Significance
A key step in bacterial cell division is the polymerization of FtsZ at midcell into a ring-like structure. This so-called Z ring forms a scaffold for the other cell division proteins. FtsA anchors the Z ring to the cell membrane; however, many bacterial species do not have FtsA. Here, we show that the conserved protein SepF, which forms large protein rings, also functions as a membrane anchor for the Z ring. We determined the molecular structure of the FtsZ-binding and ring-forming domain of SepF and show that the membrane-binding domain is located at the very beginning of the protein. These results explain why FtsA and SepF can fulfill similar functions in bacterial cell division.
Abstract
A key step in bacterial cell division is the polymerization of the tubulin homolog FtsZ at midcell. FtsZ polymers are anchored to the cell membrane by FtsA and are required for the assembly of all other cell division proteins. In Gram-positive and cyanobacteria, FtsZ filaments are aligned by the protein SepF, which in vitro polymerizes into large rings that bundle FtsZ filaments. Here we describe the crystal structure of the only globular domain of SepF, located within the C-terminal region. Two-hybrid data revealed that this domain comprises the FtsZ binding site, and EM analyses showed that it is sufficient for ring formation, which is explained by the filaments in the crystals of SepF. Site-directed mutagenesis, gel filtration, and analytical ultracentrifugation indicated that dimers form the basic units of SepF filaments. High-resolution structured illumination microscopy suggested that SepF is membrane associated, and it turned out that purified SepF not only binds to lipid membranes, but also recruits FtsZ. Further genetic and biochemical analyses showed that an amphipathic helix at the N terminus functions as the membrane-binding domain, making SepF a unique membrane anchor for the FtsZ ring. This clarifies why Bacillus subtilis grows without FtsA or the putative membrane anchor EzrA and why bacteria lacking FtsA contain SepF homologs. Both FtsA and SepF use an amphipathic helix for membrane binding. These helices prefer positively curved membranes due to relaxed lipid density; therefore this type of membrane anchor may assist in keeping the Z ring positioned at the strongly curved leading edge of the developing septum.
One of the first steps in bacterial cell division is the polymerization of the conserved protein FtsZ at midcell. FtsZ shares structural homology with eukaryotic tubulin and uses GTP to polymerize into filaments close to the cell membrane. These filaments then assemble into a ring-like structure, the Z ring, which recruits other proteins needed for the division septum (1). Several cell division proteins support the formation of a stable Z ring, such as ZapA that forms cross-links between FtsZ filaments (2, 3). In Gram-positive bacteria and cyanobacteria, the protein SepF also stimulates bundling of FtsZ polymers (4–6). Electron microscopic (EM) studies have shown that SepF assembles into large and regular protein rings with diameters of about 50 nm. In vitro, these rings are able to bundle FtsZ protofilaments into long tubular structures (4). Furthermore, SepF is essential for the synthesis of regular and smooth division septa (7–9).
The Z ring is associated with the cell membrane and the best-characterized membrane anchor is the conserved protein FtsA. This protein binds to FtsZ directly and contains a C-terminal amphipathic helix that binds to lipid bilayers in a membrane-potential-dependent manner (10–12). In Escherichia coli, FtsA is essential, but in Bacillus subtilis, ftsA can be deleted, although this affects Z-ring formation and cells become elongated. Because B. subtilis can grow without FtsA, there must be another protein that links the Z ring to the cell membrane. The essential E. coli cell division protein ZipA binds to FtsZ and contains an N-terminal transmembrane domain (13). The necessity for ZipA can be bypassed by a gain-of-function mutation in FtsA (14). Gram-positive bacteria contain EzrA, which shows a similar topology to that of ZipA, with an N-terminal transmembrane helix and a large C-terminal domain that binds to the FtsZ C terminus (15). It therefore seemed likely that EzrA functions as an alternative membrane anchor for the Z ring in B. subtilis when FtsA is absent.
Comparison of the SepF amino acid sequence against protein databases did not reveal conserved motifs that provide clues to potential molecular mechanisms. To gain insight into the residues that are important for FtsZ interaction, we used a yeast two-hybrid screen. This revealed that the conserved C-terminal part of SepF comprises the FtsZ binding site. We were able to obtain diffracting crystals of this domain that revealed a tight dimer structure. Site-directed mutagenesis indicated that the protein polymerizes as units of dimers. Although the FtsZ interaction domain constitutes only 60% of the full-length protein, it appeared to be also sufficient for the formation of the large protein rings. This raised the question of what the function is of the N-terminal domain of SepF. Interestingly, high-resolution fluorescent-light microscopy suggested that SepF associates with the cell membrane. Subsequent analyses indicated that the membrane-binding domain resides in the N-terminal domain of SepF. Biochemical experiments using purified SepF showed that the protein binds specifically to lipid membranes. These data suggested that SepF might function as a membrane anchor for the Z ring. Interestingly, we were able to delete both ftsA and the putative FtsZ-membrane anchor ezrA in B. subtilis and this did not affect viability. However, it turned SepF into an essential protein. Indeed, using purified SepF we were able to show that the protein recruits FtsZ to artificial lipid membranes. We conclude that SepF is a unique membrane anchor for FtsZ and the Z ring.
Results
Localization of FtsZ Interaction Mutants.
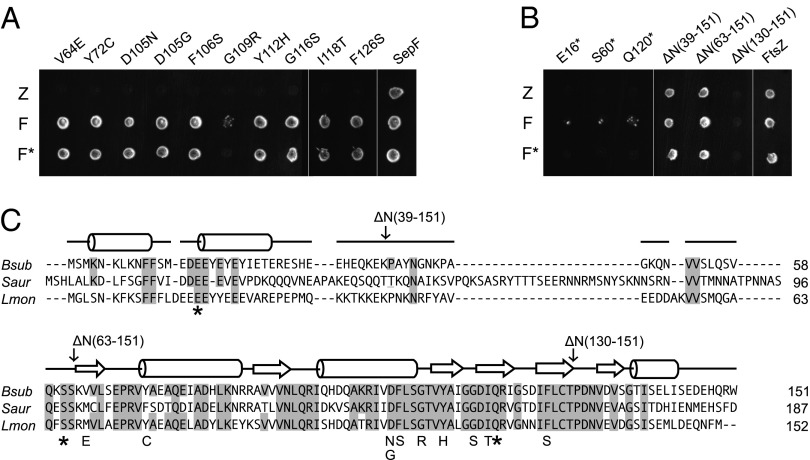
SepF binds directly to the C-terminal domain of FtsZ (6, 16). However, the protein has no apparent homology to proteins of known function and the amino acid sequence does not indicate a clear docking site for FtsZ. In earlier studies it was shown that FtsZ and SepF show a strong and specific interaction in yeast two-hybrid assays (8, 17). This assay was used to identify amino acid residues in SepF that are important for FtsZ binding. Mutations in SepF were introduced by means of error-prone PCR. SepF also shows strong self-interaction, and the mutants were tested for this as well (Fig. 1 A and B). Thirteen different mutants were isolated, of which 10 were amino acid substitutions and 3 were nonsense mutations, the latter resulting in C-terminal truncations at amino acids 16, 60, and 120, respectively. To confirm that the SepF mutants show a defect in FtsZ interaction in B. subtilis cells, the mutants were expressed in an ftsA deletion strain. Previous work has shown that sepF becomes essential when ftsA is deleted in B. subtilis (17). Indeed, all mutants were unable to support growth in a B. subtilis ftsA mutant background (Fig. S1). Fig. 1C shows an amino acid sequence alignment of SepF homologs from three different bacterial species. An extensive ClustalW alignment can be found in Fig. S2. The 10 missense mutations are all located in the conserved C-terminal part of the protein. To further analyze this, three N-terminal truncation mutants were constructed, lacking the first 39 aa, 63 aa, or 130 aa, respectively (Fig. 1B). Interestingly, it was possible to delete the complete 63-aa-long N terminus without affecting FtsZ interaction and self-interaction in our assay. This suggests that the C-terminal part of the protein constitutes a separate domain carrying the FtsZ-interaction and self-interaction functionalities.
Fig. 1.
Yeast two-hybrid screen for SepF-FtsZ interaction mutants. (A) SepF mutants were created using error-prone PCR, and FtsZ-interaction deficient mutants were selected using the yeast two-hybrid assay. Diploid colonies were obtained by mating PJ69-4α strains harboring pGBT9 derivatives containing sepF mutants, and wild-type sepF, with PJ69-4A strains that harbored pGAD424 derivatives containing ftsZ (Z), wild-type sepF (F), or the corresponding sepF mutants (F*). (B) Yeast two-hybrid interactions of the nonsense mutants and the different N-terminal deletion constructs. Remaining amino acid regions are indicated between parentheses. A and B are discontinuous images of the same plate. (C) Amino acid sequence alignment of SepF proteins, from B. subtilis (Bsub), Staphylococcus aureus (Saur), and Listeria monocytogenes (Lmon), using ClustalW. Secondary structure prediction (using PSIPRED) for B. subtilis SepF is shown above the sequences, whereby α-helices and β-sheets are indicated by rods and arrows, respectively. Conserved amino acids are shaded. Yeast two-hybrid mutations are indicated.
