Abstract
Efficient labeling of protein-based targeting ligands with various cargos (drugs, imaging agents, nanoparticles, etc.) is essential to the fields of molecular imaging and targeted therapeutics. Many common bioconjugation techniques, however, are inefficient, non-stoichiometric, not site-specific, and/or incompatible with certain classes of protein scaffolds. Additionally, these techniques can result in a mixture of conjugated and unconjugated products, which are often difficult to separate. In this study, a bacterial sortase enzyme was utilized to condense targeting ligand purification and site-specific conjugation at the C-terminus into a single step. A model was produced to determine optimal reaction conditions for high conjugate purity and efficient utilization of cargo. As proof-of-principle, the sortase-tag expressed protein ligation (STEPL) technique was used to generate tumor-specific affinity ligands with fluorescent labels and/or azide modifications at high purity (>95%) such that is was not necessary to remove unconjugated impurities. Click chemistry was then used for the highly efficient and site-specific attachment of the azide-modified targeting ligands onto nanoparticles.
Keywords: Sortase, bioconjugation, click chemistry, expressed protein ligation, nanoparticle
INTRODUCTION
Molecular imaging and targeted therapeutics rely heavily on ligands that can seek out and bind to specific cell surface biomarkers. These targeting ligands allow specific populations of cells to be tracked, quantified, and/or killed in vivo. A targeting ligand’s utility lies in its ability to direct a therapeutic and/or imaging moiety to the targeted cells. A number of methods have been developed to label ligands with cargo, including maleimide, N-hydroxysuccinimide, carbodiimide, and click chemistries 1–4. However, many of these approaches suffer from poor reaction efficiencies and indiscriminate labeling of nucleophilic residues (e.g. lysines and cysteines) on the targeting ligand. Random labeling of targeting ligands is problematic because a poorly placed cargo can greatly reduce a ligand’s affinity for its target. Additionally, each targeting ligand may be labeled with any number of functional moieties, eliminating their ability to be quantitative or stoichiometric.
Bioconjugation techniques that rely on ligases (e.g. biotin or lipoic acid) can overcome many of these shortcomings because they allow for site-specific and stoichiometric modifications of target proteins 5. However, ligases are generally limited to functionalizing proteins with their natural substrates (i.e. biotin and lipoic acid). The introduction of alternative substrates is possible, but this has generally led to a significant loss in conjugation efficiency and/or requires extensive protein engineering 6, 7. As a result, the versatility and broad applicability of these techniques is limited.
Another approach that has been used for the site-specific labeling of proteins involves fusing a protein tag to the target ligand. The protein tag is used to facilitate the covalent attachment of small-molecule substrates that can be customized with a variety of functional moieties, including imaging and protein capture ligands. Examples of protein tags include SNAP tags 8, HaloTags 9, CLIP tags 10, and acyl carrier protein domains 11. These labeling domains have been used to site-specifically attach a wide range of cargo onto target proteins. However, a noted disadvantage of this approach is that the protein tags add significant and potentially undesirable size to the final product (~10–20 kDa).
Recently, expressed protein ligation (EPL) has garnered interest as a chemoselective bioconjugation strategy that allows for site-specific coupling reactions 12. EPL refers to a native chemical ligation whereby a recombinant protein with a c-terminal thioester is ligated to a second molecule containing an N-terminal cysteine and the cargo of interest. Thioester generation is typically driven by a family of autoprocessing enzymes known as inteins, under reducing conditions. EPL has already been successfully used for the controlled conjugation between two proteins, and between proteins and drugs, imaging agents, and nanoparticles 2, 13–15. Despite the benefits of EPL, this methodology suffers from several shortcomings. Specifically, the need for reducing agents to generate thioesters, which can prevent the use of this technique with proteins that contain disulfides; a large excess and high concentration of the cysteine-containing cargo for efficient EPL, which can be costly; and it can be difficult to separate unlabeled recombinant proteins from labeled proteins.
Another recently described enzyme-based conjugation technique relies on Staphylococcus aureus Sortase A (SrtA) 16, 17. SrtA is a calcium-assisted transpeptidase that is responsible for anchoring surface proteins to the peptidoglycan cell wall of Gram-positive bacteria 18. Briefly, the enzyme cleaves the peptide bond between the amino acids threonine and glycine, within the motif, LPXTG. However, the products remain transiently attached through the active cystine residue of SrtA, until the N-terminal glycine of another protein displaces the cysteine residue and forms a new peptide bond between the two peptide chains 17, 19. This activity has been used recently for a number of protein engineering tasks, including protein purification. In this case a fusion protein was constructed with an N-terminal His-tag followed by SrtA, an LPXTG linker, and the protein of interest 20. The target protein, with only a single extra N-terminal glycine, was readily released upon the addition of Ca2+ and triglycine.
SrtA has also recently been used to site-specifically label proteins at the C-terminus with various cargos (e.g. fluorophores, haptens, etc.) 17. In these studies, the coding sequence for the LPXTG tag is simply inserted downstream of the protein of interest. The SrtA enzyme is then used to link any short peptide with an N-terminal glycine and the desired cargo onto the purified recombinant protein. Unfortunately, this conjugation technique requires the sortase enzyme, which is simply added to the sample, to be purified from the ligated protein, adding additional complexity. Further, efficient ligation requires the peptide with cargo to be used in large excess to prevent the reattachment of the liberated glycine.
