Abstract
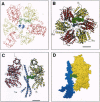
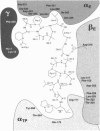
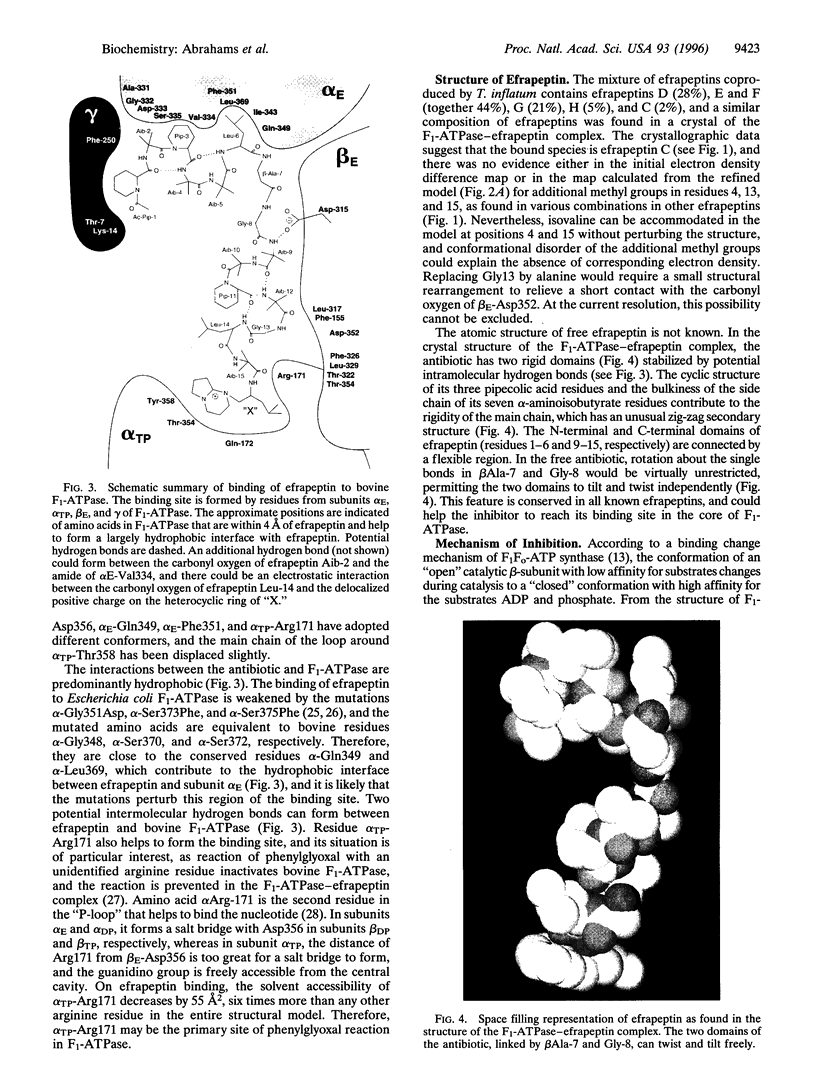
In the previously determined structure of mitochondrial F1-ATPase determined with crystals grown in the presence of adenylyl-imidodiphosphate (AMP-PNP) and ADP, the three catalytic beta-subunits have different conformations and nucleotide occupancies. AMP-PNP and ADP are bound to subunits beta TP and beta DP, respectively, and the third beta-subunit (beta E) has no bound nucleotide. The efrapeptins are a closely related family of modified linear peptides containing 15 amino acids that inhibit both ATP synthesis and hydrolysis by binding to the F1 catalytic domain of F1F0-ATP synthase. In crystals of F1-ATPase grown in the presence of both nucleotides and inhibitor, efrapeptin is bound to a unique site in the central cavity of the enzyme. Its binding is associated with small structural changes in side chains of F1-ATPase around the binding pocket. Efrapeptin makes hydrophobic contacts with the alpha-helical structure in the gamma-subunit, which traverses the cavity, and with subunit beta E and the two adjacent alpha-subunits. Two intermolecular hydrogen bonds could also form. Intramolecular hydrogen bonds probably help to stabilize efrapeptin's two domains (residues 1-6 and 9-15, respectively), which are connected by a flexible region (beta Ala-7 and Gly-8). Efrapeptin appears to inhibit F1-ATPase by blocking the conversion of subunit beta E to a nucleotide binding conformation, as would be required by an enzyme mechanism involving cyclic interconversion of catalytic sites.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abrahams J. P., Leslie A. G., Lutter R., Walker J. E. Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994 Aug 25;370(6491):621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- Boyer P. D. The binding change mechanism for ATP synthase--some probabilities and possibilities. Biochim Biophys Acta. 1993 Jan 8;1140(3):215–250. doi: 10.1016/0005-2728(93)90063-l. [DOI] [PubMed] [Google Scholar]
- Cross R. L., Kohlbrenner W. E. The mode of inhibition of oxidative phosphorylation by efrapeptin (A23871). Evidence for an alternating site mechanism for ATP synthesis. J Biol Chem. 1978 Jul 25;253(14):4865–4873. [PubMed] [Google Scholar]
- Cross R. L. The mechanism and regulation of ATP synthesis by F1-ATPases. Annu Rev Biochem. 1981;50:681–714. doi: 10.1146/annurev.bi.50.070181.003341. [DOI] [PubMed] [Google Scholar]
- Jackson P. J., Harris D. A. The mitochondrial ATP synthase inhibitor protein binds near the C-terminus of the F1 beta-subunit. FEBS Lett. 1988 Feb 29;229(1):224–228. doi: 10.1016/0014-5793(88)80832-9. [DOI] [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991 Mar 1;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kohlbrenner W. E., Cross R. L. Efrapeptin prevents modification by phenylglyoxal of an essential arginyl residue in mitochondrial adenosine triphosphatase. J Biol Chem. 1978 Nov 10;253(21):7609–7611. [PubMed] [Google Scholar]
