Abstract
Posttraumatic stress disorder (PTSD) is a common and debilitating mental disorder with a particularly high burden for women. Emerging evidence suggests PTSD may be more heritable among women and evidence from animal models and human correlational studies suggest connections between sex-linked biology and PTSD vulnerability, which may extend to the disorder’s genetic architecture. We conducted a genome-wide association study (GWAS) of PTSD in a primarily African American sample of women from the Detroit Neighborhood Health Study (DNHS) and tested for replication in an independent cohort of primarily European American women from the Nurses Health Study II (NHSII).
We genotyped 413 DNHS women - 94 PTSD cases and 319 controls exposed to at least one traumatic event - on the Illumina HumanOmniExpress BeadChip for > 700,000 markers and tested 578 PTSD cases and 1963 controls from NHSII for replication. We performed a network-based analysis integrating data from GWAS-derived independent regions of association and the Reactome database of functional interactions.
We found genome-wide significant association for one marker mapping to a novel RNA gene, lincRNA AC068718.1, for which we found suggestive evidence of replication in NHSII. Our network-based analysis indicates that our top GWAS results were enriched for pathways related to telomere maintenance and immune function. Our findings implicate a novel RNA gene, lincRNA AC068718.1, as risk factor for PTSD in women and add to emerging evidence that non-coding RNA genes may play a crucial role in shaping the landscape of gene regulation with putative pathological effects that lead to phenotypic differences.
Keywords: PTSD, GWAS, lincRNA, telomere maintenance, immune system
1. Introduction
Posttraumatic stress disorder (PTSD) is pervasive and debilitating mental disorder that develops in some persons following exposure to a traumatic event. The lifetime prevalence of PTSD is 7.6% in the United States (Kessler et al., 1995). The majority of persons who are exposed to even a severe traumatic event do not develop PTSD (Kessler et al., 1995; Breslau et al., 1999) Why some individuals are vulnerable and others are resilient remains an open question; genetic factors are hypothesized to play an important role (Cornelis et al., 2010). However, a recent review of the biology of PTSD concluded that the candidate gene studies design had not produced robust results (Pitman et al., 2012). To date, only two genome-wide association studies (GWASs) of PTSD have been published. The first identified the retinoid-related orphan receptor alpha (RORA) gene as a novel risk locus and was conducted in a primarily male (59.7%) European American sample of military veterans and their partners (Logue et al., 2012). The second was conducted in a European American sample of men and women and found a genome-wide significant hit at rs406001 with the second strongest association in the Tolloid-Like 1 gene (TLL1). The TLL1 association was replicated in an independent sample of European Americans (Xie et al., 2013).
The burden of PTSD is particularly high for women. Women in the general population are at twice the lifetime risk of PTSD as men: 1 in 9 women versus 1 in 20 men in the US will have met criteria for the diagnosis at some point in their lifetime (Resnick et al., 1993; Kessler et al., 1995; Breslau et al., 1998). PTSD in women is more likely to follow a chronic course (Breslau and Davis, 1992; Kessler et al., 1995; Breslau et al., 1998) and may be associated with greater functional impairment (McLean et al., 2011). The traumatic events associated with PTSD in women differ from those in men; women are more likely to be exposed to traumatic events such as sexual abuse, sexual assault, and rape that have high conditional risks of PTSD (Kessler et al., 1995; Roberts et al., 2012). PTSD may also be more heritable in women; the heritability of PTSD was recently estimated at 72% in a community-based sample of young adult female twins (Sartor et al., 2011), more than twice that of the 30% heritability observed in a male sample of Vietnam era veterans. Recently, animal models and human genetic studies have suggested connections between sex-linked biology and vulnerability to PTSD (Lebron-Milad and Milad, 2012). Mechanisms underlying sex-linked biology may be determined by sex-specific genetic risk factors for the disorder. For example, Ressler and colleagues reported a sex-specific association of SNP rs2267735 mapping to a putative estrogen response element within the gene PACAPR1 (encoded by ADCYAP1R1) with PTSD in females only (Ressler et al., 2011) which has been replicated in several studies (Almli et al., 2013; Uddin et al., 2013; Wang et al., 2013). However, Chang and colleagues were unable to replicate the association between rs2267735 and PTSD in either AA or EA females in these two large independent samples (Chang et al., 2012).
Taken together, these findings support the importance of investigating the genetic risk factors that specifically increase vulnerability in women. In order to identify novel risk genes for PTSD in women, we conducted a GWAS in a primarily African American sample of women from the Detroit Neighborhood Health Study (DNHS) (Goldmann et al., 2011), an epidemiologic study of adults in urban Detroit and replicated our top finding in a primarily European American sample from the Nurses Health Study II (NHSII) (Koenen et al., 2009). To better characterize the biological pathways implicated by our GWAS, we conducted an exploratory pathway enrichment analysis using a functional interaction network-based approach of top hits from our initial GWAS.
2. Materials and Methods
2.1 DNHS Sample
Participants were recruited from the DNHS (N = 1,547), a longitudinal cohort of predominately African American men and women adults (18+) living in Detroit, Michigan and screened for lifetime trauma exposure using procedures described in more detail elsewhere (Uddin et al., 2010; Goldmann et al., 2011). The analytic sample for this paper is 413 women, who signed consent to provide DNA samples that were viable for analysis and were exposed to at least one traumatic event. The self-reported ethnicity of these women is: 342 African Americans, 45 European Americans, 26 individuals of other ethnicity. Overall, 406 women identified themselves as non-Hispanic and 7 as Hispanic. 94 had a lifetime diagnosis of PTSD and 319 were trauma-exposed controls. PTSD diagnosis was assessed via structured interview administered via telephone (Breslau et al., 1998). PTSD symptoms were assessed in reference to both the traumatic event the participant regarded as their worst and one randomly selected traumatic event from the remaining traumas the participant experienced. Lifetime PTSD cases met all six DSM-IV criteria in reference to either the worst or random traumatic event. The diagnostic interview showed good validity against the Clinician Administered PTSD Scale (CAPS) (Blake et al., 1995) as described elsewhere (Uddin et al., 2010). The Institutional Review Board of the University of Michigan reviewed and approved the study protocol.
