Abstract
The hrp genes of the plant pathogen Ralstonia solanacearum are key pathogenicity determinants; they encode a type III protein secretion machinery involved in the secretion of mediators of the bacterium–plant interaction. These hrp genes are under the genetic control of the hrpB regulatory gene, expression of which is induced when bacteria are co-cultivated with plant cell suspensions. In this study, we used hrp–gfp transcriptional fusions to demonstrate that the expression of the hrpB and type III secretion genes is specifically induced in response to the bacterium–plant cell contact. This contact-dependent induction of hrpB gene expression requires the outer membrane protein PrhA, but not a functional type III secretion apparatus. Genetic evidence indicates that PrhA constitutes the first example of a bacterial receptor for a non-diffusible signal present in the plant cell wall and which triggers the transcriptional activation of bacterial virulence genes.
Keywords: Arabidopsis thaliana/green fluorescent protein/hrp genes/signal transduction/type III secretion pathway
Introduction
Many species of soil bacteria form specialized symbiotic and parasitic interactions with plant cells. To initiate an interaction, a bacterium must first recognize its appropriate host plant cell. This recognition can be used by the bacterium to activate bacterial genes whose products mediate the development of the interaction. For >10 years, it has been known that several small diffusible plant molecules play a major role in these recognition processes by inducing a variety of genes in plant-associated bacteria. This was first observed for Agrobacterium tumefaciens, which, in wounded tissues, senses a structurally diverse group of plant phenolics, including acetosyringone (Stachel et al., 1985). This perception of phenolic signal molecules leads to transcriptional activation of virulence (vir) genes necessary for the transfer of T-DNA to host cells. Certain phenolic β-glycosides also serve as signals that induce phytotoxin production by Pseudomonas syringae pv. syringae (Mo et al., 1995), while in rhizobia, specific plant flavonoids activate nodulation genes essential for the establishment of symbiosis (Peters et al., 1986). Despite these well-documented examples, little is known about how the process of bacterium–plant cell recognition occurs in many other interactions, especially for the other main groups of Gram-negative phytopathogenic bacteria.
With the exception of Agrobacterium, Gram-negative plant pathogenic bacteria possess key pathogenicity determinants called hrp genes. These hrp genes are required both for causing disease symptoms on host plants and for eliciting a defence reaction, called the hypersensitive response, on resistant or non-host plants. The molecular characterization of hrp gene clusters has revealed that several hrp genes are homologous to pathogenicity determinants of animal pathogens, which encode a type III secretion machinery (reviewed in Van Gijsegem et al., 1993; Hueck, 1998; Galan and Collmer, 1999). In animal pathogens, type III secretion pathways enable the translocation of several effector proteins directly into eukaryotic cells, where they exert their anti-host functions (see Cornelis, 1998; Galan and Collmer, 1999). In plant pathogens, multiple reports suggest that the products of avr genes encode effector proteins, which are injected into host plant cells by the Hrp type III secretion machinery. This hypothesis is supported by the fact that avr gene products are active when they are expressed, either stably or transiently, within plant cells (reviewed in Galan and Collmer, 1999). Bacterial type III secretion is thought to proceed through the formation of a pilus-like structure connecting the pathogen and host cell. The structural components of this pilus are encoded by hrp genes, hrpA for P.syringae (Roine et al., 1997) and hrpY for Ralstonia solanacearum (Van Gijsegem et al., 2000). Several other proteins have been shown to be secreted in the extracellular medium in an Hrp-dependent manner: harpins from Erwinia amylovora, P.syringae and R.solanacearum (Wei et al., 1992; He et al., 1993; Arlat et al., 1994; Charkowski et al., 1998; Kim and Beer, 1998) and a set of Avr/Dsp proteins from different plant pathogenic bacteria (Gaudriault et al., 1997; Bogdanove et al., 1998; Mudgett and Staskawicz, 1999; Rossier et al., 1999; van Dijk et al., 1999; Guéneron et al., 2000). However, the function of these diverse proteins and their contributions to pathogenicity are still poorly understood.
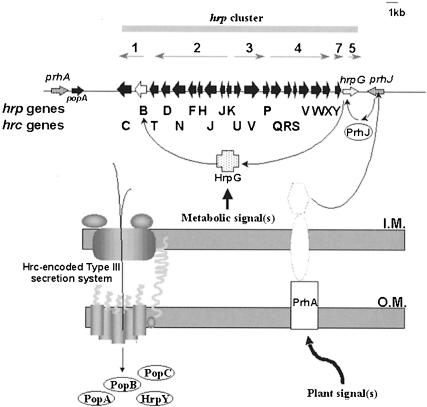
The hrp genes are induced in plants, but no plant inducers of hrp gene expression have been characterized so far. hrp genes are not expressed when bacteria are grown in rich media; rather, they are most strongly expressed in various minimal media that mimic plant apoplastic fluids (for references, see Van Gijsegem, 1997), and in several cases the level of induction found in plants is comparable to that observed in the synthetic medium. However, recent results in our laboratory provide evidence that specific plant factor(s) induce(s) R.solanacearum hrp gene expression. Upon co-culture with Arabidopsis thaliana or tomato cell suspensions, the expression of the regulatory hrpB gene is induced up to 20-fold more than in minimal medium (Marenda et al., 1998). This specific plant cell induction of hrp genes is controlled by PrhA, a protein that shows homology to outer membrane siderophore receptors (Marenda et al., 1998). PrhA was proposed to act as a receptor for a plant-derived signal and appears to activate a specific plant-dependent pathway controlling the induction of R.solanacearum hrp genes (Figure 1). Two additional components of this regulatory cascade, HrpG and PrhJ, have recently been characterized (Brito et al., 1999).
Fig. 1. Model of regulation of the R.solanacearum Hrp type III secretion system in response to plant signal(s). Above, genetic organization of the hrp gene cluster, with grey arrows showing the orientation and length of the hrp transcriptional units. A new nomenclature has distinguished hrp and hrc (for hrp-conserved) genes, the latter are homologues of type III secretion genes of animal pathogens and are predicted to encode components of the type III secretion apparatus. Four proteins, the harpin PopA, PopB, PopC and HrpY have been described to be secreted by the Hrp pathway into the extracellular medium. HrpY is the structural component of the Hrp pilus, an essential extracellular structure involved in PopA secretion. The outer membrane protein PrhA controls the plant-responsive regulatory cascade composed of PrhJ, HrpG and HrpB, the final activator of hrp transcription units 1–4 and 7. Two additional regulatory components (dotted lines) have recently been identified (B.Brito and S.Genin, unpublished). The HrpG protein integrates another hrp-inducing signal pathway, which is dependent on the nutrient/metabolic status of the bacterium. This model is based on data compiled from Arlat et al. (1994), Marenda et al. (1998), Brito et al. (1999), Guéneron et al. (2000) and Van Gijsegem et al. (2000).
We developed a green fluorescent protein (GFP) reporter gene system to monitor the in situ expression of bacterial genes upon co-cultivation of R.solanacearum with plant cells. In this paper, we show that the strong induction of hrp genes detected in the presence of plant cells specifically requires a physical contact between the bacterium and its target cell. We demonstrate that this plant cell contact-dependent induction of hrp genes is abolished in a prhA mutant strain, indicating that PrhA is the first example of a bacterial receptor for a non-diffusible plant cell wall molecule that induces the expression of pathogenicity genes.
