Abstract
We present, to our knowledge, the first quantitative evidence that music and genes may have coevolved by demonstrating significant correlations between traditional group-level folk songs and mitochondrial DNA variation among nine indigenous populations of Taiwan. These correlations were of comparable magnitude to those between language and genes for the same populations, although music and language were not significantly correlated with one another. An examination of population structure for genetics showed stronger parallels to music than to language. Overall, the results suggest that music might have a sufficient time-depth to retrace ancient population movements and, additionally, that it might be capturing different aspects of population history than language. Music may therefore have the potential to serve as a novel marker of human migrations to complement genes, language and other markers.
Keywords: music, genes, language, population structure, coevolution, Taiwan
1. Introduction
As human populations migrate to new regions of the world, their evolutionary divergence leaves its mark on both genes and culture. Correlations between cultural markers such as language and genetic markers such as mitochondrial, Y chromosome or autosomal DNA demonstrate that these features can co-migrate and coevolve over the course of thousands of years [1–3]. The same coevolutionary process has been proposed for music and genes [4–7], not least because music is a universal feature of human cultures [8,9] and because it shows quantifiable diversity both within and between populations [10,11]. However, this hypothesis has been criticized on the grounds that music evolution might occur at too rapid a rate [12] and therefore that music's time depth might be too shallow to be correlated with something as ancient and slowly evolving as genes. Although a few studies have found suggestive parallels between music and genes [5,13–15], none have demonstrated a statistically significant correlation between them. We wanted to examine this relationship quantitatively for the first time, to our knowledge, and explore whether music might have the potential to serve as a new type of marker for the study of human population history.
We decided to examine Taiwan as a test case as it has several clear advantages for such an analysis. Taiwan has a small number of well-characterized indigenous populations that are located in geographically distinct regions of the island [16]. These populations have been well studied musically, linguistically and genetically such that there exists ample material for performing correlational analyses. The indigenous musics have been extensively recorded and archived by ethnomusicologists since the 1920s [17–21], and genetic analyses of mitochondrial DNA (mtDNA) haplotypes for most of the indigenous groups have been published [22]. Therefore, the degree of musical and genetic sampling makes Taiwan an ideal case for analysis. In addition to this, Taiwan has been the focal point of theories about one of the most significant migrational events in human history, namely the expansion of the Austronesian-speaking peoples [23–31], thereby making any findings related to the population history of Taiwan relevant to the larger backdrop of the Austronesian migration.
The major objective of this study was to examine music's potential to serve as a novel marker of human population structure and to complement findings coming from genetics, linguistics and archaeology. To do so, we analysed for the first time, to our knowledge, correlations between genetics—specifically, mtDNA, a marker with a known time depth—and music—a marker of unknown time depth—for nine indigenous populations of Taiwan for which both genetic and musical data were available. We predicted that if music has a sufficient time depth to serve as a useful marker of human population history, we would observe significant correlations between musical diversity and genetic diversity. We also examined correlations with language, because gene–language correlations have been well studied in other parts of the world (although not in Taiwan) and because language shows both similarities with and differences from music at the cognitive and cultural levels [8,32,33].
2. Material and methods
(a). Musical sample
We restricted our analysis to group-level (choral) vocal songs—excluding solo songs and purely instrumental music—because we predicted that the constraints involved in coordinating musical parts among multiple singers would make this repertoire the most resistant to change over time, and hence the most stable. The songs comprised a mixture of traditional genres with a focus on ritual songs. We excluded children's songs and songs with explicit signs of borrowing, for example Christian missionary songs.
The musical sample consisted of 220 traditional, group-level vocal songs from nine indigenous populations from Taiwan (see figure 1 for the geographical location of these populations): the Amis (30 songs), Atayal (8), Bunun (30), Paiwan (30), Puyuma (24), Rukai (30), Saisiyat (22), Tao/Yami (28) and Tsou (18). These songs were obtained in consultation with an ethnomusicologist with an expertise in these musics, Ying-fen Wang. Most of the songs are available from the Taiwan National Music Archive (http://music.ncfta.gov.tw). This archive contains a variety of ethnomusicological recordings, some of them published commercially (notably [19,21]). Our sample represents all of the populations whose genetic data were published by Trejaut et al. [22], which comprises the nine populations that have been officially recognized by the government for many decades. However, it does not include five groups that were only recognized in the twenty-first century or any of the groups that are not officially recognized. Musicological characterization of a larger dataset that included musics from these former five groups has been reported in other publications from our laboratory [11,34].
Figure 1.