Ring-Forming Domain.
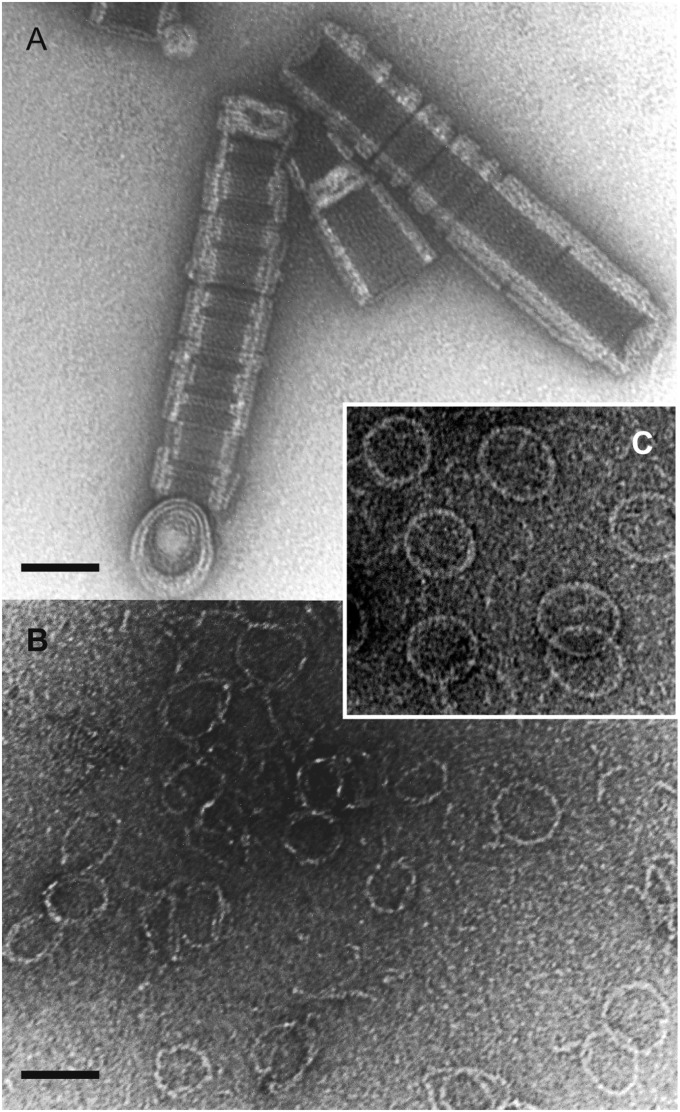
We were unable to obtain diffraction-quality crystals of full-length SepF from B. subtilis or from other bacteria. One of the reasons could be that the protein contains unstructured domains that hamper the crystallization process. Mild proteolytic treatment is able to remove such domains, and indeed a controlled incubation with α-chymotrypsin yielded a truncated protein that formed crystals, which diffracted to 2 Å resolution. Mass spectrometry and N-terminal sequencing revealed that the crystallized protein consists of residues 57–151, thereby neatly encompassing the C-terminal FtsZ-binding region identified in the yeast two-hybrid screen. EM analysis of this truncated SepF protein revealed tubular structures of ∼50 nm in diameter, which were made up of stacks of SepF rings (Fig. 2A). Very high pH conditions (∼pH 10) resolved the structures into individual rings (Fig. 2B). Thus, the C-terminal domain of SepF is responsible for the polymerization of SepF into large rings.
Fig. 2.
Negatively stained electron micrographs of purified chymotrypsin-treated SepF used for crystallization. (A) Under neutral pH conditions (50 mM Tris⋅HCl, pH 7.0) this protein forms large tubes made up of stacked 50-nm diameter rings. (B) Under basic pH conditions (50 mM glycine, pH 10.0) individual rings become visible. (C) Normal full-length SepF. (Scale bars, 50 µm.)
Crystal Structure.
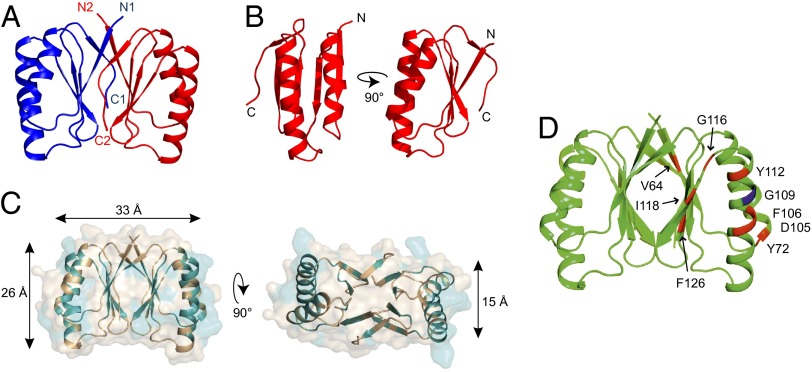
To elucidate the atomic structure, two additional structures of SepF-like proteins were solved by multiwavelength anomalous dispersion (MAD) phasing, one from Archaeoglobus fulgidus and one from Pyrococcus furiosus, and used for molecular replacement of the Bacillus protein. The resulting crystal structure of the SepF C-terminal domain is shown in Fig. 3. The structure comprises residues Ser-61 to Ser-140. Because the protein spans residues 57–151, it means that the first 4 aa and the last 11 aa are disordered in the crystal. The crystal structure shows a tight dimer, with monomers that contain a compact α/β-sandwich arrangement of two α-helices stacked against a five-stranded β-sheet (Fig. 3 A and B). The dimer interface is formed by the β-sheets. Interestingly, the C-terminal β-strand from each monomer joins the β-sheet of the other monomer, such that it is parallel to its N-terminal β-strand. The tight packing of the dimer suggests that this is the functional unit in SepF polymers. In fact, many of the conserved residues are located at the interface between the monomers on the opposing β-sheets (Fig. 3C). Moreover, the A. fulgidus and P. furiosus SepF-like proteins show the same dimer packing in their crystals (Fig. S3; structure statistics are listed in Table S1).
Fig. 3.
Crystal structure of B. subtilis SepF residues 61–140. (A) The crystal structure reveals a dimer whereby the C-terminal β-strand of one monomer (C1, C2) joins the β-sheet of the other monomer and is parallel to its N-terminal β-strand (N1, N2). Structure statistics are listed in Table S1. (B) Monomer of SepF C-terminal domain. (C) Conserved residues are highlighted in teal on the SepF dimer structure. Most conserved residues are located at the interface between monomers (β-sheets) and the interface between dimers (α-helices). Dimensions indicated by arrows are based on the protein backbone. (D) FtsZ-interaction mutants from the yeast two-hybrid screen are indicated on the crystal structure. G109 residue is indicated in blue.
Polymerization of Dimers.