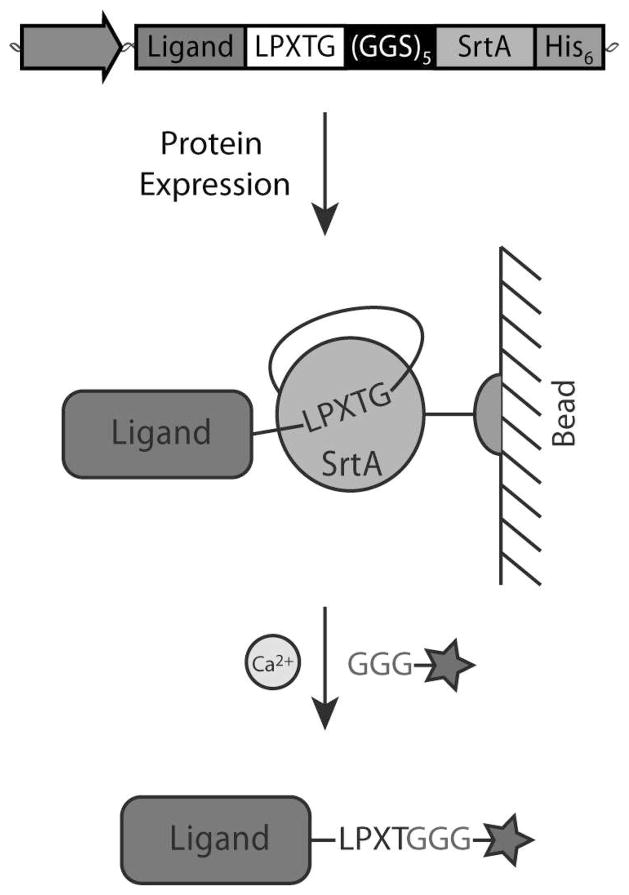
To mitigate these shortcomings, we have created a single protein construct with the amino acid sequence LPXTG, a (GGS)5 linker, SrtA, and a His-tag, respectively, fused to the C-terminal end of the protein of interest (Figure 1). This sortase-tag expressed protein ligation (STEPL) technique links protein purification and conjugation into a single step. The flexible (GGS)5 linker gives the sortase domain the conformational freedom to recognize the LPXTG in a unimolecular reaction. Addition of calcium and any protein/peptde with an N-terminal glycine (and attached cargo, if desirable) activates the sortase domain, ligating the protein of interest to the peptide while simultaneously cleaving it from the rest of the sortase chimera. Thus the conjugate is released while the sortase enzyme is retained on the column via the His-tag. By making purification and conjugation codependent, STEPL remains site-specific and stoichiometric in nature, but does not require any additional steps to remove SrtA from the purified protein sample. Further, large excesses of peptide are not essential since only correctly ligated product is released from the affinity column and conditions can be optimized to nearly exhaust any added peptide. In this study, the STEPL protocol is optimized, modeled, and used to conjugate the Her2/neu and EGFR-targeting affibody to fluorophores for imaging and/or an azide for subsequent copper-free click chemistry reactions with azadibenzocyclooctyne (ADIBO)-functionalized superparamagnetic iron oxide nanoparticles, demonstrating the system’s flexibility, efficacy, and utility.
Figure 1.
Sortase Expressed Protein Ligation Scheme. Ligands are cloned in series with the amino acid sequence LPXTG, a (GGS)5 linker, SrtA and a hexahistidine tag, respectively. The chimeric protein is expressed and isolated on a nickel column. The addition of calcium and a peptide with an N-terminal glycine (and optimally 3 glycines) allows the SrtA enzyme to simultaneously catalyze ligand release and peptide ligation. This allows any cargo (e.g. a fluorophore, azide, biotin, PEG, proteins, etc. – represented by the star) that is attached to the triglycine peptide to be site-specifically conjugated to the ligand.
EXPERIMENTAL PROCEDURES
Cloning
Sa-SrtAΔ59 20 was amplified from pGMBCS-SrtA (Addgene plasmid 21931 21) with an N-terminal (GGS)5 sequence and C-terminal H6 sequence. To facilitate blue/white screening, the Lac operon was amplified from pUC19 (Invitrogen) in an antisense orientation with a C-terminal sequence coding for the restriction site XhoI, the sortase recognition sequence LPETG, and the (GGS)5 linker. Overlap-extension PCR was used to join the Lac operon product to the Sa-SrtAΔ59 product. The full sequence was then cloned into pRSET-A (Invitrogen) via the NdeI and MluI restriction sites, creating the STEPL vector, pSTEPL. Sequences were verified by restriction analysis and sequencing. The Her2/neu affibody sequence 22 and the EGFR sequence 23 were amplified with 5′ NdeI and 3′ SalI sites and cloned into pSTEPL via its NdeI and XhoI sites. White colonies were picked and verified by restriction analysis and sequencing.
Notably, five GGS repeats were chosen for this fusion construct because the crystal structure reports a length of 26.2Å between the N-terminus of the sortase domain and its active site 24, corresponding to the length of approximately 3 GGS repeats (8.8Å each). Thus a (GGS)5 linker was expected to provide sufficient spatial flexibility for the sortase domain to recognize and bind the LPXTG motif.
Protein Expression, Cleavage, & Bioconjugation
Constructs were transformed into the BL12-derived Rosetta2 BL21(DE3) line (EMD Millipore). 50mL starter cultures were grown overnight in LB-Ampicillin. These were added to 450mL of LB and grown to an OD600 of 0.8–1 before induction with 0.5mM Isopropyl β-D-1-thiogalactopyranoside (IPTG). Cultures were allowed to express for 24hrs at 25°C. Cells were then harvested by centrifugation (6000 RPM, 15min) and resuspended in 10mL of lysis buffer (50mM NaH2PO4, 300mM NaCl, 1mg/mL Lysozyme, 1 EDTA-free cOmplete Mini protease inhibitor tablet (Roche), pH 7.5). Lysates were incubated at room temperature for 30min under gentle agitation before freezing overnight at −80°C. Samples were then thawed and incubated for 30min with DNAse I (Roche) under gentle agitation. Lysates were then sonicated and separated by centrifugation (10,000 rpm, 30min).
