Abstract
The Ski and Sno oncoproteins are components of a macromolecular complex containing the co-repressor N-CoR/SMRT, mSin3 and histone deacetylase. This complex has been implicated in the transcriptional repression exerted by a number of repressors including nuclear hormone receptors and Mad. Further more, Ski and Sno negatively regulate transforming growth factor-β (TGF-β) signaling by recruiting this complex to Smads. Here we show that loss of one copy of sno increases susceptibility to tumorigenesis in mice. Mice lacking sno died at an early stage of embryogenesis, and sno was required for blastocyst formation. Heterozygous (sno+/–) mice developed spontaneous lymphomas at a low frequency and showed an increased level of tumor formation relative to wild-type mice when challenged with a chemical carcinogen. sno+/– embryonic fibroblasts had an increased proliferative capacity and the introduction of activated Ki-ras into these cells resulted in neoplastic transformation. The B cells, T cells and embryonic fibroblasts of sno+/– mice had a decreased sensitivity to apoptosis or cell cycle arrest. These findings demonstrate that sno acts as a tumor suppressor at least in some types of cells.
Keywords: histone deacetylase/Mad/Rb/Sno/tumor suppressor
Introduction
The oncogene v-ski was originally identified in avian Sloan–Kettering viruses and was found to transform chicken embryo fibroblasts (Li et al., 1986). The chicken and human c-ski proto-oncogene products (c-Ski) are nuclear proteins of 750 and 728 amino acids, respectively (Nomura et al., 1989; Stavnezer et al., 1989; Sutrave and Hughes, 1989). The v-Ski protein lacks 292 amino acids from the C-terminal coiled-coil region of chicken c-Ski, but still contains the N-terminal cysteine-rich region (Stavnezer et al., 1989). The ski gene family consists of two members, ski and sno (ski-related novel gene) (Nomura et al., 1989), which share significant homology in their N- and C-terminal regions (Nomura et al., 1989; Nagase et al., 1993). Ski and Sno form a complex via their C-terminal regions (Nagase et al., 1993; Heyman and Stavnezer, 1994). Interestingly, ski can induce both transformation (growth) and differentiation, which is usually associated with the cessation of growth. Overexpression of either c-ski or v-ski induces transformation or muscle differentiation of quail embryo fibroblasts, depending on the growth conditions (Colmenares and Stavnezer, 1989; Colmenares et al., 1991). In addition, v-ski transgenic mice have more muscle mass because of hypertrophy of type II fast muscle fibers (Sutrave et al., 1990). The N-terminal region is responsible for both the cellular transformation and myogenic capacities of ski (Zheng et al., 1997). High levels of sno expression also cause transformation of chicken embryo fibroblasts and muscle differentiation of quail embryo fibroblasts (Boyer et al., 1993).
Recently, we demonstrated that c-Ski and Sno bind directly to the co-repressors N-CoR/SMRT and mSin3A, and are involved in the histone deacetylase (HDAC) complex (Nomura et al., 1999). N-CoR and SMRT, which show striking homology, were originally identified as co-repressors that bind to, and mediate transcriptional repression by, nuclear hormone receptors (Chen and Evans, 1995; Hörlein et al., 1995). Mammalian Sin3 orthologs (mSin3A and mSin3B) were originally shown to bind to, and mediate transcriptional repression by, basic helix–loop-helix (bHLH) proteins of the Mad family (Ayer et al., 1995), which act as transcriptional repressors after heterodimerization with Max (Ayer et al., 1993). N-CoR/SMRT forms a complex with mSin3, and the binding of mSins to HDAC suggested that transcriptional repression through N-CoR/SMRT and mSin3 involves deacetylation of nucleosomal histones (Alland et al., 1997; Hassing et al., 1997; Heinzel et al., 1997; Laherty et al., 1997; Nagy et al., 1997). The N-terminal cysteine-rich region of Ski/Sno binds to N-CoR/SMRT, while the C-terminal coiled-coil region interacts with mSin3. Ski and Sno are required for transcriptional repression by Mad and thyroid hormone receptor β (Nomura et al., 1999) and modulate the transcriptional regulation mediated by retinoid receptor α (Dahl et al., 1998). In addition to N-CoR/SMRT, mSin3 and Ski/Sno, the HDAC macromolecular complex is thought to contain many other components such as SAP30 (mSin3-associated protein) (Zhang et al., 1997; Laherty et al., 1998).
Recently, c-Ski and Sno were found to bind directly to Smad proteins, which play an important role in mediating the transforming growth factor-β (TGF-β) signal (Akiyoshi et al., 1999; Luo et al., 1999; Stroschein et al., 1999; Sun et al., 1999a,b). Both c-Ski and Sno inhibit Smad-induced transcriptional activation by recruiting the HDAC complex. These findings are consistent with the view that Ski and Sno act as oncoproteins. On the other hand, c-Ski and Sno are required for transcriptional repression not only by Mad, but also by Rb (Tokitou et al., 1999), which also interacts with HDAC (Brehm et al., 1998; Luo et al., 1998; Magnaghi-Jaulin et al., 1998). Both Mad and Rb act as tumor suppressors. A heterodimer of Myc–Max binds to the same target sequence as Mad–Max (the so-called E-box), and activates transcription. Transcriptional activation of a group of target genes by Myc–Max enhances cellular proliferation or transformation, whereas transcriptional repression of the same target genes by Mad–Max leads to suppression of proliferation or induction of terminal differentiation in a wide range of cell types (Ayer and Eisenman, 1993; Chin et al., 1995; Roussel et al., 1996). In fact, mutant mice lacking mxi1, a member of the mad gene family, were more susceptible to tumorigenesis (Schreiber-Agus et al., 1998). The Rb gene was originally identified as a tumor suppressor of retinoblastoma; the Rb protein regulates the G1–S transition in the cell cycle by silencing a group of target genes regulated by E2F transcription factors (Nevins, 1992; Weinberg, 1995). Rb binds to the activation domain of E2F and then actively represses the promoter by recruiting HDAC. Thus, c-Ski and Sno are required for the function of two tumor suppressors, Mad and Rb. v-Ski, which lacks the C-terminal region of c-Ski, blocks Mad- and Rb-induced transcriptional repression (Nomura et al., 1999; Tokitou et al., 1999). Since v-Ski lacks the C-terminal region of c-Ski, which is responsible for interaction with mSin3A, v-Ski is thought to act as a dominant-negative form. These observations suggest that ski and sno may be tumor suppressors in some types of cell, although v-ski was originally identified as an oncogene.
Here we demonstrate that loss of one copy of sno increases susceptibility to tumorigenesis in mice. Spontaneous lymphomas in heterozygous (sno+/–) mice and an increased level of tumor formation in sno+/– mice relative to wild-type mice following carcinogen treatment indicate that sno acts as a tumor suppressor. The growth properties of lymphocytes and embryonic fibroblasts suggest that sno may play different roles in proliferation control in different cell types.
