas (Szutowicz et al 1979, Turyn et al 2003). In view of all these findings that suggest that modulation of the activity of AMPK, Sirt1, GCN5 and ACL all impact on carcinogenesis, it is of interest to investigate if these effect are accomplished by changes in the PGC-1α acetylation status and PGC-1α mediated oxidative phosphorylation, thereby ultimately shifting cancer cells away from protumorigenic glycolytic pathways. If this is the case, strategies to induce PGC-1α expression and/or activity could become attractive to treat cancer and should incite the exploration of the therapeutic potential of compounds that interfere with PGC-1α activity, such as ACL inhibitors, GCN5 inhibitors, SIRT-1 activators, and AMPK agonists.
Conclusions and future perspectives
The maintenance of metabolic homeostasis is controlled by transcriptional events that enable the cell to adapt its gene expression to its energetic status. The transcriptional cofactor PGC-1α, which acts as a master regulator of mitochondrial function, acts in coherence with the energy status of the cell, being activated in times of energy demand, such as during fasting, physical activity and cold exposure, and being inhibited when energy is around. We have summarized here the wide body of evidence in support of the regulation of PGC-1α activity by changes in its acetylation status. In fact, PGC-1α activity is conversely regulated by the Sirt1 deacetylase, which activates PGC-1α, and the GCN5 acetyltransferase, which represses PGC-1α activity. GCN5 and Sirt1 seem therefore to act as the yin-yang that controls the activity of PGC-1α (see figure 3). As the activity of PGC-1α is reflected by the metabolic status of the cell, it is of interest to investigate if also the activity of GCN5 and Sirt1 is regulated in an opposite fashion by nutrients. In the case of Sirt1, cellular levels of its co-substrate NAD+ are shown to determine its activity. This seems to depend on nutrient availability, as NAD+ levels are increased in C2C12 myotubes on low glucose, whereas this is reversed after addition of glucose (Canto et al 2010). GCN5 requires acetyl-CoA as a co-substrate for its acetylation reaction. In mammals, the main metabolic enzyme providing acetyl-CoA for GCN5-catalyzed histone acetylation in the nucleus is ACL (Wellen et al 2009). The ACL-mediated histone acetylation depends on glucose availability as this was decreased in glucose-deprived conditions (Wellen et al 2009). Whether the pool of ACL-generated acetyl-CoA is also used by GCN5 for the acetylation of PGC-1α needs to be further explored. If this is the case, these observations support the idea that the co-substrates of Sirt1 and GCN5 are regulated in a yin-yang fashion by nutrients, with high levels of NAD+ and low levels of acetyl-CoA when glucose is limited, whereas this is the opposite when glucose is around.
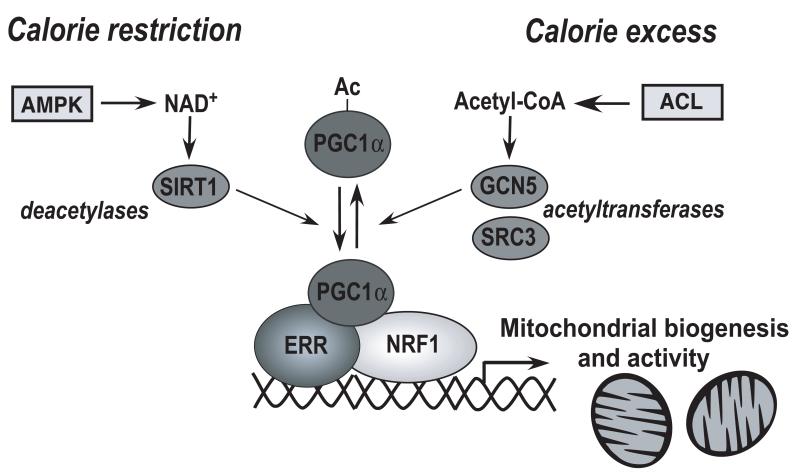
Figure 3. Energy sensing mediated by cofactors.
In times of high energy demand, an AMPK-mediated increase in NAD+ activates Sirt1 resulting in the deacetylation and activation of PGC-1α leading ultimately to mitochondrial biogenesis and improved mitochondrial function. Conversely, GCN5-mediated acetylation inactivates PGC-1α when plenty of energy is around. The acetyl-CoA necessary for this acetylation reaction is provided by ACL, which acts as rate limiting for this reaction.