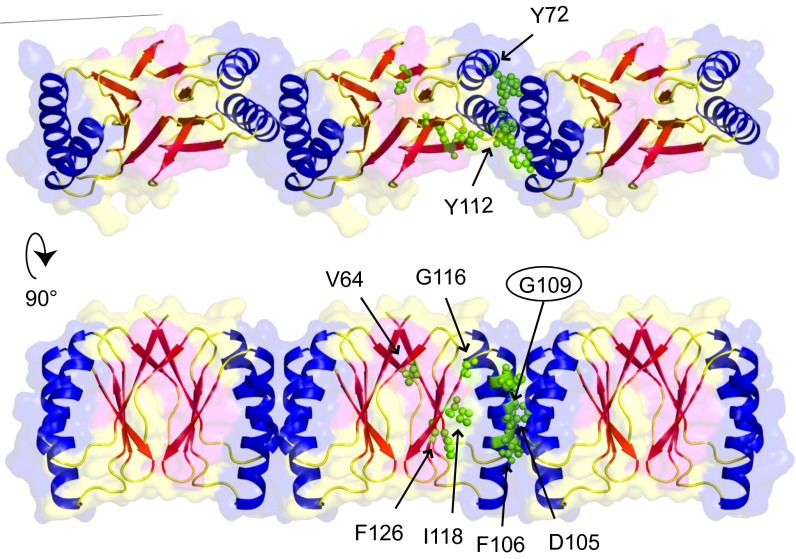
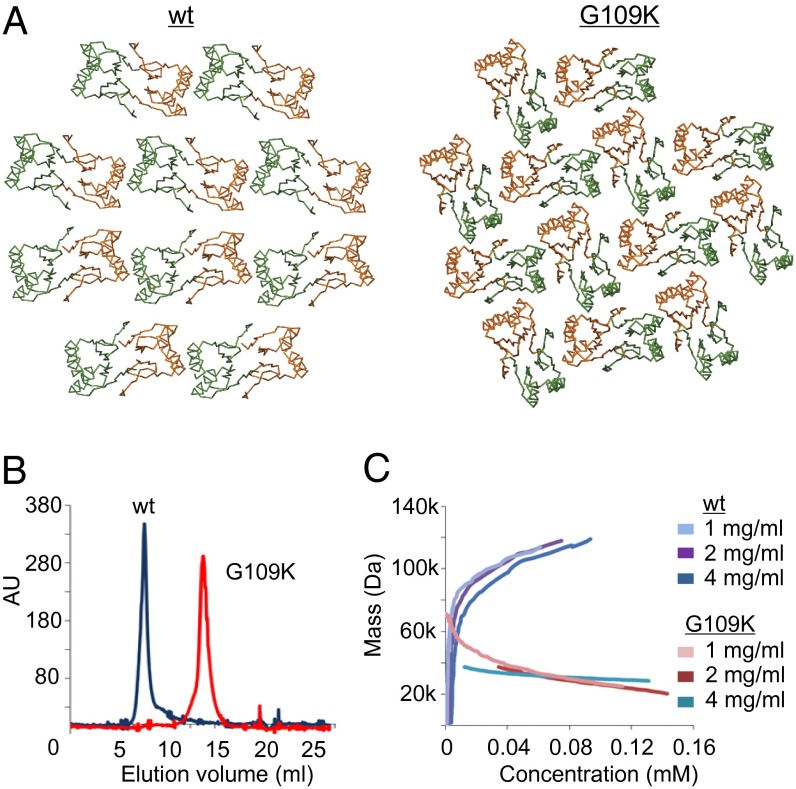
In Fig. 3D the yeast two-hybrid mutations from Fig. 1 are highlighted in the crystal structure. Several mutations that affect FtsZ interaction are located on the surface of the α-helices. This is surprising because there is no clear pocket that could function as a docking site for the C-terminal domain of FtsZ on the dimer structure. However, the crystal packing reveals a side-by-side chain arrangement of very tightly packed dimers (Fig. 4), and in this arrangement an elongated pocket exists that could accommodate the FtsZ C terminus, which most likely forms an alpha α-helix (18, 19). In the SepF polymer, the α-helices on the outside of dimers form the interfaces between the dimers. It seems highly likely that the SepF rings are polymers of SepF dimers. To determine whether the α-helices are indeed involved in multimerization, we identified residues that are important for such interactions. At the interface between adjacent dimers there are two glycine residues (Gly109) contacting each other, one contributed by each monomer (Fig. 4). The lack of side chains on this glycine leads to an unusually close packing of the two longest helices in the four-helix bundle. In fact, Gly109 is highly conserved (Fig. S2B), and the yeast two-hybrid experiments showed that an arginine substitution destroys self-interaction (Fig. 1A). Importantly, the crystal lattices of the A. fulgidus and P. furiosus proteins do not show the polymeric arrangement, and these proteins do not contain this conserved glycine residue. It should be mentioned that these proteins have currently no known function and were picked solely because of their sequence similarity to SepF. To confirm the role of Gly109 in multimerization, we replaced it with a lysine and crystallized the mutant protein. The G109K substitution has a dramatic effect, because in the new crystals the protein filaments are absent, but dimers are still present (Fig. 5A). EM analysis showed that this mutant completely abolishes ring formation. This was not a consequence of structural differences between the wild type and the G109K mutant proteins, because superposition of the two monomers shows close to no variation in the conformation of the C-α backbone between the two structures (Fig. S4B).
Fig. 4.
Polymer structure of SepF revealed by crystal packing. Crystal packing reveals a tight packing of dimers that assemble into polymers via a close interface between their outer helices. FtsZ-interaction mutants from the yeast two-hybrid screen are highlighted, as well as glycine residue G109 (encircled).
Fig. 5.
G109K mutation diminishes polymerization. (A) Crystal lattice of wild-type SepF and crystal lattice of the G109K mutant showing disruption of the interdimer interface that leads to polymerization. (B) Gel-filtration profiles of full-length SepF wild type and the G109K mutant. (C) Analytical ultracentrifugation run at 21,000 rpm. Diagram of mass against concentration indicates that the G109K mutant has a molecular weight close to the theoretical mass of a SepF dimer (34 kDa). Concentrations used are indicated by different colors.
The G109K mutation should not affect dimerization. To test this, gel filtration and analytical ultracentrifugation were performed using full-length SepF with and without the G109K substitution. The gel filtration data indicate that, unlike wild-type SepF that elutes in the void volume, the G109K mutant is considerably smaller in size (Fig. 5B). Using three different protein concentrations, analytical ultracentrifugation assays show that the calculated molecular weight of the G109K mutant ranges between 30 kDa and 40 kDa, which is close to the molecular weight of the dimer (34 kDa, Fig. 5C). These experiments support the conclusion that SepF forms polymers of dimers and that the outside α-helices form the polymerization interfaces.
One aspect that remains uncertain is the precise location of the FtsZ binding site. Differential scanning calorimetry using truncated SepF protein and FtsZ gave no conclusive results, and mixing the truncation with FtsZ did not result in tubular structures other than what is shown in Fig. 2A. Possibly, the close stacking of rings in these tubular structures interferes with FtsZ interaction. Yeast two-hybrid data show that the G109R mutation also abolishes FtsZ binding (Fig. 1A). The same was observed with the G109K mutation, using an affinity chromatography assay (Fig. S4C), suggesting that the interacting α-helices are part of the FtsZ binding site.
A Dali structural similarity search (20) revealed that the C-terminal domain of SepF shows some resemblance to the E. coli protein TusA [YhhP, Protein Data Bank (PDB) ID 1dcj] (21). Because TusA is involved in thiomodification of tRNAs, it is unlikely that this similarity has any functional relevance (22).
An ftsA ezrA Double Mutant Is Viable.
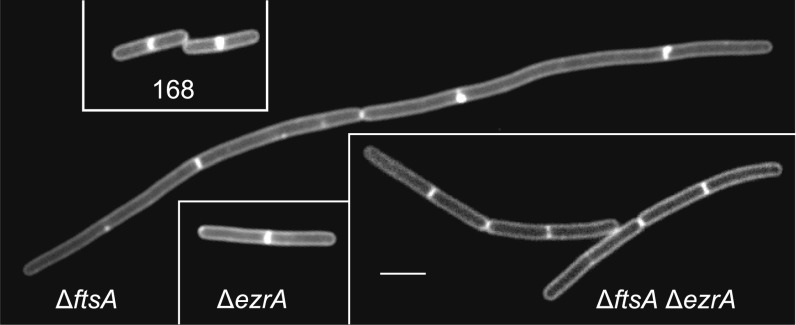
Because the C-terminal domain of SepF is involved in FtsZ interaction and ring formation, the question arises what the function is of the 60-aa-long N-terminal region. A surprising genetic result provided a possible clue. Superficially, the overall architecture of the cell division protein EzrA shows some similarity to that of the E. coli cell division protein ZipA, with an N-terminal transmembrane helix preceding a much larger cytoplasmic FtsZ-binding domain. Therefore, EzrA might be able to anchor FtsZ to the membrane, which would explain why B. subtilis cells are able to grow when the main membrane anchor of the Z ring, ftsA, is deleted. An ezrA-null mutant forms slightly longer cells (∼20%) compared with wild-type cells (23), but an ftsA null mutant has problems dividing and forms long filamentous cells (24). Interestingly, when we transformed an ftsA deletion into a ∆ezrA background, the resulting double mutant grew well and did not show a strong cell division defect (Fig. 6). We concluded that there must be another protein that links FtsZ to the cell membrane. Certain Gram-positive bacteria, such as Mycobacteria, lack FtsA but they do contain a SepF homolog. In addition, deleting both ftsA and sepF is lethal in B. subtilis, and so is the ezrA sepF double knockout (8, 17). These data might suggest that SepF can complement the function of FtsA. However, this is possible only if SepF binds to the cell membrane. Previously, it was shown that a SepF-GFP fusion binds to the Z ring and that the SepF-GFP signal becomes dispersed when FtsZ is depleted (8, 17). In fact, repetition of this experiment, but this time using high-resolution structured illumination microscopy (SIM), showed that the SepF-GFP signal clearly overlaps with the fluorescent signal of the cell membrane when FtsZ is depleted (Fig. S5), suggesting that SepF has indeed affinity for the cell membrane.
Fig. 6.
Viability of ftsA ezrA double mutant. Images show membrane-stained wild-type, ΔftsA, ΔezrA, and ΔftsA ΔezrA double-mutant B. subtilis cells. (Scale bars, 3 µm.)
Purified SepF Binds to Lipid Membranes.