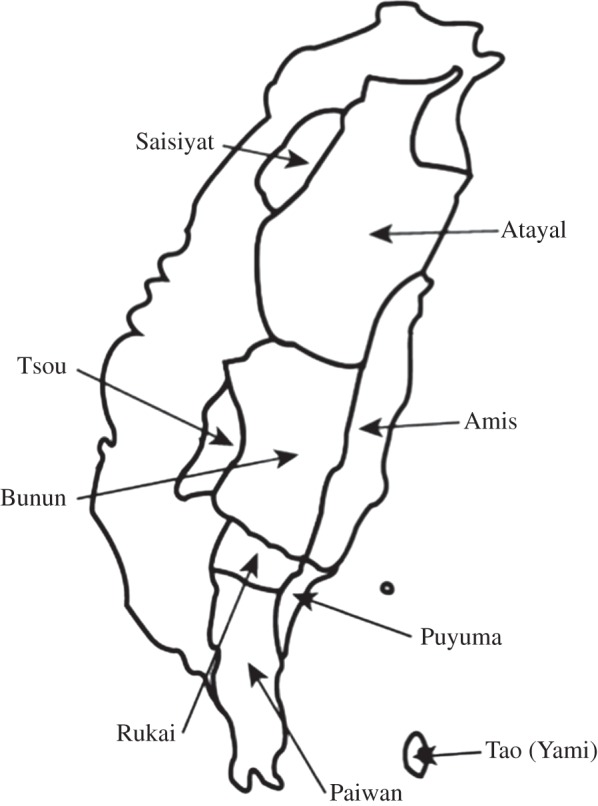
Map of Taiwan. A map of Taiwan showing the geographical locations of the nine indigenous populations included in the analyses.
The complete corpus of 220 songs was coded using two different methods of classification. P.E.S. coded all the songs acoustically using the CantoCore song-classification scheme developed in our laboratory [35], while Victor Grauer coded all of the same songs using the Cantometric coding system [36,37]. In order to avoid unreliable characters as well as character duplication between the two schemes, we chose 41 characters a priori for the analysis: all 26 structural characters from CantoCore (related to rhythm, pitch, text, texture and form) and the 15 performance-style characters from Cantometrics (related to vocal style, ornamentation and dynamics; see Savage et al. [35] for descriptions of specific characters and their coding reliability). Distances between songs were calculated using these 41 characters, accounting for ordinal, nominal and missing characters, as described in Rzeszutek et al. [11]. Raw musical codings and inter-rater reliability information are presented in the electronic supplementary material.
In the process of combining the two coding systems, 22 songs included in Rzeszutek et al. [11] were excluded from the analysis after realizing through consultation with Ying-fen Wang and Victor Grauer that they were either non-traditional songs, children's songs or duplicate recordings of the same song.
(b). Genetic sample
Genetic samples were obtained from the published data of Trejaut et al. [22]. The samples consisted of hypervariable segments 1 and 2 of the control region of the mitochondrial genome from 640 individuals. While this analysis was being performed, a newly obtained cohort of 410 mtDNA samples from these same nine Taiwanese populations became available at the Max Planck Institute in Leipzig (Ko et al. [38]). The Mantel correlation between the genetic datasets was rs = 0.45, p = 0.005. A combined dataset was prepared such that the same populations were pooled across the two datasets. The final sample sizes were: Amis (148 individuals), Atayal (159), Bunun (139), Paiwan (105), Puyuma (91), Rukai (100), Saisiat (87), Tao (113) and Tsou (108). After all sequences were aligned and edited, a 744 base-pair haplotype that encompassed the entirety of hypervariable segments 1 and 2 across a total of 1050 individuals was used for the correlational analyses.
(c). Distances between populations
For both the genetic and musical data, pairwise distances among the populations were calculated using the analysis of molecular variance (AMOVA) framework [39] in Arlequin v. 3.5.1.2, as described in detail for music in Rzeszutek et al. [11]. These distances were measured using the statistic ΦST (uncorrected for heterogeneous sites), which represents the proportion of variability among individual songs or genetic sequences that is owing to between-population differences.
Pairwise linguistic distances (patristic distances) between all nine populations were obtained from the published analysis of Gray et al. [40], which was based on lexical cognates across 210 items of basic vocabulary. Pairwise geographical distances were calculated based on the averaged sampling locations of Ko et al. [38]. Matrices of pairwise distance between the nine populations for music, genes, language and geography are presented in the electronic supplementary material, tables S1–S4, respectively.
We used the NeighborNet method [41] in order to visualize the relationships among the populations and to calculate Q-residuals (normalized to an average distance of 1 when calculating Q-residuals, as recommended by Gray et al. [42]). The analysis was performed in SplitsTree4 using standard settings [43].