Results
Construction of hrp–gfp transcriptional fusions
We assayed the functional expression of the gfp gene in R.solanacearum using a mini-transposon located on the suicide delivery plasmid pAG408 (Suarez et al., 1997), in order to generate several chromosomal fusions. One of them, GMI1600, appeared to be highly and constitutively expressed; the corresponding strain was subsequently used as a positive GFP control strain for studying bacterium–plant cell interactions. We used polymerase chain reaction (PCR) cloning techniques to construct a transcriptional fusion between a 250 bp DNA fragment containing the functional promoter of the hrpB regulatory gene (Genin et al., 1992) and the gfp gene from pAG408 (Suarez et al., 1997) (see Materials and methods). This hybrid hrpB–gfp gene fusion was expressed in R.solanacearum on the low copy number plasmid pLAFR6 (Huynh et al., 1989) between two synthetic trp terminator sequences to ensure that the expression of the cloned fragment was driven from its own promoter region. The resulting plasmid, pSG261, was transferred by conjugation into the wild-type strain GMI1000 and various hrp/prh mutants (Table I). The recombinant GMI1000/pSG261 strain gave slightly fluorescent colonies on minimal medium plates after incubation at 28°C, but only background activity was detected after growth on rich medium; this is the expression pattern expected for the hrpB gene (Genin et al., 1992). Similarly, another GFP reporter fusion was generated by PCR amplification with the hrpY gene promoter and introduced into R.solanacearum on a pLAFR6 plasmid.
Table I. Bacterial strains and plasmids.
| Designation | Relevant characteristics | Reference/source |
|---|---|---|
| R.solanacearum strains | ||
| GMI1000 | wild-type strain | Boucher et al. (1985) |
| GMI1410 | hrpY mutant | Van Gijsegem et al. (1995) |
| GMI1462 | hrcC mutant | Gough et al. (1992) |
| GMI1475 | hrpB-Tn5B20 mutant | Genin et al. (1992) |
| GMI1525 | hrpB mutant | Genin et al. (1992) |
| GMI1575 | prhA mutant | Marenda et al. (1998) |
| GMI1578 | hrpG mutant | Brito et al. (1999) |
| GMI1579 | prhJ mutant | Brito et al. (1999) |
| GMI1600 | GMI1000 with constitutive GFP expression | this study |
| GMI1601 | GMI1600 carrying a prhA mutation | this study |
| Plasmids | ||
| pAG408 | suicide plasmid containing a promotorless gfp reporter gene on a mini-transposon | Suarez et al. (1997) |
| pLAFR6 | pLAFR1 with trp terminators, carries tetracycline resistance gene | Huynh et al. (1989) |
| pSG261 | pLAFR6 carrying hrpB–gfp transcriptional fusion | this study |
| pSG282 | pLAFR6 carrying hrpY–gfp transcriptional fusion | this study |
Expression of the regulatory hrpB gene and type III secretion genes is specifically induced in response to bacterium–plant cell contact
Expression of the regulatory hrpB gene is strongly induced (15- to 20-fold) when R.solanacearum are co-cultured with A.thaliana or tomato plant cell suspensions (Marenda et al., 1998; Brito et al., 1999). The observation that conditioned medium (i.e. medium in which plant cells have been grown and then removed by filtration) is not able to induce the transcription of hrpB (Marenda et al., 1998) indicates that this induction either specifically requires plant cells or involves a very labile diffusible plant signal.
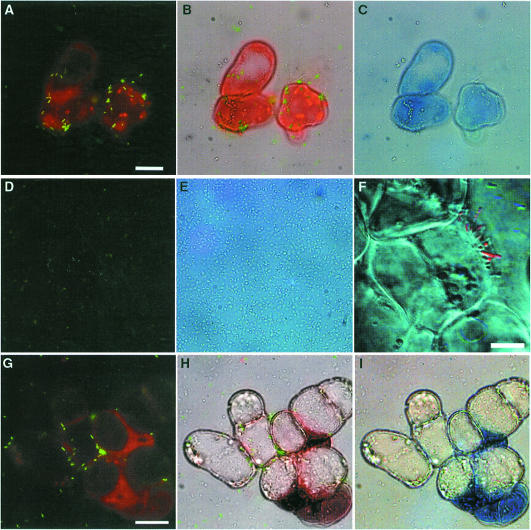
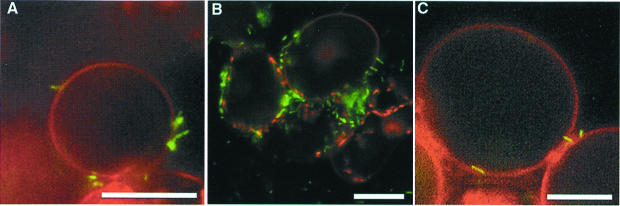
By cytological GFP fluorescence detection, we used a GMI1000/pSG261 hrpB–gfp transcriptional fusion to monitor the expression of the hrpB gene when bacteria were co-cultivated with Arabidopsis cells. Plasmid retention studies indicated that the population of bacteria present in the co-culture showed no net loss of plasmid-borne tetracycline resistance during the experiments (data not shown). Because the autofluorescence of plant cell wall material at 395 nm (the wavelength at which GFP absorbs) interferes with the observation of bacterium–plant cell interaction we used the dye Evans blue, which binds to plant cells and causes a shift in the fluorescence emission of plant cell material, which thus appears red. This method allowed easy detection of gene activation in individual bacteria co-cultivated with host plant cells. Strong fluorescence was detected when bacteria were physically associated with Arabidopsis cells (Figure 2A), whereas only a weak signal (comparable to that detected after growth in conditioned medium, Figure 2D) was observed from bacteria present in the medium, but not in close contact with plant cells. Confocal laser scanning microscopy was used to confirm that the strongly fluorescent bacteria were attached to the plant cells, often in a polar fashion (Figure 2F). Under the experimental conditions used, distinctly fluorescent bacteria in contact with plant cells were first detected after 90 min of co-cultivation. Measurements of the β-galactosidase activity of strain 1475 (carrying a chromosomal hrpB–lacZ fusion) confirmed that after 4 h the activity found in bacteria attached to plant cells was 6-fold higher than that observed in bacteria recovered from the culture medium (data not shown).
Fig. 2. Induction of hrp gene expression in response to bacterium–plant cell contact. Interaction of R.solanacearum and A.thaliana cells observed by epifluorescence microscopy (A–E and G–I) or by confocal microscopy (F) after 16 h of co-cultivation. The fluorescence and phase contrast images have been overlaid in (B) and (H) [(A) + (C) and (G) + (I), respectively] in order to co-localize induced bacteria and plant cell surfaces. Images were acquired with the same parameters to allow comparisons between the emitted fluorescence level in the different frames. The bar represents 20 µm, except in F (10 µm). (A–C) Co-cultivation of GMI1000/pSG261 with Arabidopsis cells. Strongly induced bacteria expressing the hrpB–gfp fusion are found in contact with plant cells; some uninduced bacteria present in the medium are indicated by arrowheads (B). The few strongly induced bacteria observed in the medium are assumed to have previously been in close association with plant cells. (D and E) Basal level of hrpB–gfp expression obtained when strain GMI100/pSG261 is grown in Arabidopsis-conditioned medium. (F) Detail of the interaction between strain GMI1000/pSG261 and Arabidopsis cell surface observed by confocal microscopy. The bacteria in contact with the plant cell surface exhibit a high level of GFP fluorescence (red colour); non-induced bacteria are visible at the upper right. (G–I) Co-cultivation of strain GMI1000/pSG282 (hrpY–gfp) with Arabidopsis cells, showing that the hrpY gene follows the same expression pattern as hrpB.