Teleologically, such a regulation of PGC-1α by nutrients makes sense as it inhibits mitochondrial function in situations with high energy supplies, favouring fat accumulation, whereas it solicits fat release from adipose stores and their use in oxidative metabolism in situations of energy need (Coste et al 2008). The relative level of activity of Sirt1 and GCN5, which seems to be determined by changes in the supply of their co-substrates as a function of cellular metabolic activity, hence ultimately will inform PGC1α about the energetic status of the cell. PGC-1α then will adapt cellular energy production through its role in mitochondrial biogenesis and function, so that energy requirements are exactly met. Although our review mainly focussed on the regulation of PGC-1α as a prototypical example of metabolic regulation of protein activity by reversible acetylation/deacetylation, it needs to be underscored that such regulation may be more wide-spread and affect other proteins that control metabolism, such as the FOXO family of transcription factors, which already have been shown to be controlled by Sirt1-mediated acetylation (Brunet et al 2004).
In conclusion, unravelling the mechanisms by which PGC-1α is regulated by the energy status of the cell will contribute to our understanding of how cellular metabolic processes are regulated and as such might help to design novel therapeutics strategies to combat metabolic diseases and cancer.
Acknowledgements
This work was supported by grants of the Ecole Polytechnique Fédérale de Lausanne, Swiss National Science Foundation, NIH (DK59820), and the European Research Council Ideas programme (Sirtuins; ERC-2008-AdG23118).
References
- Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature. 2003;423:181–185. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Biol Chem. 2002;277:45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- Bracha AL, Ramanathan A, Huang S, Ingber DE, Schreiber SL. Carbon metabolism-mediated myogenic differentiation. Nat Chem Biol. 2010;6:202–204. doi: 10.1038/nchembio.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown WJ, Burton NW, Rowan PJ. Updating the evidence on physical activity and health in women. Am J Prev Med. 2007;33:404–411. doi: 10.1016/j.amepre.2007.07.029. [DOI] [PubMed] [Google Scholar]
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, et al. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, et al. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Jiang LQ, Deshmukh AS, Mataki C, Coste A, Lagouge M, et al. Interdependence of AMPK and SIRT1 for Metabolic Adaptation to Fasting and Exercise in Skeletal Muscle. Cell Metab. 2010;11:213–219. doi: 10.1016/j.cmet.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste A, Louet JF, Lagouge M, Lerin C, Antal MC, Meziane H, et al. The genetic ablation of SRC-3 protects against obesity and improves insulin sensitivity by reducing the acetylation of PGC-1{alpha} Proc Natl Acad Sci U S A. 2008;105:17187–17192. doi: 10.1073/pnas.0808207105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costford SR, Bajpeyi S, Pasarica M, Albarado DC, Thomas SC, Xie H, et al. Skeletal Muscle NAMPT is Induced by Exercise in Humans. Am J Physiol Endocrinol Metab. 2009 doi: 10.1152/ajpendo.00318.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JM, Donnelly LA, Emslie-Smith AM, Alessi DR, Morris AD. Metformin and reduced risk of cancer in diabetic patients. BMJ. 2005;330:1304–1305. doi: 10.1136/bmj.38415.708634.F7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feilchenfeldt J, Brundler MA, Soravia C, Totsch M, Meier CA. Peroxisome proliferator-activated receptors (PPARs) and associated transcription factors in colon cancer: reduced expression of PPARgamma coactivator 1 (PGC-1) Cancer Lett. 2004;203:25–33. doi: 10.1016/j.canlet.2003.08.024. [DOI] [PubMed] [Google Scholar]
- Firestein R, Blander G, Michan S, Oberdoerffer P, Ogino S, Campbell J, et al. The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth. PLoS One. 2008;3:e2020. doi: 10.1371/journal.pone.0002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friis RM, Wu BP, Reinke SN, Hockman DJ, Sykes BD, Schultz MC. A glycolytic burst drives glucose induction of global histone acetylation by picNuA4 and SAGA. Nucleic Acids Res. 2009;37:3969–3980. doi: 10.1093/nar/gkp270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino T, Kondo J, Ishikawa M, Morikawa K, Yamamoto TT. Acetyl-CoA synthetase 2, a mitochondrial matrix enzyme involved in the oxidation of acetate. J Biol Chem. 2001;276:11420–11426. doi: 10.1074/jbc.M008782200. [DOI] [PubMed] [Google Scholar]