(d). Correlations
The statistical significance of the correlations between distance matrices was tested with the permutation-based Mantel test [44] using 10 000 permutations, with the threshold for significance set at p < 0.05. This test controls for the fact that the 36 pairwise distances among the nine populations are not independent of one another by randomly permuting the rows and columns of the distance matrices to construct an empirical null distribution that is used to assess the significance of the observed correlation. Partial Mantel tests [45] were used to assess correlations between two distance matrices while controlling for a third distance matrix.
One-tailed tests were used, as recommended by Legendre & Fortin [46], because coevolutionary hypotheses predict positive correlations between the various distances, because the Mantel test already has lower power to reject the null hypothesis than a standard correlation not based on distances, and in order to make our analysis comparable with published gene–language analyses that also use one-tailed tests by default [47–52]. It should be noted that the Mantel r2 is always smaller than an R2-value based on rectangular data tables in cases (unlike this one) when both types of data can be compared, and so it is not appropriate to interpret the Mantel r-value in terms of the percentage of variance accounted for [46].
3. Results
AMOVA analyses were performed for music and genes in order to examine the partitioning of variances into between-culture and within-culture components; such an analysis was not possible for language because there are no data available on intracultural linguistic diversity in Taiwan. The ΦST value for music was 0.047 (4.7% between-culture variance), whereas that for genes was 0.127 (12.7%). These between-culture components for both musical and genetic diversity were highly significant (p < 0.00001) despite their relatively small absolute magnitudes.
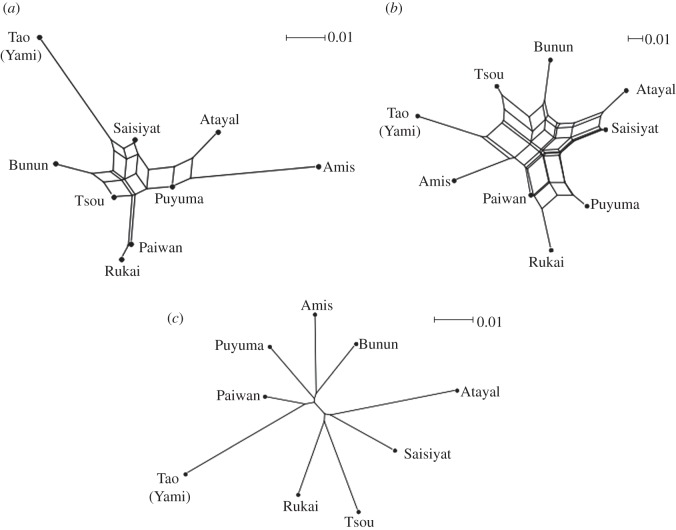
Next, we used NeighborNets to analyse population structure for music, genes and languages for the nine indigenous Taiwanese populations for which all three types of data were available (figure 2; see figure 1 for the geographical locations of the populations). In keeping with the extreme linguistic diversity of Taiwan, the NeighborNet for language was more or less star-shaped (figure 2c), implying that each language was nearly equidistant from every other language in the set. The Q-residual value for language was essentially zero (2 × 10–16), suggesting that its branching pattern was almost completely tree-like.
Figure 2.
NeighborNets for music, genes and language. The figure shows NeighborNet diagrams for (a) music, (b) genes and (c) language for the nine populations included in the analyses. The bar represents distance within each domain.
In contrast to this, the NeighborNets for both music and genes showed specific structures to them such that clustering was seen among populations. For example, neighbouring groups such as Paiwan and Rukai or Bunun and Tsou showed proximity in both networks. In addition, both of these NeighborNets showed extensive amounts of reticulation, as reflected in Q-residual values that were far greater than that for language (0.211 for music and 0.104 for genes), suggesting that the population structures for music and genes are much less tree-like than that for language.
Because patristic distances are, by definition, calculated from a tree and may thus overestimate how tree-like the linguistic data really are, we also calculated the Q-residual value for language using Hamming distances. The obtained value was 0.001, which is greater than that calculated using patristic distances but still more than 100 times smaller than the values for music and genes.
In order to look for potential coevolutionary relationships, we analysed correlations between musical, genetic, linguistic and geographical distance matrices. Table 1 presents the correlations for all analyses, and figure 3 shows the associated regression plots, with the r- and p-values presented inside each plot. The correlation between music and genes was statistically significant (r = 0.417, p = 0.015). This correlation remained significant even when geographical distance was controlled for (r = 0.385, p = 0.032). This finding suggests that these correlations reflect a branching coevolution of music and genes through shared ancestry [53] rather than a process of ‘isolation by distance’ [54], in which case the correlations would simply result from recent diffusion to geographical neighbours.
