Abstract
Secretins, a superfamily of multimeric outer membrane proteins, mediate the transport of large macromolecules across the outer membrane of Gram-negative bacteria. Limited proteolysis of secretin PulD from the Klebsiella oxytoca pullulanase secretion pathway showed that it consists of an N-terminal domain and a protease-resistant C-terminal domain that remains multimeric after proteolysis. The stable C-terminal domain starts just before the region in PulD that is highly conserved in the secretin superfamily and apparently lacks the region at the C-terminal end to which the secretin-specific pilot protein PulS binds. Electron microscopy showed that the stable fragment produced by proteolysis is composed of two stacked rings that encircle a central channel and that it lacks the peripheral radial spokes that are seen in the native complex. Moreover, the electron microscopic images suggest that the N-terminal domain folds back into the large cavity of the channel that is formed by the C-terminal domain of the native complex, thereby occluding the channel, consistent with previous electrophysiological studies showing that the channel is normally closed.
Keywords: protein secretion/PulD/pullulanase/secretins
Introduction
Gram-negative bacteria possess several different pathways to secrete proteins into the extracellular medium. In the type I and type III secretion systems, secreted proteins are transported in one step across both the inner and outer membranes. In the secreton or type II secretion pathway, substrates are transported first across the cytoplasmic membrane by the Sec system, and then across the outer membrane via a specialized secretion apparatus (the secreton) composed of or assembled by 12 or more different proteins (Pugsley, 1993; Possot et al., 2000). One of the most interesting features of the secreton pathway is that it contains only one integral outer membrane protein, protein D, which is consequently the only protein that could conceivably form the translocation channel by which proteins are transported across the outer membrane (Nouwen et al., 1999).
Protein D belongs to a large superfamily of homologous proteins, the secretins (Genin and Boucher, 1994). Besides the D proteins, this family includes components of the type III secretion pathway, proteins involved in DNA uptake (natural competence) and proteins needed for the assembly and secretion of filamentous phages, type IV pili and S-layers (Genin and Boucher, 1994). Secretins show particularly high sequence similarity in their C-terminal halves. This region (called the β domain in Guilvout et al., 1999) is predicted to contain several amphipathic β-strands similar to the transmembrane segments of porins and, consequently, is likely to be embedded in the outer membrane (Bitter et al., 1998; Guilvout et al., 1999). The N-terminal half of secretins is conserved only in proteins from related secretion pathways (Genin and Boucher, 1994) and is presumed to face the periplasm (Guilvout et al., 1999). This domain might be involved in substrate recognition (Shevchik et al., 1997), interaction with other components of the secretion machinery (Feng et al., 1999) or gating of the proposed secretin channel (Guilvout et al., 1999). In addition, some secretins contain a small C-terminal domain to which a secretin-specific pilot protein binds (Daefler et al., 1997; Daefler and Russel, 1998). This pilot protein protects secretins from proteolysis and is essential for their insertion in the outer membrane (Hardie et al., 1996a,b; Daefler and Russel, 1998; Shevchik and Condemine, 1998).
To investigate whether secretins do indeed contain different structural domains, as suggested by sequence comparisons, we analysed the structure of secretin biochemically. As a representative of the secretin family, we studied the domain structure of the 633 amino acid PulD protein, the secretin of the pullulanase-specific secreton of Klebsiella oxytoca (d’Enfert et al., 1989; Nouwen et al., 1999).
Results
Limited proteolysis of PulD
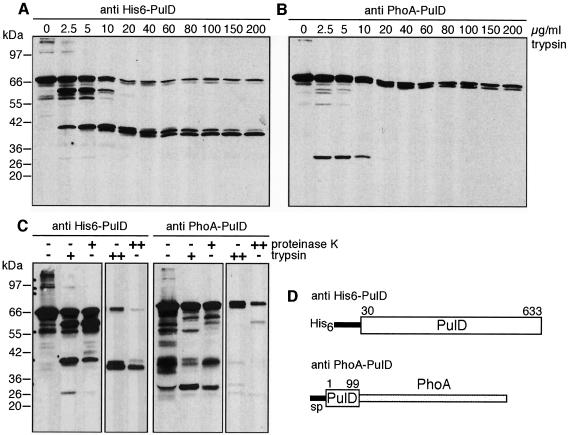
On the basis of sequence comparisons, secretins are presumed to contain different structural domains. To investigate whether there is biochemical evidence for this presumed domain structure, we treated outer membranes containing secretin PulD with limited amounts of trypsin. PulD forms very stable high molecular weight complexes in vivo that are not dissociated by prolonged heating at 100°C in SDS. To facilitate the analysis, samples were treated with phenol, which converts PulD into monomers, before loading onto an SDS–polyacrylamide gel. Immunoblot analysis using an antibody raised against almost complete PulD (His6-PulD) (Hardie et al., 1996a) (Figure 1D) showed that low concentrations of trypsin gave rise to proteolytic fragments of ∼60 and ∼40 kDa (Figure 1A). These proteolytic fragments did not react with an antibody raised against the 99 N-terminal amino acids of mature PulD (PulD-PhoA) (d’Enfert et al., 1987) (Figure 1B), indicating that they do not contain the extreme N-terminal region of PulD. Instead, a proteolytic fragment of ∼28 kDa was detected with this antibody (Figure 1B). Interestingly, the sum of the sizes of the 40 and 28 kDa fragments is close to that of full-length PulD. At higher trypsin concentrations, the 60 and 28 kDa fragments were completely degraded whereas the 40 kDa fragment was trimmed to a stable fragment of ∼38 kDa (Figure 1A and B). The latter proteolysis product will be referred to hereafter as the stable C-terminal fragment.