Results
Targeted mutation of the sno gene
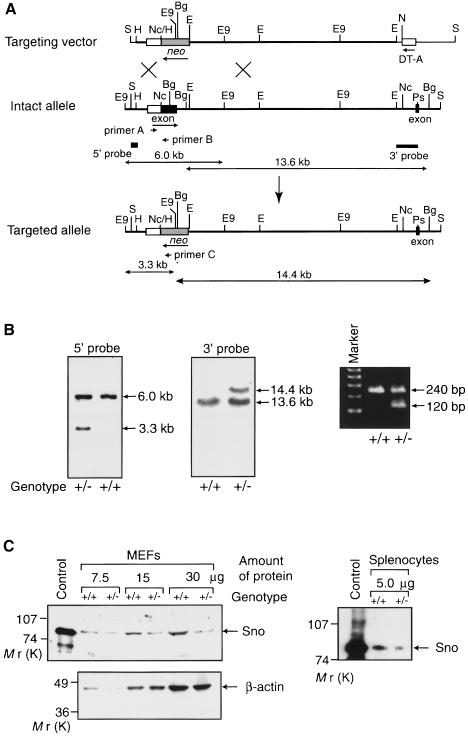
Mutant mice were generated by homologous recombination in TT2 embryonic stem (ES) cells. Genomic sno DNA clones were isolated from a TT2 genomic library and used to construct a gene-targeting vector. The exon encoding amino acids 1–362 of SnoN, which is common to all of the Sno protein species generated by alternative splicing, was replaced with a neo cassette in the gene-targeting vector (Figure 1A). We selected this large exon as a target because replacement of the exon containing the initiator methionine may severely affect the biosynthesis of full-length, normal Sno protein. Homologous recombinants were characterized by the appearance on Southern blots of a 3.3 kb EcoO109I fragment hybridizing to the 5′ probe, and a 14.4 kb BglI fragment detected by the 3′ probe (Figure 1B). Of 248 G418-resistant ES cell colonies, 12 (5%) were found to have a mutated target allele. Chimeras were obtained from two independent mutant ES clones using standard techniques and were mated with C57BL/6 females to generate F1 heterozygous mutant mice. Heterozygous mice were identified by Southern blotting and PCR analysis using genomic DNA isolated from tail biopsies (Figure 1B). To confirm that the expression level of full-length Sno protein in the sno+/– mice was reduced to about half of the wild-type level, Western blot analysis was performed using mouse embryonic fibroblasts (MEFs). The expression level of the 85 kDa Sno protein in sno+/– MEFs was reduced to about half of the wild-type level; a similar decrease in the level of Sno protein was observed in sno+/– splenocytes (Figure 1C). The protein fragment containing the C-terminal half of Sno, which can act as a dominant-negative form of Sno, was not detected by Western blotting using the anti-Sno monoclonal antibodies, which recognize the C-terminal half of SnoN.
Fig. 1. Generation of sno-deficient mice. (A) Diagrammatic representations of the mouse sno locus and targeting construct. The protein-coding regions of the two exons containing the N-terminal region of SnoN (amino acids 1–362) and the following 33 amino acid region are shown as closed boxes, while the 5′-untranslated region is indicated by an open box. The thick line indicates the genomic DNA, whereas the thin line shows the vector backbone. The neomycin resistance gene (neo) was inserted into the exon encoding the N-terminal region (amino acids 1–362). The probes used for Southern blot analyses are shown together with the predicted sizes of the hybridizing fragments. The three primers used for PCR are also indicated. B, BamHI; Bg, BglI; E, EcoRI; E9, EcoO109I; H, HindIII; Nc, NcoI; Ps, PstI; S, SalI. (B) Examples of genomic Southern blot (left and center panel) and PCR (right panel) analyses of wild-type (+/+) and heterozygous (+/–) mutant mice. Genomic DNA was isolated from the tails of mice and digested with EcoO109I or BglI for Southern blot analysis using the 5′ or 3′ probe, respectively. PCR was performed using the primers shown in (A). (C) Immunodetection of Sno protein. Extracts from wild-type (+/+) or sno+/– (+/–) MEFs or splenocytes were used for Western blotting with anti-Sno antibodies (above left). Extract from 293T cells transfected with the Sno expression vector was used as a control. To confirm equal loading of total protein in all lanes for MEFs, Western blotting was also carried out with an anti-actin antibody (below left). The loading of protein from splenocytes was adjusted by measuring protein concentrations using a dye-binding method.
Phenotype of sno–/– mice
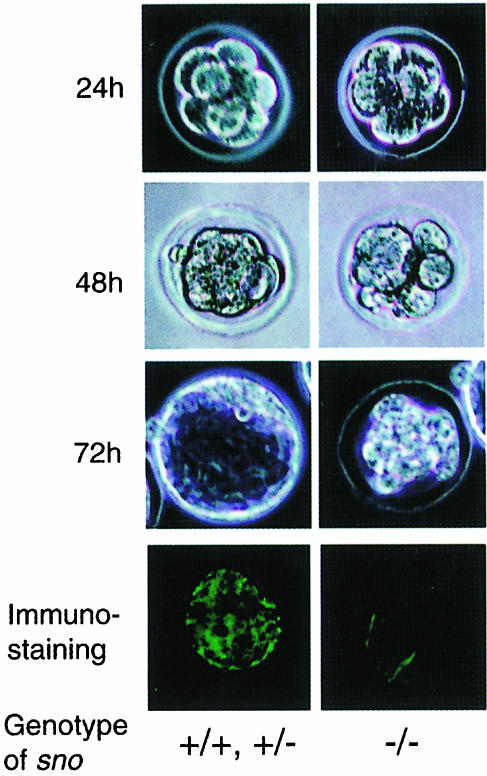
Heterozygous mice were intercrossed and the genotypes of resulting 3-week-old offspring were determined. Of the 211 offspring derived from two independent germline chimeras, none were sno–/– homozygotes; the ratio of sno+/+ to sno+/– genotypes was 1:2 (data not shown). In crosses of sno+/– and sno+/+ mice (in either gender combination), the ratio of sno+/+ and sno+/– genotypes in the progeny population was 1:1, indicating that sno– germ cells were not eliminated during spermatogenesis or oogenesis. Since 99 of the progeny were from intercrosses between two independent ES cell line-derived heterozygotes, the lack of sno-null progeny cannot be due to homozygosity of an undetected linked mutation inadvertently generated in either of the targeted ES cell lines. These results indicate that homozygosity for the sno-null allele is lethal during embryonic development. To determine when sno–/– animals died, the genotypes of post-implantation embryos [embryonic days (E) 3.5–16.5] from heterozygous matings were determined. No sno–/– embryos were obtained after E3.5 among 96 embryos examined (data not shown), suggesting that sno–/– embryos die before implantation. Under optimal growth conditions, blastocysts grown in vitro develop to the egg cylinder stage and can even be taken to the eight-somite stage (Gonda and Hsu, 1980; Wu et al., 1981). The series of developmental changes that occur during this process have been clearly delineated. To characterize the pre-implantation development of the sno–/– embryos, two-cell embryos derived from heterozygous intercrosses were recovered at E1.5, cultured individually and examined. About 25% of the embryos (n = 20) exhibited no significant staining with anti-Sno antibodies, and had a visible defect in blastocyst formation (Figure 2, right-hand panels). These embryos divided normally up to the eight-cell stage during the first 24 h in culture, but became decompacted during the second and third days and failed to divide beyond approximately the 16-cell stage or to form a blastocoel. The remaining embryos were strongly labeled by anti-Sno antibodies and developed into normal blastocysts (Figure 2, left-hand panels). These results indicate that sno is required for blastocyst formation. Since ski-deficient mice were alive at E18.5 (Berk et al., 1997), ski and sno appear to play different roles during development.
Fig. 2. sno is required for blastocyst formation. Embryos were recovered from (sno+/– × sno+/–) intercrosses at E1.5 (two-cell stage) and cultured for different lengths of time, as indicated. Photographs in the right-hand column show an abnormal embryo, which decompacts and fails to form a blastocyst, whereas those in the left-hand column show a normal embryo. Embryos at the eight-cell stage are also shown stained with Sno-specific monoclonal antibodies (bottom panels).