To determine whether the expression of type III secretion genes followed the same expression pattern, we monitored expression of an hrpY–gfp transcriptional fusion in a wild-type strain. The hrpY gene was recently shown to encode a component of an essential extracellular pilus structure of the secretion apparatus, required for the secretion of PopA (Van Gijsegem et al., 2000). We observed that this hrpY–gfp fusion was also strongly induced upon contact of bacteria with plant cells (Figure 2G), but was not expressed in an hrpB mutant strain (data not shown), confirming the previous report that hrpY is HrpB regulated. From these observations, we conclude that transcription of the hrpB gene and of type III secretion genes is strongly induced in R.solanacearum only after contact with plant cells has been established, indicating that the pathogen is able to recognize a cell surface ligand and responds accordingly.
Contact-dependent induction of hrpB gene expression requires the outer membrane protein PrhA
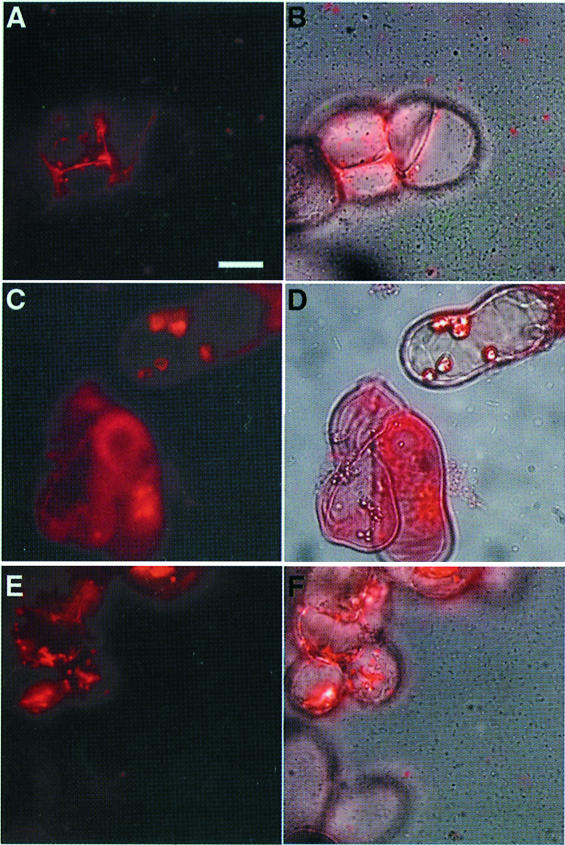
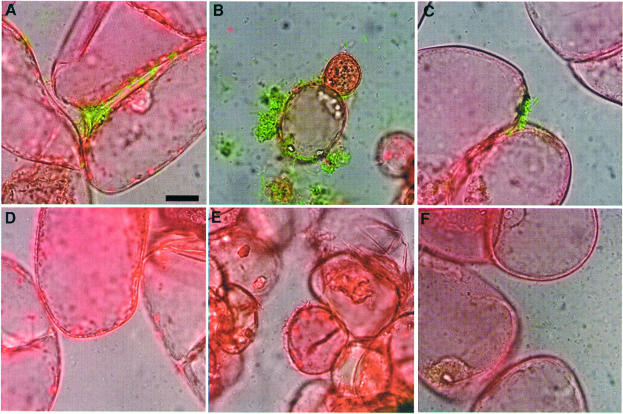
The high level of induction of hrpB gene transcription observed when bacteria are co-cultivated with Arabidopsis cells depends on a functional prhA gene, the product of which has been proposed to act as a receptor for plant-specific signal(s) (Marenda et al., 1998). Genetic evidence indicates that the PrhA protein is at the top of a regulatory cascade involved in the transduction of plant-dependent signal(s) to the hrpB gene (Marenda et al., 1998; Brito et al., 1999). In order to determine whether the contact-dependent induction of hrpB expression was also controlled by PrhA, the hrpB–gfp fusion carried by the plasmid pSG261 was introduced into strain GMI1575 (prhA mutant). After growth of the resulting strain in the presence of Arabidopsis cells, only a weak level of fluorescence, corresponding to the basal level of hrpB expression, could be detected after 16 h, even when bacteria were found in the vicinity of plant cells (Figure 3A and B). These observations demonstrate that PrhA is specifically involved in the recognition of the bacterium–plant cell contact, confirming that the plant signal that induces hrpB expression is a non-diffusible ligand molecule.
Fig. 3. Contact-dependent induction of hrpB gene expression requires components of the plant-responsive regulatory cascade. Epifluorescence microscopy study after 16 h of co-cultivation of R.solanacearum and Arabidopsis cells. The fluorescence and phase contrast images have been overlaid in (B), (D) and (F). The bar represents 20 µm. (A and B) Strain GMI1575/pSG261 carrying a prhA mutation. (C and D) Strain GMI1579/pSG261 carrying a prhJ mutation. (E and F) Strain GMI1425/pSG261 carrying an hrpG mutation.
Plasmid pSG261 was also introduced into strains carrying disruptions of regulatory genes encoding other components of the plant signal transduction pathway: GMI1578 (hrpG::Ω) and GMI1579 (prhJ::Ω), and into strain GMI1525 (hrpB::Ω). In a prhJ mutant, a basal level of fluorescence was observed, but no transcriptional induction could be detected upon bacterium–plant cell contact, as described above in a prhA mutant background (Figure 3C and D). Expression of the hrpB–gfp fusion was undetectable in an hrpG mutant co-cultivated with plant cells, whether bacteria were in contact with plant cells or not (Figure 3E and F). As expected, plant cell contact-dependent expression of the hrpB–gfp fusion was observed in the hrpB mutant strain (data not shown), confirming the specificity of the upstream components of the PrhA-dependent regulatory cascade in transducing the signal triggered by the bacterium–plant cell contact.
PrhA is required for recognition, but not for attachment to plant cells
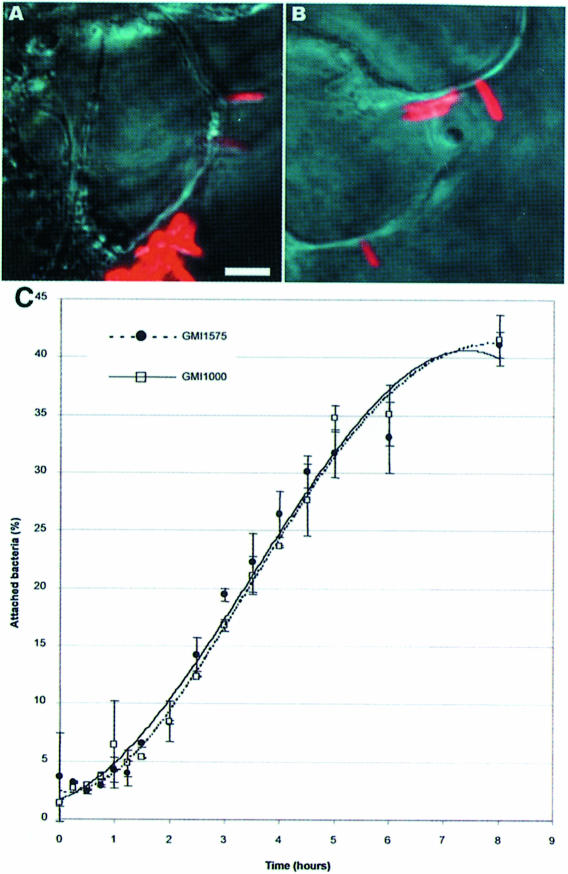
The above data indicate that the putative PrhA receptor recognizes a ligand upon contact with the plant cell surface, which leads to a specific up-regulation of the hrpB gene promoter. Because cytological studies revealed that R.solanacearum are tightly attached to plant cells, we wanted to find out whether PrhA is required for bacteria to become attached to plant cells. Strain GMI1575 (prhA mutant) bearing the pSG261 plasmid was only weakly fluorescent and therefore not suitable for accurate study of the nature of the bacterium–plant cell interaction, so a prhA mutation was introduced into strain GMI1600, which constitutively expresses GFP activity. We observed by confocal laser microscopy (Figure 4A and B) that mutant strain GMI1601 interacts qualitatively with plant cells, like the wild-type strain GMI1000 (Van Gijsegem et al., 2000). To confirm this finding, we determined the kinetics of attachment of 32P-radiolabelled bacterial populations to Arabidopsis cells. Figure 4C shows that strain GMI1575 (prhA mutant) was still able to bind to plant cells as efficiently as the parental strain. In both cases, only 5% of the bacteria present in the co-culture were attached to plant cells after 1 h (when cultures were shaken), but this proportion increased gradually so that after 7 h, 40% of the bacterial population were bound to plant cell surfaces. In conclusion, PrhA does not appear to be involved in the attachment of bacteria to plant cells, supporting the view that it acts instead as a receptor for a non-diffusible plant signal.