Table 1.
Correlations between music, genes and language. (Values in italic are significant at the 0.05 level.)
| r | p-value | |
|---|---|---|
| full correlations | ||
| music–genes | 0.417 | 0.015 |
| music–language | 0.411 | 0.085 |
| music–geography | 0.174 | 0.248 |
| language–genes | 0.492 | 0.006 |
| language–geography | 0.540 | 0.014 |
| genes–geography | 0.468 | 0.003 |
| partial correlations | ||
| music–genes (geography) | 0.385 | 0.032 |
| music–genes (language) | 0.271 | 0.054 |
| music–language (geography) | 0.382 | 0.101 |
| language–genes (geography) | 0.321 | 0.071 |
| language–genes (music) | 0.387 | 0.031 |
Figure 3.
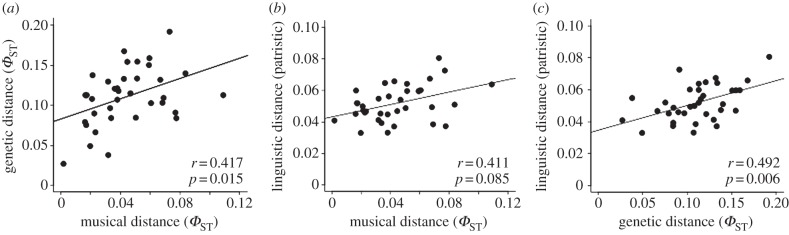
Regression plots for music, genes and language. Regression plots are shown for (a) genes versus music, (b) language versus music and (c) language versus genes. The Mantel statistic r and the associated p-value are shown inside each plot. The significance of the p-values is calculated using a Mantel permutation test to account for the fact that the 36 pairwise distances among the nine populations are not independent of one another.
The correlation between languages and genes was also significant (r = 0.492, p = 0.006), although this correlation became non-significant when geographical distance was taken into account (r = 0.321, p = 0.071). Interestingly, despite the fact that both music and language showed significant correlations with genes, the correlation between the two of them was not statistically significant (r = 0.411, p = 0.085), suggesting that these two cultural markers might be capturing, at least in part, distinct facets of genetic population history. Finally, correlations with geographical distance were significant for genes (r = 0.468, p = 0.003) and language (r = 0.540, p = 0.014) but not for music (r = 0.174, p = 0.248).
4. Discussion
These results provide, to our knowledge, the first quantitative evidence that music might be useful as a novel marker to study human population history by demonstrating statistically significant correlations between musical and genetic diversity for nine indigenous populations of Taiwan. Our results provide to the best of our knowledge, the first empirical support for the proposals of Lomax [10], Grauer [4,5] and Jordania [6] that music—particularly polyphonic group singing—might serve as a useful marker to study human migrations and human origins more generally. The fact that music can be shown to be correlated with a robust genetic marker like mtDNA suggests that it might have a sufficient time-depth to track population movements, although it is impossible at the present time to determine whether the correlations between musics, genes and languages date back to the initial peopling of Taiwan thousands of years ago or to more recent population movements within the last few centuries.
The magnitude of the music/gene correlation was quite high (r = 0.417). It is important to note that the value of this first-reported music/gene correlation is comparable to the language/gene correlation measured in this study as well as to most published language/gene correlations, which generally report maximum r-values in the range of 0.3–0.5 [47–52]. The observation that both music and language were significantly correlated with genes but not with one another suggests that these two cultural markers might be capturing partially distinct components of human population history, a contention that is supported by the fact that the musical and genetic data were much less tree-like than the linguistic data. This strengthens the case for using music as a complementary and informative marker for the study of population history.
We compared AMOVA analyses for music and genes, something made possible by our ability to measure both within- and between-culture diversity for music—just as is routinely done for genes—but something not readily possible for languages [55]. We recently performed, to our knowledge, the first AMOVA analysis of a cultural trait, namely music [11]. Using a subset of the songs from that study, the current analysis found that the vast majority of musical diversity was accounted for by the within-cultural component. However, while the between-cultural component had a small absolute magnitude of 4.7%, this component was highly statistically significant (p < 0.00001). This suggests that there is ample between-culture musical diversity for performing cross-cultural comparisons for music.