Tumor susceptibility of sno+/– mice
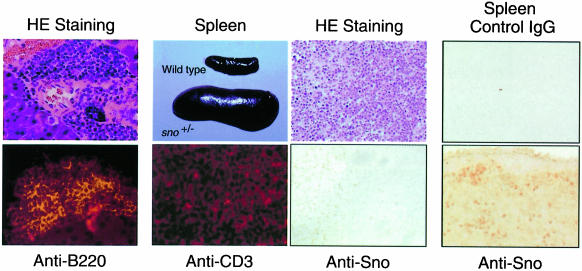
Although sno+/– heterozygotes appeared to be healthy, two of 104 sno+/– heterozygotes over the age of 4 months displayed severe wasting. Histological analysis of one of these mice (female, aged 4 months) revealed tumors in the pancreas, and immunostaining indicated that the tumors expressed the B-cell marker B220 (B lymphomas) (Figure 3, left-most panels). The other mouse, a 7-month-old female, developed tumors in the spleen, and immunostaining showed that the tumors expressed the T-cell marker CD3 (T lymphomas) (Figure 3, second panels). Although the frequency of spontaneous tumorigenesis in sno+/– heterozygotes was low (2/104), no tumors were observed in 220 wild-type mice in the same colony. Most of the tumor suppressors examined to date fulfill Knudson’s ‘two-mutation’ criterion (Knudson, 1971, 1997) and the wild-type allele is lost or expression of the protein is impaired in tumors related to these genes. Studies of various tumor suppressor genes indicated that loss of expression from the wild-type allele is caused by multiple mechanisms, including rearrangement or deletion of the gene, point mutation and methylation. To determine directly whether expression of Sno protein from the wild-type allele in sno+/– mice was lost in tumors, we immunostained Sno protein in tumor cells. Although we examined only one spontaneously developed tumor, Sno protein was not detected in this tumor (Figure 3, third panels), suggesting that sno may be a canonical ‘two-hit’ tumor suppressor.
Fig. 3. Histological analysis of a spontaneously arising tumor in sno+/– mouse. Left-most panels: B lymphoma in pancreas. Hematoxylin and eosin (HE) staining of a tumor in the pancreas (above). The tumor is positive for the B-cell marker B220 (below). Second panels: T lymphomas in spleen. Splenomegaly appeared in the tumor-bearing sno+/– mouse (above). The tumor is positive for the T-cell marker CD3 (below). Third panels: T lymphomas in lymph node. Staining with HE (above) or anti-Sno antibody (below). Note that nuclei of normal lymphocytes were stained with anti-Sno antibody, but those of tumor cells were not. Right-most panels: T-cell-rich region in wild-type spleen as a control. Staining with control IgG (above) or anti-Sno antibody (below).
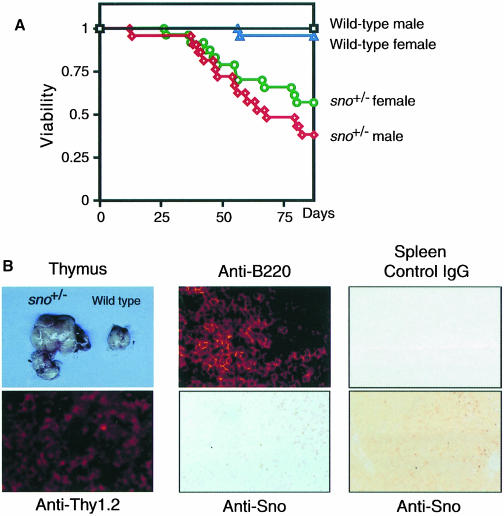
In light of the spontaneous development of malignant lymphomas of B- or T-cell origin in sno+/– heterozygotes, we further examined the possibility that sno might act as a tumor suppressor by comparing the tumor susceptibility of wild-type and sno+/– mice during a course of intraperitoneal injections of the carcinogen 9,10-dimethyl-1,2-benzanthracene (DMBA). Over an observation period of 80 days, most of the 43 wild-type mice, but only 56.5% of the 23 sno+/– female mice and 42.9% of the 21 sno+/– male mice survived this treatment and remained free of clinically apparent tumors (Figure 4A; p <0.001). In total, 22 sno+/– mice died with clinically apparent tumors, most of which were malignant lymphomas: 19 mice developed lymphomas (six of which were T-cell, four B-cell and nine of indeterminate origin; the latter were negative for T- and B-cell markers of CD3, Thy1.2, B220 and IgM), two mice developed both T lymphoma and fibroma, and one mouse developed both lymphoma of indeterminate origin and fibroma. Histological analysis of a typical tumor is shown in Figure 4B. Lymphomas in sno+/– mice homogeneously expressed the T-cell marker Thy-1.2 (T lymphomas) (Figure 4B, left panels). Among the seven DMBA-induced tumors examined, expression of Sno protein was completely lost in three tumors (42%) (Figure 4B, middle panels). Thus, sno fulfills Knudson’s ‘two-mutation’ criterion for a tumor suppressor gene (Knudson, 1971). We could not analyze further the remaining four tumors that expressed Sno because we could not isolate enough tumor cells. Therefore, we cannot exclude the possibility that a mutated Sno protein is expressed in these tumors.
Fig. 4. Chemical carcinogen-induced tumor formation in sno+/– mice. (A) Survival curves of DMBA-treated wild-type and sno+/– mice. For all informative cases, the cause of death was the development of a clinically apparent and histologically confirmed tumor. (B) Histopathological analysis of tumors developed in DMBA-treated sno+/– mice. Left panels: T lymphoma. The enlarged thymus of a tumor-bearing sno+/– mouse is shown (above). The tumor was positive for the T-cell marker Thy-1.2 (below). Center panels: B lymphoma in lymph node. The tumor was positive for B220 (above). Immunostaining with anti-Sno antibody (below). Note that nuclei of normal lymphocytes were stained with anti-Sno antibody, but those of tumor cells were not. Right panels: germinal center in wild-type spleen as a control. Staining with control IgG (above) or anti-Sno antibody (below).
Growth properties of sno+/– lymphocytes
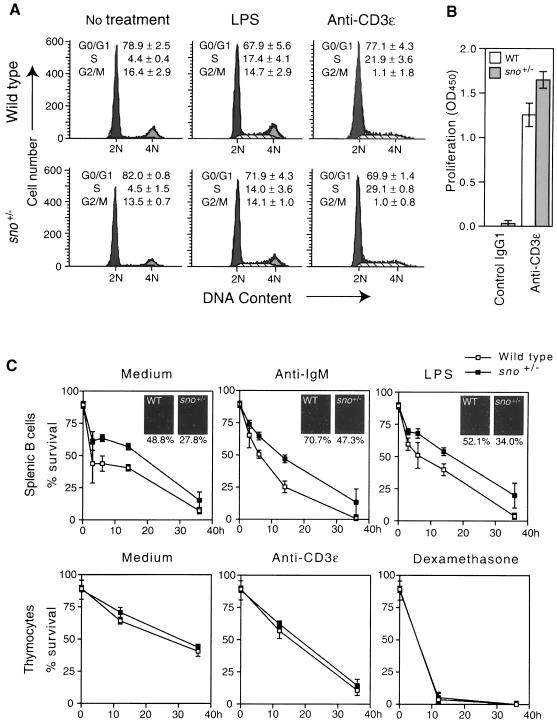
To investigate the role of sno in cellular growth control, we examined the effects of loss of one copy of sno on the growth of specific mouse cell types, including lymphocytes and embryonic fibroblasts. Loss of sno expression in three out of seven tumors induced by DMBA suggests that complete loss of Sno leads to neoplastic transformation. However, we examined the possibility that loss of one copy of the sno gene might predispose cells to tumor formation, partly because sno–/– mice die at an early stage of development, which renders tumor formation studies in these mice impossible. To evaluate the proliferative capacity of sno+/– and control lymphocytes, we cultured lymph node cells from 6- to 8-week-old mice in the presence or absence of B- or T-cell-specific mitogens [bacterial lipopolysaccharide (LPS) or anti-CD3 antibody], and assayed the cell cycle profile. In anti-CD3-treated sno+/– cells there was an increase in exit from the G1 phase of the cell cycle (Figure 5A, right panels; ∼7% decrease in the G1 population of anti-CD3-treated sno+/– cells), whereas no decrease in the G1 population of LPS-treated sno+/– samples was observed (Figure 5A, middle panels). Furthermore, anti-CD3-treated sno+/– cells incorporated more bromodeoxyuridine (BrdU) than wild-type cells (Figure 5B), supporting the increase in S-phase population of anti-CD3-treated sno+/– cells. There was, however, a significant decrease in the percentage of apoptotic cells in the LPS-treated sno+/– samples (data not shown). To confirm this, we examined the rate of cell death in B and T cells. As measured by viability dye staining, sno+/– B cells were slightly, but significantly, more resistant to apoptosis induced by medium, anti-IgM or LPS (Figure 5C, upper panels). The results with viability dye staining were confirmed by detecting apoptotic cells using TUNEL labeling (Figure 5C, small panels). In contrast, there was no difference in the sensitivity to apoptosis induced by medium, anti-CD3 or dexamethasone between wild-type and sno+/– T cells (Figure 5C, lower panels). These results suggest that sno+/– B cells are less sensitive to apoptosis, while sno+/– T cells have a defect in cell cycle arrest.