2.2 GWAS Genotyping
DNA was isolated from whole blood or saliva. Whole-blood derived genomic DNA was isolated from 400μl of whole blood using either the DNA Mini Kit (Qiagen, Valencia, CA) or the QuickGene DNA Blood Kit using the Quickgene Mini80 system (Fujifilm, Tokyo, Japan) following the manufacturer’s recommended protocol. Saliva-derived genomic DNA was isolated from 4ml of saliva, which was then processed using the Oragene DNA OG-500 kit (Orasure Technologies, Bethlehem, PA) following the manufacturer’s recommended protocol. 0.25 μg of each DNA sample was sent to the Applied Genomics Technology Facility (Wayne State University, Detroit, MI) for genotyping using the HumanOmniExpress BeadChips (Illumina Inc, San Diego, CA). The BeadChips were run on an Illumina iScan system using a combination of the Infinium HD Assay Super Manual Protocol and the Infinium HD Assay Super Automated Protocol, switching protocols from manual to automated at the XStain step on Day 3. The GenomeStudio Genotyping (GT) Module (Illumina Inc, San Diego, CA) was used for data normalization and genotype calling.
2.3 Genome-wide association analysis
We genotyped 730,525 SNPs. Genotyping was performed on 471 women. A total of 4 women had samples with call rate < 95% and were removed from the study. Other samples were removed due to duplicate issues, missing phenotype data or because they were not exposed to at least one traumatic event with a remaining sample of 413 women. A total of 688,890 SNPs passed quality control filters (call rate > 95%, minor allele frequency > 0.01, Hardy-Weinberg disequilibrium p > 1×10−6). Genotype reports generated using Illumina GenomeStudioGT v. 1.8.4 software were used to generate PLINK input files (i.e. lgen, map and fam files). GWAS was conducted using logistic regression for genotypes coded additively (0,1,2 copies of the minor allele) and PTSD case status. Two Multidimensional Scaling (MDS) components, described in detail in the next section, were used in the regression model as covariates to adjust for population stratification. All analyses were performed using PLINK software package (v 1.07, October 2009) (Purcell et al., 2007). The study had limited power to detect a disease-associated allele at the genome-wide threshold. Simulations showed that in a sample of 94 cases with a control/case ratio of 3.4 like ours, we would have 48% statistical power to detect an associated allele with a population frequency of 0.20 and an odds ratio of 2.5 (at P = 5 × 10−8 under a multiplicative genetic model).
2.4 Population stratification
We used the MDS analysis of genome-wide identity-by-state data implemented in PLINK to determine ancestry in the whole sample. The analysis was conducted using the 688,890 SNPs that passed quality control filters previously described. The first two components from the MDS analysis identified clearly separated clusters that correlate with self-report ethnicity identification. The first two MDS components distinguished African American from European American participants and others and the second component distinguished Hispanic and non-Hispanic subjects. Both components were used to adjust for population stratification in the association analysis.
2.5 Nurses Health Study II sample
We used data from the PTSD diagnostic subsample (N=3,013) of the NHSII. The ascertainment of the subsample has been described in detail previously (Roberts et al., 2012) and the protocol has been published elsewhere (Koenen et al., 2009). See the Supplementary Material for further demographic information.
2.6 Diagnostic Interviews
Lifetime PTSD diagnosis was assessed in relation to participant’s worst event via structured telephone interview similar to that used for the DNHS (Roberts et al., 2012).
2.7 Replication genotyping
All samples were genotyped using the ABI PRISM 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA), in 384-well format or using the TaqMan OpenArray™ SNP Genotyping Platform (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions. TaqMan® assays were ordered using either Assays-on-Demand or using the ABI Assays-By-Design service.
2.8 Statistical analysis of rs10170218 in the NHSII replication sample
We attempted to replicate our top GWAS finding from the primary PTSD analysis in the female DNHS subsample in an independent cohort of 578 PTSD cases and 1963 trauma-exposed controls from the NHSII. To avoid population stratification biases, we confined the analysis to the subsample of European American females who self-reported as non-Hispanic. After checking for Hardy Weinberg Equilibrium, the SNP with genome-wide significant p-value in the primary analysis was tested for evidence of association in the replication sample using a logistic regression model for genotypes coded additively (0,1,2 copies of the minor allele) and PTSD case status. In consideration of potential differences in the allelic distribution between the primary and the replication samples due to different ancestry-related genetic background, we also tested the association of the SNP with PTSD status using different genetic models, namely the genotypic (2df), dominant and recessive models (1df) using PLINK.
2.9 In-silico analysis
To further characterize genes and their potential predicted function, we conducted an in-silico computational analysis by integrating the current genome annotations available at UCSC Genome Browser (Rosenbloom et al., 2012). We used the ENCODE/GENCODE (version 7, May 2011) annotation through Ensembl to annotate the region that harbors our top significant SNPs. The potential transposable elements and interspersed repeats overlapping with the DNA sequence of the SNPs/gene found associated were searched for by using the annotation created with Repeatmasker (Smit et al., 1996–2010) program on the reference genome sequence, assembly February 2009, GRCh37/hg19.
2.10 Pathway analysis
To better characterize the top findings of our GWAS, we set out to identify independent regions of association using a clumping algorithm and test pathway enrichment analysis of the genes mapping to these regions through a network-based approach. First, for each SNP with p-value < 5 × 10−4, we looked for any SNPs in linkage disequilibrium (at least above r2=0.2) and associated with PTSD at p-value < 0.05 within 250 Kb distance from the index SNP. Then, we used the Reactome Functional Interaction (FI) network to reconstruct a FI sub-network based on the set of genes mapping on the non-desert genomic regions of association derived from the GWAS. The FI sub-network mines the Reactome manually curated pathway-based protein functional interaction network, which covers close to 50% of human proteins. To reconstruct the network, the algorithm used genes from our list and “linker” genes necessary to identify patterns of functional interactions. We ran a network-clustering algorithm based on spectral partition (Newman, 2006). Pathway enrichment analysis was performed for each module using only genes provided in the original list without considering “linker” genes in the enrichment evaluation. Reactome FI uses the False Discovery Rate approach to determine the adjusted p-values for each enriched pathway. We considered a pathway to be significantly enriched at FDR < 1 × 10−3.