Fig. 1. Release of fragments from PulD by limited proteolysis with trypsin. (A and B) Outer membranes were incubated for 15 min with the indicated amounts of trypsin on ice. After phenol treatment to dissociate the multimers, proteins were precipitated with acetone and resuspended in SDS–PAGE sample buffer. Samples were analysed by SDS–PAGE (10% acrylamide) and immunoblotting using anti-His6-PulD (almost complete PulD) or anti-PulD-PhoA (N-terminal 99 amino acids of mature PulD. (C) Outer membranes were incubated with 5 µg/ml (+) or 100 µg/ml (++) trypsin or proteinase K. After 15 min on ice, samples were treated and analysed as described above. (D) Schematic representation of proteins used to raise antibodies against different regions of PulD. sp, PulD signal peptide.
To determine whether the different proteolytic fragments were generated by cleavage at unique trypsin cleavage sites or at sites that are highly accessible to proteases, the experiments were repeated with proteinase K. At both low and high proteinase K concentrations, exactly the same proteolytic fragments were generated as with trypsin (Figure 1C). Therefore, the proteolytic fragments are likely to correspond to structural domains in PulD.
Both N- and C-terminal fragments are integrated into the outer membrane
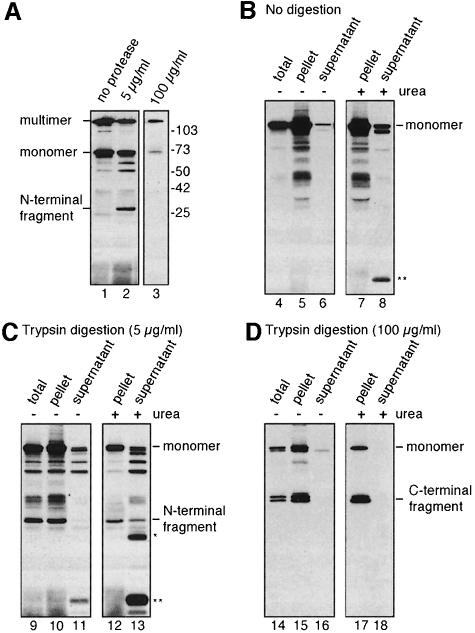
Without phenol treatment, PulD migrates as a large multimeric complex on SDS–PAGE (Hardie et al., 1996a). However, the N-terminal 28 kDa fragment migrated as a monomer on SDS–PAGE, indicating that, once excised from the stable multimer, it does not retain its multimeric state in SDS (Figure 2A). In contrast, the stable C-terminal fragment remained multimeric in SDS (Figure 2A, lane 3; see below). To analyse whether the N- and stable C-terminal fragments remained associated with the outer membrane, outer membranes were treated with 5 M urea and then centrifuged to separate extracted proteins from membranes. The C-terminal fragment could not be extracted with urea, indicating that it is integrated firmly into the outer membrane (Figure 2D). Urea treatment seemed to extract ∼20% of the N-terminal fragment from the membrane, whereas without this treatment it remained entirely in the membrane fraction. However, the urea extract also contained novel proteolytic fragments (* and ** in Figure 2B and C) that were much more abundant than the 28 kDa fragment. Their appearance might have been due to denaturation of the protein and degradation by endogenous proteases during sample preparation. Thus, the band at 28 kDa might not be derived from membrane-associated N-terminal fragment. The fact that the majority of the N-terminal fragment was not extracted by urea indicates that this domain of PulD, like the C-terminal domain, is integrated firmly into the outer membrane.
Fig. 2. Multimeric state and susceptibility to urea extraction of proteolytic fragments derived from outer membrane-associated PulD. (A) Outer membranes were treated with the indicated amount of trypsin and analysed without phenol treatment by SDS–PAGE (11% acrylamide) and immunoblotting with antibodies against PulD-PhoA (lanes 1 and 2) or His6-PulD (lane 3). Note that in lane 3, the multimer composed of proteolysed monomers co-migrates with multimers composed of full-length monomer and that 40 kDa monomers are not detected. (B–D) Outer membranes were treated with the indicated amount of trypsin. After proteolysis, the membranes were extracted with 5 M urea and/or pelleted. Pellet and supernatant fractions were treated with phenol to dissociate PulD multimers and analysed by SDS–PAGE and immunoblotting using antibodies raised against PulD-PhoA (lanes 1 and 2, and 4–13) or His6-PulD (lanes 3 and 14–18).