Ross et al. [56] recently performed an AMOVA analysis of another cultural trait, namely 700 variants of a single folktale across 31 populations in Europe. Their measured ΦST value of 0.091 is comparable to but somewhat higher than our own value for music. However, these authors argued—along with Bell et al. [57] in their analysis of the World Values Survey—that cultural differences between populations are far greater than genetic differences (the latter measured in Europe as a ΦST value less than 0.01 [58,59]), whereas we found the opposite relationship. Also, whereas Ross et al. found geographical distance to be a strong predictor of variation in folktales across cultures, our correlational analyses showed far less of an effect of geographical distance on music. mtDNA ΦST values for Taiwan (0.127) are quite large for a region of this size and are more comparable to values obtained from world surveys, which tend to range from 0.05 to 0.30 [60–63]. Therefore, we argue that it is premature to make generalizations about the relative sizes of cultural versus genetic ΦST, because each one might show substantial variation across world regions and because cultural ΦST might vary strongly as a function of the trait being measured and the classification tool used to measure it.
Recently, Pamjav et al. [15] performed a similar analysis to our own, examining the relationship between genetic distance and musical distance across Eurasia for 42 cultures for Y-chromosome analysis and 56 cultures for mtDNA analysis. Their automated computer analyses of the music were based on one-line song notations (as opposed to recordings in our case) and relied on a single musical feature related to melody. While clustering methods were employed for both the genes and the music, no significance testing or correlations were performed. However, their analysis lent support for a relationship between musical distance and genetic distance. Other qualitative findings have suggested similar relationships that span large distances in both geographical extent and in the amount of time that the populations are thought to have been isolated from one another, including that between central African Pygmies and southern African San ‘Bushmen’ [4,64], Bantu-speaking populations throughout sub-Saharan Africa [14] and Arctic cultures on both sides of the Bering Strait [13]. The fact that independent analyses of a variety of different regions using a variety of different methods and cultural samples coalesce on a similar conclusion lends support to the idea that music truly has a substantial time-depth that can aid in the study of human history. Our own findings are also, to the best of our knowledge, the first to demonstrate a relationship between music and genes over such a geographically restricted area, which is essential for understanding the mechanisms by which large-scale correlations might arise [49].
Unlike Pamjav et al. [15], we focused our analysis on choral songs based on the idea that the coordination involved in group singing should provide important constraints on song evolution. The observation of extensive polyphonic singing among most of the Taiwanese indigenous people creates an important cultural link between Taiwan and a majority of the extant Austronesian-speaking populations, including most ethnic groups throughout Island Southeast Asia, Oceania, coastal Papua New Guinea and Madagascar. It also creates a link to the southern region of China from which the proto-Austronesian peoples supposedly emanated [65,66], as polyphonic singing is ubiquitous among the ethnic minorities of southern China [67,68]. It therefore may be possible to use music to investigate the status of Taiwan versus other areas as the source-population for the expansion of the Austronesian-speaking people [3,23–31,40].
Overall, music showed greater similarities to genes than did languages with regard to population structure and degree of reticulation. This might reflect intrinsic similarities between music and genes in their mechanisms of evolution, migration and cross-cultural contact compared with languages. This work therefore opens the door to using music—a universal yet highly diverse feature of human cultures—as a novel marker of human history, one that provides complementary information to the more established cultural marker of language. The correlations we observed between musical and genetic diversity support the contention that music and genes may have been coevolving for a significant time period and that music might possess the capacity to track population changes occurring on the time scale of perhaps thousands of years. To the extent that migrational models are validated by the concordance of results across multiple markers [1–3,69], music may well contribute to a richer understanding of human evolution.
Acknowledgements
We thank Tom Rzeszutek for his work on the distance matrices and correlational analyses, and Ying-fen Wang for ethnographic and musicological advice on song selection for the Taiwanese musical sample. We thank Victor Grauer for the Cantometric coding of all of the songs in this study, Emily Merritt for the inter-rater reliability codings, Simon Greenhill for providing linguistic data and Marie Lin for her support of the collaborative arrangement with the Taipei group.
Funding statement
This work was supported by a grant to S.B. from the Social Sciences and Humanities Research Council of Canada, by an Amherst College Roland Wood Fellowship and a Japanese Ministry of Education, Culture, Sports, Science, and Technology Scholarship to P.E.S., and by the Max Planck Society for the research of A.M.-S.K. and M.S. The GenBank accession numbers for the Max Planck Taiwan sample are KF540506-KF541055.
References
- 1.Cavalli-Sforza LL, Menozzi P, Piazza A. 1994. The history and geography of human genes. Princeton, NJ: Princeton University Press [Google Scholar]
- 2.Bellwood P, Renfrew C. Eds. 2002. Examining the farming/language dispersal hypothesis. Cambridge, UK: McDonald Institute for Archaeological Research [Google Scholar]
- 3.Diamond J, Bellwood P. 2003. Farmers and their languages: the first expansions. Science 300, 597–603 (doi:10.1126/science.1078208) [DOI] [PubMed] [Google Scholar]
- 4.Grauer V. 2006. Echoes of our forgotten ancestors. World of Music 48, 5–59 [Google Scholar]
- 5.Grauer VA. 2011. Sounding the depths: tradition and the voices of history CreateSpace See http://soundingthedepths.blogspot.com/.