Fig. 5. Role of sno in apoptosis and cell cycle arrest of lymphocytes. (A) Cell cycle analysis. Lymph node cells were treated with LPS or anti-CD3, labeled with propidium iodide, and analyzed by FACScan. The average population and standard deviation of triplicate measurements are shown. (B) BrdU incorporation. Purified lymph node T cells were plated into a plate coated with anti-CD3ε antibody or control IgG1, and cell proliferation was assessed by incorporation of BrdU. The average of triplicate measurements is shown with the standard deviation. (C) Decreased sensitivity of sno+/– B cells to apoptosis. B cells purified from spleen or thymocytes were treated with the reagents indicated, and cell viability was determined by negative staining with 7-amino-actinomycin D. Experiments were repeated three times; average values are indicated together with standard deviations. Insets: cells were also treated with the indicated reagents for 15 h, and apoptotic cells were detected by TUNEL labeling. The percentage ratio of the number of apoptotic cells to the total number of cells is indicated.
Growth properties of sno+/– MEFs
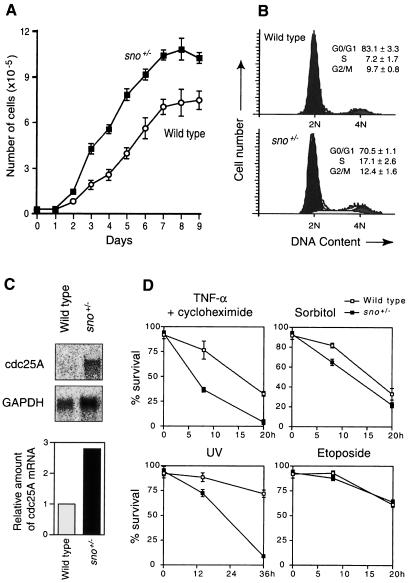
We next assayed the proliferative properties of cultured MEFs using growth curves and flow cytometry of exponentially growing cultures. Although sno+/+ and sno+/– cultures were morphologically indistinguishable at low density, sno+/– MEFs grew more quickly than wild-type MEFs (Figure 6A). In addition, sno+/– MEF monolayers achieved 30% higher cell densities than wild-type MEFs (Figure 6A). Analysis of cell cycle profiles indicated that there was a 9.9% increase in the S-phase population of sno+/– cells relative to the wild type (Figure 6B). These results suggest that sno+/– MEFs have a defect in cell cycle arrest. We speculated that decreased activity of Mad or Rb in sno+/– MEFs may cause a defect in cell cycle arrest. One of the most famous target genes that is down-regulated by Rb and Mad is the cdc25A gene (Galaktionov et al., 1996; Neufeld et al., 1998). We examined the cdc25A mRNA levels in wild-type and sno+/– MEFs (Figure 6C). The level of cdc25A mRNA in the sno+/– mutant was 2.8-fold higher than that in wild type.
Fig. 6. Growth advantage conferred on MEFs by the loss of one copy of sno. (A) Growth curves of MEFs. The average and standard deviation of triplicate measurements are shown for each time point. (B) Cell cycle analysis. Exponentially growing MEFs were stained with propidium iodide and analyzed by FACScan. The average population and standard deviation of triplicate measurements are shown. (C) Expression of cdc25A. Poly(A)+ RNA was isolated from confluent MEFs and expression of cdc25A and GAPDH was analyzed by Northern blotting. The amount of cdc25A mRNA was normalized with respect to the GAPDH mRNA level, and the relative amount is indicated as a bar graph. (D) Analysis of apoptosis of MEFs. Apoptosis was induced and cells were assayed at the indicated time points. TNF-α (10 ng/ml) and cycloheximide (5 µg/ml); sorbitol (0.4 M); UV irradiation (390 J/m2); etoposide (100 µM). The average and standard deviation of triplicate measurements are shown for each time point.
We also examined the rate of cell death in MEFs (Figure 6D). sno+/– MEFs were more sensitive to apoptosis induced by tumor necrosis factor-α (TNF-α) and cycloheximide, UV or sorbitol than wild-type cells. In contrast, the two types of cell did not differ in their sensitivity to apoptosis induced by etoposide. These results suggest that sno+/– MEFs have an increased sensitivity to apoptosis induced by specific stimuli.
As a further measure of proliferative potential, we compared the abilities of sno+/– and wild-type MEFs to produce visible foci following low-density seeding; sno+/– cells were better at generating actively growing colonies of clonal origin (Table I, left column). We then measured colony formation in methylcellulose after infection with murine leukemia virus (MuLV) carrying the v-K-ras oncogene. sno+/– MEFs generated 32–85 colonies per 103 cells, whereas wild-type MEFs generated no colonies (Table I, middle column). Under the same conditions, p53-deficient MEFs produced 230 colonies per 103 cells. MEFs expressing the v-K-ras oncogene were injected subcutaneously into nude mice, and tumor formation was examined. sno+/– MEFs expressing v-K-ras were significantly more tumorigenic in nude mice than wild-type MEFs, although their tumor-forming capacity was weaker than that of p53-deficient MEFs (Table I, right column). These results indicate that loss of one copy of the sno gene facilitated transformation by activated ras in cell culture.
Table I. Loss of one copy of sno predisposes MEFs to tumorigenic transformation.
| MEF | Number of focia | Number of colonies/103 cellsb | Tumors/number of injectionsc | |
|---|---|---|---|---|
| |
|
No virus |
v-K-ras |
|
| Wild type | ||||
| E14-1 | 0.3 ± 0.6 | 0 | 0.0 ± 0.0 | ND |
| E14-2 | 0.7 ± 1.2 | 0 | 0.7 ± 0.6 | ND |
| E14-3 | 0.7 ± 0.6 | 0 | 0.3 ± 0.6 | 0/12 |
| E14-6 | 0.7 ± 1.2 | 0 | 0.3 ± 0.5 | ND |
| Sno+/– | ||||
| E14-4 | 6.0 ± 2.7 | 0 | 72.3 ± 6.3 | ND |
| E14-7 | 21.0 ± 3.6 | 0 | 85.3 ± 8.5 | ND |
| E15-1 | 22.7 ± 6.2 | 0 | 57.7 ± 9.8 | 3/12 |
| E15-2 | 10.7 ± 0.6 | 0 | 31.6 ± 7.9 | ND |
| p53–/– | ND | 0 | 230.0 ± 26.1 | 12/12 |
Focus formation and neoplastic transformation were assayed using MEFs. Four independent preparations of MEFs from wild-type or sno+/– embryos were used.
aThe number of foci arising 12 days after low-density seeding. The average population and standard deviation of triplicate measurements are indicated.
bThe number of colonies generated in methylcellulose gel after infection with MuLV carrying the activated K-ras oncogene. MEFs prepared from p53-deficient mice were used as a control. Experiments were repeated three times, and the average number of colonies formed per 103 cells is indicated together with the standard deviation.
cNumber of tumors formed when cells expressing v-K-ras were injected into nude mice. ND, not determined.