N-terminal sequencing of the stable proteolytic fragment
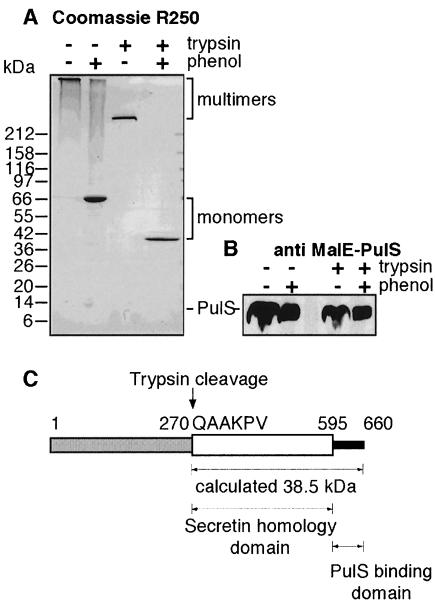
The trypsin cleavage site that generates the stable C-terminal fragment could not be determined by proteolysis of outer membranes because of the low amount of PulD present and because the 38 kDa band was contaminated by other proteins that co-migrated upon SDS–PAGE. Therefore, we generated sufficient stable C-terminal fragment for sequencing by treating purified PulD complex with immobilized trypsin. Proteolytic fragments were separated on a 4–20% acrylamide gel. Proteolysis led to the formation of a multimer of ∼500 kDa that could be dissociated with phenol to give a monomer of ∼38 kDa (Figure 3A). Since this 38 kDa fragment was multimeric, did not react with antibodies against the N-terminal region of PulD and was exactly the same size as the stable fragment obtained after proteolysis of outer membranes (data not shown), we conclude that they are identical. The determined N-terminal sequence of the fragment was QAAKPV, indicating that cleavage had occurred before amino acid 270 in the mature sequence of PulD. Interestingly, this site is just before the start of the region that is highly conserved in all secretins.
Fig. 3. Proteolysis of purified PulD–PulS complex. (A) Purified, detergent-solubilized PulD–PulS complex was proteolysed using trypsin-coated agarose beads. Stable proteolytic fragments were separated by SDS–PAGE (4–20% acrylamide) and then stained with Coomassie Brilliant Blue. (B) Samples from (A) were separated by SDS–PAGE (12% acrylamide), electroblotted on nitrocellulose and analysed for the presence of PulS using antibodies raised against MalE–PulS (only that part of the blot displaying PulS is shown). (C) Schematic representation of the mature part of PulD protein showing the N-terminal domain (shaded), secretin homology (β) domain (white) and PulS-binding domain (black). The trypsin cleavage site and the calculated molecular size of the resulting fragment are also indicated.
Stable C-terminal fragment lacks the PulS-binding domain
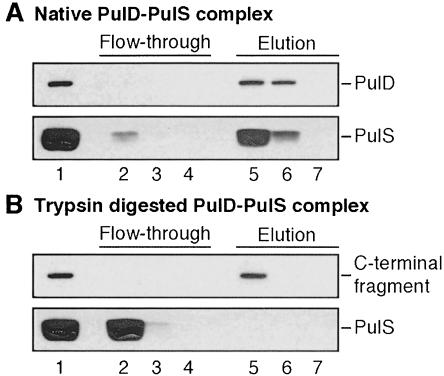
PulS binds to the 65 C-terminal amino acids of PulD and protects it from degradation. Proteolysis of the complex with trypsin does not change the size of PulS (Figure 3B). To investigate whether PulS was still associated with the C-terminal fragment, we loaded the proteolysed complex on an anion-exchange column. Normally, PulS co-elutes with PulD on such a column (Figure 4A). However, after proteolysis, PulS did not bind to the column whereas the stable C-terminal fragment did (Figure 4B). Since trypsin does not degrade PulS (see above), this result indicates that the C-terminal fragment lacks the PulS-binding domain (i.e. the extreme C-terminus of PulD).
Fig. 4. The stable C-terminal fragment lacks the PulS-binding domain. Native or trypsin-proteolysed PulD–PulS complex was loaded on a miniQ™ column. Flow-through fractions (lanes 2–4) and fractions eluted with NaCl (lanes 5–7) were treated with phenol to dissociate PulD multimers and then analysed by SDS–PAGE and immunoblotting. (A) Native PulD–PulS complex. (B) Trypsin-proteolysed PulD–PulS complex.