Fig. 4. PrhA is not involved in the attachment of bacteria to plant cell surfaces. (A and B) Confocal analysis of the interaction between Arabidopsis cells and (A) strain GMI1600 carrying a constitutive gfp reporter fusion or (B) strain GMI1601, which carries, in addition, a prhA mutation. In both cases, adherence and polar attachment of the bacteria (in red) to the plant cell surface can be observed, as has been described for the wild-type strain GMI1000 (Van Gijsegem et al., 2000). The bar represents 15 µm. (C) Time course of attachment of 32P-radiolabelled bacteria (wild-type strain GMI1000 and prhA mutant strain GMI1575) to Arabidopsis cells grown under continuous agitation.
The up-regulation of hrpB in response to pathogen–plant cell contact does not require a functional type III secretion system
The hrp regulon controlled by HrpB comprises at least 20 genes encoding components of the type III secretion machinery, along with a secreted protein (PopA) and other candidate proteins exported by the Hrp apparatus. In Yersinia, the expression of virulence genes induced by the sensing of target cell contact depends on the secretion of the negative regulator LcrQ, through the type III secretion pathway (Pettersson et al., 1996). This strategy, based on the sensing of secretion itself, allows the coupling of the transcription and secretion processes. We investigated whether the Hrp pilus of R.solanacearum could influence the transcription of hrpB, either by being directly involved in the sensing of the host plant cell or by promoting the translocation of proteins into plant cells. The hrpB–gfp fusion carried on pSG261 was introduced into strain GMI1410, which carries a Tn5 insertion within the hrpY gene coding sequence. Microscopic detection of fluorescent bacteria grown in co-culture with Arabidopsis cell suspensions clearly revealed that the absence of the Hrp pilus in this strain does not affect the contact-dependent up-regulation of the hrpB gene (Figure 5A). This also confirmed the observation that bacterial attachment is not impaired in this hrpY mutant strain (Van Gijsegem et al., 2000). A contact-dependent induction of the hrpB gene was also observed when the pSG261 plasmid was introduced into another hrp secretion mutant strain, GMI1462, carrying a disruption of the hrcC gene, which encodes the R.solanacearum secretin (Figure 5B). An hrpY mutant strain carrying an hrpY–gfp transcriptional fusion was also up-regulated upon contact of the bacteria with plant cells; up-regulation thus still occurred in a strain unable to synthesize the Hrp pilus (Figure 5C). From these data, we conclude that the Hrp pilus is not involved in the sensing of host plant cells and that the inability to secrete virulence factors does not modulate the transcriptional activation of the hrpB and hrpY genes in response to contact with the target cell.
Fig. 5. Transcriptional induction of hrpB gene expression in response to plant cell contact is not type III secretion dependent. Observations by epifluorescence microscopy after 16 h of co-cultivation of Arabidopsis cells with the R.solanacearum hrpY mutant strain GMI1410/pSG261 (A) and the hrcC mutant strain GMI1462/pSG261 (B), both carrying an hrpB–gfp reporter fusion. The same experimental conditions were used for strain GMI1410/pSG282, carrying an hrpY–gfp reporter fusion (C). The bar represents 20 µm.
The signal that induces hrp gene expression during pathogen–plant cell contact is a ubiquitous and non-diffusible molecule present in the Arabidopsis cell wall
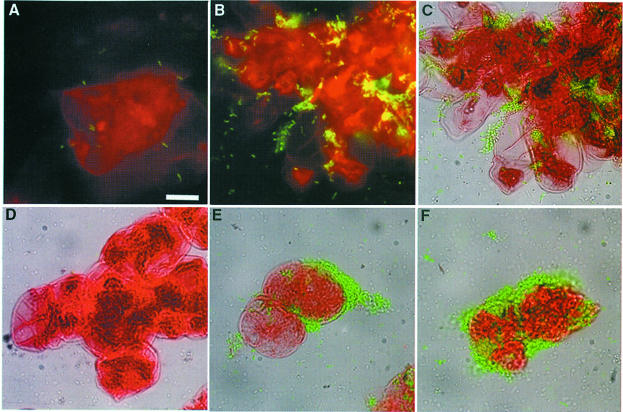
In order to define the nature of the hrp-inducing plant signal, we investigated whether it could be found in plant species other than A.thaliana. We monitored the expression level of an hrpB–gfp gene fusion in individual bacteria co-cultivated with plant cell suspensions from tomato, which is a host plant for R.solanacearum, and from tobacco and Medicago truncatula, two non-host plants in which the bacterium is unable to cause disease. In order to standardize the culture conditions, these different plant cell suspensions were maintained and grown for experiments in the same defined culture medium (Gamborg medium). Results shown in Figure 6 indicate that contact-dependent induction of hrpB expression was observed in the presence of all the plant cell types tested and that this induction was, in each case, dependent on a functional prhA gene. This latter observation differs from our previous results in which hrpB gene expression was only reduced in a prhA mutant strain co-cultivated with tomato cells (Marenda et al., 1998). This discrepancy is probably due to the difference between the plant cell culture media used in the two studies (T-MSMO was used in the earlier study and Gamborg medium in this one). In the presence of tomato, tobacco or Medicago cells, the induction kinetics of hrpB expression were comparable to those observed in the presence of Arabidopsis cells and reached a maximum after 4 h of co-cultivation. None of the conditioned media prepared from these different plant cell cultures induced hrpB gene expression (data not shown).
Fig. 6. Different plant cell species induce hrpB gene expression in a PrhA-dependent manner. Illustrations are an overlay of the fluorescence and visible images. The bar represents 20 µm. (A and D) Co-cultivation of tobacco cells with R.solanacearum strain GMI1000/pSG261 (A) or the prhA mutant strain GMI1575/pSG261 (D). (B and E) Co-cultivation of M.truncatula cells with GMI1000/pSG261 (B) or GMI1575/pSG261 (E). (C and F) Co-cultivation of tomato cells with GMI1000/pSG261 (C) or GMI1575/pSG261 (F).
To localize the active plant signal in plant cells, cell wall material was prepared from Arabidopsis cell suspensions and incubated at 28°C with R.solanacearum. Under these conditions, bacteria carrying the hrpB–gfp fusion bound to this plant cell wall material started to fluoresce after 4 h (Figure 7A) and were intensely fluorescent after 16 h (Figure 7B and C). The low fluorescence detected with a prhA mutant strain confirmed that the transcriptional induction of the hrpB gene observed in the presence of cell wall material follows the same expression pattern as in the presence of plant cells (Figure 7D), and indicated that wild-type bacteria were able to recognize a non-diffusible ligand molecule concentrated in the cell wall fraction. This signal appears to be a constitutive component of the plant cell wall since it was present in the Arabidopsis cell wall material prepared from plant cells grown in the absence of bacteria. Furthermore, this Arabidopsis cell wall material still activated hrpB gene expression after heating (10 min at 100°C) and proteinase K treatment before incubation with bacteria (Figure 7E and F).