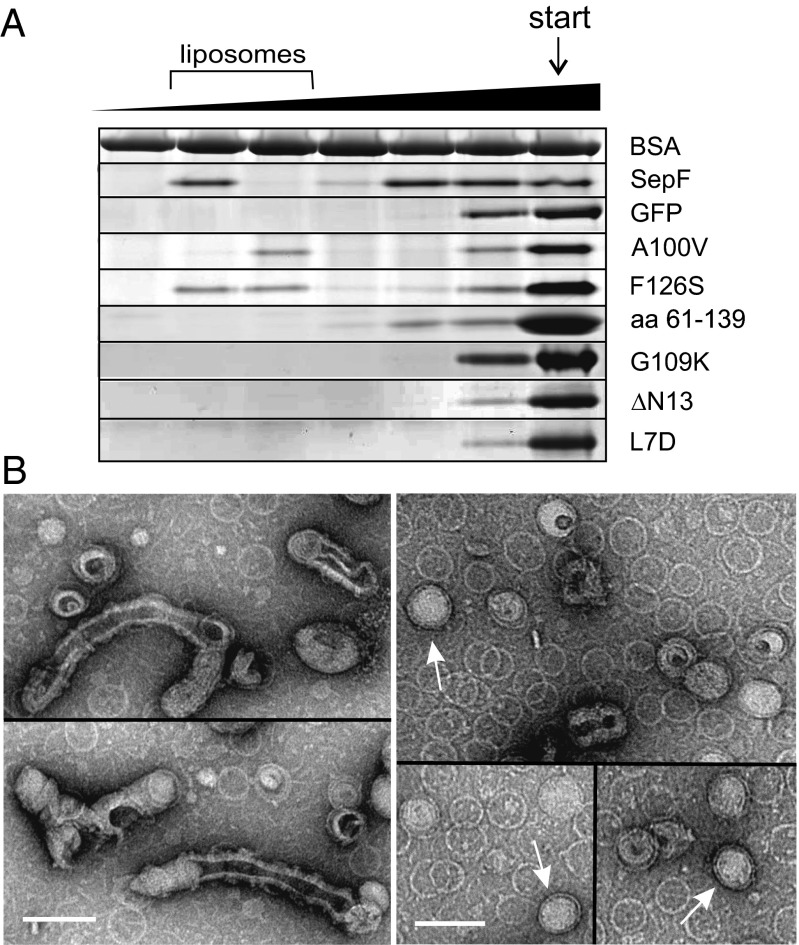
SepF might interact with a membrane protein or SepF is a peripheral membrane protein and has an affinity for lipid bilayers itself. To test this, purified SepF was mixed with liposomes and after centrifugation the amount of SepF in the pellet and supernatant was determined. Excess BSA and 200 mM KCl were included to reduce nonspecific interactions. Approximately four times more SepF is pelleted when liposomes are present, suggesting that SepF has some affinity for lipid membranes (Fig. S6A). However, even without liposomes, a substantial amount of SepF is spun down, presumably through polymerization. Therefore, a more suitable sucrose flotation experiment was performed. SepF and liposomes were loaded at the bottom of a sucrose gradient, and during centrifugation the liposomes migrated to a lower density, upward (lower sucrose concentration). Again BSA and 200 mM KCl were included throughout the gradient to increase specificity. As shown in Fig. 7A, a fraction of SepF clearly comigrates with liposomes, whereas purified GFP does not, lending further support to the idea that SepF can bind directly and specifically to the cell membrane. Another compelling indication was the observation that fluorescently labeled liposomes mixed with purified SepF started to aggregate and often fused into larger vesicles (Fig. S6C).
Fig. 7.
SepF binds to lipid membranes. (A) Flotation assay using sucrose density gradients. SepF with or without liposomes was loaded at the bottom of the gradient (indicated by “start”), and after centrifugation the different gradient fractions were analyzed by SDS/PAGE gel. The sucrose gradient and the liposome-containing fractions are indicated on top of the protein gels. BSA was present throughout the sucrose gradient. The various SepF mutants are described in the main text. (B) EM images of negatively stained SepF rings mixed with small liposomes. (Left) SepF deforms liposomes into extended structures. The SepF rings can be seen in the background. (Right, open arrows) Some SepF rings contain a liposome inside. Liposomes were made from E. coli polar lipid extract. (Scale bar, 100 nm.)
SepF forms remarkably large and uniform protein rings. To examine whether lipid membranes would influence these structures, SepF was mixed with small liposomes, negatively stained, and examined by EM. The presence of SepF resulted in large amorphous clumps of liposomes, as was observed with fluorescence light microscopy, but isolated liposomes were often strongly deformed, giving rise to elongated structures (Fig. 7B). Deformation of liposomes is regularly observed with peripheral membrane proteins, because insertion of the membrane-binding domain displaces lipid molecules that induce local curvature of the lipid bilayer (25). Interestingly, some SepF rings contained a liposome inside (Fig. 7B).
Membrane-Binding Domain.
The SIM images suggested that binding to FtsZ is not required for the association of SepF with the membrane. To corroborate this, we tested the binding of two previously described mutations, A100V and F126S, which impair FtsZ binding but do not block protein ring formation (4). Indeed, both proteins comigrated with the liposome fraction in sucrose density gradients and induced liposome clustering (Fig. 7A and Fig. S6C).
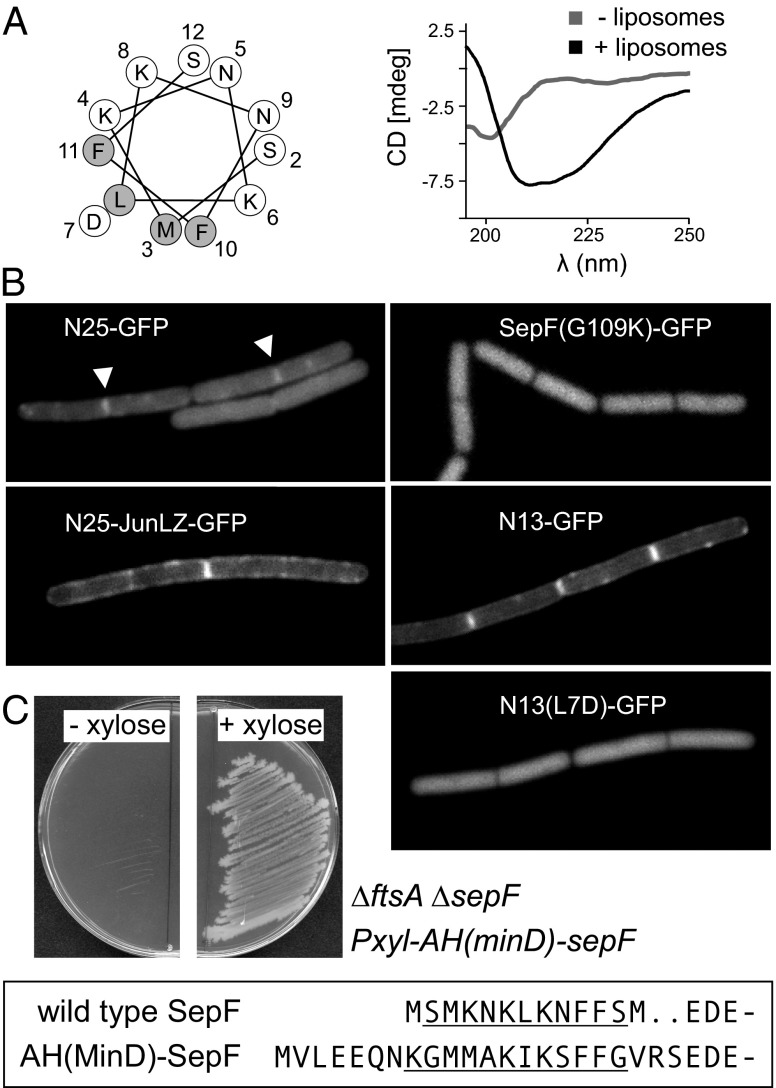
To determine whether the membrane-binding domain is located in the main ring-forming part of SepF, we examined whether the SepF truncation used for crystallography showed any affinity for liposomes. As shown in Fig. 7A, the C-terminal domain (amino acids 61–139) does not comigrate with liposomes in sucrose gradients, and is not able to cluster liposomes (Fig. S6C). This implies that the membrane-binding domain resides in the 60-aa-long N-terminal region of SepF. In fact, prediction using AmphipaSeek (26) suggested that amino acids 2–12 form a putative amphipathic helix (Fig. 8A), and this region is highly conserved in the N-terminal domain (Fig. S2A). Such helical structures are known to insert into lipid membranes and are used by many peripheral membrane proteins as membrane-binding domains, including the cell division proteins FtsA, MinD, and MreB (11, 27–29). To examine whether the first 13 aa of SepF form an amphipathic helix upon interaction with lipid bilayers, this small peptide was synthesized and CD spectra with and without liposomes were determined. As shown in Fig. 8A, SepF1–13 is structurally unfolded in solution but exhibits a clear shift to an α-helical fold in the presence of liposomes, a transition that is characteristic of membrane-binding amphipathic helices (30, 31).
Fig. 8.
SepF N terminus contains a membrane-binding domain. (A) (Left) Helical wheel representation of the N-terminal amphipathic helix (AH). Amino acid positions, L7D mutation, and hydrophobic (shaded) amino acids are indicated. (Right) Far-UV circular dichroism spectra of SepF1-13 in the absence (shaded line) and presence (solid line) of liposomes. Liposomes were made from synthetic phospholipids (natural mixture of PG, PE, and CL). (B) In vivo membrane binding of different fragments of the SepF N terminus and the nonpolymerizing SepFG109K mutant. See main text for description of the mutants. (C) Functional replacement of SepF amphipathic helix with an amphipathic helix of MinD. The amino acid sequence of the N terminus of wild-type SepF and the SepF mutant with the MinD membrane-binding amphipathic helix [AH(MinD)-SepF] are boxed and the amphipathic helices are underlined. Strain HS242 [∆ftsA ∆sepF Pxyl-AH(mind)-sepF] grows only in the present of xylose, indicating that the MinD amphipathic helix can replace the SepF amphipathic helix.