- Fulco M, Cen Y, Zhao P, Hoffman EP, McBurney MW, Sauve AA, et al. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev Cell. 2008;14:661–673. doi: 10.1016/j.devcel.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, et al. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci U S A. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8:774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- Hatzivassiliou G, Zhao F, Bauer DE, Andreadis C, Shaw AN, Dhanak D, et al. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell. 2005;8:311–321. doi: 10.1016/j.ccr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Hawley SA, Boudeau J, Reid JL, Mustard KJ, Udd L, Makela TP, et al. Complexes between the LKB1 tumor suppressor, STRAD alpha/beta and MO25 alpha/beta are upstream kinases in the AMP-activated protein kinase cascade. J Biol. 2003;2:28. doi: 10.1186/1475-4924-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig S, Long F, Jhala US, Hedrick S, Quinn R, Bauer A, et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- Houtkooper RH, Canto C, Wanders RJ, Auwerx J. The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr Rev. 2010;31:194–223. doi: 10.1210/er.2009-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Jager S, Handschin C, St-Pierre J, Spiegelman BM. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc Natl Acad Sci U S A. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang WG, Douglas-Jones A, Mansel RE. Expression of peroxisome proliferator activated receptor-gamma (PPARgamma) and the PPARgamma co-activator, PGC-1, in human breast cancer correlates with clinical outcomes. Int J Cancer. 2003;106:752–757. doi: 10.1002/ijc.11302. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kals M, Natter K, Thallinger GG, Trajanoski Z, Kohlwein SD. YPL.db2: the Yeast Protein Localization database, version 2.0. Yeast. 2005;22:213–218. doi: 10.1002/yea.1204. [DOI] [PubMed] [Google Scholar]
- Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- Kelly TJ, Lerin C, Haas W, Gygi SP, Puigserver P. GCN5-mediated transcriptional control of the metabolic coactivator PGC-1beta through lysine acetylation. J Biol Chem. 2009;284:19945–19952. doi: 10.1074/jbc.M109.015164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 2000;19:1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Agarwal S, Heyman JA, Matson S, Heidtman M, Piccirillo S, et al. Subcellular localization of the yeast proteome. Genes Dev. 2002;16:707–719. doi: 10.1101/gad.970902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerin C, Rodgers JT, Kalume DE, Kim SH, Pandey A, Puigserver P. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1alpha. Cell Metab. 2006;3:429–438. doi: 10.1016/j.cmet.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Li X, Monks B, Ge Q, Birnbaum MJ. Akt/PKB regulates hepatic metabolism by directly inhibiting PGC-1alpha transcription coactivator. Nature. 2007;447:1012–1016. doi: 10.1038/nature05861. [DOI] [PubMed] [Google Scholar]
- Libby G, Donnelly LA, Donnan PT, Alessi DR, Morris AD, Evans JM. New users of metformin are at low risk of incident cancer: a cohort study among people with type 2 diabetes. Diabetes Care. 2009;32:1620–1625. doi: 10.2337/dc08-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- Luong A, Hannah VC, Brown MS, Goldstein JL. Molecular characterization of human acetyl-CoA synthetase, an enzyme regulated by sterol regulatory element-binding proteins. J Biol Chem. 2000;275:26458–26466. doi: 10.1074/jbc.M004160200. [DOI] [PubMed] [Google Scholar]
- Malavasi F, Deaglio S, Funaro A, Ferrero E, Horenstein AL, Ortolan E, et al. Evolution and function of the ADP ribosyl cyclase/CD38 gene family in physiology and pathology. Physiol Rev. 2008;88:841–886. doi: 10.1152/physrev.00035.2007. [DOI] [PubMed] [Google Scholar]
- Nagy Z, Tora L. Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene. 2007;26:5341–5357. doi: 10.1038/sj.onc.1210604. [DOI] [PubMed] [Google Scholar]
- Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha} J Biol Chem. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- Olson BL, Hock MB, Ekholm-Reed S, Wohlschlegel JA, Dev KK, Kralli A, et al. SCFCdc4 acts antagonistically to the PGC-1alpha transcriptional coactivator by targeting it for ubiquitin-mediated proteolysis. Genes Dev. 2008;22:252–264. doi: 10.1101/gad.1624208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortolan E, Vacca P, Capobianco A, Armando E, Crivellin F, Horenstein A, et al. CD157, the Janus of CD38 but with a unique personality. Cell Biochem Funct. 2002;20:309–322. doi: 10.1002/cbf.978. [DOI] [PubMed] [Google Scholar]