Discussion
The susceptibility of sno+/– mice to spontaneous and carcinogen-induced tumors indicates that sno acts as a tumor suppressor, although ski and sno have been considered oncogenes to date. We also observed that ski-heterozygous mice showed an increased level of tumor formation relative to wild-type mice when challenged with a chemical carcinogen (our unpublished results), suggesting that ski and sno act as tumor suppressors in a similar fashion. Similar observations have been made concerning E2F-1 and PML. Overexpression of E2F-1 in tissue culture cells stimulates cell proliferation and is oncogenic, but mice lacking E2F-1 develop a broad spectrum of tumors (Field et al., 1996; Yamasaki et al., 1996). The PML gene was originally identified as a PML–retinoic acid receptor (RAR) fusion oncogene causing leukemia; however, later studies showed it to be a tumor suppressor gene (Doucas and Evans, 1996; Wang et al., 1998). These results are consistent with the observations that overexpression of ski or sno is accompanied by myogenic differentiation, which is associated with growth inhibition (Colmenares and Stavnezer, 1989; Sutrave et al., 1990; Boyer et al., 1993) and that v-Ski acts in a dominant-negative fashion (Nomura et al., 1999; Tokitou et al., 1999). Our observations regarding lymphoma in sno+/– mice may be consistent with the fact that v-ski can transform chicken bone marrow cells (Larsen et al., 1993). Although we found differences in growth properties between sno+/– and wild-type cells (cell cycle arrest or sensitivity to apoptosis, depending on the cell type), these differences were small in comparison with the reported effects of other tumor suppressors. However, it should be noted that most studies to date have used homozygous mutant cells completely lacking the tumor suppressor protein. In contrast, we used heterozygous sno mutant cells that retain one wild-type allele. Since we observed that expression of Sno protein was completely lost in three of seven DMBA-induced tumors, it appears that complete loss of Sno may be needed for tumor formation. Although we could not analyze the remaining four tumors that expressed Sno, we cannot exclude the possibility that the mutated Sno protein is expressed in these tumors. It is likely that that complete loss of Sno would generate a much more striking difference in growth properties than loss of one copy of the gene, although we could not confirm this because of difficulty in obtaining sno–/– cells.
It is likely that v-Ski blocks the repressor activity of Rb and Mad in a dominant-negative fashion and induces transformation. However, the mechanism by which overexpression of normal c-Ski or Sno also transforms chicken embryo fibroblasts remains unknown. There appear to be two possible mechanisms. First, overexpression of wild-type c-Ski or Sno could lead to an imbalance between the components of the HDAC complex and could abrogate transcriptional repression mediated by Mad or Rb, rather than potentiating transcriptional repression. In fact, we observed that overexpression of c-Ski partly abrogates Mad- and Rb-mediated transcriptional repression (Nomura et al., 1999; Tokitou et al., 1999). In addition, the punctate pattern of N-CoR localization in the nuclei was disrupted by co-expression of a high level of c-Ski relative to that of N-CoR (Nomura et al., 1999). The importance of the balance between the components of the HDAC complex is also suggested by the observation that transcriptional repression by RAR can be either positively or negatively regulated by changes in the levels of N-CoR expression (Söderström et al., 1997). The second possibility is that ski and sno may act as either tumor suppressors or oncogenes, depending on the cell type. In addition to Rb, Mad and nuclear hormone receptors, many other repressors may also require Ski/Sno for their regulatory activity. Since these putative repressors may be expressed in various cells at different levels, the effect of loss of Ski/Sno on cellular proliferation may differ depending on the cell type. Recently, it was reported that c-Ski binds directly to Smad proteins, which are key regulators in TGF-β signaling (Akiyoshi et al., 1999; Luo et al., 1999; Stroschein et al., 1999; Sun et al., 1999a,b). c-Ski inhibits TGF-β-induced transcriptional activation by recruiting the HDAC complex to Smad proteins. These findings are consistent with the previous observation that the Smad-binding site mediates transcriptional repression by Ski (Nicol and Stavnezer, 1998). Therefore, overexpression of c-Ski/Sno may enhance proliferation by inhibiting TGF-β signaling in TGF-β-responsive cells.
The role of sno in proliferation control appears to depend on the cell type: loss of one copy of sno affected cell cycle arrest in T cells and MEFs, and apoptosis in B cells and MEFs. This suggests that different transcriptional repressors may mediate the effect of Sno on proliferation control in different cell types. Ski and Sno are required for the repressor function of the tumor suppressor gene products, Rb and Mad (Nomura et al., 1999; Tokitou et al., 1999). Since both Rb and Mad appear to be positive regulators of cell cycle arrest (Goodrich and Lee, 1992; Leone et al., 1997; Zhang et al., 1999), the defect in cell cycle arrest observed in sno+/– T cells and MEFs could be a result of decreased Rb and/or Mad activity. In fact, the cdc25A gene is upregulated in sno+/– MEFs (Figure 6C). Loss of one copy of sno decreases sensitivity to apoptosis in B cells, whereas it leads to increased sensitivity to apoptosis in MEFs. The mechanism by which Sno affects apoptosis in B cells and MEFs is unclear.
The sno gene is located between 3q26.31 and 3q26.32 (Shinagawa et al., unpublished results), a region to which one of the highest frequencies of loss of constitutional heterozygosity in human osteosarcoma has also been mapped (Kruzelock et al., 1997). The human c-ski gene maps to 1p36 (T.Shinagawa et al., unpublished results), close to the reported location of multiple uncharacterized tumor suppressor genes for neuroblastoma (Cheng et al., 1995). Together with our results, these observations indicate that the human ski gene family may be important for tumor susceptibility.
Many transcriptional regulators act through shared transcriptional mediators (co-activators and co-repressors), which are thought to be available in limited amounts. This is consistent with the observation that loss of one copy of the co-activator CBP gene causes abnormal pattern formation (Petrij et al., 1995; Tanaka et al., 1997). Similarly, we have shown that loss of one copy of sno increases susceptibility to tumor formation. This is consistent with the fact that multiple transcriptional repressors utilize the HDAC complex, which contains Ski/Sno. These results suggest that mutation of other genes encoding transcriptional mediators may also be responsible for some autosomal-dominant genetic diseases.
Materials and methods
Generation of sno-deficient mutant mice
Genomic clones of sno were isolated from a library derived from TT2 cells by a standard hybridization procedure. A 15.0 kb HindIII–EcoRI genomic DNA subfragment was used to generate the targeting vector. A 1.5 kb NcoI–EcoRI fragment containing the exon encoding the N-terminal 362 amino acids of Sno was deleted and replaced with a neomycin (neo) cassette driven by the phosphoglycerate kinase gene promoter. To increase the frequency of gene targeting, the diphtheria toxin poly(A) signal (DT-A) cassette for negative selection was fused to the short arm (Yagi et al., 1993a). The ES cells used were TT2 cells derived from an F1 embryo resulting from a cross between C57BL/6 and CBA mice (Yagi et al., 1993b). Isolation of ES clones containing the sno mutation, generation of chimeras and production of heterozygous mutants were performed as described elsewhere (Tanaka et al., 1997). The homologous nature of recombination was confirmed by Southern blot analysis using several restriction enzymes and several probes located either inside or outside of the targeting vector (Figure 1). In addition, three different primers were used to amplify a 240 bp fragment from the wild-type allele or a 120 bp fragment from the mutant allele. PCR (35 cycles) was carried out using the following primers: forward sno primer, 5′-GCATCTCCATAGTCATCTAA-3′; reverse sno primer, 5′-AGT TAAGGGCCAGCTCATTC-3′; neo primer, 5′-GATTAGGAACTG AGAACAAC-3′. One PCR cycle consisted of 94°C for 45 s, 55°C for 30 s and 72°C for 1 min. The mice were maintained by the Division of Experimental Animal Research of RIKEN Institute.