To test whether this amphipathic helix is able to recruit proteins to the membrane in vivo, we constructed a C-terminal GFP fusion with the first 25 aa of SepF (N25-GFP) and analyzed the localization of the fusion protein in B. subtilis cells. As shown in Fig. 8B, the fluorescence signal is primarily diffuse although in some cells a weak membrane signal can be observed at division sites (arrowheads). Because SepF forms large polymers, it is likely that a weak membrane affinity of the monomers is compensated by multimerization. In fact, the G109K mutation that blocks polymerization also abolishes comigration with liposomes in sucrose gradients and clustering of liposomes (Fig. 7A and Fig. S6C). Indeed, introduction of this mutation into a SepF-GFP reporter fusion [SepF(G109K)-GFP] resulted in a diffuse fluorescent signal (Fig. 8B). Previously, Szeto et al. (32) have shown that the 12-aa membrane-binding amphipathic helix of E. coli MinD could target GFP to the cell membrane only when they fused this domain to the c-Jun leucine zipper. When the c-Jun leucine zipper dimerization domain was introduced between the first 25 aa of SepF and GFP (N25-JunLZ-GFP), membrane binding became very clear (Fig. 8B). Interestingly, when only the first 13 aa of SepF were fused to GFP (N13-GFP), the fusion product also localized to the cell membrane, even without the presence of a dimerization domain (Fig. 8B). Mutating the hydrophobic leucine residue of the amphipathic helix to an aspartate [N13(L7D)-GFP] abolished membrane binding (Fig. 8B). These data strongly suggest that the amphipathic helix at amino acid positions 1–12 constitutes the membrane targeting domain of SepF. To further corroborate this conclusion, a SepF variant was purified, lacking the first 13 aa (∆N13). As expected, this mutant showed no lipid interaction (Fig. 7A). When the L7D mutation was introduced into full-length SepF, lipid binding was also abolished (Fig. 7A).
To support these findings, the importance of the amphipathic helix for the function of SepF in cell division was tested. It has been shown that SepF expressed in trans is able to support growth in the absence of both ftsA and wild-type sepF (17). However, we were unable to delete both sepF and ftsA when a SepF mutant was expressed that lacked the first 13 aa, indicating that the membrane binding of SepF is essential to support cell division. This result was not a consequence of an increased instability of the mutant protein (Fig. S6D). Interestingly, the replacement of the native amphipathic helix with the membrane-binding amphipathic helix of B. subtilis MinD restored growth in a strain lacking ftsA and wild-type sepF (Fig. 8C), demonstrating the key membrane-binding function of the N terminus of SepF. It should be mentioned that the in vitro binding experiments were performed using E. coli polar lipid extract. However, SepF also binds to liposomes formed from B. subtilis lipid extract (Fig. S6E).
SepF Recruits FtsZ to Lipid Membranes.
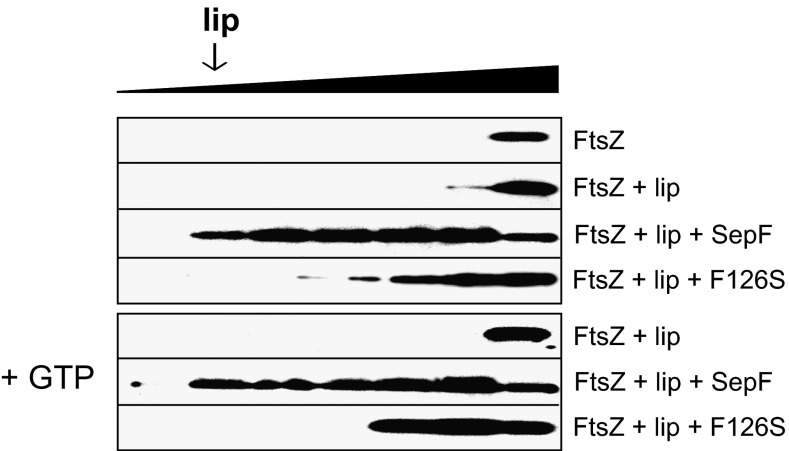
If SepF functions as a membrane anchor for the Z ring, then the protein should be able to recruit FtsZ to lipid membranes. To test this, FtsZ was included in the sucrose gradient centrifugation assays. As shown in Fig. 9, FtsZ remains in the bottom fraction of the gradient when mixed with liposomes. However, when SepF is included, FtsZ migrates with the liposomes. As a control, the experiment was repeated with mutant F126S that shows a reduced affinity for FtsZ but binds normally to liposomes. Indeed, this SepF variant is unable to recruit FtsZ to the liposome-containing gradient fraction (Fig. 9). FtsZ polymerizes into long protofilaments in the presence of GTP. However, previous work has shown that the binding of FtsZ to SepF does not depend on GTP (4, 6), and when the flotation experiments were performed in the presence of GTP, similar results were obtained (Fig. 9).
Fig. 9.
SepF recruits FtsZ to lipid membranes. Recruitment of FtsZ to liposomes was analyzed using sucrose gradient centrifugation. FtsZ was detected using Western blotting. The F126S mutation reduces SepF-FtsZ binding (4). The presence of GTP (+GTP) has no effect on lipid interaction. Liposomes were made from E. coli polar lipid extract.
It has been shown that SepF rings assemble FtsZ protofilaments into very long 50-nm-wide tubular structures (4). In fact, these tubular structures were easily observed on EM grids when liposomes were added to a mixture of purified SepF and FtsZ, (Fig. S7). The main difference is that there are many more FtsZ-SepF tubules when liposomes are present. Charged surfaces, including lipid bilayers, tend to stimulate polymerization of FtsZ (33, 34), and it is likely that this effect facilitates the assembly of FtsZ-SepF tubules.
Discussion
SepF Is a Unique FtsZ Membrane Anchor.
Until now, FtsA has been considered the key membrane anchor of the Z ring. However, not all bacterial species contain an FtsA homolog. We demonstrate here that SepF displays three of the known FtsA activities: polymerization, FtsZ binding, and membrane binding (19). This could explain why bacterial species that do not contain FtsA, such as the Mycobacteria and Streptomycetes, contain SepF homologs. It should be stressed that Mycobacteria and Streptomycetes species also lack EzrA homologs.
Alternative Model for SepF.
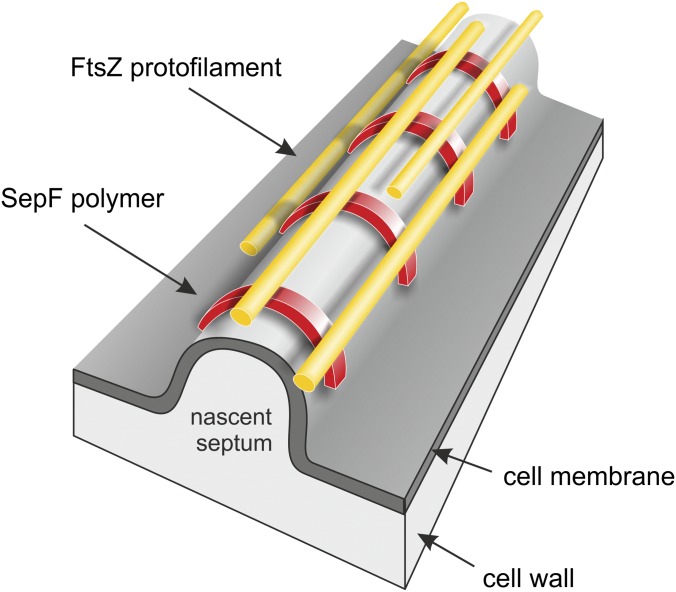
Purified SepF rings will align FtsZ polymers into long and regular tubular structures. This bundling of FtsZ polymers seems to be necessary to synthesize a regular and smooth division septum, because sepF mutants of B. subtilis and Streptococcus pneumonia contain strongly deformed septa (7, 8). However, how are the SepF rings positioned in the cell when they also bind to the cell membrane? We postulate that SepF does not make rings but instead forms arcs that lay perpendicular over the leading edge of the division septa, on top of which the FtsZ protofilaments assemble (Fig. 10). This model is based on the following observations. First, EM analyses showed SepF rings that contained liposomes inside the rings, and we never observed liposomes attached to the outside of rings, strongly suggesting that the membrane-binding domain is located on the inside of SepF rings. In this way, membrane binding and FtsZ binding are possible at the same time, because previous EM observations have suggested that the FtsZ polymers are attached to the outside of SepF rings (4). Second, the strong and stable curvature of SepF polymers makes it hard to envision how these structures can anchor to a flat lipid membrane. However, the cell membrane is strongly curved at the leading edge of the growing septum, and it might be that SepF polymerizes as semicircles over this convex (positively) curved membrane area. In fact, there is an intriguing correlation between the size of SepF rings and the width of division septa. The mean inner diameter of SepF rings varies roughly around 40 nm (Fig. S8). The average thickness of septa measures ∼33 nm, according to transmission EM images of dividing B. subtilis cells (Fig. S8). If we assume that the cell membrane thickness approximates to ∼5 nm, then the average width of septa is more in the range of 43 nm, which comes close to the inner diameter of SepF rings. This would mean that arcs of SepF polymers would fit neatly on top of the leading edge of the developing septum (Fig. 10). This model also better accommodates the fact that FtsZ protofilaments become curved upon GTP hydrolyses. In previous work it was shown that this bending of protofilaments separates the ends of FtsZ-SepF tubules, resulting in splayed tips containing highly curved FtsZ filaments (4).