Fig. 7. The plant signal recognized by PrhA that induces hrpB gene expression is present in the Arabidopsis cell wall. Epifluorescence microscopy observations of R.solanacearum strains grown in the presence of Arabidopsis cell wall material. Illustrations (C–F) are overlays of the fluorescence and visible light microscopy images. The bar represents 20 µm. (A) Strain GMI1000/pSG261 co-cultivated for 4 h with cell wall material. (B–D) Strain GMI1000/pSG261 (B and C) and the prhA mutant strain GMI1575/pSG261 (D) co-cultivated for 16 h with cell wall material. (E) Strain GMI1000/pSG261 with heat-treated cell wall material (15 min, 100°C). (F) Strain GMI1000/pSG261 with proteinase K-treated cell wall material.
Discussion
Bacterial virulence genes are integrated into sophisticated regulatory networks that allow their control at the transcriptional level in response to environmental changes encountered by the pathogen in its various habitats. Expression of the hrp genes of phytopathogenic bacteria can be induced in vitro by a variety of environmental signals, such as pH, osmolarity, and the carbon and nitrogen sources used in the medium. In this study, we demonstrate for the first time that contact between R.solanacearum and host plant cells is a signal triggering maximal hrp gene expression. It has already been shown that contact of bacteria with mammalian cells is an important signal, stimulating the production of several effector proteins translocated by type III secretion pathways (Ménard et al., 1994; Rosqvist et al., 1994; Watarai et al., 1995; Zierler and Galan, 1995) and also results in increased transcription of bacterial virulence genes (Pettersson et al., 1996; Zhang and Normark, 1996; Jacobi et al., 1998; Taha et al., 1998). Here, we present evidence that this is also the case for plant-pathogenic bacteria. In R.solanacearum, the transcription of two essential pathogenicity determinants, namely the hrpB regulatory gene (controlling both Hrp type III secretion genes and genes encoding type III-secreted proteins such as PopA) and hrpY (encoding the structural component of the Hrp pilus), is strongly induced when the bacterium comes into contact with plant cells. We show that the plant signal that triggers hrp gene expression upon bacterium–plant cell contact is a component of the Arabidopsis plant cell wall. To our knowledge, this is the first description of a non-diffusible plant signal inducing bacterial gene expression. The induction of R.solanacearum hrp genes detected in response to contact with diverse plant cell species from three different dicotyledonous plant families (Solanaceae, crucifers and legumes) and including both host and non-host plants strongly suggests that this inducing signal involves a ubiquitous component of the plant cell wall. Two additional arguments support this hypothesis: (i) the elicitation of the hypersensitive response on non-host plants by phytopathogenic bacteria is specifically dependent upon hrp genes, which argues against host-specific hrp gene induction; and (ii) the biochemical composition of the plant cell wall is widely conserved among dicotyledonous plants (Varner and Lin, 1989). This hrp-inducing signal appears to be heat stable and resistant to proteinase K treatment. This suggests the involvement of a complex carbohydrate macromolecule present in the hemicellulosic/pectic fraction, although the involvement of a proteinaceous component protected in the pectic gel matrix cannot be ruled out completely.
This work provides evidence that the recently characterized plant-responsive regulatory cascade that induces hrp gene expression in R.solanacearum in the presence of plant cells (Marenda et al., 1998; Brito et al., 1999) is activated in response to pathogen–plant cell contact. Accordingly, contact-dependent induction of hrpB gene expression is strongly reduced or abolished, respectively, in mutant strains disrupted in prhA or hrpG, which both encode components of this regulatory cascade. Genetic evidence and structural features of PrhA indicate that PrhA defines a novel class of receptor proteins involved in the sensing of plant cell contact by the recognition of a non-diffusible ligand molecule present in the plant cell wall. PrhA is homologous to siderophore receptors belonging to the FecA/PupB subclass (Marenda et al., 1998). FecA and PupB are the receptor components of the so-called ‘three-compartment signal transduction systems’, which are involved in transcriptional regulation of, respectively, the Escherichia coli and Pseudomonas putida iron transport genes (Koster et al., 1994; Härle et al., 1995). This newly discovered signal transduction system allows Gram-negative bacteria to rapidly modulate transcription in response to a stimulus from the cell surface. Preliminary data suggest that in R.solanacearum PrhA might, along with two other components, form an analogous three-compartment signal transduction system, which specifically induces the expression of downstream genes in the presence of plant cells (our unpublished data). The phylogenetic relationship between PrhA and hydroxamate siderophore receptors suggests that PrhA can recognize a ligand molecule related to ferrichrome derivatives. Nethertheless, it has been shown that co-evolution of siderophore receptor structures is not necessarily correlated with substrate specificity (Koebnik et al., 1993). Finally, it should be noted that no bacterial sensor of target cell contact has yet been identified in mammalian pathogens. It will therefore be of prime interest to determine whether pathogenic bacteria have evolved different classes of external receptors able to recognize contact with their host cells and to generate a specific signal triggering virulence functions.
Our observations indicate the existence of two distinct steps in the interaction between bacteria and plant cells. First, bacteria attach to the plant cell surface (often in a polar manner); this first step does not require a functional PrhA protein and is not dependent on hrp-encoded functions (since this attachment process is still observed with an hrpB mutant strain). Once this attachment is established, the PrhA receptor is able to detect an accessible ligand; this event leads to increased transcription of the hrpB regulatory gene. This two-step interaction is reminiscent of the situation described during the interaction of enteropathogenic E.coli with its target cell, which requires initial adhesion, through a bundle-forming pilus, followed by intimate adhesion mediated by intimin (Rosenshine et al., 1996). Since the R.solanacearum Hrp pilus is not involved in the sensing of plant cells (this study) or in the attachment process, but rather in the translocation of Hrp-dependent secreted products (Van Gijsegem et al., 2000), this suggests that specific determinants involved in bacterial attachment to the host cell surface remain to be identified.
This study shows that the contact-dependent induction of R.solanacearum hrp genes can be observed even in the absence of a functional type III secretion apparatus. We have also shown that transcription of the type III-secreted protein HrpY is not regulated by the secretion status, as is also probably the case for PopA, expression of which is hrpB dependent (Arlat et al., 1994). This situation is clearly different from that observed in Yersinia, where export of the negative regulator LcrQ via the type III secretion pathway is a prerequisite for achieving full induction of yop gene expression (Pettersson et al., 1996). In Shigella flexneri, transcription of the secreted protein VirA also requires the integrity of the type III secretion machinery (Demers et al., 1998). In R.solanacearum, control of the type III secretion process appears to be regulated differently, suggesting that this regulation operates mainly at a post-transcriptional level, probably by ensuring the coupling of translation and secretion at the mRNA level (Anderson and Schneewind, 1997). This observation, along with the fact that contact-dependent induction of hrpB expression occurs with a wide range of host and non-host plant cell cultures, favours the view that the specificity of the plant–bacterium interaction does not reside in the transcriptional activation of type III secretion genes, but rather at the level of the Hrp-dependent secreted products.