3. Results
3.1 PTSD GWAS in the DNHS
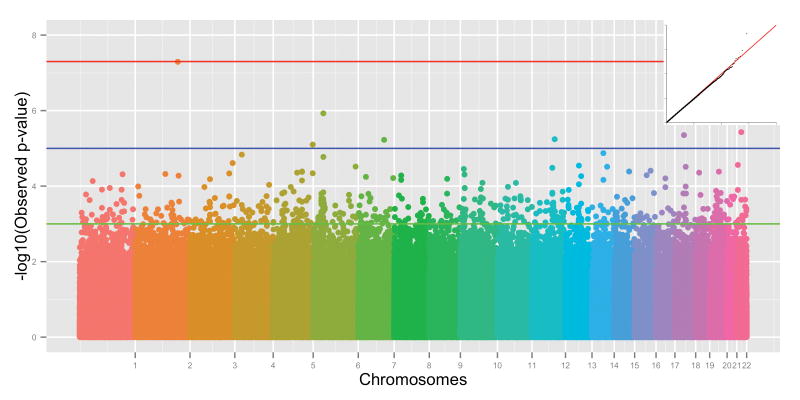
We performed a GWAS in DNHS women. As DNHS is ethnically diverse, all our association analysis used principal components of genetic variation inferred from the whole genome data as covariates to account for ethnicity. The q–q plot of the results shows no departures of the overall distribution of the observed p-values from the expected distribution, with the only exception of the extreme tail of the distribution of the test statistics (Figure 1). The genomic inflation factor (lambda) of 0.99 further confirms that population stratification is not biasing association results. The Manhattan plot illustrates the results of the genome-wide association analysis with PTSD, with 201 SNPs (0.03%) showing evidence of association at p-value < 5 × 10−4 (Figure 1). Table 2 lists the 7 SNPs that yielded p-values < 1 × 10−5 (Table 2). The strongest signal was at SNP rs10170218 with a genome-wide significant p-value (p-value = 5.09 × 10−8; OR = 2.88, 95% CI = 1.97–4.23). The Bonferroni corrected p-value is 0.035. SNP rs10170218 is located within AC068718.1, a novel long intergenic non-coding RNA (lincRNA) gene located at 2q32.1. The total length of the gene is 578.8 Kb. The region containing AC068718.1 showed a very mild degree of linkage disequilibrium (LD) (Figure 2). Of the 92 SNPs mapping to AC068718.1, 11 additional SNPs were significant with p-value < 0.05. Among these, the strongest signals were at SNPs rs10181512 and rs10497691, located ~9Kb downstream of SNP rs10170218, and rs6713963, located in the same 28-Kb LD block as rs10170218, with p-values at 10−3 threshold of significance.
Figure 1.
Manhattan plot and Q–Q plot for the GWAS in the female sample from DNHS. The chromosomes are presented in order and color coded for ease of identification. The individual SNP p-values are represented by circles and are plotted according to –log10 scale. Three lines identify different significance threshold: green for p-value < 0.001, blue for p-value < 1 × 10−5 and red for p-value < 5 × 10−8 (i.e., genome-wide significance threshold). In the upper right corner, Q–Q plot of GWAS analyses with correction for population stratification (y-axis: observed –log10(p-value); x-axis: expected –log10(p-value). Lambdagc = 0.99, indicating adequate control of bias. For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.
Table 2.
Loci with strongest evidence of association (p-value < 1 × 10−5) with PTSD in the DNHS.
| Gene Annotation |
Variant Annotation |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | SNP | Coordinate annotated | Alleles (Minor/Major) | Minor Allele Frequency | Beta | p-value | Odds Ratio (95% CI) | Gene Symbol | Gene Type | Gene Status | Variant type | Variant distance (bp) to gene |
| 2 | rs10170218 | 188,814,444 | C/A | 0.22 | 5.45 | 5.10E-08 | 2.89 (1.97–4.23) | AC068718.1 | lincRNA | Novel | intronic | 0 |
| 6 | rs1611133 | 29,809,382 | A/G | 0.17 | 4.47 | 1.18E-06 | 3.56 (2.04–6.22) | HLA-G3 | Protein coding | Known | intergenic | 10,480 |
| 22 | rs2413187 | 33,864,817 | G/A | 0.21 | 4.86 | 3.71E-06 | 2.78 (1.84–4.19) | LARGE | Protein coding | Known | intronic | 0 |
| 18 | rs10502692 | 36,040,352 | A/G | 0.22 | −4.53 | 4.46E-06 | 0.35 (0.22–0.55) | Intergenic | NA | NA | intergenic | NA |
| 12 | rs7302717 | 95,119,816 | A/G | 0.39 | −4.54 | 5.72E-06 | 0.41 (0.28–0.60) | TMCC3 | Protein coding | Known | intergenic | −75,478 |
| 7 | rs11767398 | 112,130,009 | C/A | 0.29 | 4.59 | 5.95E-06 | 2.54 (1.71–3.78) | C7orf53 | Protein coding | Known | 3′ UTR | 0 |
| 5 | rs4701170 | 178,897,590 | A/G | 0.09 | 4.63 | 7.92E-06 | 2.41 (1.66–3.51) | RP11–798K23.1 | lincRNA | Putative | intergenic | 14,494 |
Figure 2.