For optimization experiments, 8mL of clarified lysate for each condition was incubated for 1 hr with 0.6mL Talon resin (Clontech, equilibrated in lysis buffer). The lysate and beads were then added to a column and beads were washed with 6mL STEPL buffer (20mM Tris-base, 50mM NaCl, pH 7.5). Washed beads were resuspended in a total of 1.2mL of STEPL buffer containing the indicated amounts of CaCl2 and triglycine (Sigma-Aldrich) and aliquoted into three 1.5mL microcentrifuge tubes. Samples were shaken at 1,000 rpm, the indicated temperature, and protected from light. At each timepoint, samples were spun down at 3,000 rpm for 5min. Absorbance spectra were taken of the supernatants from 400 – 600nm using a Cary 100 Bio UV-Visible Spectrophotometer (Varian) and the sample was returned to shaking. At the end of the timecourse, beads were washed three times with 1mL STEPL buffer and incubated for 30 min in 100mM EDTA. Stripped beads were spun down as before and the absorbance spectra were taken of the supernatants from 400 – 600nm.
For bioconjugation experiments, 8mL of clarified lysate was incubated for 1hr with 0.5mL Talon resin (equilibrated in lysis buffer). The lysate and beads were added to a column and beads were washed with 5mL STEPL buffer. 400μL of STEPL buffer containing 150μM synthetic peptide (Table 2) or 5mM triglycine (for labeled and unlabeled preparations, respectively) and 100μM CaCl2 was flowed over the beads until it replaced the wash buffer. Columns were protected from light and reacted for 6hrs at 37°C. 1mL of STEPL buffer was added to the column and the flow through collected. To remove unreacted peptide, flowthrough was dialyzed three times against 4L of STEPL buffer at 4°C while protected from light (Slide-A-Lyzer2 cassettes, 3.5K cutoff, Thermo Scientific).
Table 2.
Synthetic Peptides
| Peptide | Molecular Weight (kDa) | λex/λem (nm) |
|---|---|---|
| NH2-Gly-Gly-Gly-Lys(HiLyteFlour 750)-NH2 | 1,327 | 754/778 |
| NH2-Gly-Gly-Gly-Lys(5-FAM)-Gly-Gly-Ser-Lys(N3)-NH2 | 1,030 | 492/518 |
EGFP Release Analysis & Model Design
Absorbance spectra obtained from optimization samples were baseline corrected and the EGFP concentration was determined using the Beer-Lambert law (ε(EGFP, 488nm) = 55,000 M−1cm−1 25. EGFP concentrations were fit to the sum of equations 4 and 5 in the following system of ODEs using non-linear least squares:
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
| (7) |
Where the model fits for A1, A2, , and . Temperatures are on the Kelvin scale. Initial EGFP·SrtA concentration was determined by adding the concentration of EGFP in the final timepoint to the concentration of EGFP from the stripped beads. Initial triglycine concentration and temperature were varied experimentally. Initial conditions for SrtA, EGFP, and EGFP·GGG were zero. Model predictions were produced with the following initial conditions: 100μM Ca2+, 100μM EGFP·SrtA, 1μM – 1mM triglycine, 4° – 37°C.
Cell Culture
NIH/3T3 and T6-17 cells (i.e. NIH/3T3 cells engineered to stably express the Her2/neu receptor; kindly provided by Dr. Mark Greene, University of Pennsylvania) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin at 37°C and 5% CO2. H1666 cells expressing pLKO.shCTRL and pLKO.shEGFR 26 (kindly provided by Dr. Matthew Lazzara, University of Pennsylvania) were maintained in RPMI supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin at 37°C and 5% CO2.
Fluorescence Analysis of Cell Targeting
NIH/3T3 and T6-17 cells were incubated with 1μM Her2/neu affibody conjugated to HiLyte Fluor 750 for 4 hours in full media with and without a 10-fold excess of unlabeled Her2/neu affibody. Cells were washed 3 times with affibody-free media before being imaged in serum-free DMEM. Imaging was performed with an Olympus IX81 inverted fluorescent microscope with a back-illuminated EMCCD camera (Andor) and a SOLA excitation source (Lumencor). Images of HiLyte Fluor 750 were acquired using the filter set (HQ710/75, HQ810/90, Q750LP) (Chroma). A LUC PLAN FLN 40X objective (NA 0.6) was used for all imaging studies. ImageJ was used to merge the fluorescent images and equalize levels. After optical imaging, the plate was scanned by an Odyssey Imaging System (Li-Cor). User-defined regions of interest were drawn within each well and fluorescence within the 800-channel was quantified to determine relative Her2/neu expression.
MR Relaxation Measurements of Cell Targeting and MR Imaging
Azodibenzocyclooctyne (ADIBO)-functionalized superparamagnetic iron oxide (SPIO) nanoparticles were synthesized as previously described 27. Azide-modified Her2/neu-targeted affibody were conjugated to ADIBO-SPIO nanoparticles by combining 5mg Fe/mL SPIO nanoparticles with 30μM affibody. Reactions were mixed overnight at room temperature and affibody-SPIO conjugates were purified on PD-10 columns (GE Healthcare).
T6-17 and NIH/3T3 cells were incubated with 125 μg Fe/mL of Her2/neu-targeted SPIO for 45 minutes in full media with and without a 100-fold excess of unlabeled Her2/neu affibody in triplicate. Cells were transferred to 1.5mL microcentrifuge tubes and washed with 500μL PBS three times before being resuspended in 300μL RIPA Lysis Buffer (Millipore). T2 measurements were taken using a benchtop relaxometer (Bruker mq60). Following relaxation measurements, cell lysates were combined 100μL transferred to wells of a 364-well plate. Images of the cells were taken on a 9.4-T magnet interfaced to a Varian INOVA console using a 70 mm inner diameter volume coil for radiofrequency transmission and reception. T2-weighted gradient echo (GRE) MR images were collected using parameters as follows: repetition time (TR) = 200 ms, echo time (TE) = 5 ms, flip angle = 20°, slice thickness = 0.5 mm, number of acquisitions = 8.
RESULTS
Optimization of STEPL
A valuable feature of the STEPL system is that it allows for the site-specific labeling of recombinantly expressed proteins without requiring any steps in addition to what is normally required for protein purification. Under optimal conditions, all of the recombinant protein that is released from the affinity column would be labeled with the desired cargo as a result of the SrtA-mediated ligation reaction. To evaluate the efficiency of this ligation reaction and to assess the extent of any non-specific cleavage of the LPXTG motif, in the absence of ligation, a model system was designed with EGFP as the “ligand” (EGFP-STEPL). This allowed for quantitative monitoring of protein release from the affinity column in the presence and absence of triglycine and calcium. Notably, peptides with two or more glycines are typically preferred for SrtA-mediated ligations since they exhibit significantly improved binding and catalysis 20.