The plant cell contact-dependent induction of hrp gene expression is probably very rapid: hrpB gene induction is clearly detected within 90 min, which is shorter than the generation time of R.solanacearum grown under optimal conditions and also represents the time required for attachment of bacteria to plant cells (Figure 4C) and the synthesis of a detectable amount of GFP protein under our experimental conditions. In Yersinia, the up-regulation of yopE gene expression is also very rapid, being detected within 30 min after infection of eukaryotic cells (Jacobi et al., 1998). It is noteworthy that a basal level of GFP activity can be detected in bacteria before the contact with plant cells. This basal level is comparable to the level of hrpB expression observed after growth of the bacteria in minimal medium in vitro and it is known that protein secretion via the Hrp type III secretion pathway can be observed in these conditions (Arlat et al., 1994). This observation indicates that contact of the bacteria with plant cells does not trigger de novo expression of hrp genes, but rather triggers a strong (full?) induction of hrp gene expression. Several recent reports also support the view that, even for mammalian pathogens, type III secretion can occur in the absence of contact with eukaryotic cells (Blocker et al., 1999; Daefler, 1999; Lee and Schneewind, 1999) and that some environmental factors, such as pH of the growth medium, strongly influence the secretion process. A contact-dependent signal leading to full and rapid activation of hrp genes could serve to produce more Hrp proteins and Hrp-dependent effectors in the vicinity of plant cells, ensuring the release of these pathogenicity factors at the appropriate time and place. Another attractive possibility is that this contact-dependent signal could result in a differential control of virulence gene expression, suggesting the existence of a hierarchy in the type III secretion process (Collazo and Galan, 1996; Demers et al., 1998). Our observations are consistent with the hypothesis that this contact-dependent signal could contribute to triggering the polarized translocation of some specific type III effector proteins into the eukaryotic cell, even though this translocation of bacterial effectors has not yet been formally demonstrated in the case of plant pathogens (Galan and Collmer, 1999). As mentioned above, however, this translocation step is certainly a finely tuned process that also involves additional controls at the post-transcriptional level.
In summary, we have identified a new type of non-diffusible plant signal inducing bacterial virulence gene expression and we have presented genetic evidence that prhA encodes the candidate receptor protein of this contact-dependent signal. Our next goal is to characterize the nature of the PrhA ligand, which was shown to be a component of the plant cell wall, in order to demonstrate that PrhA is the specific receptor of this ligand. Since this hrp-inducing signal was found to be present in a wide range of plant species, it is plausible that it may induce the expression of genes encoding type III secretion systems in the main groups of non-tumorigenic Gram-negative bacteria.
Materials and methods
Bacterial strains and conjugation procedure
Bacterial strains used in this study are listed in Table I. Ralstonia solanacearum strains were grown in B medium or minimal medium (Boucher et al., 1985) supplemented with glutamate (20 mM final concentration). When needed, antibiotics were added to the media at the following concentrations (mg/l): kanamycin, 50; spectinomycin, 40; gentamycin, 25; tetracycline, 10. Plasmids were introduced in R.solanacearum by triparental mating using the helper strain E.coli K12 carrying plasmids pRK2013 or pRK2073 as conjugative helper plasmids (references and method in Brito et al., 1999).
Construction of hrp–gfp transcriptional fusions
Escherichia coli DH5α (Bethesda Research Laboratory) was used as a host for plasmid constructs. Transcriptional fusions between the R.solanacearum hrpB and hrpY genes and gfp were constructed by a PCR cloning procedure using the Expand Long Template PCR kit (Boehringer). A 727 bp fragment of the gfp gene was amplified from the plasmid pAG408 (Suarez et al., 1997) using the forward primer 5′-CATATGAGTAAAGGAGAAG-3′ and the reverse primer 5′-GAA TTCATTATTTGTAGAGC-3′. The hrpB gene promoter was amplified with the forward primer 5′-CAGGTCAAGGGTACGCTC-3′ and the reverse primer 5′-GGAATTCCATATGAATGCTCCTGAAGCGTCA-3′. The resulting 236 bp amplified fragment was cloned upstream of the gfp gene, resulting in plasmid pSG257. This hrpB–gfp transcriptional fusion was then cloned in the low copy number plasmid pLAFR6 (Huynh et al., 1989), generating plasmid pSG261.
The hrpY promoter was amplified with the following pair of primers: 5′-GAATTCGAGCAGCGCGCC-3′ and 5′-GCTAGCCATGATAGT TTCCTTTGATGG-3′. The resulting 249 bp amplified fragment was cloned upstream of the gfp gene, creating plasmid pSG278. This hrpY–gfp transcriptional fusion was cloned in the pLAFR6 vector to generate plasmid pSG282.
Detailed descriptions of plasmid constructs can be obtained on request from S.G.
Construction of an R.solanacearum strain with constitutive GFP expression
We used the promoter–probe gfp-based mini-transposon pAG408 located on a suicide delivery plasmid (Suarez et al., 1997) to generate chromosomal fusions. Escherichia coli S17-1 (λpir) cells (Suarez et al., 1997) transformed with pAG408 were used as donor in mating experiments to transfer the suicide plasmid into the R.solanacearum wild-type strain GMI1000. Transconjugants were recovered on minimal medium plates supplemented with gentamycin. Several colonies that fluoresced bright green were examined after growth on both complete and minimal media, and strain GMI1600 was selected on the basis of its constitutive high GFP expression. The pathogenicity of strain GMI1600 was investigated by its ability to induce the hypersensitive response on tobacco leaves and to cause wilting symptoms in tomato plants as described by Brito et al. (1999). Plant tests showed that, under our experimental conditions, strain GMI1600 behaved in the same way as the wild-type strain GMI1000, indicating that the random chromosomal insertion of the pAG408 mini-transposon did not alter the pathogenicity of the wild-type strain.
Strain GMI1601 was generated after transformation of strain GMI1600 by total genomic DNA of strain GMI1575, which carries a prhA::Ω mutation (Marenda et al., 1998). The occurrence of a double recombination event in strain GMI1601 leading to disruption of the prhA gene was checked by Southern analysis.
Plant cell cultures and co-cultivation procedure
Cell suspensions of A.thaliana At-202 (accession Col 0), tomato (Lycopersicon esculentum) MSK8, Nicotiana tabacum L. var. Wisconsin no. 38 and M.truncatula were grown in Gamborg B5 medium (ICN Biomedicals, Aurora, OH) supplemented with 20 g/l sucrose and 1 µM α-naphthalene acetic acid at 23°C under continuous illumination and shaking. Samples of 0.5 g of plant cell suspensions in the exponential phase were inoculated with R.solanacearum as described previously (Marenda et al., 1998; Brito et al., 1999) and grown at 28°C on a rotary shaker.
The term ‘conditioned medium’ refers to culture medium from a 1-week-old plant cell suspension from which plant cells have been removed by filtration; the medium is then sterilized by filtration through 0.22 µm Millipore filters.
Time course of attachment of bacteria to plant cells
Bacteria were labelled by growth overnight in a phosphate-depleted liquid medium supplemented with 100 µCi of 32P-radiolabelled NaPO4 (NEN Life Science Products, Boston, MA). Bacteria were washed four or five times to eliminate unincorporated radioactivity before co-culture with an Arabidopsis cell suspension. Independent 200 µl aliquots taken from co-cultures were filtered through 20 µm pore size nylon membranes and washed thoroughly three times with sterile culture medium to eliminate bacteria that were not specifically bound to plant cells. The radioactivity of this washed plant cell fraction was measured by Cerenkov determination (1 min) on a liquid scintillation analyser (Tri-Carb 2100TR; Packard Instrument Co., Meriden, USA).
Cytological analysis of the plant cell–bacterium interactions
Microscopic examination of the plant cell–bacteria co-culture was performed after the addition of the dye Evans blue at a final concentration of 0.001% (w/v). Plant cells treated with Evans blue emit a red fluorescence when observed in the GFP excitation wavelength range (480 ± 30 nm) with a 520 nm long-pass glass emission filter. This procedure decreases the plant cell wall autofluorescence and facilitates the discrimination between GFP-tagged bacteria and plant cells. Observations were performed using 10 µl of the co-culture mounted on a glass slide previously coated with poly-l-lysine (Sigma, St Louis, MO).