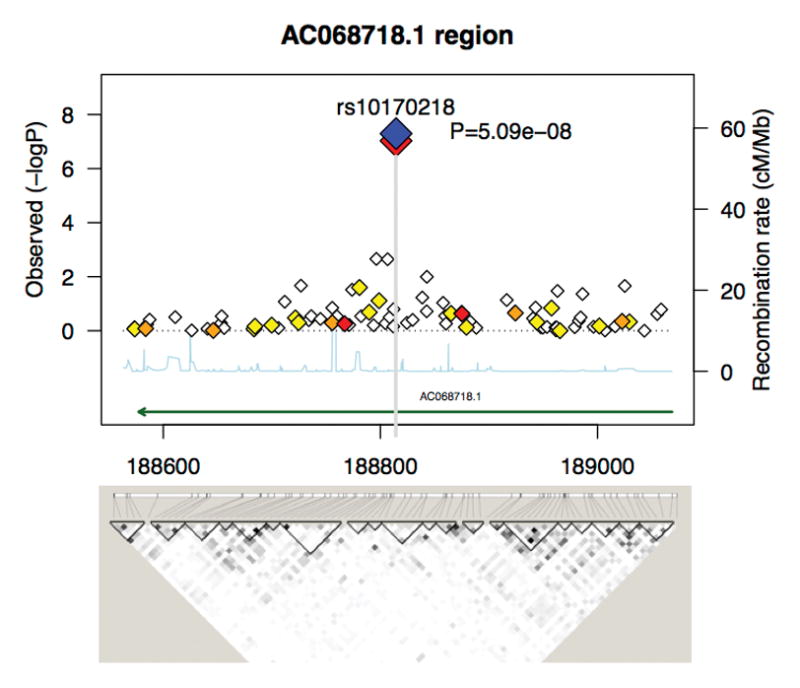
Case-control association results and linkage disequilibrium (LD) structure at 2q32.1. Results are reported for the most significant SNP rs10170218 and SNPs genotyped across 800 Kb as part of the original GWAS in DNHS. The color of each diamond is based on r2 (white for r2=0, red for r2>0.8) with SNP rs10170218. The recombination rates are based on YRI HapMap data and are shown in light grey along the x axis. The green arrows indicate gene location. For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.
The next strongest signals were at SNP rs1611133 (p-value = 1.18 × 10−6; OR = 3.56, 95% CI = 2.04–6.22) located in a region of high LD on chromosome 6, 10-Kb from the gene HLA-G, which encodes for the major histocompatibility complex, class I, G and SNP rs2413187 maps to an intron of LARGE, a gene that encodes acetyl-glucosaminyl-transferase-like protein. Of note, SNP rs1611133 was associated with rheumatoid arthritis with p-value = 4 × 10−6 (OR = 0.73) in the opposite direction (Plenge et al., 2007). Both SNP rs7302717 and rs10502692 map to an intergenic region. The first of these two regions is gene desert, while the second harbors the gene TMCC3, which encodes the transmembrane and coiled-coil domain family 3, and is located ~75-Kb from the associated SNP. SNP rs11767398 maps to the 3′ UTR of gene C7orf53, which encodes a coiled-coil domain-containing transmembrane protein. SNP rs4701170 maps in an intergenic region, and is located 14-Kb from the gene RP11–798K23.1, a putative lincRNA.
3.2 Top hit SNP rs10170218 replication in NHS-II Sample
We tested the association between SNP rs10170218 and PTSD in the NHSII cohort for replication in an independent female sample. SNP rs10170218 was in HWE and the minor allele C frequency (MAF) was 0.27 in the European American females sample from NHSII (MAF of allele C was 0.22 in DNHS). The association test based on the additive genetic model gave a marginally significant p-value for association with PTSD (p-value = 0.07), with stronger evidence for the genotypic model (p-value = 0.03), and the dominant model (p-value = 0.01).
3.3 In-silico analysis of the genome annotations for lincRNA AC068718.1
Given the novelty of lincRNAs and the relative SNP found associated in our study, we decided to further characterize the lincRNA AC068718.1 performing an in silico computational analysis by integrating the current genome annotations available at UCSC Genome Browser (Rosenbloom et al., 2012). The ENCODE/GENCODE (version 7, May 2011) annotation defines AC068718.1 as a novel lincRNA. We found that ~ 52.38% of the DNA sequence contains transposable elements (TEs) as previously reported elsewhere for other lincRNAs (Jendrzejewski et al., 2012). TEs are discrete pieces of DNA that can move within genomes, inserting themselves into new genomic sites and generating interspersed repeats of the original TEs, and have been recently implicated in the origin of lincRNAs (Ponting et al., 2009). Consulting the database of TEs, we found that 7.14% of the reference sequence of the lincRNA AC068718.1 overlaps with short interspersed nuclear elements (SINEs), 26.51% with long interspersed nuclear elements (LINEs), of which 71% are of Family L1, and 16.23% with long terminal repeat (LTR) transposable elements from the endogenous retrovirus (ERV) family. We found that exons 1, 2, and 3 are located within repetitive elements, SINE (Family Alu) and LTR (Family Erv1), respectively, while exon 4 seems to be a unique sequence in the genome.
3.4 Pathway analysis of functional interactions network reconstructed in silico based on independent genomic regions of association
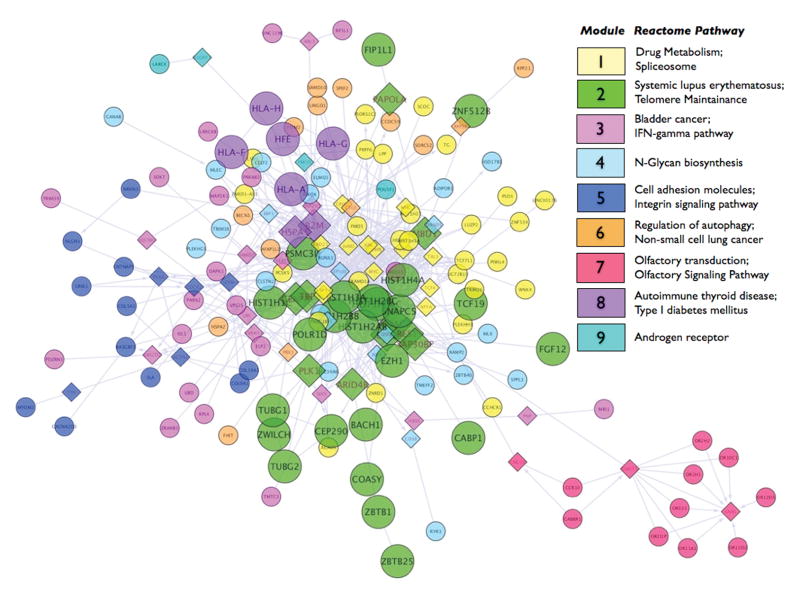
The clumping algorithm identified 133 independent genomic regions of association harboring genes (Table S1). 98 of these harbored more than one SNP, with region sizes ranging from 0.8 Kb (i.e., 2 SNPs clumped) to 346 Kb (i.e., 49 SNPs clumped). Then, we used the Reactome Functional Interaction (FI) network to reconstruct a FI subnetwork based on the set of genes mapping on the 133 non-desert genomic regions of association derived from the GWAS (Figure 3). The network-clustering algorithm based on spectral partition (Newman, 2006) identified 9 different modules of highly interacting genes (Table S2). Pathway enrichment analysis within each module revealed enrichment of the Telomere Maintenance pathway (fdr < 2 × 10−3), and Systemic Lupus Erythematosus pathway (fdr < 1 × 10−3) in module 2 and a set of signaling pathways pertinent to immunity in module 8, including the Antigen processing and presentation pathway, Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell, Type I diabetes mellitus and Autoimmune thyroid disease pathways with fdr < 3 × 10−4.