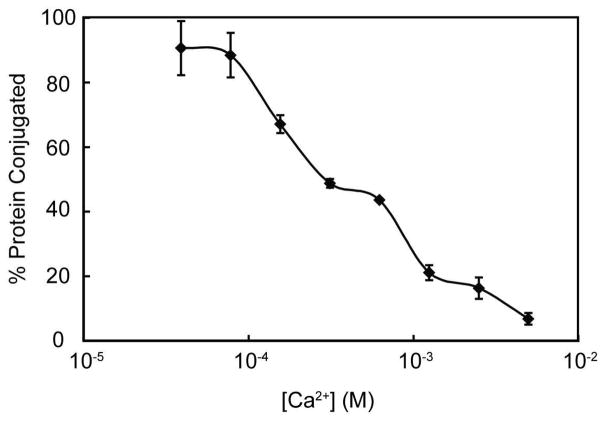
Initial studies with the EGFP-STEPL system, in the presence and absence of Ca2+ (5mM) and triglycine ([GGG], 25 μM or 100 μM), revealed that release of the GFP from the affinity column increased with triglycine concentration (Figure 2). However, it was also observed that Ca2+ alone could lead to some non-specific release of GFP, albeit at lower levels than when triglycine was also present. In the absence of Ca2+, no GFP was released from the affinity column, with or without triglycine. These results suggested that it was important to identify an optimal Ca2+ concentration that would maximize the ratio of ligated (i.e. GFP-triglycine conjugates) to unligated recombinant protein. Therefore, GFP-STEPL was performed in the presence of a fixed concentration of triglycine (25μM) and increasing concentrations of Ca2+ (0 to 5 mM). These experiments provided a measure of the total amount of ligated and unligated product released from the affinity column for each Ca2+ concentration. The amount of unligated product was determined by performing analogous experiments in the absence of triglycine. The maximum percent of ligated product occurred at Ca2+ concentrations below 100μM (Figure 3). Therefore, a Ca2+ concentration of 100μM was used for all subsequent experiments.
Figure 2.
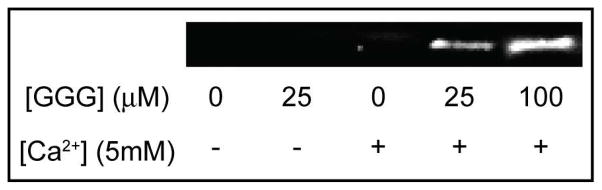
Western Blot of GFP released from an affinity column under various conditions. A GFP-STEPL fusion protein was expressed and washed on an affinity column. The column was then treated with various concentrations of triglycine and Ca2+. Released GFP was detected by western blot.
Figure 3.
Effect of calcium on the efficiency of protein ligation. A GFP-STEPL fusion protein was expressed and washed on an affinity column. The column was then treated with various concentrations of Ca2+, in the presence of triglycine, at 37°C for 4hrs. These experiments provided a measure of the total amount of ligated and unligated product ([GFP]Total) released from the affinity column for each Ca2+ concentration. The amount of unligated product ([GFP]unligated) was determined by performing analogous experiments in the absence of triglycine. The percent protein conjugated ([GFP]ligated) was then calculated calculated as ([GFP]Total − [GFP]unligated)/([GFP]Total)*100.
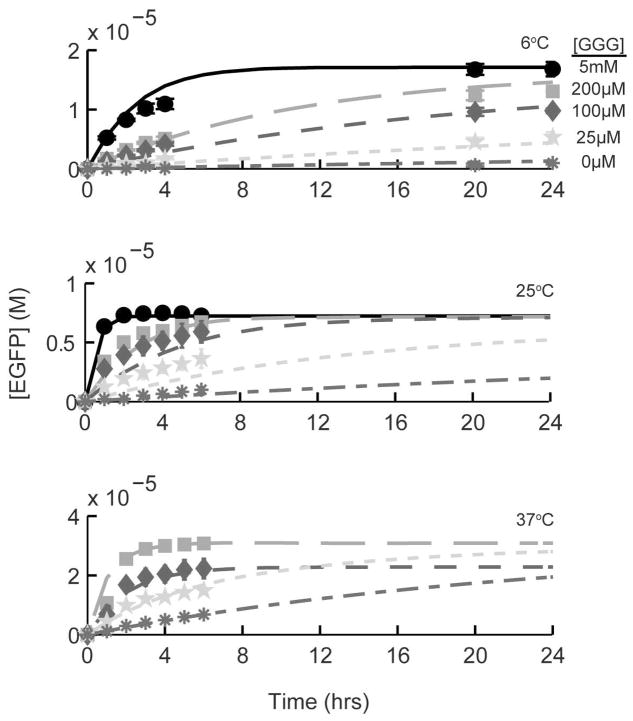
In order to further optimize the GFP-STEPL procedure, a systematic study on the effect of triglycine concentration, reaction temperature, and reaction time on the amount of recombinant protein released from the affinity column was conducted (Figure 4). As expected, the rate of protein release increased with triglycine concentration and reaction temperature. All experimental data was fit with a kinetic model of the reaction, detailed in Scheme 1. This model assumes that transpeptidation is the rate-limiting step of the glycine-dependent pathway and therefore collapses reversible peptide binding and transpeptidation into a single second-order rate constant. To include temperature dependence, the model also assumes that the rate constants can be modeled by the Arrhenius equation. Thus, the model’s parameters are the preexponential constants and activation energies of the two pathways (Table 1). As shown in Figure 4, the model provides an acceptable fit to the observed data.
Figure 4.