- Paine PL, Moore LC, Horowitz SB. Nuclear envelope permeability. Nature. 1975;254:109–114. doi: 10.1038/254109a0. [DOI] [PubMed] [Google Scholar]
- Picard F, Gehin M, Annicotte J, Rocchi S, Champy MF, O’Malley BW, et al. SRC-1 and TIF2 control energy balance between white and brown adipose tissues. Cell. 2002;111:931–941. doi: 10.1016/s0092-8674(02)01169-8. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Adelmant G, Wu Z, Fan M, Xu J, O’Malley B, et al. Activation of PPARgamma coactivator-1 through transcription factor docking. Science. 1999;286:1368–1371. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Rhee J, Donovan J, Walkey CJ, Yoon JC, Oriente F, et al. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature. 2003;423:550–555. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc Natl Acad Sci U S A. 2007;104:12861–12866. doi: 10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- Shore D, Squire M, Nasmyth KA. Characterization of two genes required for the position-effect control of yeast mating-type genes. EMBO J. 1984;3:2817–2823. doi: 10.1002/j.1460-2075.1984.tb02214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelman BM, Heinrich R. Biological control through regulated transcriptional coactivators. Cell. 2004;119:157–167. doi: 10.1016/j.cell.2004.09.037. [DOI] [PubMed] [Google Scholar]
- Suwa M, Nakano H, Kumagai S. Effects of chronic AICAR treatment on fiber composition, enzyme activity, UCP3, and PGC-1 in rat muscles. J Appl Physiol. 2003;95:960–968. doi: 10.1152/japplphysiol.00349.2003. [DOI] [PubMed] [Google Scholar]
- Szutowicz A, Kwiatkowski J, Angielski S. Lipogenetic and glycolytic enzyme activities in carcinoma and nonmalignant diseases of the human breast. Br J Cancer. 1979;39:681–687. doi: 10.1038/bjc.1979.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, McCaffery JM, Irizarry RA, Boeke JD. Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Mol Cell. 2006;23:207–217. doi: 10.1016/j.molcel.2006.05.040. [DOI] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- Trievel RC, Rojas JR, Sterner DE, Venkataramani RN, Wang L, Zhou J, et al. Crystal structure and mechanism of histone acetylation of the yeast GCN5 transcriptional coactivator. Proc Natl Acad Sci U S A. 1999;96:8931–8936. doi: 10.1073/pnas.96.16.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turyn J, Schlichtholz B, Dettlaff-Pokora A, Presler M, Goyke E, Matuszewski M, et al. Increased activity of glycerol 3-phosphate dehydrogenase and other lipogenic enzymes in human bladder cancer. Horm Metab Res. 2003;35:565–569. doi: 10.1055/s-2003-43500. [DOI] [PubMed] [Google Scholar]
- van den Berg MA, de Jong-Gubbels P, Kortland CJ, van Dijken JP, Pronk JT, Steensma HY. The two acetyl-coenzyme A synthetases of Saccharomyces cerevisiae differ with respect to kinetic properties and transcriptional regulation. J Biol Chem. 1996;271:28953–28959. doi: 10.1074/jbc.271.46.28953. [DOI] [PubMed] [Google Scholar]
- Vega RB, Huss JM, Kelly DP. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor alpha in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol Cell Biol. 2000;20:1868–1876. doi: 10.1128/mcb.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins G, Douglas-Jones A, Mansel RE, Jiang WG. The localisation and reduction of nuclear staining of PPARgamma and PGC-1 in human breast cancer. Oncol Rep. 2004;12:483–488. [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- Xu J, Wu RC, O’Malley BW. Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nat Rev Cancer. 2009;9:615–630. doi: 10.1038/nrc2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin PH, Lee HC, Chau GY, Wu YT, Li SH, Lui WY, et al. Alteration of the copy number and deletion of mitochondrial DNA in human hepatocellular carcinoma. Br J Cancer. 2004;90:2390–2396. doi: 10.1038/sj.bjc.6601838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo EJ, Chung JJ, Choe SS, Kim KH, Kim JB. Down-regulation of histone deacetylases stimulates adipocyte differentiation. J Biol Chem. 2006;281:6608–6615. doi: 10.1074/jbc.M508982200. [DOI] [PubMed] [Google Scholar]
- Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413:131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- Zakikhani M, Dowling R, Fantus IG, Sonenberg N, Pollak M. Metformin is an AMP kinase-dependent growth inhibitor for breast cancer cells. Cancer Res. 2006;66:10269–10273. doi: 10.1158/0008-5472.CAN-06-1500. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ba Y, Liu C, Sun G, Ding L, Gao S, et al. PGC-1alpha induces apoptosis in human epithelial ovarian cancer cells through a PPARgamma-dependent pathway. Cell Res. 2007;17:363–373. doi: 10.1038/cr.2007.11. [DOI] [PubMed] [Google Scholar]