- 6.Jordania J. 2006. Who asked the first question? The origins of human choral singing, intelligence, language and speech. Tblisi, Georgia: Logos [Google Scholar]
- 7.Savage PE, Brown S. 2013. Toward a new comparative musicology. Anal. Approaches World Music 2, 148–198 [Google Scholar]
- 8.Wallin NL, Merker B, Brown S. (eds) 2000. The origins of music. Cambridge, MA: MIT Press [Google Scholar]
- 9.Brown S, Jordania J. 2013. Universals in the world's musics. Psychol. Music 41, 229–248 (doi:10.1177/0305735611425896) [Google Scholar]
- 10.Lomax A. 1968. Folk song style and culture. Washington, DC: American Association for the Advancement of Science [Google Scholar]
- 11.Rzeszutek T, Savage PE, Brown S. 2012. The structure of cross-cultural musical diversity. Proc. R. Soc. B 279, 1606–1612 (doi:10.1098/rspb.2011.1750) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stock JPJ. 2006. Clues from our present peers? A response to Victor Grauer. World of Music 48, 73–91 [Google Scholar]
- 13.Nattiez J-J. 1999. Inuit throat-games and Siberian throat singing: a comparative, historical, and semiological approach. Ethnomusicology 43, 399–418 (doi:10.2307/852555) [Google Scholar]
- 14.Callaway E. 2007. Music is in our genes. Nat. News. See http://www.nature.com/news/2007/071210/full/news.2007.359.html (doi:10.1038/news.2007.359) [Google Scholar]
- 15.Pamjav H, Juhasz Z, Zalan A, Nemeth E, Damdin B. 2012. A comparative phylogenetic study of genetics and folk music. Mol. Genet. Genomics 287, 337–349 (doi:10.1007/s00438-012-0683-y) [DOI] [PubMed] [Google Scholar]
- 16.Council of Indigenous Peoples 2013. The tribes in Taiwan See http://www.apc.gov.tw/ (accessed 30 April 2013).
- 17.Kurosawa T. 1973. Taiwan takasagozoku no ongaku music of the Taiwan Aborigines. Tokyo, Japan: Oyamakaku [Google Scholar]
- 18.Loh I-T. 1982. The tribal music of Taiwan: with special reference to the Ami and Puyuma tribes. PhD dissertation, University of California, Los Angeles, CA, USA [Google Scholar]
- 19.Wu R-S. Ed. 1992–1995. The music of the aborigines on Taiwan island (set of 8 CDs). Taipei, Taiwan: China Publishing Company [Google Scholar]
- 20.Hsu T-H. 2002. Taiwan: music of the Taiwan aborigines. In The Garland encyclopedia of world music, volume 7. East Asia: China, Japan and Korea (eds Provine RC, Tokumaru Y, Witzleben JL.), pp. 523–529 New York, NY: Routledge [Google Scholar]
- 21.Wang Y-F. Ed. 2008. Sounds from wartime Taiwan 1943 (set of 2 CDs). Taipei, Taiwan: National Taiwan University Press SJL-78 & SJL-79 [Google Scholar]
- 22.Trejaut JA, Kivisild T, Loo JH, Lee CL, He CL, Hsu CJ, Li ZY, Lin M. 2005. Traces of archaic mitochondrial lineages persist in Austronesian-speaking Formosan populations. PLoS Biol. 3, 1362–1372 (doi:10.1371/journal.pbio.0030247) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirch PV. 2000. On the road of the winds: an archaeological history of the Pacific islands before European contact. Berkeley, CA: University of California Press [Google Scholar]
- 24.Diamond J. 2000. Taiwan's gift to the world. Nature 403, 709–710 (doi:10.1038/35001685) [DOI] [PubMed] [Google Scholar]
- 25.Gray RD, Jordan FM. 2000. Language trees support the express-train sequence of Austronesian expansion. Nature 405, 1052–1055 (doi:10.1038/35016575) [DOI] [PubMed] [Google Scholar]
- 26.Oppenheimer S, Richards M. 2001. Fast trains, slow boats, and the ancestry of the Polynesian islanders. Sci. Prog. 84, 157–181 (doi:10.3184/003685001783238989) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bellwood P, Dizon E. 2005. The Batanes archaeological project and the ‘Out of Taiwan’ hypothesis for Austronesian dispersal. J. Austronesian Stud. 1, 1–33 [Google Scholar]
- 28.Bellwood P, Dizon E. 2008. Austronesian cultural origins: out of Taiwan, via the Batanes Islands, and onwards to Western Polynesia. In Past human migrations in East Asia: matching archaeology, linguistics and genetics (eds Sanchez-Mazas A, Blench R, Ross M, Peiros I, Lin M.), pp. 23–39 London, UK: Routledge [Google Scholar]