Electron microscopic analysis of the stable C-terminal fragment
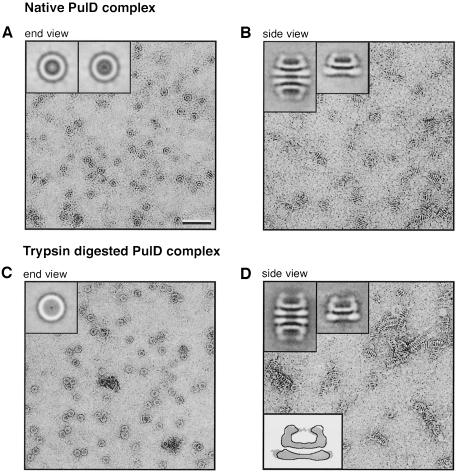
Recent electron microscopic analysis of negatively stained samples of purified PulD–PulS complex showed that it forms ring-like structures with 12 peripheral radial spokes, giving a total radius of 20–21 nm (Nouwen et al., 1999). These previous analyses did not resolve structures inside the ring. The new data presented here clearly show that there is protein structure in the centre of the ring of the PulD complex. Principal components analysis of 1276 images revealed two major classes of 621 and 576 images whose averages are presented in the insets of Figure 5A. Both average images show the ring-like structure of 14 nm diameter surrounded by the 12 peripheral radial spokes. The more abundant class average (left inset) reveals a plug in the centre of the complex, while the second average (right inset) shows a smaller, ring-like structure with a 2.6 nm diameter central hole.
Fig. 5. Electron microscopic analysis of native and trypsin-digested PulD complex. A sample of purified native or trypsin-proteolysed PulD complex was applied to air- or pentylamine-glow-discharged thin carbon-coated grids. (A) Uranyl formate-stained images of native PulD complex on air-glow-discharged grids; insets, averaged images of an end-on view of two major classes (n = 621 and 576). (B) Uranyl formate-stained images of native PulD complex on pentylamine-glow-discharged grids; insets, averaged image of double multimeric (n = 150) and single multimeric (n = 217) side views of native PulD complex. (C) Uranyl formate-stained images of proteolysed PulD complex on air-glow-discharged grids; inset, averaged image of an end view (n = 276). (D) Uranyl formate-stained images of proteolysed PulD complex on pentylamine-glow-discharged grids; top insets, averaged images of double complex (n = 150) and single complex (n = 217) side views of proteolysed PulD complex; bottom inset, comparison of undigested (grey shading) and trypsin-digested (outline) secretin complexes. The number of particles (n) used for constructing the averaged image is indicated in parentheses. The scale bar corresponds to 50 nm and the inset baseline corresponds to 25 nm.
Only end views of PulD–PulS complexes were clearly distinguished when air-glow-discharged carbon films were used, preventing the analysis of side views. However, to our surprise, when carbon films were glow discharged in the presence of pentylamine, the most abundant structures seen were side views of single or stacked multimers (Figure 5B). These side views suggest that the PulD complex consists of two ring-shaped structures that are 3.5 nm apart. The lower ring is 1.5 nm thick at its narrowest, central point in the side views, and 20 nm in diameter, consistent with the outer diameter of the ring, including spokes, in the end views. The upper ring is thicker (2.3 nm) than the lower ring and has a diameter of 14 nm. This upper ring appears to form a cavity via protrusions that fold back towards the centre of the ring, thereby closing the cavity that it forms. Many of the stacked multimers were doublets that resembled rugby balls and consisted of two PulD complexes associated via their lower rings.
Proteolysis of the complex removes PulS and >40% of mature PulD. Thus, the structure of the remaining complex should be drastically different from that of the original complex. Examination of negatively stained samples of the purified stable C-terminal fragment revealed that the peripheral spokes and the structure in the centre of the end views were essentially absent (Figure 5C). The end views displayed ring-shaped structures filled with homogeneously distributed material and a small central cavity of ∼1.6 nm.
The purified C-terminal fragment had a strong tendency to form long ‘ladder-like’ structures on pentylamine-glow-discharged carbon grids. In these structures, single complexes associated not only via their lower rings but also via their upper rings. This observation clearly indicates that proteolysis changed the rim of the upper ring. Averages of both single multimeric and double multimeric side views exhibit major changes in both rings (Figure 5D, bottom inset). The lower ring is narrower (16.7 nm compared with 20 nm in the native complex), consistent with the removal of the spokes seen in the end views. The lateral projections on the upper ring are longer (5.3 nm compared with 4.5 nm in the native complex). Moreover, faint protrusions emanating from the rim of the extensions towards the centre appeared to be shorter and less well ordered in the proteolysed sample, consistent with the small pore seen in the end views.