Figure 3.
Visualization of the Functional Interactions network constructed on the genes mapping in GWAS-derived independent regions of association. Each circle represents a node of the network and each node is a gene. Diamonds represent “linker” genes, i.e. nodes necessary to reconstruct the network but not present in the original list of genes. Nodes are connected by directional arrows that indicate the target gene. Nodes in different network modules are shown in different colors. For each module, the top two associated biological pathways from the Reactome database are reported in the legend. Larger circle size for nodes in modules 2 and 8 indicate modules with biological pathways associated with FDR < 0.05 (see Supplementary Table 2). For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article
4. Discussion
We have completed the largest and most comprehensive GWAS of PTSD to date in a female sample involving more than 400 women using a dense panel of > 700,000 markers. Our results implicate a novel risk locus associated with PTSD (rs10170218) mapping to a novel RNA gene, the lincRNA AC068718.1 that reached genome-wide significance. We found suggestive evidence for replication in a female European-American sample from the NHSII. Our exploratory pathway enrichment analysis using a functional interaction network-based approach of top hits from our initial GWAS, albeit preliminary, revealed molecular signatures implicating pathways involved in telomerase maintenance and immune functioning. We considered our findings in reference to the two other published PTSD GWASs (Logue et al., 2012; Xie et al., 2013). With regard to the RORA gene, the strongest association was found for rs2433025 (p-value = 8 × 10−4). This SNP maps ~ 117 Kb upstream from the SNPs found associated in the first GWAS by Logue and colleagues. As for the second, we were not able to evaluate the association of SNP rs6812849 (TLL1) as it was not present in our set of available genotyped SNPs; however, our results revealed a marginal association of SNP rs10031332 with PTSD (p-value = 5.2 × 10−3). This SNP maps ~ 27 Kb downstream from the top hit by Xie and colleagues.
The DNHS and the NHSII have different ancestry-related genetic background, mainly African American for the DNHS and European American for the NHSII. To be consistent with the analysis of the GWAS in the DNHS sample, we tested the association of SNP rs10170218 using the additive genetic model. The frequency of the putative risk allele of SNP rs10170218 differed somewhat in the two samples (MAFNHSII = 0.27 versus MAFDNHS = 0.22). This allelic difference should be considered in the context of the potential heterogeneity in the genetic architecture of the disease in the independent cohorts used in this study. It is well known that various forces, including genetic drift, gene flow, mutation, selection, and admixture, shape the population frequencies at any given locus (Adeyemo and Rotimi, 2010). These factors play a major role in determining the evolutionary history of loci associated to the disease and may lead to a non-homogeneous distribution of genetic risk variants across populations with a different ancestry-related background (Marigorta et al., 2011). The genotypic and the dominant model show a progressive increase in statistical significance, which is consistent with the idea that a more specific genetic model, i.e. the dominant, when it is in fact true, is more powerful that a more generic one, i.e. the genotypic.
Our genome-wide significant hit maps to a novel RNA gene, the lincRNA AC068718.1. Our in silico analyses using current genome annotations available at UCSC suggest our finding is consistent with what has been reported elsewhere about the possible origin of exons of noncoding RNA genes by retrotransposable elements. SNP rs1070218 maps to intron 3–4, 236 Kb upstream from the start of exon 4, and overlaps with a repeat sequence of class LTR8B (Family Erv1). Emerging literature supports a functional role of transposable elements such as transposons or retrotransposons in gene regulation (Cordaux and Batzer, 2009). The presence of mobile genetic elements in this lincRNA structure seems to be consistent with a noncoding RNA gene with putative regulatory functions (Mariner et al., 2008; Ponicsan et al., 2010). The absence of information about lincRNA function does not allow studying them in the context of any curated known functional pathways, which makes difficult to fully evaluate their role in the etiology of PTSD.
lincRNAs have been classified as RNA molecules longer than 200 nucleotides that are capped, polyadenylated and often spliced, not overlapping with protein-coding genes or other non-coding RNA genes previously identified (Guttman et al., 2011). Emerging literature suggests a regulatory role of lincRNA on other genes in terms of protein expression, DNA binding, and transcriptional function (Guttman and Rinn, 2012). Further experimental analyses are needed to elucidate the biological mechanism underlying lincRNAs such as AC068718.1.
We conducted an exploratory analysis to assess whether our list of protein-coding genes in the independent regions of association that harbored hits with p-value < 5 × 10−4 was significantly enriched for gene ontology categories, and whether these genes were implicated in known functional pathways. We used a network-based approach to reconstruct the overall network of functional interactions between the genes in our list. Our clustering analysis of highly interacting genes within our reconstructed network identified two sets of pathways of interest in two of the nine modules. The molecular signatures revealed by pathway enrichment analysis are consistent with recent developments related to the pathophysiology of PTSD. In module 2, we found association with the Telomerase Maintenance and the Systemic Lupus Erythematosus pathways, which apply to four genes and six genes respectively, encoding for histone proteins mapping on 6p22.1. Telomeres are nucleoprotein structures that protect chromosome ends from degradation. Telomerase maintenance refers to the enzyme telomerase-dependent mechanisms of maintenance of the telomere length. Histone proteins, associated in our GWAS, are included in this pathway because of their central role for DNA repair and chromosomal stability. Oxidative and psychological stress, including PTSD, has been associated with accelerated telomere shortening (Epel et al., 2004; Danese et al., 2009; O’Donovan et al., 2011). Interestingly, one of the emerging themes among many lincRNAs is the formation of RNA-protein interactions between the ncRNAs and proteins, such as the telomerase, used to carry out their function. Apparently the telomerase activity requires a specific telomerase RNA component, i.e., gene TERC, for telomeric regulation (Guttman and Rinn, 2012). Specific TERC domains seem to interact with the telomerase bringing specific regulatory components into proximity with each other, which results in the formation of a unique functional RNA-protein functional complex ultimately responsible to telomerase repair.