Modeled and actual EGFP release from an affinity column as a function of temperature, triglycine concentration, and time. A GFP-STEPL fusion protein was expressed and washed on an affinity column. The column was then treated with 0mM (asterisk), 25mM (star), 100mM (diamond), 200mM (square), or 5mM (circle) triglycine and 100mM Ca2+. GFP release was monitored as a function of time. Protein release experiments were conducted at 6°C (top), 25°C (middle), and 37°C (bottom). All data was fit using a kinetic model of EGFP cleavage that takes into account both triglycine-dependent and triglycine-independent pathways. Modeled GFP release (lines) has been superimposed onto the recorded data (symbols).
Scheme 1.
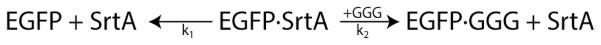
Kinetic model of EGFP cleavage via triglycine-dependent and triglycine-independent pathways.
Table 1.
Model Parameters. The kinetics model determined Ahrenneous pre-exponential constants and activation energies for the glycine-free (k1) and glycine-dependent pathways (k2).
| Pathway | Pre-exponential Constant (A) | Activation Energy (ΔG‡) | Rate Constant at 37°C |
|---|---|---|---|
| Glycine-free | 4.0568 × 102 s−1 | 3.8463 × 1010 J/mol | 0.0419 s−1 |
| Glycine-dependent | 2.9246 × 1010 s−1 M−1 | 5.4958 × 104 J/mol | 5.0352 × 103 s−1 M−1 |
The kinetic model was used to predict the effect of various reaction conditions on three outcomes: (i) the percentage of released protein that is ligated to the peptide, (ii) the percentage of peptide consumed in the reaction, and (iii) the yield of ligated protein (i.e. the amount of ligated protein normalized by the total amount of protein initially bound to the affinity column) (Figure 5). The value for each of the desired outcomes was determined for reaction times of 2, 4, 6, and 24 h, with 100μM CaCl2, initial triglycine peptide concentrations of 1 to 1000 μM, reaction temperatures of 4, 25, or 37°C, and assuming 100μM initial protein concentration on the affinity column. It was determined that the purity of the ligated protein was independent of reaction time, as it is simply a ratio of the two rate constants. As a result, adding excess peptide could be used to drive the ligation reaction and overwhelm the basal cleavage rate. Overall, if a highly pure ligated product is desirable, >95% purity can be achieved by simply using 2-fold or greater molar excess of triglycine-containing peptide compared with the concentration of total column-bound protein (at 37°C, Figure 5C). This is significantly lower than the 10-fold excess of peptide typically required for efficient intein-based EPL 14.
Figure 5.
Model predictions of STEPL-ligation efficiency, triglycine peptide utilization, and the percent yield of expressed GFP that is recovered from the affinity column. The kinetic model of GFP cleavage from the STEPL system was evaluated with initial conditions of 100μM Ca2+, 100μM GFP-SrtA, and 1μM to 1mM triglycine at 4°, 25°, and 37°C for 0 to 24hrs. The percentage of GFP that has been ligated to triglycine (time independent) and the percentage of triglycine peptide consumed during a 6 hr incubation was determined for reaction temperatures of (A) 4°C, (B) 25°C, and (C) 37°C. Dotted lines at 90% and 95% are included for reference. The percentage of ligated GFP recovered after a 2, 4, 6, and 24 hour incubation was determined as a function of excess triglycine (relative to the total amount of GFP-SrtA) for reaction temperatures of (D) 4°C, (E) 25°C, and (F) 37°C.
If it is more desirable to exhaust all of the triglycine peptide than achieve high purity of the ligated product, perhaps because the peptide is functionalized with a cargo that is cost-prohibitive, then >90% peptide consumption can be achieved by adopting reaction conditions whereby the recombinantly expressed protein is in 4-fold or greater molar excess over the peptide (at 37°C, Figure 5C). However, this comes at the cost of reduced purity of the ligated product and will likely require additional purification to remove unligated targeting ligands.
Higher reaction temperatures can be used to speed up the reaction and improve peptide utilization, particularly at lower triglycine peptide concentrations, but purity of the ligated product is only marginally improved. When yield is considered, the STEPL system clearly favors higher temperatures regardless of whether high purity of the ligated protein or high peptide utilization is desirable. (Figure 5D–F). Of course, some proteins may be unstable at high temperatures, requiring longer reactions to be performed at room temperature or in a cold room.
Fluorophore Ligation
To demonstrate the utility of STEPL as a general methodology for the site-specific labeling of tumor targeting ligands with imaging agents, the coding sequence for a Her2/neu-targeting affibody (Her2-affibody) was cloned into the STEPL vector. The affibody was expressed and conjugated to a triglycine peptide containing the near-infrared dye HiLyte Fluor™ 750 (Table 2) using conditions that were expected to result in >95% purity of the fluorescent labeled affibody, based on the kinetic model established above (2-fold excess peptide, 100μM Ca2+, 37°C, 6hr). Efficient ligation between the affibody and the fluorescently-labeled peptide was confirmed by SDS-PAGE (Figure 6). The major band in the 700nm channel (protein stain) co-localized with the single fluorescent band in the 800nm channel (HiLyte Fluor™ 750), following removal of excess free peptide by dialysis. Only a very faint signal stemming from the unligated protein was observed in the 700 nm channel, at a slightly lower molecular weight than the ligated product. To confirm that the affibody remained functional following the ligation reaction, it was incubated with T6-17 and NIH-3T3 cells in vitro, which are positive and negative for the Her2/neu receptor, respectively (Figure 7). As expected, the affibody labeled the T6-17 cells exclusively, with no observable labeling of the NIH-3T3 cells. Further, the addition of excess unlabeled affibody (i.e. cleaved with triglycine) competitively inhibited the binding of the fluorescently labeled affibody to T6-17 cells, suggesting that binding was specific for the Her2/neu receptor. Quantification using an in-cell western assay (Figure 7B) corroborated the fluorescence microscopy findings. Similar results were obtained by applying the epidermal growth factor receptor (EGFR)-targeted affibody to EGFR-positive and negative cells (Supplementary Figure S1).