Discussion
PulD is a member of the superfamily of secretins, outer membrane proteins that mediate the transport of macromolecules across the outer membrane of many Gram-negative bacteria. On the basis of sequence comparisons, secretins were proposed to consist of two main domains (Genin and Boucher, 1994; Guilvout et al., 1999). The C-terminal domain (also called the β domain) is highly conserved in all members of the superfamily and is presumably embedded in the outer membrane. The N-terminal domain is thought to extend into the periplasm and to interact with other components of the secreton. In addition, some secretins contain a small C-terminal domain to which a secretin-specific pilot protein binds (in the case of PulD, this is the lipoprotein PulS). Our biochemical analyses of secretin PulD are largely consistent with such a domain organization. Limited proteolysis of outer membranes containing PulD led to the formation of an N-terminal 28 kDa fragment that did not retain the SDS-resistant multimeric organization typical of native PulD on SDS–PAGE. Under the same conditions, the C-terminal fragments generated all retained the stable multimeric state of PulD. This observation is in line with those of previous studies showing that peptide insertions in the C-terminal (β) domain of PulD had a dramatic effect on the formation and stability of multimers. A fragment of PulD corresponding to the stable C-terminal region of PulD, as defined here, but with its PulS-binding site intact and associated with PulS did not form multimers in vivo (Guilvout et al., 1999). However, multimers were observed when an N-terminal fragment was produced in trans. These multimers contained only the C-terminal region of PulD (the N-terminal region either did not associate with the C-terminal region or dissociated from it in SDS) and contained <12 copies of the PulD fragment (Guilvout et al., 1999). These results show unambiguously that the N-terminal region does not remain oligomeric in SDS when removed from the dodecameric C-terminal region of PulD but is required for multimerization.
Interestingly, the 28 kDa fragment could not be extracted with 5 M urea, indicating that it is firmly integrated into the outer membrane. High amounts of protease degraded the N-terminal 28 kDa fragment, but multimers of an ∼38 kDa C-terminal fragment appeared to be very resistant to proteolysis, as would be expected for a domain of PulD that is embedded in the outer membrane. Using purified, detergent-solubilized PulD–PulS complex, we demonstrated that the fragment corresponds to a C-terminal segment of mature PulD starting at amino acid position 270 and lacking the PulS-binding domain as a result of a second, undetermined proteolysis event close to the C-terminus of PulD. Thus, the stable proteolytic fragment corresponds almost exactly to the region (the β domain) that is highly conserved in the secretin superfamily.
We recently analysed the structure of negatively stained native PulD–PulS complex by electron microscopy (Nouwen et al., 1999). In these studies, as in other studies of different secretins (Koster et al., 1997; Bitter et al., 1998; Crago and Koronakis, 1998), we only obtained clear images of the end view of the complex. Here we present an averaged image of the side views of the PulD–PulS complex. It consists of a 20 nm diameter, 2.3 nm thick ring or disc that supports a smaller cup-like structure that is 14 nm in diameter and 8.5 nm high. We hypothesize that the broader of the two rings is integrated into the outer membrane. This hypothesis is supported by the following: (i) the broader ring has a strong tendency to self-associate to form doublets, suggesting that it is very hydrophobic; (ii) the ring is ∼2.3 nm thick at its outer limits, which is sufficient to span the outer membrane; and (iii) proteolysis of the complex removes the region to which the outer membrane-associated lipoprotein PulS binds, coincident with a narrowing of the ring. In this model, the majority of the complex would protrude so far into the periplasm that the ends of the top rim of the cup would almost touch the cytoplasmic membrane, and could therefore come into contact with other secreton components that are located in this membrane. In addition, folded proteins would not have to pass the peptidoglycan layer to reach the translocation channel since they might be captured within the funnel of the channel close to the surface of the cytoplasmic membrane. However, the secretin channel must itself penetrate through the peptidoglycan layer. Even the bulk of the β domain might not be fully integrated into the membrane, since less than half of the amino acids in this domain are predicted to be in potential transmembrane β-strands (Guilvout et al., 1999). The loops between most of these predicted β-strands are longer than in other outer membrane proteins that have been analysed (Guilvout et al., 1999) and could extend into the medium as well as into the periplasm.
In the averaged images of the side views of both the single and double multimers, the ends of the rim of the cup seem to fold back into the cavity of the channel. This structure is likely to be part of the N-terminal domain of PulD that forms the ‘plug’ in the centre of the ring that is seen in approximately half of the end views. In agreement with this idea, the ‘plug’ is totally absent in all of the end views of the stable C-terminal fragment. Moreover, side views of this C-terminal fragment show that, in contrast to the native complex, the upper rims of the cup have a strong tendency to self-associate. This indicates that drastic changes have occurred in this region of the protein as a result of proteolysis by trypsin. It is worth noting, however, that almost half of the end views of the native complex appear to have an open centre (right hand inset in Figure 5A). This observation is difficult to interpret at present but one possibility is that some of the native secretin particles examined were in a partially open configuration when examined end-on.
The ‘plug’ that could be formed by the N-terminal domain of PulD might be analogous to a protein domain that folds backwards into the channels formed by the outer membrane ferrisiderophore transporters FhuA and FepA (Ferguson et al., 1998; Locher et al., 1998; Buchanan et al., 1999). In these transporters, the N-terminal ‘plug’ or ‘cork’ domain controls channel opening and subsequent transport of the ferrisiderophore. However, complete displacement of the central plug would be necessary to allow proteins to transit the secretin channel, whereas removal of the plug is not necessary to create a channel sufficiently large to permit siderophore transport through FepA and FhuA. In previous studies, we showed that the PulD secretin complex exhibited only very low level electrical conductance when incorporated into planar lipid bilayers (Nouwen et al., 1999). Thus, we concluded that these secretin complexes were in the uniformly closed configuration, which we now propose corresponds to the structure with the plug in the centre of the barrel or cup-like structure. Therefore, one would predict that the C-terminal fragment should be in the open configuration and would have very high conductance when incorporated into lipid bilayers. This was not found to be the case, however, apparently because it is not possible to incorporate the stable C-terminal fragment into lipid bilayers (N.Nouwen and A.Ghazi, unpublished observations). We are currently exploring other ways to manipulate the opening and closure of the secretin channel.