- 29.Bellwood P. 2011. Holocene population history in the Pacific region as a model for worldwide food producer dispersals. Curr. Anthropol. 52, S363–S378 (doi:10.1086/658181) [Google Scholar]
- 30.Meacham W. 1984. On the improbability of Austronesian origins in South China. Asian Perspect. 26, 89–106 [Google Scholar]
- 31.Blust R. 1999. Subgrouping, circularity and extinction: some issues in Austronesian comparative linguistics. In Selected papers from the Eighth International Conference on Austronesian linguistics (eds Zeitoun E, Li R.), pp. 31–94 Taipei, Taiwan: Institute of Linguistics, Academia Sinica [Google Scholar]
- 32.Feld S, Fox AA. 1994. Music and language. Annu. Rev. Anthropol. 23, 25–53 (doi:10.1146/annurev.an.23.100194.000325) [Google Scholar]
- 33.Patel AD. 2008. Music, language, and the brain. Oxford, UK: Oxford University Press [Google Scholar]
- 34.Savage PE, Brown S. In press Mapping music: cluster analysis of song-type frequencies within and between cultures. Ethnomusicology 58 [Google Scholar]
- 35.Savage PE, Merritt E, Rzeszutek T, Brown S. 2012. CantoCore: a new cross-cultural song classification scheme. Anal. Approaches World Music 2, 87–137 [Google Scholar]
- 36.Lomax A, Grauer V. 1968. The Cantometric coding book. In Folk song style and culture (ed. Lomax A.), pp. 34–74 Washington, DC: American Association for the Advancement of Science [Google Scholar]
- 37.Lomax A. 1976. Cantometrics: an approach to the anthropology of music. Berkeley, CA: University of California Extension Media Center [Google Scholar]
- 38.Ko AM-S, Chen C-Y, Fu O, Delfin F, Li M, Chiu H-L, Aximu-Petri A, Stoneking M, Ko Y-C. Submitted. Early Austronesians: into and out of Taiwan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Excoffier L, Smouse P, Quattro J. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131, 479–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gray RD, Drummond AJ, Greenhill SJ. 2009. Language phylogenies reveal expansion pulses and pauses in Pacific settlement. Science 323, 479–483 (doi:10.1126/science.1166858) [DOI] [PubMed] [Google Scholar]
- 41.Bryant D, Moulton V. 2004. Neighbor-Net: an agglomerative method for the construction of phylogenetic networks. Mol. Biol. Evol. 21, 255–265 (doi:10.1093/molbev/msh018) [DOI] [PubMed] [Google Scholar]
- 42.Gray RD, Bryant D, Greenhill SJ. 2010. On the shape and fabric of human history. Phil. Trans. R. Soc. B 365, 3923–3933 (doi:10.1098/rstb.2010.0162) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267 (doi:10.1093/molbev/msj030) [DOI] [PubMed] [Google Scholar]
- 44.Mantel N. 1967. The detection of disease clustering and a generalized regression approach. Cancer Res. 27, 209–220 [PubMed] [Google Scholar]
- 45.Smouse PE, Long JC, Sokal RR. 1986. Multiple regression and correlation extensions of the Mantel test of matrix correspondence. Syst. Zool. 35, 627–632 (doi:10.2307/2413122) [Google Scholar]
- 46.Legendre P, Fortin M-J. 2010. Comparison of the Mantel test and alternative approaches for detecting complex multivariate relationships in the spatial analysis of genetic data. Mol. Ecol. Resour. 10, 831–844 (doi:10.1111/j.1755-0998.2010.02866.x) [DOI] [PubMed] [Google Scholar]
- 47.Sokal RR. 1988. Genetic, geographic, and linguistic distances in Europe. Proc. Natl Acad. Sci. USA 85, 1722–1726 (doi:10.1073/pnas.85.5.1722) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen J, Sokal RR, Ruhlen M. 1995. Worldwide analysis of genetic and linguistic relationships of human populations. Hum. Biol. 67, 595–612 [PubMed] [Google Scholar]