Detection of Sno protein
To induce the expression of Sno protein, MEFs were cultured for a further 7 days after reaching confluency, then washed in phosphate-buffered saline (PBS) and resuspended in 0.3 ml of lysis buffer [45 mM Tris–HCl pH 7.4, 270 mM NaCl, 0.9% Triton X-100, 0.9% sodium deoxycholate, 0.09% sodium dodecyl sulfate (SDS), 22 mM EDTA and 1 mM phenylmethylsulfonyl fluoride (PMSF)]. After rotation for 1 h at 4°C, lysates were centrifuged at 15 000 r.p.m. for 20 min, and the supernatant was mixed with 0.2 vols of 6× SDS sample buffer. Proteins in the supernatant were resolved on 10% SDS–polyacrylamide gels, transferred on to PVDF membrane (Immobilon, Millipore) and probed with anti-Sno monoclonal antibodies (mixtures of four kinds of monoclonal antibody) (Tokitou et al., 1999) or anti-actin antibody (I-19, Santa Cruz). Western blots were developed using ECL detection reagents (Amersham). Similar analysis was performed using nuclear extracts from splenocytes prepared as described elsewhere (Tanaka et al., 1999).
In vitro culture of two-cell embryos and immunostaining
M2 and M16 media were prepared as described by Hogan et al. (1986). Embryos were obtained by mating sno+/– males and females. The day on which a vaginal plug was found was considered to be day 0 post-coitus (0 d.p.c.). Embryos were flushed from the oviducts at 1.5 d.p.c. using M2 medium, cultured individually in drops of M16 medium under paraffin oil at 37°C in a humidified atmosphere containing 5% CO2 and examined periodically with a dissecting microscope. For immunostaining, embryos were fixed at room temperature for 30 min in freshly prepared 2.5% paraformaldehyde in saline buffer (pH 7.5) and then treated with methanol for 30 min followed by 4% hydrogen peroxide for 30 min. Anti-Sno monoclonal antibodies were used to stain the cells and primary antibody–antigen complexes were detected using fluorescein isothiocyanate (FITC)-conjugated secondary antibodies and confocal microscopy.
Histological analysis and immunohistochemistry
Tissues were fixed in 4% paraformaldehyde, dehydrated and embedded in paraffin. Sections (5 µm) were stained with hematoxylin and eosin according to standard procedures. Paraffin sections of 4 µm were used for immunohistochemistry. Phycoerythrin-conjugated anti-Thy-1.2, anti-CD3-ε and anti-B220 antibodies (30-H12, 500A2 and RA3-6B2, respectively; all from PharMingen) were used for immunostaining. For Sno protein staining, deparaffinized sections were incubated in 3% hydrogen peroxide for 10 min to block endogenous peroxidase. Specimens were microwaved for 10 min in the presence of 10 mM sodium citrate pH 6.0, treated with 0.3% Triton X-100 in PBS for 10 min at room temperature and then incubated overnight at 37°C with anti-Sno monoclonal antibody 3′-1 (1:10). Horseradish peroxidase-conjugated anti-mouse IgG1 antibody was used as secondary antibody and 3,3′-diaminobenzine tetrahydrochloride (DAB) as a chromogen. Isotype-matched mouse antibody (IgG1) was used for negative controls.
DMBA tumorigenic treatment
A lipid emulsion of DMBA (0.5% w/w, Sigma) was injected intraperitoneally. The first dose, 35 mg/kg body weight, was given 2–3 months after birth, followed by successive injections of 17.5 mg/kg at 2 week intervals. Mice were monitored regularly and killed when their overall health was compromised. A full histopathological survey was conducted on the first 8–10 mice in each cohort; thereafter, detailed pathological analysis was restricted to the site of tumor involvement and organs of potential metastatic involvement.
Analysis of sno+/– lymphocytes
For cell cycle analysis, lymph node cells (2 × 105 cells/well) were activated with LPS (10 µg/ml) or plate-bound anti-CD3-ε (10 µg/ml) in culture medium. After 6 h with LPS or 66 h with anti-CD3-ε, cells were suspended in 50 µg/ml propidium iodide (Sigma), 4 mM sodium citrate, 150 mM NaCl, 0.5 mg/ml RNase A and 0.05% Nonidet P-40, and were analyzed by FACScan (Becton Dickinson). Cell cycle distributions were analyzed using the CellQuest and ModFit LT software packages. For analysis of BrdU incorporation, T cells (2 × 105) purified by using MACS CD45R (B220) and CD11b (Mac-1) MicroBeads (Miltenyi Biotec) were plated in 96-well plates coated with anti-CD3ε antibody (10 µg/ml) or control hamster IgG1 (10 µg/ml) and grown for 60 h. BrdU was added to the cells for an additional 6 h. Fixed cells were treated using the Biotrak cell proliferation ELISA system (Amersham) and the resultant color developed by the anti-BrdU peroxidase-labeled immune complexes was read at 450 nm in an OPTImax microplate reader (Molecular Devices). For analysis of apoptotic profiles, B cells were purified from spleen by negative selection using anti-Thy1.2 and anti-CD11b magnetic beads (Milteni Biotec). B cells or thymocytes were seeded at 2 × 105 cells/well and treated with anti-IgM antibody (10 µg/ml), LPS (10 µg/ml), anti-CD3 antibody (10 µg/ml) or dexamethasone (10 nM). Cell viability was determined by staining with 7-amino-actinomycin D (Sigma) and flow cytometry as described by Philpott et al. (1996).
Analysis of embryonic fibroblasts
MEFs were isolated from embryos at 14.5 d.p.c. For growth curves, cells were grown from a starting density of 3 × 104 cells/well in 24-well plates in Dulbecco’s modifed Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Nine independent experiments were carried out from four different MEF preparations. For cell cycle analysis, exponentially growing cells were stained with propidium iodide and analyzed by FACScan as described above. Cell viability was determined as described above. To determine the efficiency of focus formation, cells were seeded into 6 cm dishes at 100 cells/dish and incubated in DMEM plus 10% FBS. After 12 days, samples were stained with Giemsa and the number of visible colonies was scored. For colony formation assays, MEFs were infected with MuLV carrying the activated K-ras oncogene (at a multiplicity of infection of 1). After 48 h, 3 × 104 cells were suspended in 1.3% methylcellulose gel dissolved in culture medium and overlaid on an agarose bed (0.53% agarose in culture medium). Colonies were scored 3 weeks after plating. To test the tumorigenicity of the cell clones, nude mice (BALB/c nu/nu; Clea Japan Inc.) were injected subcutaneously with 106 cells resuspended in 200 µl of DMEM without FCS. Cells were scored as tumorigenic if a visible nodule >2 mm in diameter appeared at the site of injection 10 days after injection.
Northern analysis
MEFs were seeded at 3 × 106 cells/10 cm dish and harvested 3 days later. Total RNA was isolated by Isogen (Nippon Gene, Japan) and poly(A)+ RNA was isolated using a mRNA purification kit (Amersham Pharmacia). Five micrograms of poly(A)+ RNA were electrophoresed in a 1% agarose–formaldehyde gel and transferred on to Hybond-N+ membrane (Amersham). The blot was hybridized with a 32P-labeled cdc25A probe or a GAPDH probe.
Acknowledgments
Acknowledgements
We thank M.Noda for the kind gift of MuLV, and C.Colmenares and E.Stavnezer for helpful discussion.