The second set of pathways in module 8 pointed to immunity-related molecular signature with nominal association of antigen processing and presentation, immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell, Type I diabetes mellitus and autoimmune thyroid disease pathways, which all apply to the same set of genes encoding for the major histocompatibility complex (HLA) class I heavy chain paralogues, i.e. HLA-A, -G and -F, mapping as well on 6p22.1. Both sets of genes in the most enriched biological pathways map in the two largest LD regions identified by the independent regional association analysis. The first LD region contains 33 and the second LD region contains 49 SNPs with p-value < 0.05 in LD with one of the top nominally associated SNP with p-value < 5 × 10−4.
Evidence from a variety of sources suggests an association between dysfunction of the innate immune inflammatory system in the pathophysiology of PTSD. Previous work by the our group (Uddin et al., 2010) and others (Smith et al., 2011) has shown that immune system functions were significantly overrepresented among the annotations associated with genes uniquely unmethylated among those with PTSD. In a female sample, childhood abuse related PTSD has been associated with enhanced inflammatory system activity and decreased immune cell glucocorticoid sensitivity (Pace et al., 2012). PTSD in veteran samples has been associated with increased risk of Rheumatoid Arthritis (RA) onset and, among RA patients, greater pain, more functional impairment, and less well-being (Mikuls et al., 2012; Pace et al., 2012). This combined evidence suggests a potential role of dysfunction of the innate immune inflammatory system in the pathophysiology of PTSD.
Our findings must be considered in the context of several limitations. Telephone interviews, used to ascertain PTSD status, may result in PTSD misclassification, biasing our results towards the null. We were powered only to find very strong genetic effects. Therefore, the marginally significant results in this cohort should be considered with caution. Results from GWAS of other psychiatric disorders including schizophrenia, bipolar disorder and major depression suggest these disorders are highly polygenic, characterized by potentially hundreds of variants with weak effects (Purcell et al., 2009; Middeldorp et al., 2011). Whether the genetic architecture of PTSD is similar remains to be seen.
Genome-wide association studies have revealed a number of new genetic variants for common diseases (Hindorff et al., 2009). The identification of genetic variants of susceptibility to PTSD is still relatively in its infancy. Although there have been some notable successes, candidate gene association studies have often failed to deliver robust results. Our results suggest the potential role of a novel RNA gene, the lincRNA AC068718.1, in the pathophysiology of PTSD. We also found several additional SNPs with highly significant evidence of association (p-value < 1 × 10−5), none of which have previously been reported in association with PTSD. Moreover, the possibility of sex-specific genetic effects in PTSD merit further investigation in much larger samples that include both men and women.
Supplementary Material
Table 1.
Demographic characteristics of GWAS subjects
| Variable b | PTSD Status |
||
|---|---|---|---|
| Cases n (%)/ Mean (SD) | Controls n (%)/ Mean (SD) | Group Comparison P valuea |
|
| Age | 52.2 (13.5) | 54.3 (15.9) | 0.2325 |
| Race | 0.4080 | ||
| White | 10 (10.6%) | 35 (11.0%) | . |
| Black | 81 (86.2%) | 261 (82.1%) | . |
| Other | 3 (3.2%) | 22 (6.9%) | . |
| Ethnicity | 1.0000 | ||
| not Hispanic | 93 (98.9%) | 313 (98.4%) | . |
| Hispanic | 1 (1.1%) | 5 (1.6%) | . |
| Education | 0.9289 | ||
| Less than high school | 12 (12.8%) | 44 (13.8%) | . |
| High school grad/GED | 25 (26.6%) | 80 (25.1%) | . |
| some college | 37 (39.4%) | 114 (35.7%) | . |
| college degree | 14 (14.9%) | 58 (18.2%) | . |
| grad degree | 6 (6.4%) | 23 (7.2%) | . |
| Marital Status | 0.3417 | ||
| married | 22 (23.4%) | 65 (20.4%) | . |
| divorced or separated | 33 (35.1%) | 88 (27.6%) | . |
| widowed | 14 (14.9%) | 58 (18.2%) | . |
| never married | 25 (26.6%) | 108 (33.9%) | . |
| Number of traumatic events | 9.0 (4.5) | 5.5 (3.8) | <.0001 |
P-values for continuous variables are from Wilcoxon exact test, while Monte Carlo estimate for Pearson chi-square exact test for categorical variables
Counts may not add up to 449 due to missing values from some variables
Acknowledgments
We acknowledge the Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School for its management of The Nurses’ Health Study II.
Role of the funding Source
The Detroit Neighborhood Health Study (PI: Allison Aiello) is funded by DA022720, DA 022720-S1, and MH 088283. Funding for this study was also provided by National Institute on Mental Health awards R01 MH093612, MH078928 to Karestan C. Koenen. The Nurses’ Health Study II is funded in part by NIH CA50385.
Footnotes
Contributors
Drs. Guffanti and Yan undertook the statistical analysis. Drs. Roberts and Solovieff contributed with quality control analysis and phenotypic variables preparation for the two datasets involved DNHS and NHSII. Dr. Wildman performed the genotyping in DNHS and Dr. Ranu the genotyping in NHSII. Dr. Koenen supervised the overall project. Drs. Guffanti and Koenen wrote the first draft of the manuscript. All authors contributed to and have approved the final manuscript.
Conflict of interest statement
All authors declare that they have no conflicts of interest.