- 49.Lansing JS, et al. 2007. Coevolution of languages and genes on the island of Sumba, eastern Indonesia. Proc. Natl Acad. Sci. USA 104, 16 022–16 026 (doi:10.1073/pnas.0704451104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hunley K, Dunn M, Lindström E, Reesink G, Terrill A, Healy ME, Koki G, Friedlaender FR, Friedlaender JS. 2008. Genetic and linguistic coevolution in Northern Island Melanesia. PLoS Genet. 4, e1000239 (doi:10.1371/journal.pgen.1000239) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tishkoff SA, et al. 2009. The genetic structure and history of Africans and African Americans. Science 324, 1035–1044 (doi:10.1126/science.1172257) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.HUGO Pan-Asian SNP Consortium 2009. Mapping human genetic diversity in Asia. Science 326, 1541–1545 (doi:10.1126/science.1177074) [DOI] [PubMed] [Google Scholar]
- 53.Cavalli-Sforza LL, Piazza A, Menozzi P, Mountain J. 1988. Reconstruction of human evolution: bringing together genetic, archaeological, and linguistic data. Proc. Natl Acad. Sci. USA 85, 6002–6006 (doi:10.1073/pnas.85.16.6002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wright S. 1943. Isolation by distance. Genetics 28, 114–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Levinson SC, Gray RD. 2012. Tools from evolutionary biology shed new light on the diversification of languages. Trends Cogn. Sci. 16, 167–173 (doi:10.1016/j.tics.2012.01.007) [DOI] [PubMed] [Google Scholar]
- 56.Ross MR, Greenhill SJ, Atkinson QD. 2013. Population structure and cultural geography of a folktale in Europe. Proc. R. Soc. B 280, 20123065 (doi:10.1098/rspb.2012.3065) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bell AV, Richerson PJ, McElreath R. 2009. Culture rather than genes provides greater scope for the evolution of large-scale human prosociality. Proc. Natl Acad. Sci. USA 106, 17 671–17 674 (doi:10.1073/pnas.0900137106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lao O, et al. 2008. Correlation between genetic and geographic structure in Europe. Curr. Biol. 18, 1241–1248 (doi:10.1016/j.cub.2008.07.049) [DOI] [PubMed] [Google Scholar]
- 59.Novembre J, et al. 2008. Genes mirror geography within Europe. Nature 456, 98–101 (doi:10.1038/nature07331) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lewontin RC. 1972. The apportionment of human diversity. In Evolutionary biology (eds Dobzhansky T, Hecht MK, Steer CW.), pp. 391–398 New York, NY: Appleton-Century-Crofts [Google Scholar]
- 61.Jorde LB, Watkins WS, Bamshad MJ, Dixon ME, Ricker CE, Seielstad MT, Batzer MA. 2000. The distribution of human genetic diversity: a comparison of mitochondrial, autosomal, and Y-chromosome data. Am. J. Hum. Genet. 66, 979–988 (doi:10.1086/302825) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW. 2002. Genetic structure of human populations. Science 298, 2381–2385 (doi:10.1126/science.1078311) [DOI] [PubMed] [Google Scholar]
- 63.Li JZ, et al. 2008. Worldwide human relationships inferred from genome-wide patterns of variation. Science 319, 11 000–11 004 (doi:10.1126/science.1153717) [DOI] [PubMed] [Google Scholar]
- 64.Grauer V. 2007. New perspectives on the Kalahari debate: a tale of two ‘genomes’. Before Farming 2, 1–14 [Google Scholar]
- 65.Li H, et al. 2008. Paternal genetic affinity between western Austronesians and Daic populations. BMC Evol. Biol. 15, 146 (doi:10.1186/1471-2148-8-146) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mirabal S, Cadenas AM, Garcia-Bertrand R, Herrera RJ. 2013. Ascertaining the role of Taiwan as a source for the Austronesian expansion. Am. J. Phys. Anthropol. 150, 551–564 (doi:10.1002/ajpa.22226) [DOI] [PubMed] [Google Scholar]
- 67.Zhang X. 2001. Alili: multi-part folksongs of Yunnan‘s ethnic minorities. PAN 2038CD. Leiden, The Netherlands: PAN Records [Google Scholar]
- 68.Shen Q. 2002. National minorities in China's south and southwest: ethnic groups and musical styles. In East Asia: China, Japan and Korea. Garland encyclopedia of world music, vol. 7, pp. 485–493 London, UK: Routledge [Google Scholar]
- 69.Sanchez-Mazas A, Blench R, Ross M, Peiros I, Lin M. (eds) 2008. Past human migrations in East Asia: matching archaeology, linguistics and genetics. London, UK: Routledge [Google Scholar]