Fig. 10.
Schematic representation of SepF binding model. SepF polymers (red) bind as arcs onto the convex leading edge of the nascent division septum and bundle FtsZ protofilaments (yellow). See main text for explanation of the model.
Localization by Curved Membrane Area.
Finally, both SepF and FtsA use an amphipathic helix to bind to the cell membrane. These protein structures bind stronger to convex (positively curved) membrane surfaces because curvature creates space in between lipid molecules (25). By far the most prominent convex membrane area in a bacterial cell appears at the leading edge of a developing division septum (Fig. 10). Thus, it seems likely that both FtsA and SepF have an intrinsic binding preference for this area in the cell. This characteristic might help to keep the Z ring positioned at the edge of the growing septum during cell division.
Materials and Methods
Strains and General Methods.
Yeast, E. coli, and B. subtilis strains and primers used in this study are listed in Tables S2 and S3, respectively. Molecular cloning, PCR reactions, yeast transformation and mating experiments, and E. coli transformations were carried out using standard techniques. B. subtilis chromosomal DNA for transformation and PCR reactions was purified as described before (35). Transformation of competent B. subtilis cells was performed using an optimized two-step starvation procedure based on the method of Anagnostopoulos and Spizizen (36, 37). Selection of transformants was carried out on nutrient agar supplemented with 100 μg/mL ampicillin for E. coli and, for B. subtilis, 4 μg/mL erythromycin, 50 μg/mL spectinomycin, 5 μg/mL tetracycline, or 0.5 µg/mL phleomycin.
Construction of Plasmids for Protein Purification.
pMal-C2X (New England Biolabs) was used to clone and purify variants of SepF used in this study as N-terminal MBP-SepF fusions. Using B. subtilis 168 genomic DNA as template, the ΔN13 SepF truncation (oligos inc22/inc6) was amplified, digested with SmaI/XbaI, and ligated into pMal-C2 linearized with XmnI/XbaI, resulting in plasmid pHJS106 (MBP-SepF∆N13). The L7D and G109K exchanges were introduced into pNC12 with quick-change mutagenesis, using oligos HS492/HS493 (L7D) and oligos inc40/inc41, resulting in plasmids pHJS107 and pNC13.
Construction of Plasmids and Strains for Complementation Assays.
For the construction of the B. subtilis ftsA mutant containing different SepF mutants, sepF genes were amplified from the pGBT9-sepF derivatives with oligos sepF-mutIn-f2 and pGBT9-mutIn-r. The upstream region, containing the 5′ end of the sepF coding sequence, and the downstream region of sepF were amplified from B. subtilis 168 genomic DNA, using the oligo sets sepF-mutF-f/sepF-mutF-r2 and ylmG-rPCR-f/sepF-mutB-r, respectively. The chloramphenicol-resistant cassette was amplified from pDLT3 (38) with oligos rPCR-CmF2 and rPCR-CmR(-ter). The four DNA fragments were then fused together by recombinant PCR, using the oligos sepF-mutF-f and sepF-mutB-r. The sepF mutations were introduced into B. subtilis MD194, which contains a complete deletion of the sepF gene, by transformation with the PCR products. To test functionalities of the SepF mutants, the sepF mutations were then introduced into a B. subtilis MD197 mutant strain, which contains a complete sepF deletion, and a xylose-inducible ftsA (amyE::Pxyl-ftsA, ΔftsA::erm, ΔsepF::spec), by transformation with the genomic DNA extracted from the resultant sepF mutant strains.
The strains encoding SepF without amphipathic helix (∆AH, strain HS230) and the native AH replaced with AH of B. subtilis MinD (strain HS232) were constructed by PCR amplification of Pxyl promoter (oligos HS495/HS497 for ∆AH, oligos HS495/HS498 for AHMinD) and SepF fragments (oligos HS500/HS501 for ∆AHSepF, oligos HS500/HS503 for AHMinD-SepF) followed by insertion into aprE spec delivery vector, using the Stargate cloning system (IBA) and direct transformation into B. subtilis 168. The correct integration in the aprE locus and the sequence of the constructs were verified by PCR amplification followed by sequencing.
Construction of Plasmids and Strains for Microscopy.
The strains encoding N-terminal fragments of SepF fused to GFP were constructed by amplification of sepF1-39 (N13) and sepF1-75 (N25), using oligos HS478/HS480 and HS478/HS481, respectively. The amplified products were fused with the plasmid pSG1154 linearized with oligos HS09/HS479, using the InFusion HD cloning kit (Clontech), resulting in plasmids pHJS108 (N13-GFP) and pHJS109 (N15-GFP). The homologous sequences required for InFusion cloning were designed so that a flexible SGSGSGS linker is positioned between the SepF fragments and GFP. This linker also contains a unique BamHI restriction site, which was used to insert junLZ amplified with oligos HS484/HS485 from plasmid pTS37 into pHJS109, resulting in plasmid pHJS110. The plasmid pHJS111 encoding N13-GFP with L7D exchange was constructed with quick-change mutagenesis of plasmid pHJS108, using primers HS492/HS493. The plasmid pNC14 encoding SepFG109K-GFP was constructed with quick-change mutagenesis of plasmid pFG1 (8), using primers inc40/inc41. The plasmids were transformed into B. subtilis 168, resulting in the corresponding strains HS206, HS207, HS208, HS223, and NC21.
Yeast Two-Hybrid Assay.
For library screening, Saccharomyces cerevisiae AH109 was used as a host, and S. cerevisiae PJ69-4A and PJ69-4α were used for mating experiments. To construct a sepF mutant library in pGBT9 (Clontech), the sepF gene was amplified from pENTR-sepF (17) with oligos pDONR-F and pDONR-R to attach attL1 and attL2 sequences at the 5′ and 3′ ends, respectively, necessary for cloning with the Gateway cloning technique (Life Technologies, previously named Clontech). Random mutations were introduced by using Ex Taq DNA polymerase (Takara) (17, 39). The PCR product was then recombined with pBDGW, using the LR Gateway cloning reaction, and the reaction mixture was transformed to E. coli. Plasmids from ∼1.2 × 105 transformants were collected. S. cerevisiae AH109 harboring pGAD424-ftsZ was transformed with the plasmid library, and transformants were selected on synthetic defined medium without tryptophan and leucine in the presence of X-α-gal, to monitor interactions between FtsZ and SepF. The pGBT9 derivatives containing mutant sepF genes were selected using the E. coli tryptophan auxotroph KC8 strain and the mutations were identified by sequencing.
To check the N-terminal deletions of SepF in the yeast-two hybrid assay, sepF encoding ΔN(39–151), ΔN(63–151), and ΔN(130–151) regions were amplified with the oligo sets sepF-ΔN1-gwF/sepFgwR, sepF-ΔN2-gwF/sepFgwR, and sepF-ΔN3-gwF/sepFgwR, respectively, and then recombined with pDONR201 by the BP Gateway reaction. The resultant plasmids were then recombined with pGBT9 or pGAD424, using the LR Gateway reaction.
Interactions of the SepF mutants with wild-type SepF and self-interactions were tested by mating experiments, using the following plasmids. The mutated sepF genes from selected pGBT9 derivatives (pGBT9-sepFm01 to -sepFm15) were amplified with oligos Adapter-attB1 and Adapter-attB2, and the products were integrated into pDONR201 (Life Technologies), using the BP Gateway cloning technology. The inserts were then transferred into pGAD424 by LR Gateway reaction.
Cloning and Protein Purification for Crystallography.
The genes encoding B. subtilis SepF (GI: 238054369) and residues 37–122 of A. fulgidus SepF-like protein (GI: 11498388) and residues 49–134 of P. furiosus SepF-like protein (GI: 33359575) were amplified from genomic DNA by PCR, using forward oligos containing an NdeI restriction site and reverse oligos carrying a BamHI restriction site. The PCR product was digested with NdeI and BamHI before ligation with plasmid pHis17. A single point mutation, Ile-105 to Met, was introduced in the sequence of the A. fulgidus protein, using a QuikChange Mutagenesis Kit (Stratagene) for phasing.