Supplementary Material is available at the Psychoneuroendocrinology website
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adeyemo A, Rotimi C. Genetic variants associated with complex human diseases show wide variation across multiple populations. Public Health Genomics. 2010;13:72–79. doi: 10.1159/000218711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almli LM, Mercer KB, Kerley K, Feng H, Bradley B, Conneely KN, Ressler KJ. ADCYAP1R1 genotype associates with post-traumatic stress symptoms in highly traumatized African-American females. Am J Med Genet B Neuropsychiatr Genet. 2013;162B:262–272. doi: 10.1002/ajmg.b.32145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake DD, Weathers FW, Nagy LM, Kaloupek DG, Gusman FD, Charney DS, Keane TM. The development of a Clinician-Administered PTSD Scale. J Trauma Stress. 1995;8:75–90. doi: 10.1007/BF02105408. [DOI] [PubMed] [Google Scholar]
- Breslau N, Chilcoat HD, Kessler RC, Peterson EL, Lucia VC. Vulnerability to assaultive violence: further specification of the sex difference in post-traumatic stress disorder. Psychol Med. 1999;29:813–821. doi: 10.1017/s0033291799008612. [DOI] [PubMed] [Google Scholar]
- Breslau N, Davis GC. Posttraumatic stress disorder in an urban population of young adults: risk factors for chronicity. Am J Psychiatry. 1992;149:671–675. doi: 10.1176/ajp.149.5.671. [DOI] [PubMed] [Google Scholar]
- Breslau N, Kessler RC, Chilcoat HD, Schultz LR, Davis GC, Andreski P. Trauma and posttraumatic stress disorder in the community: the 1996 Detroit Area Survey of Trauma. Arch Gen Psychiatry. 1998;55:626–632. doi: 10.1001/archpsyc.55.7.626. [DOI] [PubMed] [Google Scholar]
- Chang SC, Xie P, Anton RF, De Vivo I, Farrer LA, Kranzler HR, Oslin D, Purcell SM, Roberts AL, Smoller JW, Uddin M, Gelernter J, Koenen KC. No association between ADCYAP1R1 and post-traumatic stress disorder in two independent samples. Mol Psychiatry. 2012;17:239–241. doi: 10.1038/mp.2011.118. [DOI] [PubMed] [Google Scholar]
- Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis MC, Nugent NR, Amstadter AB, Koenen KC. Genetics of post-traumatic stress disorder: review and recommendations for genome-wide association studies. Curr Psychiatry Rep. 2010;12:313–326. doi: 10.1007/s11920-010-0126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danese A, Moffitt TE, Harrington H, Milne BJ, Polanczyk G, Pariante CM, Poulton R, Caspi A. Adverse childhood experiences and adult risk factors for age-related disease: depression, inflammation, and clustering of metabolic risk markers. Arch Pediatr Adolesc Med. 2009;163:1135–1143. doi: 10.1001/archpediatrics.2009.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epel ES, Blackburn EH, Lin J, Dhabhar FS, Adler NE, Morrow JD, Cawthon RM. Accelerated telomere shortening in response to life stress. Proc Natl Acad Sci U S A. 2004;101:17312–17315. doi: 10.1073/pnas.0407162101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmann E, Aiello A, Uddin M, Delva J, Koenen K, Gant LM, Galea S. Pervasive exposure to violence and posttraumatic stress disorder in a predominantly African American Urban Community: the Detroit Neighborhood Health Study. J Trauma Stress. 2011;24:747–751. doi: 10.1002/jts.20705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jendrzejewski J, He H, Radomska HS, Li W, Tomsic J, Liyanarachchi S, Davuluri RV, Nagy R, de la Chapelle A. The polymorphism rs944289 predisposes to papillary thyroid carcinoma through a large intergenic noncoding RNA gene of tumor suppressor type. Proc Natl Acad Sci U S A. 2012;109:8646–8651. doi: 10.1073/pnas.1205654109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler RC, Sonnega A, Bromet E, Hughes M, Nelson CB. Posttraumatic stress disorder in the National Comorbidity Survey. Arch Gen Psychiatry. 1995;52:1048–1060. doi: 10.1001/archpsyc.1995.03950240066012. [DOI] [PubMed] [Google Scholar]
- Koenen KC, De Vivo I, Rich-Edwards J, Smoller JW, Wright RJ, Purcell SM. Protocol for investigating genetic determinants of posttraumatic stress disorder in women from the Nurses’ Health Study II. BMC Psychiatry. 2009;9:29. doi: 10.1186/1471-244X-9-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebron-Milad K, Milad MR. Sex differences, gonadal hormones and the fear extinction network: implications for anxiety disorders. Biol Mood Anxiety Disord. 2012;2:3. doi: 10.1186/2045-5380-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logue MW, Baldwin C, Guffanti G, Melista E, Wolf EJ, Reardon AF, Uddin M, Wildman D, Galea S, Koenen KC, Miller MW. A genome-wide association study of post-traumatic stress disorder identifies the retinoid-related orphan receptor alpha (RORA) gene as a significant risk locus. Mol Psychiatry. 2012 doi: 10.1038/mp.2012.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marigorta UM, Lao O, Casals F, Calafell F, Morcillo-Suarez C, Faria R, Bosch E, Serra F, Bertranpetit J, Dopazo H, Navarro A. Recent human evolution has shaped geographical differences in susceptibility to disease. BMC Genomics. 2011;12:55. doi: 10.1186/1471-2164-12-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, Goodrich JA. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell. 2008;29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- McLean CP, Asnaani A, Litz BT, Hofmann SG. Gender differences in anxiety disorders: prevalence, course of illness, comorbidity and burden of illness. J Psychiatr Res. 2011;45:1027–1035. doi: 10.1016/j.jpsychires.