Observations were made using an epifluorescence microscope (Leitz DMIRBE; Leica, Wetzlar, Germany; Fluotar objective 63×, n.a. 0.7) or a confocal laser scanning microscope (Axiovert; Carl Zeiss, Oberkochen, Germany) with a Neofluar objective (63×, n.a. 1.3) equipped with dual detectors and an argon–helium laser for simultaneous scanning of fluorescence and DIC-like images. Fluorescence images were acquired using a CCD camera (Colour Coolview; Photonic Science, Robertsbridge, UK) connected to the Leitz microscope. Images in the same figure were acquired with the same parameters to allow comparisons between the emitted fluorescence level in the different samples. Figures were edited using Adobe Photoshop 5.0 software.
Preparation of cell wall material from Arabidopsis cell suspensions
A 1-week-old Arabidopsis cell culture removed from the medium by filtration was placed in 90% cold ethanol to inactivate degradative enzymes. Cell walls were isolated according to the methods described by Selvendran and O’Neill (1987) and Morvan et al. (1991). Briefly, cells were homogenized with an Ultraturrax for 10 min. The resulting suspension was washed twice with distilled water and insoluble material was collected by centrifugation (10 min, 2000 g). The pellet was resuspended in 5 vols of chloroform–methanol 1:1 (v/v), washed three times on the sinter with this mixture and then twice more with acetone. The resulting material was called ‘cell wall material’. The ability of this cell wall material to trigger hrpB–gfp gene expression was tested after growth of the bacteria for 16 h in minimal medium containing cell wall material at a final concentration of 1% (w/v). Similar conditions were used with cell wall material that had been heat treated (15 min at 100°C) or treated with proteinase K (50 µg/ml for 90 min at 40°C).
Acknowledgments
Acknowledgements
We thank A.Jauneau and P.Cochard for expert technical assistance with microscopy, C.Guzman for providing the plasmid pAG408, S.Charlier and J.Vasse for their contribution in developing GFP in R.solanacearum, P.Barberis for technical assistance, and M.Arlat. We are indebted to M.-C.Roland and C.Gough for critical reading of the manuscript. This work was funded by projects BIO4-CT-97-2244 from the European Commission and AIP-188 Microbiologie from the Institut National de la Recherche Agronomique. B.B. was the recipient of a Marie Curie TMR postdoctoral grant from the European Commission.
References
- Anderson D.M. and Schneewind,O. (1997) A mRNA signal for the type III secretion of Yop proteins in Yersinia enterocolitica. Science, 278, 1140–1143. [DOI] [PubMed] [Google Scholar]
- Arlat M., Van Gijsegem,F., Huet,J.C., Pernollet,J.C. and Boucher,C.A. (1994) PopA1, a protein which induces a hypersensitive-like response on specific petunia genotypes, is secreted via the Hrp pathway of Pseudomonas solanacearum. EMBO J., 13, 543–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blocker A., Gounon,P., Larquet,E., Niebuhr,K., Cabiaux,V., Parsot,C. and Sansonetti,P. (1999) The tripartite type III secreton of Shigella flexneri inserts IpaB and IpaC into host membranes. J. Cell Biol., 147, 683–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanove A.J., Bauer,D.W. and Beer,S.V. (1998) Erwinia amylovora secretes DspE, a pathogenicity factor and functional AvrE homolog, through the Hrp (type III secretion) pathway. J. Bacteriol., 180, 2244–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher C.A., Barberis,P., Trigalet,A.P. and Démery,D.A. (1985) Transposon mutagenesis of Pseudomonas solanacearum: isolation of Tn5-induced avirulent mutants. J. Gen. Microbiol., 131, 2449–2457. [Google Scholar]
- Brito B., Marenda,M., Barberis,P., Boucher,C. and Genin,S. (1999) prhJ and hrpG, two new components of the plant signal-dependent regulatory cascade controlled by PrhA in Ralstonia solanacearum. Mol. Microbiol., 31, 237–251. [DOI] [PubMed] [Google Scholar]
- Charkowski A.O., Alfano,J.R., Preston,G., Yuan,J., He,S.Y. and Collmer,A. (1998) The Pseudomonas syringae pv. tomato HrpW protein has domains similar to harpins and pectate lyases and can elicit the plant hypersensitive response and bind to pectate. J. Bacteriol., 180, 5211–5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collazo C.M. and Galan,J.E. (1996) Requirement for exported proteins in secretion through the invasion-associated type III system of Salmonella typhimurium. Infect. Immun., 64, 3524–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis G.R. (1998) The Yersinia deadly kiss. J. Bacteriol., 180, 5495–5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daefler S. (1999) Type III secretion by Salmonella typhimurium does not require contact with a eukaryotic host. Mol. Microbiol., 31, 45–51. [DOI] [PubMed] [Google Scholar]
- Demers B., Sansonetti,P.J. and Parsot,C. (1998) Induction of type III secretion in Shigella flexneri is associated with differential control of transcription of genes encoding secreted proteins. EMBO J., 17, 2894–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan J.E. and Collmer,A. (1999) Type III secretion machines: bacterial devices for protein delivery into host cells. Science, 284, 1322–1328. [DOI] [PubMed] [Google Scholar]
- Gaudriault S., Malandrin,L., Paulin,J.-P. and Barny,M.-A. (1997) DspA, an essential pathogenicity factor of Erwinia amylovora showing homology with AvrE of Pseudomonas syringae, is secreted via the Hrp secretion pathway in a Dsp-dependent way. Mol. Microbiol., 26, 1057–1069. [DOI] [PubMed] [Google Scholar]
- Genin S., Gough,C.L., Zischek,C. and Boucher,C.A. (1992) Evidence that the hrpB gene encodes a positive regulator of pathogenicity genes from Pseudomonas solanacearum. Mol. Microbiol., 6, 3065–3076. [DOI] [PubMed] [Google Scholar]
- Gough C.L., Genin,S., Zischek,C. and Boucher,C.A. (1992) hrp genes of Pseudomonas solanacearum are homologous to pathogenicity determinants of animal pathogenic bacteria and are conserved among plant pathogenic bacteria. Mol. Plant Microbe Interact., 5, 384–389. [DOI] [PubMed] [Google Scholar]
- Guéneron M., Timmers,A.C.J., Boucher,C. and Arlat,M. (2000) Two novel proteins, PopB, which has functional nuclear localisation signals, and PopC, which has a large leucine-rich repeat domain, are secreted through the Hrp-secretion apparatus of Ralstonia solanacearum. Mol. Microbiol., 36, 261–277. [DOI] [PubMed] [Google Scholar]
- Härle C., Kim,I., Angerer,A. and Braun,V. (1995) Signal transfer through three compartments: transcription initiation of the Escherichia coli ferric citrate transport system from the cell surface. EMBO J., 14, 1430–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S.Y., Huang,H.-C. and Collmer,A. (1993) Pseudomonas syringae pv. syringae harpin Pss: a protein that is secreted via the Hrp pathway and elicits the hypersensitive response in plants. Cell, 73, 1255–1266. [DOI] [PubMed] [Google Scholar]
- Hueck C.J. (1998) Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Rev., 62, 379–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh T.V., Dahlbeck,D. and Staskawicz,B.J. (1989) Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science, 245, 1374–1377. [DOI] [PubMed] [Google Scholar]