All proteins were expressed in E. coli strain C41. Following introduction of plasmids into cells by electroporation, cells were first grown overnight, at 37 °C, on three nutrient agar plates, containing 100 µg/mL ampicillin. All cells from the agar plates were then transferred to 12 L of 2× tryptone-yeast extract medium containing 100 µg/mL ampicillin and grown at 37 °C. For B. subtilis SepF and SepF-G109K, protein expression was induced at OD600 by the addition of 1 mM IPTG, at 25 °C for 6 h. In the case of the A. fulgidus and P. furiosus proteins, synthesis was induced with 1 mM IPTG, at 37 °C for 3–4 h. For purification, cells expressing B. subtilis SepF or SepF-G109K were resuspended in 25 mM Tri⋅HCl, pH 8.5, 1 mM EDTA, and 1 mM NaN3 and subsequently lysed using a cell disruptor (Constant Systems). The cell lysate was clarified by ultracentrifugation at 25.171 × g, which was followed by precipitation of the protein from the lysate supernatant by gradual addition of ammonium sulfate, to a final concentration of 25% (saturation), while incubating at 4 °C. The precipitated protein was pelleted at 17.590 × g; and the pellet was resuspended in 25 mM Tris⋅HCl, pH 8.5, 1 mM EDTA, 1 mM NaN3, and 100 mM NaCl; concentrated; and loaded onto a HiPrep Sephacryl S200 (GE Healthcare) gel filtration column. To obtain the truncated forms of B. subtilis SepF or SepF-G109K, before the gel filtration step, the protein concentrations were measured using a NanoDrop ND-1000 Spectrophotometer (Thermo), and the sample was incubated with α-chymotrypsin in a 1:1,000 concentration ratio between protease and SepF protein. Limited proteolysis, at 37 °C for 1 h, was immediately followed by gel filtration on a Sephadex S200 column. Both SepF and SepF-G109K were concentrated to 10 mg/mL for crystallization trials.
For expression of selenomethionine-derivatized A. fulgidus and P. furiosus proteins, chemically competent E. coli B834 (DE3) cells (Novagen) were transformed with the relevant plasmids; grown overnight at 37 °C on agar plates containing 100 µg/mL ampicillin; and then transferred to sterile M9 minimal media containing 2 mM magnesium sulfate, 0.4% (wt/vol) glucose, 25 μg/mL ferrous sulfate heptahydrate, 1 μg/mL each of a mixture of vitamins (riboflavin, niacinamide, pyridoxine monohydrochloride, and thiamine), 40 μg/mL amino acid Complete Supplement Mixture (CSM, −Met), 40 μg/mL of seleno-l-methionine (Fisher 259960025), and 100 µg/mL ampicillin. Protein synthesis was induced with 1 mM IPTG, at 22 °C for 10 h. Pelleted cells were resuspended in 50 mM Hepes (pH 7.5) and 5 mM DTT and lysed using a cell disruptor, and lysate was clarified by ultracentrifugation at 25.171 × g. The lysate supernatant was loaded onto a 10-mL cation-exchange column (Pharmacia), and protein was eluted with a linear gradient of 0–1 M NaCl. Relevant fractions were pooled, concentrated, and loaded onto a Sephacryl S100 gel filtration column preequilibrated in 25 mM Tris⋅HCl, pH 7.5, 1 mM EDTA, 1 mM NaN3, and 5 mM DTT. For crystallization trials, P. furiosus protein was concentrated to 15 mg/mL and A. fulgidus protein to 10 mg/mL.
Protein Crystallization.
Crystallization experiments were carried out using the sitting-drop vapor diffusion technique, with incubation at 19 °C. P. furiosus protein crystals grew in 0.2 M ammonium sulfate, 0.1 M Hepes, pH 7.5, and 25% (wt/vol PEG 3350) and were cryo-protected in mother liquor containing 20% (vol/vol) ethylene glycol. A. fulgidus protein crystals appeared in 0.2 M lithium sulfate, 0.1 M sodium acetate, pH 4.5, and 30% (wt/vol PEG 8000) and were cryo-protected with 25% (vol/vol) glycerol in mother liquor. Truncated B. subtilis SepF was crystallized in 30% (wt/vol) PEG 8000 and 0.2 M ammonium sulfate and for data collection crystals were cryo-protected with 20% (vol/vol) glycerol in mother liquor. Crystals of truncated B. subtilis SepF-G109K grew in 15% (vol/vol) ethanol and 0.1 M Tris, pH 7.0, and were cryo-protected with 20% (vol/vol) ethylene glycol in mother liquor.
Structure Determination.
The crystal structures of the A. fulgidus and P. furiosus proteins (P6522 crystal form) were solved by multiple-wavelength anomalous diffraction, using selenomethionine-derivatized crystals. Selenium sites were located and phases obtained with SOLVE/RESOLVE (40). Clear electron density maps revealed the presence of three dimers in the asymmetric unit of the A. fulgidus crystal and two monomers packed as a dimer in that of P. furiosus. The structures of B. subtilis wild-type SepF and the P2 crystal form of the P. furiosus protein were solved by molecular replacement, with Phaser 2.1 (41), using the P6522 form of the P. furiosus protein as template. The search revealed one dimer in the asymmetric unit of the B. subtilis SepF crystal and six monomers packed as three dimers for the P. furiosus protein (P2). B. subtilis SepF-G109K was also solved by molecular replacement, using the truncated B. subtilis wild-type SepF monomer as template, which revealed one monomer in the asymmetric unit. Model building was done with Coot (42), and refinement was carried out using Refmac (43) and Phenix (44).
Analytical Ultracentrifugation and Gel Filtration Assays.
Gel filtration assays were carried out by loading 1.5 mg of protein onto a Superdex S-200 gel filtration column (GE Healthcare), preequilibrated in buffer containing 25 mM Tris⋅HCl, pH 7.5, 1 mM EDTA, 1 mM NaN3, and 50 mM NaCl, and eluting at a flow rate of 0.5 mL/min.
Sedimentation equilibrium experiments were carried out in a Beckman Optima XL-I analytical ultracentrifuge with an An-60 Ti rotor, using interference and absorbance at 280 nm. Samples were spun at 20 °C, 21,000 rpm, in two-sector 12-mm path-length cells, until they reached equilibrium, as judged by the lack of changes in subsequent scans. The three different concentrations used were 1 mg/mL, 2 mg/mL, and 4 mg/mL. The data were analyzed using Ultraspin (http://www.mrc-lmb.cam.ac.uk/dbv/ultraspin2/ultraspin1/). Partial-specific volumes were calculated from the amino acid compositions of the proteins, using SEDNTERP, which was also used for estimation of the density and viscosity of buffers (45).
Electron Microscopy.
Negative staining electron microscopy images of truncated SepF were collected on a Philips 208 TEM operating at 80 kV and at a magnification of 180,000×. Protein was diluted to a concentration of 0.05 mg/mL in 50 mM Tris⋅HCl, pH 7.0, and was applied to carbon-coated grids that had been made hydrophilic by glow discharging for 10 s. Grids were stained with 2% uranyl acetate. To image SepF with liposomes, the preformed liposomes were diluted to 5 mg/mL in 10 mM Tris⋅HCl, pH 7.4, 50 mM KCl, and 2 mM MgCl2 and sonicated for 15 min in a bath sonicator. SepF (0.3 mg/mL) was incubated with the sonicated liposome suspension (25 μL final volume) for 10 min at room temperature. In the case that FtsZ was included (0.1 mg/mL) the buffer also contained 4 mM GTP. After incubation, 20 µL of sample was applied to glow-discharged 200-mesh carbon-coated grids, which were negatively stained with 100 µL uranyl-acetate (2% wt/vol). The grids were imaged using a Phillips CM100 electron microscope.
For the measurement of septum widths, B. subtilis wild-type (strain 168) cells were grown in LB at 37 °C until the midexponential phase and pelleted. Fixation of cells was done in 2% glutaraldehyde, and then cells were washed with sodium cacodylate buffer, followed by secondary fixation in 1% osmium tetroxide. Fixed cells were washed with distilled water. After fixation, cells were dehydrated with gradual addition of acetone and embedded in resin at 60 °C for 24 h. Resin blocks were sectioned to 70-nm samples with a Leica EM UC7, using a diamond knife. Ultrathin sections were collected on copper mesh grids and stained with uranyl acetate and lead citrate, using an automatic stainer (Leica EM AC20). Grids were examined using a Philips CM100 Compustage (FEI) transmission electron microscope.
FtsZ Interaction Assay.
The interaction of SepF with FtsZ was analyzed by a coelution experiment as described before (4). In brief, MBP-SepF and MBP-SepFG109K were expressed in E. coli, and cell extracts were mixed with amylose resin and washed with 50 mM Tris⋅HCl, pH 7.4, 200 mM KCl, 1 mM EDTA, and 0.5 mM DTT. Resin was mixed with purified B. subtilis FtsZ at 4 °C for 2 h, followed by washing and elution with 10 mM Maltose and 1% SDS. Elution fractions were analyzed by SDS/PAGE and Coomassie staining.
Fluorescence Microscopy.