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middeldorp CM, de Moor MH, McGrath LM, Gordon SD, Blackwood DH, Costa PT, Terracciano A, Krueger RF, de Geus EJ, Nyholt DR, Tanaka T, Esko T, Madden PA, Derringer J, Amin N, Willemsen G, Hottenga JJ, Distel MA, Uda M, Sanna S, Spinhoven P, Hartman CA, Ripke S, Sullivan PF, Realo A, Allik J, Heath AC, Pergadia ML, Agrawal A, Lin P, Grucza RA, Widen E, Cousminer DL, Eriksson JG, Palotie A, Barnett JH, Lee PH, Luciano M, Tenesa A, Davies G, Lopez LM, Hansell NK, Medland SE, Ferrucci L, Schlessinger D, Montgomery GW, Wright MJ, Aulchenko YS, Janssens AC, Oostra BA, Metspalu A, Abecasis GR, Deary IJ, Raikkonen K, Bierut LJ, Martin NG, Wray NR, van Duijn CM, Smoller JW, Penninx BW, Boomsma DI. The genetic association between personality and major depression or bipolar disorder. A polygenic score analysis using genome-wide association data. Transl Psychiatry. 2011;1:e50. doi: 10.1038/tp.2011.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikuls TR, Padala PR, Sayles HR, Yu F, Michaud K, Caplan L, Kerr GS, Reimold A, Cannon GW, Richards JS, Lazaro D, Thiele GM, Boscarino JA. A prospective study of posttraumatic stress disorder (PTSD) and disease activity outcomes in U.S. veterans with Rheumatoid Arthritis. Arthritis Care Res (Hoboken) 2012 doi: 10.1002/acr.21778. [DOI] [PubMed] [Google Scholar]
- Newman ME. Modularity and community structure in networks. Proc Natl Acad Sci U S A. 2006;103:8577–8582. doi: 10.1073/pnas.0601602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donovan A, Epel E, Lin J, Wolkowitz O, Cohen B, Maguen S, Metzler T, Lenoci M, Blackburn E, Neylan TC. Childhood trauma associated with short leukocyte telomere length in posttraumatic stress disorder. Biol Psychiatry. 2011;70:465–471. doi: 10.1016/j.biopsych.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace TW, Wingenfeld K, Schmidt I, Meinlschmidt G, Hellhammer DH, Heim CM. Increased peripheral NF-kappaB pathway activity in women with childhood abuse-related posttraumatic stress disorder. Brain Behav Immun. 2012;26:13–17. doi: 10.1016/j.bbi.2011.07.232. [DOI] [PubMed] [Google Scholar]
- Pitman RK, Rasmusson AM, Koenen KC, Shin LM, Orr SP, Gilbertson MW, Milad MR, Liberzon I. Biological studies of post-traumatic stress disorder. Nat Rev Neurosci. 2012;13:769–787. doi: 10.1038/nrn3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plenge RM, Seielstad M, Padyukov L, Lee AT, Remmers EF, Ding B, Liew A, Khalili H, Chandrasekaran A, Davies LR, Li W, Tan AK, Bonnard C, Ong RT, Thalamuthu A, Pettersson S, Liu C, Tian C, Chen WV, Carulli JP, Beckman EM, Altshuler D, Alfredsson L, Criswell LA, Amos CI, Seldin MF, Kastner DL, Klareskog L, Gregersen PK. TRAF1-C5 as a risk locus for rheumatoid arthritis--a genomewide study. N Engl J Med. 2007;357:1199–1209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponicsan SL, Kugel JF, Goodrich JA. Genomic gems: SINE RNAs regulate mRNA production. Curr Opin Genet Dev. 2010;20:149–155. doi: 10.1016/j.gde.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick HS, Kilpatrick DG, Dansky BS, Saunders BE, Best CL. Prevalence of civilian trauma and posttraumatic stress disorder in a representative national sample of women. J Consult Clin Psychol. 1993;61:984–991. doi: 10.1037//0022-006x.61.6.984. [DOI] [PubMed] [Google Scholar]
- Ressler KJ, Mercer KB, Bradley B, Jovanovic T, Mahan A, Kerley K, Norrholm SD, Kilaru V, Smith AK, Myers AJ, Ramirez M, Engel A, Hammack SE, Toufexis D, Braas KM, Binder EB, May V. Post-traumatic stress disorder is associated with PACAP and the PAC1 receptor. Nature. 2011;470:492–497. doi: 10.1038/nature09856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AL, Dohrenwend BP, Aiello AE, Wright RJ, Maercker A, Galea S, Koenen KC. The stressor criterion for posttraumatic stress disorder: does it matter? J Clin Psychiatry. 2012;73:e264–270. doi: 10.4088/JCP.11m07054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom KR, Dreszer TR, Long JC, Malladi VS, Sloan CA, Raney BJ, Cline MS, Karolchik D, Barber GP, Clawson H, Diekhans M, Fujita PA, Goldman M, Gravell RC, Harte RA, Hinrichs AS, Kirkup VM, Kuhn RM, Learned K, Maddren M, Meyer LR, Pohl A, Rhead B, Wong MC, Zweig AS, Haussler D, Kent WJ. ENCODE whole-genome data in the UCSC Genome Browser: update 2012. Nucleic Acids Res. 2012;40:D912–917. doi: 10.1093/nar/gkr1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor CE, McCutcheon VV, Pommer NE, Nelson EC, Grant JD, Duncan AE, Waldron M, Bucholz KK, Madden PA, Heath AC. Common genetic and environmental contributions to post-traumatic stress disorder and alcohol dependence in young women. Psychol Med. 2011;41:1497–1505. doi: 10.1017/S0033291710002072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P. RepeatMasker Open-3.0 1996–2010 [Google Scholar]
- Smith AK, Conneely KN, Kilaru V, Mercer KB, Weiss TE, Bradley B, Tang Y, Gillespie CF, Cubells JF, Ressler KJ. Differential immune system DNA methylation and cytokine regulation in post-traumatic stress disorder. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:700–708. doi: 10.1002/ajmg.b.31212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin M, Aiello AE, Wildman DE, Koenen KC, Pawelec G, de Los Santos R, Goldmann E, Galea S. Epigenetic and immune function profiles associated with posttraumatic stress disorder. Proc Natl Acad Sci U S A. 2010;107:9470–9475. doi: 10.1073/pnas.0910794107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin M, Chang SC, Zhang C, Ressler K, Mercer KB, Galea S, Keyes KM, McLaughlin KA, Wildman DE, Aiello AE, Koenen KC. Adcyap1r1 genotype, posttraumatic stress disorder, and depression among women exposed to childhood maltreatment. Depress Anxiety. 2013;30:251–258. doi: 10.1002/da.22037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Cao C, Wang R, Qing Y, Zhang J, Zhang XY. PAC1 receptor (ADCYAP1R1) genotype is associated with PTSD’s emotional numbing symptoms in Chinese earthquake survivors. J Affect Disord. 2013;150:156–159. doi: 10.1016/j.jad.2013.01.010. [DOI] [PubMed] [Google Scholar]
- Xie P, Kranzler HR, Yang C, Zhao H, Farrer LA, Gelernter J. Genome-wide Association Study Identifies New Susceptibility Loci for Posttraumatic Stress Disorder. Biol Psychiatry. 2013 doi: 10.1016/j.biopsych.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.