References
- Akiyoshi S., Inoue,H., Hanai,J., Kusanagi,K., Nemoto,N., Miyazono,K. and Kawabata,M. (1999) c-Ski acts as a transcriptional co-repressor in transforming growth factor-β signaling through interaction with smads. J. Biol. Chem., 274, 35269–35277. [DOI] [PubMed] [Google Scholar]
- Alland L., Muhle,R., Hou,H.,Jr, Potes,J., Chin,L., Schreiber-Agus,N. and DePinho,R.A. (1997) Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature, 387, 49–55. [DOI] [PubMed] [Google Scholar]
- Ayer D.E. and Eisenman,R.N. (1993) A switch from Myc:Max to Mad:Max heterocomplexes accompanies monocyte/macrophage differentiation. Genes Dev., 7, 2110–2119. [DOI] [PubMed] [Google Scholar]
- Ayer D.E., Kretzner,L. and Eisenman,R.N. (1993) Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell, 72, 211–222. [DOI] [PubMed] [Google Scholar]
- Ayer D.E., Lawrence,Q.A. and Eisenman,R.N. (1995) Mad–Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell, 80, 767–776. [DOI] [PubMed] [Google Scholar]
- Berk M., Desai,S.Y., Heyman,H.C. and Colmenares,C. (1997) Mice lacking the ski proto-oncogene have defects in neurulation, craniofacial patterning, and skeletal muscle development. Genes Dev., 11, 2029–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer P.L., Colmenares,C., Stavnezer,E. and Hughes,S.H. (1993) Sequence and biological activity of chicken snoN cDNA clones. Oncogene, 8, 457–466. [PubMed] [Google Scholar]
- Brehm A., Miska,E.A., McCance,D.J., Reid,J.L., Bannister,A.J. and Kouzarides,T. (1998) Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature, 391, 597–601. [DOI] [PubMed] [Google Scholar]
- Chen J.D. and Evans,R.M. (1995) A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature, 377, 454–457. [DOI] [PubMed] [Google Scholar]
- Cheng N.C., Van Roy,N., Chan,A., Beitsma,M., Westerveld,A., Speleman,F. and Versteeg,R. (1995) Deletion mapping in neuroblastoma cell lines suggests two distinct tumor suppressor genes in the 1p35-36 region, only one of which is associated with N-myc amplification. Oncogene, 10, 291–297. [PubMed] [Google Scholar]
- Chin L., Schreiber-Agus,N., Pellicer,I., Chen,K., Lee,H.-W., Dudast,M., Cordon-Cardo,C. and Depinho,R.A. (1995) Contrasting roles for Myc and Mad proteins in cellular growth and differentiation. Proc. Natl Acad. Sci. USA, 92, 8488–8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmenares C. and Stavnezer,E. (1989) The ski oncogene induces muscle differentiation in quail embryo cells. Cell, 59, 293–303. [DOI] [PubMed] [Google Scholar]
- Colmenares C., Sutrave,P., Hughes,S.H. and Stavnezer,E. (1991) Activation of the c-ski oncogene by overexpression. J. Virol., 65, 4929–4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl R., Kieslinger,M., Beug,H. and Hayman,M.J. (1998) Transformation of hematopoietic cells by the Ski oncoprotein involves repression of retinoic acid receptor signaling. Proc. Natl Acad. Sci. USA, 95, 11187–11192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucas V. and Evans,R.M. (1996) The PML nuclear compartment and cancer. Biochim. Biophys. Acta, 1288, 25–29. [DOI] [PubMed] [Google Scholar]
- Field S.J., Tsai,F.-Y., Kuo,F., Zubiaga,A.M., Kaelin,W.G.,Jr, Livingston,D.M., Orkin,S.H. and Greenberg,M.E. (1996) E2F-1 functions in mice to promote apoptosis and supresses proliferation. Cell, 85, 549–561. [DOI] [PubMed] [Google Scholar]
- Galaktionov K., Chen,X. and Beach,D. (1996) Cdc25 cell-cycle phosphatase as a target of c-myc. Nature, 382, 511–517. [DOI] [PubMed] [Google Scholar]
- Gonda M.A. and Hsu,Y.-C. (1980) Correlative scanning electron, transmission electron, and light microscopic studies of the in vitro development of mouse embryos on a plastic substrate at the implantation stage. J. Embryol. Exp. Morphol., 56, 23–39. [PubMed] [Google Scholar]
- Goodrich D.W. and Lee,W.H. (1992) Abrogation by c-myc of G1 phase arrest induced by RB protein but not by p53. Nature, 360, 177–179. [DOI] [PubMed] [Google Scholar]
- Hassing C.A., Fleischer,T.C., Billin,A.N., Schreiber,S.L. and Ayer,D.E. (1997) Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell, 89, 341–347. [DOI] [PubMed] [Google Scholar]
- Heinzel T. et al. (1997) A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature, 387, 43–48. [DOI] [PubMed] [Google Scholar]
- Heyman H.C. and Stavnezer,E. (1994) A carboxyl-terminal region of the Ski oncoprotein mediates homodimerization as well as heterodimerization with the related protein, SnoN. J. Biol. Chem., 269, 26996–27003. [PubMed] [Google Scholar]
- Hogan B., Constantini,F. and Lacy,E. (1986) Manipulating the Mouse Embryo: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Hörlein A.J. et al. (1995) Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature, 377, 397–404. [DOI] [PubMed] [Google Scholar]
- Knudson A.G. Jr (1971) Mutation and cancer: statistical study of retinoblastoma. Proc. Natl Acad. Sci. USA, 68, 820–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudson A.G. Jr (1997) Hereditary predisposition to cancer. Ann. N. Y. Acad. Sci., 833, 58–67. [DOI] [PubMed] [Google Scholar]
- Kruzelock R.P., Murphy,E.C., Strong,L.C., Naylor,S.L. and Hansen,M.F. (1997) Localization of a novel tumor suppressor locus on human chromosome 3q important in osteosarcoma tumorigenesis. Cancer Res., 57, 106–109. [PubMed] [Google Scholar]
- Laherty C.D., Yang,W.-M., Sun,J.-M., Davie,J.R., Seto,E. and Eisenman,R.N. (1997) Histone deacetylase associated with the mSin3 corepressor mediates Mad transcriptional repression. Cell, 89, 349–356. [DOI] [PubMed] [Google Scholar]
- Laherty C.D. et al. (1998) SAP30, a component of the mSin3 corepressor complex involved in N-CoR-mediated repression by specific transcription factors. Mol. Cell, 2, 33–42. [DOI] [PubMed] [Google Scholar]
- Larsen J., Meyer,S., Steinlein,P., Beug,H. and Hayman,M.J. (1993) Transformation of chicken bone marrow cells by the v-ski oncogene. Oncogene, 8, 3221–3228. [PubMed] [Google Scholar]
- Leone G., DeGregori,J., Sears,R., Jakoi,L. and Nevins,J.R. (1997) Myc and Ras collaborate in inducing accumulation of active cyclin E/Cdk2 and E2F. Nature, 387, 422–426. [DOI] [PubMed] [Google Scholar]
- Li Y., Turck,C.M., Teumer,J.K. and Stavnezer,E. (1986) Unique sequence, ski, in Sloan–Kettering avian retroviruses with properties of a new cell-derived oncogene. J. Virol., 57, 1065–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K., Stroschein,S.L., Wang,W., Chen,D., Martens,E., Zhou,S. and Zhou,Q. (1999) The Ski oncoprotein interacts with the Smad proteins to repress TGFβ signaling. Genes Dev., 13, 2196–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R.X., Postigo,A.A. and Dean,D.C. (1998) Rb interacts with histone deacetylase to repress transcription. Cell, 92, 463–473. [DOI] [PubMed] [Google Scholar]