Figure 6.
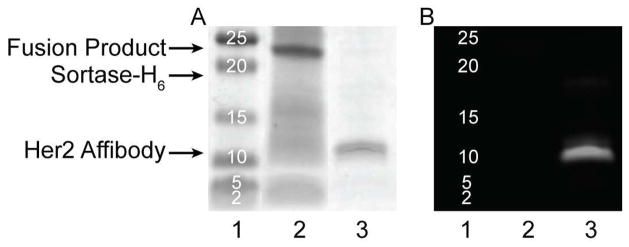
Her2/neu affibody expression and ligation. An SDS-PAGE gel was run with (1) marker, (2) raw lysate of bacterially expressed STEPL-Her2 affibody, and (3) Her2 affibody purified using a 2-fold excess of HiLyte 750-labeled triglycine peptide, 100μM Ca2+ at 37°C for 6hr. (A) SimplyBlue SafeStain protein stain. (B) HiLyte 750 peptide fluorescence.
Figure 7.
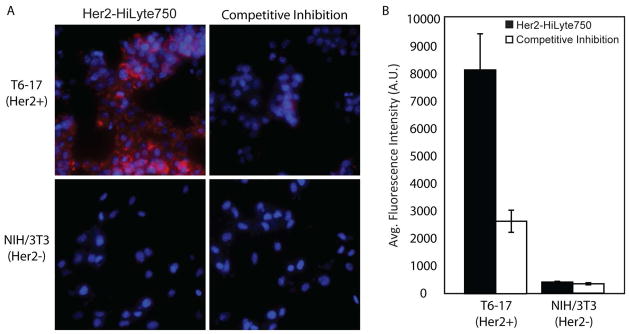
Functional evaluation of the Her2 affibody-HiLyte Fluor™ 750 conjugate. (A) Her2/neu positive and negative cells were incubated with Her2/neu-targeted affibodies that were conjugated to HiLyte Fluor™ 750 (red) using the STEPL system. Cells were also stained with Hoechst 33342 (nuclear stain, blue). (B) In-cell western quantification of HiLyte Fluor™ 750 fluorescence.
Azide Ligation and Nanoparticle Synthesis
In addition to imaging agents, STEPL can also be used to conjugate various other functional moieties including bio-orthogonal reactive groups (e.g. azide) onto the C-terminus of targeting ligands. The site-specific introduction of azides onto recombinant proteins provides a very favorable approach for the efficient coupling of targeting ligands to nanoparticles using click chemistry. In particular, this approach allows tight control over both ligand orientation and density on the nanoparticle surface. We have previously shown that both of these factors can have a dramatic impact on nanoparticle avidity 27. As proof-of-principle, the Her2/neu affibody was conjugated to a synthetic peptide containing a green fluorophore (5-FAM) as well as an azide group. This conjugate was then reacted with superparamagnetic iron oxide (SPIO) nanoparticles functionalized with azadibenzocyclooctyne (ADIBO). ADIBO is a dibenzocyclooctyne derivative capable of copper-free click reactions with azides. The resulting Her2/neu affibody-SPIO conjugates were incubated with T6-17 and NIH/3T3 cells. Cell labeling was then assessed by acquiring T2 relaxation times and T2*-weighted images of cell lysates (Figure 8). The Her2-positive cells exhibited a marked decrease in T2-relaxation times, consistent with the presence of SPIO, in comparison to Her2-negative cells. An observable negative contrast was also observed upon MR imaging of the Her2-positive cells. Competitive inhibition, using an excess of free-unlabeled Her2-affibody, led to a loss in MR contrast, indicative of receptor-specific binding. Therefore, these results provide clear evidence that STEPL can be combined with click chemistry for the site-specific attachment of targeting ligands onto nanoparticles.
Figure 8.
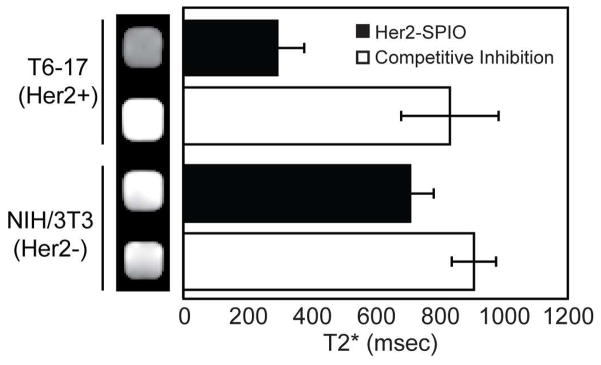
Functional evaluation of the Her2 affibody-SPIO conjugates. Her2/neu-positive and Her2/neu-negative cells were incubated with Her2-SPIO conjugates in the presence and absence of excess free affibody. Free affibody served as a competitive inhibitor to confirm specific binding of the Her2/neu receptor. Relaxivity measurements and T2*-weighted MR images of each cell suspension were acquired.
DISCUSSION
STEPL offers a number of features that make it a very favorable approach for bioconjugation reactions. First and foremost, STEPL combines release of recombinant proteins from the affinity column and bioconjugation into a single step. This greatly simplifies the entire bioconjugation procedure since no subsequent labeling and purification steps are required, saving time, money, and complexity. Second, STEPL allows for the site-specific conjugation of cargo. Site-specific functionalization has been shown to be beneficial in a number of applications including the preparation of protein-drug conjugates, which often exhibit higher efficacy than randomly labeled targeting ligands 28. It has also been shown that the site-specific attachment of targeting ligands to nanoparticles can improve nanoparticle avidity 14. Third, STEPL conjugates the peptide-to-ligand in a 1:1 stoichiometric manner. This can be important when labeling targeting ligands with imaging agents, since it allows for precise quantitative imaging. It is also beneficial for characterizing nanoparticle bioconjugations. Fourth, the conditions used to release protein from the affinity column can be manipulated to ensure that essentially all of the recovered protein is conjugated with the desired cargo. This eliminates the often-difficult process of purifying conjugated products from unconjugated proteins. Since in many applications a large protein is labeled with low molecular weight drugs or imaging agents, the conjugated and unconjugated forms of the protein can differ by as little as a few hundred to a few thousand Da, potentially without any significant change to hydrophobicity or charge. A slight excess of peptide is required to achieve complete ligation; however, excess peptide is easily removed via dialysis or gel chromatography. This purification step is analogous to the removal of imidazole from His-tagged protein samples that have been affinity purified using a nickel column. Fifth, construction of the STEPL system as a single expressible protein removes the additional step of removing sortase from the conjugated product, a common feature of current sortase conjugation systems 17, 29, 30. Additionally, in systems where unconjugated ligands are easily separable from conjugated ligands, the reaction conditions could be altered to ensure that expensive synthetic peptides can be exhausted in the ligation reaction. Thus, STEPL is a single-chain, self-cleavable system where high-cost components can be fully utilized; traits highly desirable in industrial protein production as they reduce overall cost and time 31.