- Kohlbrenner W. E., Cross R. L. The mode of inhibition of oxidative phosphorylation by efrapeptin (A23871): measurement of substrate effects on rates of inactivation by a tight-binding inhibitor. Arch Biochem Biophys. 1979 Dec;198(2):598–607. doi: 10.1016/0003-9861(79)90536-8. [DOI] [PubMed] [Google Scholar]
- Lardy H., Reed P., Lin C. H. Antibiotic inhibitors of mitochondrial ATP synthesis. Fed Proc. 1975 Jul;34(8):1707–1710. [PubMed] [Google Scholar]
- Linnett P. E., Beechey R. B. Inhibitors of the ATP synthethase system. Methods Enzymol. 1979;55:472–518. doi: 10.1016/0076-6879(79)55061-7. [DOI] [PubMed] [Google Scholar]
- Lucero H. A., Ravizzine R. A., Vallejos R. H. Inhibition of spinach chloroplasts photophosphorylation by the antibiotics leucinostatin and efrapeptin. FEBS Lett. 1976 Sep 15;68(1):141–144. doi: 10.1016/0014-5793(76)80423-1. [DOI] [PubMed] [Google Scholar]
- Lucero H., Lescano W. I., Vallejos R. H. Inhibition of energy conservation reactions in chromatophores of Rhodospirillum rubrum by antibiotics. Arch Biochem Biophys. 1978 Feb;186(1):9–14. doi: 10.1016/0003-9861(78)90457-5. [DOI] [PubMed] [Google Scholar]
- Lutter R., Abrahams J. P., van Raaij M. J., Todd R. J., Lundqvist T., Buchanan S. K., Leslie A. G., Walker J. E. Crystallization of F1-ATPase from bovine heart mitochondria. J Mol Biol. 1993 Feb 5;229(3):787–790. doi: 10.1006/jmbi.1993.1081. [DOI] [PubMed] [Google Scholar]
- Maggio M. B., Pagan J., Parsonage D., Hatch L., Senior A. E. The defective proton-ATPase of uncA mutants of Escherichia coli. Identification by DNA sequencing of residues in the alpha-subunit which are essential for catalysis or normal assembly. J Biol Chem. 1987 Jul 5;262(19):8981–8984. [PubMed] [Google Scholar]
- Orriss G. L., Runswick M. J., Collinson I. R., Miroux B., Fearnley I. M., Skehel J. M., Walker J. E. The delta- and epsilon-subunits of bovine F1-ATPase interact to form a heterodimeric subcomplex. Biochem J. 1996 Mar 1;314(Pt 2):695–700. doi: 10.1042/bj3140695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saishu T., Kagawa Y., Shimizu R. Resistance of thermophilic ATPase (TF1) to specific F1-atpase inhibitors including local anesthetics. Biochem Biophys Res Commun. 1983 May 16;112(3):822–826. doi: 10.1016/0006-291x(83)91691-1. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J. E. The regulation of catalysis in ATP synthase. Curr Opin Struct Biol. 1994 Dec;4(6):912–918. doi: 10.1016/0959-440x(94)90274-7. [DOI] [PubMed] [Google Scholar]
- Wise J. G., Duncan T. M., Latchney L. R., Cox D. N., Senior A. E. Properties of F1-ATPase from the uncD412 mutant of Escherichia coli. Biochem J. 1983 Nov 1;215(2):343–350. doi: 10.1042/bj2150343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise J. G., Latchney L. R., Ferguson A. M., Senior A. E. Defective proton ATPase of uncA mutants of Escherichia coli. 5'-Adenylyl imidodiphosphate binding and ATP hydrolysis. Biochemistry. 1984 Mar 27;23(7):1426–1432. doi: 10.1021/bi00302a014. [DOI] [PubMed] [Google Scholar]
- van Raaij M. J., Abrahams J. P., Leslie A. G., Walker J. E. The structure of bovine F1-ATPase complexed with the antibiotic inhibitor aurovertin B. Proc Natl Acad Sci U S A. 1996 Jul 9;93(14):6913–6917. doi: 10.1073/pnas.93.14.6913. [DOI] [PMC free article] [PubMed] [Google Scholar]