Secretins have highly conserved C-terminal regions, thus, other secretins probably have a similar structure to that of the C-terminal fragment of PulD. Indeed, both end and side views of this C-terminal fragment resemble images of secretin pIV of filamentous phage f1 (Linderoth et al., 1997). The major difference between PulD and pIV is that the side views of single complexes of pIV have only one ring (Linderoth et al., 1997). However, pIV might have a second ring that is not visible on the images, since Linderoth et al. (1997) observed that the contour length of the side views of the pIV double multimer was much greater than twice that of the single multimer. The lower disc and the upper cup very probably correspond to a 12-fold symmetrical structure that encircles the central channel. Recently reported studies demonstrated that a protease-resistant, membrane-associated C-terminal fragment similar to that which we report here can be obtained by proteolysis of the secretin XcpQ, a PulD homologue from Pseudomonas aeruginosa (Brok et al., 1999). Like the stable C-terminal fragment of PulD, this domain of XcpQ retained its oligomeric structure (Brok et al., 1999). Thus, the intrinsically multimeric C-terminal, ring-shaped β domain is probably a feature inherent to all secretins
Stacked rings are also seen in electron microscopic images of the flagellar basal body (L- and P-ring) (Macnab, 1996) and, most interestingly, of the complete structure of two different type III protein secretion systems (Kubori et al., 1998; Blocker et al., 1999). In the latter case, it would be very interesting to see whether these stacked rings correspond to the secretin component.
Materials and methods
Bacterial strain and growth conditions
The Escherichia coli K-12 strain NN001 (lamB ompR::Tn10) carrying plasmid pCHAP231, encoding PulD and all other secreton proteins needed for pullulanase secretion (d’Enfert et al., 1987), was used for isolation of outer membranes and purification of the PulD–PulS complex. Cells were grown at 30°C in Luria–Bertani medium (Miller, 1992) buffered with 2% M63 medium (Miller, 1992) and containing 100 µg/ml ampicillin. The medium was supplemented with 0.4% maltose to induce the pul operon.
SDS–PAGE and immunoblotting
Proteins were separated by SDS–PAGE in 10, 11 or 4–20% acrylamide gels, stained with Coomassie Brilliant Blue or transferred to nitrocellulose by semi-dry electroblotting. Immunoblots were incubated with the antibodies indicated and then with horseradish peroxidase-coupled anti-rabbit immunoglobulin G (IgG). Immunoblots were developed by enhanced chemiluminescence (ECL kit; Amersham). The primary antibodies used were a 1:4000 dilution of PulD–PhoA (d’Enfert et al., 1989), a 1:200 dilution of an affinity-purified antibody raised against His6-PulD (which lacks the first 30 amino acids of mature PulD) (Hardie et al., 1996a) and a 1:10 000 dilution of MalE–PulS (Hardie et al., 1996b).
Limited proteolysis
Outer membranes were prepared by breaking bacteria in a French press, removing unbroken cells by centrifugation at 3000 g for 15 min, and pelleting the outer membrane by centrifugation for 1 min at 165 000 g. Outer membranes were washed with 50 mM Tris pH 8.0, 1 mM EDTA, pelleted, and resuspended in 50 mM Tris pH 8.0 at a protein concentration of 50 mg/ml. Proteolysis was initiated by addition of the indicated amount of trypsin or proteinase K to a solution containing 2 mg/ml protein. After 15 min on ice, the protease was inactivated by addition of Pefabloc™ (Interchim; final concentration 100 µg/ml). Unless otherwise indicated, PulD multimers were dissociated by phenol treatment (Hardie et al., 1996a), precipitated overnight by addition of 2 vols of acetone at –20°C and dissolved in SDS–PAGE sample buffer.
To test the membrane association of the proteolytic fragments, samples were extracted with 5 M urea and then centrifuged at 100 000 g for 15 min. Membrane and supernatant fractions were treated with phenol to dissociate multimeric PulD as above and analysed for the presence of PulD by SDS–PAGE and immunoblotting.
Protein sequence determination
Purified PulD–PulS complex (1 mg/ml) was proteolysed using trypsin-coated agarose beads (Sigma; 1:5 ratio of bead slurry to protein solution). After 3 h on ice, the agarose beads were removed by centrifugation and the clarified mixture was treated with phenol to dissociate the multimeric complex. Proteins were precipitated by addition of 2 vols of acetone, resuspended in SDS–PAGE sample buffer and heated to 100°C for 5 min. After electrophoresis, proteins were electroblotted onto Immobilon P membranes (Millipore) and the proteolytic fragment was sequenced using an Applied Biosystems 473A protein sequencer according to the standard procedure of the manufacturer.