One identified shortcoming of Sortase A is that it exhibits some cleavage even in the absence of glycine. Previous studies have addressed this problem by making a destabilizing mutation to Trp-194. However, we hypothesized that reducing the calcium concentration would have a similar effect with finer control. This was found to be true, as sub-millimolar calcium concentrations provided a sharp, dose dependent drop in background cleavage. Due to the presence of cytosolic calcium in the bacterium, there is a possibility that background cleavage occurs during protein expression; however, the calcium level inside E. coli is estimated to be between 0.1 – 1 μM 32. Therefore, undesirable cleavage of the fusion protein within E. coli is likely to be minimal and could always be further discouraged by introducing the aforementioned mutation. The size of the sortase domain is also not expected to have much effect on the yield of the fusion protein. Sortase is only 147 amino acids, less than half the size of a maltose binding domain (male), which is commonly used for affinity purification, and sortase is highly soluble.
To further optimize and understand the cleavage reaction, a kinetic model was established and its parameters (time, temperature, and initial triglycine concentration) were systematically varied. To simplify the model, the reversible binding of peptide to the enzyme and product conversion were condensed into one second-order rate constant. This is justifiable because the applicable peptide concentrations do not appear to saturate the binding curve and the determined rate constants are well below the diffusion limit 33, 34, implying that product conversion is rate limiting.
The model reveals a fundamental conflict between conjugate purity and peptide utilization. The glycine-independent pathway can be easily overwhelmed by adding a large excess of peptide to drive the glycine-dependent pathway. On the other hand, the peptide can be fully utilized by making it the limiting reactant. Therefore, if product purity is required, it is optimal to use excess peptide. If peptide utilization is the primary concern, the optimal conditions are 37°C with a 1:1 ratio (or less) of synthetic peptide to STEPL protein, although it is important to note that in this latter case additional purification is needed to purify conjugated product from unconjugated proteins. Therefore, this approach is only amenable to systems where the conjugated and unconjugated products are separable. Systems where the peptide enables the conjugate to be immobilized onto a surface or particle are ideal candidates for peptide exhaustion.
In this study, STEPL was used to functionalize affibodies with chemical groups useful for molecular imaging. A near-IR fluorophore was utilized to optically differentiate between cells expressing and lacking the proto-oncogene Her2/neu. The NIR-dyed affibody was used to quantify Her2/neu expression differences between the T6-17 and NIH/3T3 cells, which demonstrates STEPL’s utility for in-cell western techniques. Additionally, the STEPL was used to conjugate a bio-orthogonal reactive group (an azide) to the Her2/neu affibody. The azide readily reacted to a strained alkyne on the surface of superparamagnetic iron oxide nanoparticles. Due to the site-specific nature of STEPL, the affibody was linked in a specific orientation, which greatly increases the particle’s efficacy in distinguishing between cells expressing and lacking Her2/neu. STEPL has the potential to conjugate many other moieties to its target protein, such as biotin, poly(ethylene-glycol), antibiotics, metal chelates, and photocrosslinkers, all of which have been proven compatible with the sortase enzyme 17.
CONCLUSION
STEPL has proven to be a flexible and efficient system for molecular imaging and targeted therapeutics. This study validated and optimized the system for ligand purity and peptide-cargo utilization. STEPL was then used to visualize and quantify Her2/neu and EGFR expression in vitro. Moreover, because it has the ability to link virtually any bacterially expressible protein with any cargo that can be attached to a triglycine peptide, STEPL has potential applications in many fields.
Supplementary Material
Acknowledgments
This work was supported in part by the National Institute of Health (NIH) NIBIB/R01-EB012065, NCI/R01-CA157766, and NIBIB/R21-EB013226.
Footnotes
SUPPORTING INFORMATION AVAILABLE. Functional evaluation of the EGFR affibody-FAM conjugate (Figure S1).