- Magnaghi-Jaulin L., Groisman,R., Naguibneva,I., Robin,P., Lorain,S., Le Villain,J.P., Troalen,F., Trouche,D. and Harel-Bellan,A. (1998) Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature, 391, 601–605. [DOI] [PubMed] [Google Scholar]
- Nagase T., Nomura,N. and Ishii,S. (1993) Complex formation between proteins encoded by the ski gene family. J. Biol. Chem., 268, 13710–13716. [PubMed] [Google Scholar]
- Nagy L., Kao,H.-Y., Chakravarti,D., Lin,R.J., Hassig,C.A., Ayer,D.E., Schreiber,S.L. and Evans,R.M. (1997) Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell, 89, 373–380. [DOI] [PubMed] [Google Scholar]
- Neufeld T.P., de la Cruz,A.F., Johnston,L.A. and Edgar,B.A. (1998) Coordination of growth and cell division in the Drosophila wing. Cell, 93, 1183–1193. [DOI] [PubMed] [Google Scholar]
- Nevins J.R. (1992) E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science, 258, 424–429. [DOI] [PubMed] [Google Scholar]
- Nicol R. and Stavnezer,E. (1998) Transcriptional repression by v-Ski and c-Ski mediated by a specific DNA binding site. J. Biol. Chem., 273, 3588–3597. [DOI] [PubMed] [Google Scholar]
- Nomura N., Sasamoto,S., Ishii,S., Date,T., Matsui,M. and Ishizaki,R. (1989) Isolation of human cDNA clones of ski and ski-related gene sno. Nucleic Acids Res., 17, 5489–5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T., Khan,M.M., Kaul,S.C., Dong,H.-D., Wadhwa,R., Colmanares,C., Kohno,I. and Ishii,S. (1999) Ski is a component of the histone deacetylase complex required for transcriptional repression by Mad and thyroid hormone receptor. Genes Dev., 13, 412–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrij F. et al. (1995) Rubinstein–Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature, 376, 348–351. [DOI] [PubMed] [Google Scholar]
- Philpott N.J., Turner,A.J., Scopes,J., Westby,M., Marsh,J.C., Gordon-Smith,E.C., Dalgleish,A.G. and Gibson,F.M. (1996) The use of 7-amino actinomycin D in identifying apoptosis: simplicity of use and broad spectrum of application compared with other techniques. Blood, 87, 2244–2251. [PubMed] [Google Scholar]
- Roussel M.F., Ashmun,R.A., Sher,C.J., Eisenman,R.N. and Ayer,D.E. (1996) Inhibition of cell proliferation by the Mad1 transcriptional repressor. Mol. Cell. Biol., 16, 2796–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber-Agus N., Meng,Y., Hoang,T., Hou,T.,Jr, Chen,K., Greenberg,R., Cordon-Cardo,C., Lee,H.-W. and DePinho,R.A. (1998) Role of Mxi1 in aging organ systems and the regulation of normal and neoplastic growth. Nature, 393, 483–487. [DOI] [PubMed] [Google Scholar]
- Söderström M., Vo,A., Heinzel,T., Lavinisky,R.M., Yang,W.-M., Seto,E., Peterson,D.A., Rosenfeld,M.G. and Glass,C.K. (1997) Differential effects of nuclear receptor corepressor (N-CoR) expression levels on retinoic acid receptor-mediated repression support the existence of dynamically regulated corepressor complexes. Mol. Endocrinol., 11, 682–692. [DOI] [PubMed] [Google Scholar]
- Stavnezer E., Brodeur,D. and Brennan,L.A. (1989) The v-ski oncogene encodes a truncated set of c-ski coding exons with limited sequence and structural relatedness to v-myc. Mol. Cell. Biol., 9, 4038–4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroschein S.L., Wang,W., Zhou,S., Zhou,Q. and Luo,K. (1999) Negative feedback regulation of TGF-β signaling by the SnoN oncoprotein. Science, 286, 771–774. [DOI] [PubMed] [Google Scholar]
- Sun Y., Liu,X., Eaton,E.N., Lane,W.S., Lodish,H.F. and Weinberg,R.A. (1999a) Interaction of the Ski oncoprotein with Smad3 regulates TGF-β signaling. Mol. Cell, 4, 499–509. [DOI] [PubMed] [Google Scholar]
- Sun Y., Liu,X., Ng-Eaton,E., Lodish,H.F. and Weinberg,R.A. (1999b) SnoN and ski protooncoproteins are rapidly degraded in response to transforming growth factor β signaling. Proc. Natl Acad. Sci. USA, 96, 12442–12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutrave P. and Hughes,S.H. (1989) Isolation and characterization of three distinct cDNAs for the chicken c-ski gene. Mol. Cell. Biol., 9, 4046–4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutrave P., Kelly,A.M. and Hughes,S.H. (1990) Ski can cause selective growth of skeletal muscle in transgenic mice. Genes Dev., 4, 1462–1472. [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Naruse,I., Maekawa,T., Masuya,H., Shiroishi,T. and Ishii,S. (1997) Abnormal skeletal patterning in embryos lacking a single Cbp allele: a partial similarity with Rubinstein–Taybi syndrome. Proc. Natl Acad. Sci. USA, 94, 10215–10220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Patestos,N.P., Maekawa,T. and Ishii,S. (1999) B-myb is required for inner cell mass formation at an early stage of development. J. Biol. Chem., 274, 28067–28070. [DOI] [PubMed] [Google Scholar]
- Tokitou F., Nomura,T., Khan,M.M., Kaul,S.C., Wadhwa,R., Yasukawa,T., Kohno,I. and Ishii,S. (1999) Viral Ski inhibits Rb-mediated transcriptional repression in a dominant negative fashion. J. Biol. Chem., 274, 4485–4488. [DOI] [PubMed] [Google Scholar]
- Wang Z.G., Delva,L., Gaboli,M., Rivi,R., Giorgio,M., Cordon-Cardo,C., Grosveld,F. and Pandolfi,P.P. (1998) Role of PML in cell growth and the retinoic acid pathway. Science, 279, 1547–1551. [DOI] [PubMed] [Google Scholar]
- Weinberg R.A. (1995) The retinoblastoma protein and cell cycle control. Cell, 81, 323–330. [DOI] [PubMed] [Google Scholar]
- Wu T.-C., Wan,Y.-J. and Damjanov,I. (1981) Positioning of inner cell mass determines the development of mouse blastocysts in vitro. J. Embryol. Exp. Morphol., 65, 105–117. [PubMed] [Google Scholar]
- Yagi T., Nada,S., Watanabe,N., Tamemoto,H., Kohmura,N., Ikawa,Y. and Aizawa,S. (1993a) A novel negative selection for homologous recombinants using diphtheria toxin A fragment gene. Anal. Biochem., 214, 77–86. [DOI] [PubMed] [Google Scholar]
- Yagi T., Tokunaga,T., Furuta,Y., Nada,S., Yoshida,M., Tsukada,T., Saga,Y., Takeda,T., Ikawa,Y. and Aizawa,S. (1993b) A novel ES cell line, TT2, with high germline-differentiating potency. Anal. Biochem., 214, 70–76. [DOI] [PubMed] [Google Scholar]
- Yamasaki L., Jacks,T., Bronson,R., Goillot,E., Harlow,E. and Dyson,N.J. (1996) Tumor induction and tissue atrophy in mice lacking E2F-1. Cell, 85, 537–548. [DOI] [PubMed] [Google Scholar]
- Zhang H.S., Postigo,A.A. and Dean,D.C. (1999) Active transcriptional repression by the Rb-E2F complex mediates G1 arrest triggered by p16INK4a, TGFβ, and contact inhibition. Cell, 97, 53–61. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Iratni,R., Erdjument-Bromage,H., Tempst,P. and Reinberg,D. (1997) SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Cell, 89, 357–364. [DOI] [PubMed] [Google Scholar]
- Zheng G., Teumer,J., Colmenares,C., Richmond,C. and Stavnezer,E. (1997) Identification of a core functional and structural domain of the v-Ski oncoprotein responsible for both transformation and myogenesis. Oncogene, 15, 459–471. [DOI] [PubMed] [Google Scholar]