Protein purification and analysis
PulD–PulS complex was purified as described (Nouwen et al., 1999) with the modification that PulD–PulS fractions from the size exclusion column that contained the complex were pooled and concentrated by chromatography on a miniQ™ column on a Smart™-system (Pharmacia).
Incubating purified PulD–PulS complex with trypsin-coated agarose beads (see above) generated the stable proteolytic fragment. After proteolysis, the agarose beads were removed by centrifugation and the clarified mixture was loaded on a miniQ™ column using a Smart™-system. After loading and washing of the column, the proteolytic fragment was eluted with 20 mM Bis-Tris-propane pH 7.0, 0.3% Zwittergen 3–14 (Fluka), 625 mM NaCl. Fractions were analysed by SDS–PAGE and immunoblotting for the presence of the proteolysed fragment and PulS. Fractions containing the C-terminal fragment were pooled and stored at –70°C.
Electron microscopy
PulD–PulS complex and purified C-terminal proteolytic fragment in 20 mM Bis-Tris-propane pH 7.0, 0.3% SB3-14, 625 mM NaCl were adsorbed for 1 min to air- or pentylamine-glow-discharged thin carbon films mounted on a thick fenestrated carbon layer on copper grids. After three washes with distilled water, the sample was stained with uranyl formate pH 4.2 for 10 s, blotted and air dried. Images of negatively stained samples were recorded on a Hitachi H7000 transmission electron microscope at 100 kV, with a nominal magnification of 50 000× at 500 electrons/nm2. Negatives were digitized by using a Leafscan 45 scanner (Leaf Systems, Westborough, MA) at 0.2 nm/pixel on the specimen plane. The SPIDER program (Frank et al., 1996) was used to select individual particles as 128 × 128 pixel (end views) and 256 × 256 pixel (side views) subframes. End views were reference-free aligned (Penczek et al., 1992), classified and the clearly visible 12-fold rotational symmetry applied. Side views were aligned using one particle as an initial reference and then averaged.
Acknowledgments
Acknowledgements
We are grateful to all members of the secretion laboratory of the Institut Pasteur for their interest and encouragement and to J.D’Alayer of the Institut Pasteur Microsequencing Laboratory for protein sequence analysis. N.N. thanks all members of the Maurice E.Müller Institut for their hospitality and help during his stay in Basel. This work was supported by the European Union (Training and Mobility in Research grant number FMRX-CT96-0004) and by a French Research Ministry grant in the Programme fondamental en Microbiologie et Maladies infectieuses et parasitaires and by the Maurice E.Müller foundation of Switzerland.
References
- Bitter W., Koster,M., Latijnhouwers,M., de Cock,H. and Tommassen,J. (1998) Formation of oligomeric rings by XcpQ and PilQ, which are involved in protein transport across the outer membrane of Pseudomonas aeruginosa. Mol. Microbiol., 27, 209–219. [DOI] [PubMed] [Google Scholar]
- Blocker A., Gounon,P., Larquet,E., Niebuhr,K., Cabiaux,V., Parsot,C. and Sansonetti,C. (1999) The tripartite type III secreton of Shigella flexneri inserts IpaB and IpaC into host membranes. J. Cell Biol., 147, 683–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brok R., Van gelder,P., Winterhalter,M., Ziese,U., Koster,A.J., de Cock,H., Koster,M., Tommassen,J. and Bitter,W. (1999) The C-terminal domain of the Pseudomonas secretin XcpQ forms oligomeric rings with pore activity. J. Mol. Biol., 294, 1169–1179. [DOI] [PubMed] [Google Scholar]
- Buchanan S.K., Smith,B.S., Venkatramani,L., Xia,D., Esser,L., Palnitkar,M., Chakraborty,R., van der Helm,D. and Deisenhofer,J. (1999) Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nature Struct. Biol., 6, 56–63. [DOI] [PubMed] [Google Scholar]
- Crago A.M. and Koronakis,V. (1998) Salmonella InvG forms a ring-like multimer that requires the InvH lipoprotein for outer membrane localization. Mol. Microbiol., 30, 47–56. [DOI] [PubMed] [Google Scholar]
- Daefler S. and Russel,M. (1998) The Salmonella typhimurium InvH protein is an outer membrane lipoprotein required for the proper localization of InvG. Mol. Microbiol., 28, 1367–1380. [DOI] [PubMed] [Google Scholar]
- Daefler S., Guilvout,I., Hardie,K.R., Pugsley,A.P. and Russel,M. (1997) The C-terminal domain of the secretin PulD contains the binding site for its cognate chaperone, PulS and confers PulS dependence on pIVf1 function. Mol. Microbiol., 24, 465–475. [DOI] [PubMed] [Google Scholar]