References
- 1.Rana S, Yeh YC, Rotello VM. Curr Opin Chem Biol. 2010;14:828–834. doi: 10.1016/j.cbpa.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sapsford KE, Algar WR, Berti L, Gemmill KB, Casey BJ, Oh E, Stewart MH, Medintz IL. Chem Rev. 2013;113:1904–2074. doi: 10.1021/cr300143v. [DOI] [PubMed] [Google Scholar]
- 3.Stephanopoulos N, Francis MB. Nat Chem Biol. 2011;7:876–884. doi: 10.1038/nchembio.720. [DOI] [PubMed] [Google Scholar]
- 4.Thorek DL, Elias DR, Tsourkas A. Mol Imaging. 2009;8:221–229. [PubMed] [Google Scholar]
- 5.Stephanopoulos N, Francis MB. Nat Chem Biol. 2011;7:876–884. doi: 10.1038/nchembio.720. [DOI] [PubMed] [Google Scholar]
- 6.Fernandez-Suarez M, Baruah H, Martinez-Hernandez L, Xie KT, Baskin JM, Bertozzi CR, Ting AY. Nat Biotechnol. 2007;25:1483–1487. doi: 10.1038/nbt1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slavoff SA, Chen I, Choi YA, Ting AY. J Am Chem Soc. 2008;130:1160–1162. doi: 10.1021/ja076655i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K. Nat Biotechnol. 2003;21:86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- 9.Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohana RF, Urh M, Simpson D, Mendez J, Zimmerman K, Otto P, Vidugiris G, Zhu J, Darzins A, Klaubert DH, Bulleit RF, Wood KV. ACS Chem Biol. 2008;3:373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 10.Gautier A, Juillerat A, Heinis C, Correa IR, Jr, Kindermann M, Beaufils F, Johnsson K. Chem Biol. 2008;15:128–136. doi: 10.1016/j.chembiol.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 11.Meier JL, Mercer AC, Rivera H, Jr, Burkart MD. J Am Chem Soc. 2006;128:12174–12184. doi: 10.1021/ja063217n. [DOI] [PubMed] [Google Scholar]
- 12.Chong S, Mersha FB, Comb DG, Scott ME, Landry D, Vence LM, Perler FB, Benner J, Kucera RB, Hirvonen Ca, Pelletier JJ, Paulus H, Xu MQ. Gene. 1997;192:271–281. doi: 10.1016/s0378-1119(97)00105-4. [DOI] [PubMed] [Google Scholar]
- 13.Dawson PE, Kent SB. Annu Rev Biochem. 2000;69:923–960. doi: 10.1146/annurev.biochem.69.1.923. [DOI] [PubMed] [Google Scholar]
- 14.Elias DR, Cheng Z, Tsourkas A. Small. 2010;6:2460–2468. doi: 10.1002/smll.201001095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erathodiyil N, Ying JY. Acc Chem Res. 2011;44:925–935. doi: 10.1021/ar2000327. [DOI] [PubMed] [Google Scholar]
- 16.Leung MKM, Hagemeyer CE, Johnston APR, Gonzales C, Kamphuis MMJ, Ardipradja K, Such GK, Peter K, Caruso F. Angew Chem, Int Ed. 2012;51:7132–7136. doi: 10.1002/anie.201203612. [DOI] [PubMed] [Google Scholar]
- 17.Proft T. Biotechnol Lett. 2010;32:1–10. doi: 10.1007/s10529-009-0116-0. [DOI] [PubMed] [Google Scholar]
- 18.Spirig T, Weiner EM, Clubb RT. Mol Microbiol. 2011;82:1044–1059. doi: 10.1111/j.1365-2958.2011.07887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Race PR, Bentley ML, Melvin JA, Crow A, Hughes RK, Smith WD, Sessions RB, Kehoe MA, McCafferty DG, Banfield MJ. J Biol Chem. 2009;284:6924–6933. doi: 10.1074/jbc.M805406200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mao H. Protein Expres Purif. 2004;37:253–263. doi: 10.1016/j.pep.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 21.Kobashigawa Y, Kumeta H, Ogura K, Inagaki F. J Biomol NMR. 2009;43:145–150. doi: 10.1007/s10858-008-9296-5. [DOI] [PubMed] [Google Scholar]
- 22.Orlova A, Magnusson M, Eriksson TL, Nilsson M, Larsson B, Hoiden-Guthenberg I, Widstrom C, Carlsson J, Tolmachev V, Stahl S, Nilsson FY. Cancer Res. 2006;66:4339–4348. doi: 10.1158/0008-5472.CAN-05-3521. [DOI] [PubMed] [Google Scholar]
- 23.Qi S, Miao Z, Liu H, Xu Y, Feng Y, Cheng Z. Bioconjug Chem. 2012;23:1149–1156. doi: 10.1021/bc200596a. [DOI] [PubMed] [Google Scholar]
- 24.Zong Y, Bice TW, Ton-That H, Schneewind O, Narayana SV. J Biol Chem. 2004;279:31383–31389. doi: 10.1074/jbc.M401374200. [DOI] [PubMed] [Google Scholar]
- 25.McRae SR, Brown CL, Bushell GR. Protein Expres Purif. 2005;41:121–127. doi: 10.1016/j.pep.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 26.Offterdinger M, Georget V, Girod A, Bastiaens PI. J Biol Chem. 2004;279:36972–36981. doi: 10.1074/jbc.M405830200. [DOI] [PubMed] [Google Scholar]
- 27.Elias DR, Poloukhtine A, Popik V, Tsourkas A. Nanomed-Nanotechnol. 2013;9:194–201. doi: 10.1016/j.nano.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Junutula JR, Raab H, Clark S, Bhakta S, Leipold DD, Weir S, Chen Y, Simpson M, Tsai SP, Dennis MS, Lu Y, Meng YG, Ng C, Yang J, Lee CC, Duenas E, Gorrell J, Katta V, Kim A, McDorman K, Flagella K, Venook R, Ross S, Spencer SD, Lee Wong W, Lowman HB, Vandlen R, Sliwkowski MX, Scheller RH, Polakis P, Mallet W. Nat Biotechnol. 2008;26:925–932. doi: 10.1038/nbt.1480. [DOI] [PubMed] [Google Scholar]
- 29.Bellucci JJ, Amiram M, Bhattacharyya J, McCafferty D, Chilkoti A. Angew Chem, Int Ed. 2013:1–7. doi: 10.1002/anie.201208292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steinhagen M, Zunker K, Nordsieck K, Beck-Sickinger AG. Bioorg Med Chem. 2013;21:3504–3510. doi: 10.1016/j.bmc.2013.03.039. [DOI] [PubMed] [Google Scholar]
- 31.Fong Ba, Wu W-Y, Wood DW. Trends Biotechnol. 2010;28:272–279. doi: 10.1016/j.tibtech.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 32.Norris V, Grant S, Freestone P, Canvin J, Sheikh FN, Toth I, Trinei M, Modha K, Norman RI. J Bacteriol. 1996;178:3677–3682. doi: 10.1128/jb.178.13.3677-3682.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alberty R, Hammes G. J Phys Chem. 1958;62:154–159. [Google Scholar]
- 34.Zhou GQ, Zhong WZ. Eur J Biochem. 1982;128:383–387. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.