- Jacobi C.A., Roggenkamp,A., Rakin,A., Zumbihi,R., Leitritz,L. and Heesemann,J. (1998) In vitro and in vivo expression studies of yopE from Yersinia enterocolitica using the gfp reporter gene. Mol. Microbiol., 30, 865–882. [DOI] [PubMed] [Google Scholar]
- Kim J.F. and Beer,S.V. (1998) HrpW of Erwinia amylovora, a new harpin that contains a domain homologous to pectate lyases of a distinct class. J. Bacteriol., 180, 5203–5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koebnik R., Hantke,K. and Braun,V. (1993) The TonB-dependent ferrichrome receptor FcuA of Yersinia enterocolitica: evidence against a strict co-evolution of receptor structure and substrate specificity. Mol. Microbiol., 7, 383–393. [DOI] [PubMed] [Google Scholar]
- Koster M., van Klompenburg,W., Bitter,W., Leong,J. and Weisbeek,P. (1994) Role of the outer membrane ferric siderophore receptor PupB in signal transduction across the bacterial cell envelope. EMBO J., 13, 2805–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee V.T. and Schneewind,O. (1999) Type III machines of pathogenic yersiniae secrete virulence factors into the extracellular milieu. Mol. Microbiol., 31, 1619–1629. [DOI] [PubMed] [Google Scholar]
- Marenda M., Brito,B., Callard,D., Genin,S., Barberis,P., Boucher,C. and Arlat,M. (1998) PrhA controls a novel regulatory pathway required for the specific induction of Ralstonia solanacearum hrp genes in the presence of plant cells. Mol. Microbiol., 27, 437–453. [DOI] [PubMed] [Google Scholar]
- Ménard R., Sansonetti,P. and Parsot,C. (1994) The secretion of the Shigella flexneri Ipa invasins is activated by epithelial cells and controlled by IpaB and IpaD. EMBO J., 13, 5293–5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo Y.-Y., Geibel,M., Bonsall,R.F. and Gross,D.C. (1995) Analysis of sweet cherry (Prunus avium L.) leaves for plant signal molecules that activate the syrB gene required for synthesis of phytotoxin syringomycin by Pseudomonas syringae pv. syringae. Plant Physiol., 107, 603–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morvan C., Abdul Hafez,M., Jauneau,A., Thoiron,B. and Demarty,M. (1991) Incorporation of d-[U-14C]glucose in the cell wall of Linum plantlets during the first step of growth. Plant Cell Physiol., 32, 609–621. [Google Scholar]
- Mudgett M.B. and Staskawicz,B.J. (1999) Characterization of the Pseudomonas syringae pv. tomato AvRpt2 protein: demonstration of secretion and processing during bacterial pathogenesis. Mol. Microbiol., 32, 927–941. [DOI] [PubMed] [Google Scholar]
- Peters N.K., Frost,J. and Long,S.R. (1986) A plant flavone, luteolin, induces expression of Rhizobium meliloti nodulation genes. Science, 233, 977–980. [DOI] [PubMed] [Google Scholar]
- Pettersson J., Nordfelth,R., Dubinina,E., Bergman,T., Gustafsson,M., Magnusson,K.E. and Wolf-Watz,H. (1996) Modulation of virulence factor expression by pathogen target cell contact. Science, 273, 1231–1233. [DOI] [PubMed] [Google Scholar]
- Roine E., Wei,W., Yuan,J., Nurmiaho-Lassila,E.-L., Kalkinen,N., Romantschuk,M. and He,S.Y. (1997) Hrp pilus: an hrp-dependent bacterial surface appendage produced by Pseudomonas syringae pv. tomato DC3000. Proc. Natl Acad. Sci. USA, 94, 3459–3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenshine I., Ruschkowski,S., Stein,M., Reinscheid,D.J., Mills,S.D. and Finlay,B.B. (1996) A pathogenic bacterium triggers epithelial signals to form a functional bacterial receptor that mediates actin pseudopod formation. EMBO J., 15, 2613–2624. [PMC free article] [PubMed] [Google Scholar]
- Rosqvist R., Magnusson,K.E. and Wolf-Watz,H. (1994) Target cell contact triggers expression and polarized transfer of Yersinia YopE cytotoxin into mammalian cells. EMBO J., 13, 964–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossier O., Wengelnik,K., Hahn,K. and Bonas,U. (1999) The Xanthomonas Hrp type III system secretes proteins from plant and mammalian bacterial pathogens. Proc. Natl Acad. Sci. USA, 96, 9368–9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvendran R.R. and O’Neill,M.A. (1987) Isolation and analysis of cell walls from plant material. Methods Biochem. Anal., 32, 25–153. [DOI] [PubMed] [Google Scholar]
- Stachel S.E., Messens,E., Van Montagu,M. and Zambryski,P.C. (1985) Identification of the signal molecules produced by wounded plant cells that activate T-DNA transfer in Agrobacterium tumefaciens. Nature, 318, 624–629. [Google Scholar]
- Suarez A., Güttler,A., Strätz,M., Staendner,L.H., Timmis,K.N. and Guzman,C.A. (1997) Green fluorescent protein-based reporter systems for genetic analysis of bacteria including monocopy applications. Gene, 196, 69–74. [DOI] [PubMed] [Google Scholar]
- Taha M.-K., Morand,P.C., Pereira,Y., Eugène,E., Giorgini,D., Larribe,M. and Nassif,X. (1998) Pilus-mediated adhesion of Neisseria meningitidis: the essential role of cell-contact-dependent transcriptional upregulation of the PilC1 protein. Mol. Microbiol., 28, 1153–1163. [DOI] [PubMed] [Google Scholar]
- Van Dijk K., Fouts,D.E., Rehm,A.H., Hill,A.R., Collmer,A. and Alfano,J.R. (1999) The Avr (effector) proteins HrmA (HopPsyA) and AvrPto are secreted in culture from Pseudomonas syringae pathovars via the Hrp (type III) protein secretion system in a temperature- and pH-sensitive manner. J. Bacteriol., 181, 4790–4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gijsegem F. (1997) In planta regulation of phytopathogenic bacteria virulence genes: relevance of plant-derived signals. Eur. J. Plant Pathol., 103, 291–301. [Google Scholar]
- Van Gijsegem F., Genin,S. and Boucher,C. (1993) Conservation of secretion pathways for pathogenicity determinants of plant and animal bacteria. Trends Microbiol., 1, 175–180. [DOI] [PubMed] [Google Scholar]
- Van Gijsegem F. et al. (1995) The hrp gene locus of Pseudomonas solanacearum, which controls the production of a type III secretion system, encodes eight proteins related to components of the bacterial flagellar biogenesis complex. Mol. Microbiol., 15, 1095–1114. [DOI] [PubMed] [Google Scholar]
- Van Gijsegem F., Vasse,J., Camus,J.C., Marenda,M. and Boucher,C. (2000) Ralstonia solanacearum produces Hrp-dependent pili that are required for PopA secretion but not for attachment of bacteria to plant cells. Mol. Microbiol., 36, 249–260. [DOI] [PubMed] [Google Scholar]
- Varner J.E. and Lin,L.-S. (1989) Plant cell wall architecture. Cell, 56, 231–239. [DOI] [PubMed] [Google Scholar]
- Watarai M., Tobe,T., Yoshikawa,M. and Sasakawa,C. (1995) Contact of Shigella with host cells triggers release of Ipa invasins and is an essential function of invasiveness. EMBO J., 14, 2461–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z.-M., Laby,R.J., Zumoff,C.H., Bauer,D.W., He,S.Y., Collmer,A. and Beer,S.V. (1992) Harpin, elicitor of the hypersensitive response produced by the plant pathogen Erwinia amylovora. Science, 257, 85–88. [DOI] [PubMed] [Google Scholar]
- Zhang J.P. and Normark,S. (1996) Induction of gene expression in Escherichia coli after pilus-mediated adherence. Science, 273, 1234–1236. [DOI] [PubMed] [Google Scholar]
- Zierler M.K. and Galan,J.E. (1995) Contact with cultured epithelial cells stimulates secretion of Salmonella typhimurium invasion protein InvJ. Infect. Immun., 63, 4024–4028. [DOI] [PMC free article] [PubMed] [Google Scholar]