- d’Enfert C., Ryter,A. and Pugsley,A.P. (1987) Cloning and expression in Escherichia coli of the Klebsiella pneumoniae genes for production, surface localization and secretion of the lipoprotein pullulanase. EMBO J., 6, 3531–3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Enfert C., Reyss,I., Wandersman,C. and Pugsley,A.P. (1989) Protein secretion by Gram-negative bacteria: characterization of two membrane proteins required for pullulanase secretion by Escherichia coli K-12. J. Biol. Chem., 264, 17462–17468. [PubMed] [Google Scholar]
- Feng J.N., Model,P. and Russel,M. (1999) A trans-envelope protein complex needed for filamentous phage assembly and export. Mol. Microbiol., 34, 745–755. [DOI] [PubMed] [Google Scholar]
- Ferguson A.D., Hofmann,E., Coulton,J.W., Diederichs,K. and Welte,W. (1998) Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide. Science, 282, 2215–2220. [DOI] [PubMed] [Google Scholar]
- Frank J., Radermacher,M., Penczek,P., Zhu,J., Li,Y., Ladjadj,M. and Leith,A. (1996) SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol., 116, 190–199. [DOI] [PubMed] [Google Scholar]
- Genin S. and Boucher,C.A. (1994) A superfamily of proteins involved in different secretion pathways in Gram-negative bacteria: modular structure and specificity of the N-terminal domain. Mol. Gen. Genet., 243, 112–118. [DOI] [PubMed] [Google Scholar]
- Guilvout I., Hardie,K.R., Sauvonnet,N. and Pugsley,A.P. (1999) Genetic dissection of the outer membrane secretin PulD: are there distinct domains for multimerization and secretion specificity? J. Bacteriol., 181, 7212–7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie K.R., Lory,S. and Pugsley,A.P. (1996a) Insertion of an outer membrane protein in Escherichia coli requires a chaperone-like protein. EMBO J., 15, 978–988. [PMC free article] [PubMed] [Google Scholar]
- Hardie K.R., Seydel,A., Guilvout,I. and Pugsley,A.P. (1996b) The secretin-specific, chaperone-like protein of the general secretory pathway: separation of proteolytic protection and piloting functions. Mol. Microbiol., 22, 967–976. [DOI] [PubMed] [Google Scholar]
- Koster M., Bitter,W., de Cock,H., Allaoui,A., Cornelis,G. and Tommassen,J. (1997) The outer membrane component, YscC, of the Yop secretion machinery of Yersinia enterocolitica forms a ring-shaped multimeric complex. Mol. Microbiol., 26, 789–797. [DOI] [PubMed] [Google Scholar]
- Kubori T., Matsushima,Y., Nakamura,D., Uralil,J., Lara-Tejero,M., Sukhan,A., Galan,J.E. and Aizawa,S.-I. (1998) Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science, 280, 602–605. [DOI] [PubMed] [Google Scholar]
- Linderoth N.A., Simon,M.N. and Russel,M. (1997) The filamentous phage pIV multimer visualized by scanning transmission electron microscopy. Science, 278, 1635–1638. [DOI] [PubMed] [Google Scholar]
- Locher K.P., Rees,B., Koebnik,R., Mitschler,A., Moulinier,L., Rosenbusch,J.P. and Moras,D. (1998) Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes. Cell, 95, 771–778. [DOI] [PubMed] [Google Scholar]
- Macnab R.M. (1996) Flagella and motility. In Neidhardt,F.C. (ed.), Escherichia coli and Salmonella. ASM Press, Washington, DC, pp. 123–145. [Google Scholar]
- Miller J.H. (1992) A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Nouwen N., Ranson,N., Saibil,H., Wolpensinger,B., Engel,A., Ghazi,A. and Pugsley,A.P. (1999) Secretin PulD: association with pilot protein PulS, structure and ion-conducting channel formation. Proc. Natl Acad. Sci. USA, 96, 8173–8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penczek P., Radermacher,M. and Frank,J. (1992) Three-dimensional reconstitution of single particles embedded in ice. Ultramicroscopy, 40, 33–53. [PubMed] [Google Scholar]
- Possot O., Vignon,G., Bomchil,N., Ebel,F. and Pugsley,A.P. (2000) Multiple interactions between pullulanase secreton components involved in stabilization and cytoplasmic membrane association of PulE. J. Bacteriol., 182, 2142–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugsley A.P. (1993) The complete general secretory pathway in Gram-negative bacteria. Microbiol. Rev., 57, 50–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchik V.E. and Condemine,G. (1998) Functional characterization of the Erwinia chrysanthemi OutS protein, an element of a type II secretion system. Microbiology, 144, 3219–3228. [DOI] [PubMed] [Google Scholar]
- Shevchik V.E., Robert-Badouy,J. and Condemine,G. (1997) Specific interaction between OutD, an Erwinia chrysanthemi outer membrane protein of the general secretory pathway and secreted proteins. EMBO J., 16, 3007–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]