Overnight cultures were diluted 1:50 in fresh LB and grown to exponential phase at 30 °C. The cell membrane was stained with FM-95 (1 μg/mL). SepF-GFP was induced with 0.5% xylose. N-terminal fragments of SepF fused to GFP were induced on nutrient agar plates supplemented with 1% xylose. Cells were immobilized on microscope slides covered with 1.2% agarose in water. Epifluorescence images were acquired with Nikon Ti-E and Zeiss 200M fluorescence microscopes, using Metamorph 6 (Molecular Devices). Two-dimensional SIM images were obtained with a Nikon N-SIM microscope and NIS-Elements 4.1 (Nikon).
Protein Purification for Lipid Interactions.
SepF mutants were purified as Maltose binding protein (MBP) fusions on amylose resin columns, as described before (4). For purification, overnight cultures of E. coli BL21 (DE3) with the relevant pMal-SepF plasmids were diluted 1:50 into fresh LB containing 100 µg/mL ampicillin. Cells were grown until an OD600 of 0.5 was reached, and protein expression was induced with 0.5 mM IPTG for 3 h. Cells were resuspended in 20 mM Tris⋅HCl, pH 7.5, 200 mM KCl, and 5 mM EDTA with complete EDTA-free protease inhibitor tablets (Roche), followed by lysis with a French press using 20 kpsi pressure. MBP-SepF was purified using an amylose column and eluted with 20 mM Tris⋅HCl, pH 7.5, and 10 mM Maltose. MBP-SepF fusion was cleaved with Factor Xa (1 µg enzyme for 100 µg MBP-fusion protein), and SepF was separated from MBP with a HiTrap Q HP column (GE Healthcare), followed by concentration with Millipore Amicon Ultra protein concentration tubes (3-kDa cutoff). Small aliquots were frozen in liquid N2 and stored at −80 °C. B. subtilis FtsZ was purified as described previously (46, 47).
Lipid Interaction Assays.
Lipid binding experiments were carried out with large unilamellar vesicles prepared from E. coli polar lipid extract or synthetic phospholipids (Avanti), essentially as described in ref. 48. Dry lipid extract was solubilized in chloroform followed by evaporation of chloroform under an argon stream. Dry lipids were resolubilized in 50 mM Tris⋅HCl, pH 7.5, 2 mM β-mercaptoethanol, and 1.5% octylglycoside (20 mg/mL lipids) under an argon stream and dialyzed against 50 mM Tris⋅HCl, pH 7.5, and 2 mM β-mercaptoethanol. The preformed liposomes were aliquoted and stored at −80 °C.
B. subtilis lipids were extracted using the Bligh/Dyer extraction method (49). In brief, 1 L B. subtilis midlog culture grown in LB at 37 °C was harvested, washed with 170 mM NaCl, and resuspended in 10 mL ice-cold PBS. Cells were broken with a French press and cell debris was removed by centrifugation. Organic extraction (chloroform, methanol, water 1:2:0.8) was carried out for 2 h at room temperature under argon, followed by addition of chloroform and water (1:1:0.9) to induce phase separation. Chloroform phase was collected after centrifugation and evaporated under a dry argon stream. Lipids were solubilized by addition of 50 mM Tris⋅HCl, pH 7.5, 2 mM β-mercaptoethanol, and 1.5% octylglycoside. Insoluble material was removed by filtering. Liposomes were formed as described for E. coli lipid extracts.
For pelleting experiments, the preformed liposomes (20 mg/mL, polar lipid extract) were extruded through a 0.4-µm pore size membrane (Avanti Polar Lipids) in SepF-binding buffer (20 mM Tris⋅HCl, pH 8.0, 2 mM MgCl2, 200 mM KCl, 0.5 mM DTT, 1 mg/mL BSA). Purified SepF (0.25–0.5 mg/mL) and liposomes (5 mg/mL) were mixed in a total volume of 120 µL and incubated for 10 min at room temperature. The mixture was centrifuged in 0.8-mL ultraclear centrifuge tubes at 40,000 rpm for 10 min at 20 °C, in a Beckman TLA100 Rotor. Pellets and supernatants were analyzed by SDS/PAGE and stained with Coomassie Brilliant Blue G.
For sucrose gradient density (flotation) experiments, preformed liposomes (1 mg/mL, polar lipid extract) were diluted in SepF-binding buffer containing a final concentration of 45% sucrose. After extrusion through a 0.1-µm pore membrane, the liposomes (80 µL) were mixed with SepF (0.25–0.5 mg/mL), resulting in a final volume of 120 µL and 30% sucrose. BSA (1 mg/mL) was included and the mixture was incubated at room temperature for 10 min. A sucrose gradient was prepared using SepF-binding buffer containing 1 mg/mL BSA and 10% (100 µL), 15% (100 µL), 20% (200 µL), or 25% (200 µL) sucrose, loaded on top of the SepF–liposome mixture. Centrifugation was carried out for 2 h (25,000 rpm, 25 °C) in a Beckman Rotor MLS50, using 0.8-mL tubes (5 × 41 mm) and suitable adapters, followed by collection of 100-µL samples from top to bottom. Samples were analyzed by SDS/PAGE and Coomassie Brilliant Blue G staining. Interaction of FtsZ (3 µg/mL) with SepF (0.25–0.5 mg/mL) and liposomes (1 mg/mL, 0.1 µm diameter) was tested with and without GTP (2 mM) in 10 mM Tris⋅HCl, pH 7.4, 50 mM KCl, 2 mM MgCl2, and 1 mg/mL BSA. The reaction was performed at room temperature for 10 min. Gradient samples were analyzed by Western blotting, using FtsZ primary antibody and goat anti-rabbit HRP (Sigma).
For the microscopic liposome binding assay, liposomes (5 mg/mL, polar lipid extract) were sonicated for 15 min in a bath sonicator, followed by incubation with SepF (0.25–0.5 mg/mL) in binding buffer for 10 min at room temperature. Liposomes were stained with 0.1 mg/mL Bodipy FL C16 (Invitrogen). A Nikon Ti equipped with a spinning-disk confocal module and a 488-nm solid-state laser light source was used to visualize the fluorescently labeled liposomes. Images were acquired with Metamorph 6 and analyzed with ImageJ 1.46 (National Institutes of Health).
CD spectroscopy was carried out with a Jasco J-810 spectropolarimeter at 25 °C, using a synthetic peptide corresponding to SepF1-13 (Biomatik), and liposomes (phosphatidylglycerol, phosphatidylethanolamine, cardiolipin = 60:38:2 mol%) formed from synthetic pure phospholipids as described above (Avanti). Before the experiment, the preformed liposomes were sized to 30 nm through extrusion and mixed with the synthetic peptide, resulting in final concentrations of 2.5 mg/mL lipids and 250 µg/mL peptide (21:1 molar ratio) in 6 mM Tris⋅HCl, pH 7.5. Spectra are smoothed background subtracted averages of nine scans acquired using a 20-nm/min scan rate, a 4-s response time, and a 0.2-mm path length cuvette (Helma).
Supplementary Material
Acknowledgments
We thank the members of the Centre for Bacterial Cell Biology for helpful discussions and experimental support and the Newcastle University Microscopy Research Centre for electron microscopy support. We acknowledge C. Johnson (Medical Research Council Laboratory of Molecular Biology) for help with analytical ultra-centrifugation and beamline support at ID23eh1, ID29 (European Synchrotron Radiation Facility, France), and I03 (Diamond Light Source, United Kingdom) and M. Yoshimura (Nara Institute of Science and Technology, Graduate School of Information Science Functional Genomics, Ikoma, Japan) for gifts of B. subtilis strains. This research was supported by a Newcastle University Overseas Research Studentship Award (to L.W.H.), Biotechnology, Netherlands Technology Foundation Vici Grant 12128 (to L.W.H.), Biological Sciences Research Council Grant BB/I01327X/1 (to L.W.H.), Medical Research Council Grant U105184326 (to J.L.), Wellcome Trust Grant 095514/Z/11/Z (to J.L.), and a Grant-in-Aid for Scientific Research (C) (to S.I.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3ZIE).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1313978110/-/DCSupplemental.
References
- 1.de Boer PA. Advances in understanding E. coli cell fission. Curr Opin Microbiol. 2010;13(6):730–737. doi: 10.1016/j.mib.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gueiros-Filho FJ, Losick R. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 2002;16(19):2544–2556. doi: 10.1101/gad.1014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Small E, et al. FtsZ polymer-bundling by the Escherichia coli ZapA orthologue, YgfE, involves a conformational change in bound GTP. J Mol Biol. 2007;369(1):210–221. doi: 10.1016/j.jmb.2007.03.025. [DOI] [PubMed] [Google Scholar]
- 4.Gündoğdu ME, et al. Large ring polymers align FtsZ polymers for normal septum formation. EMBO J. 2011;30(3):617–626. doi: 10.1038/emboj.2010.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marbouty M, Saguez C, Cassier-Chauvat C, Chauvat F. Characterization of the FtsZ-interacting septal proteins SepF and Ftn6 in the spherical-celled cyanobacterium Synechocystis strain PCC 6803. J Bacteriol. 2009;191(19):6178–6185. doi: 10.1128/JB.00723-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh JK, Makde RD, Kumar V, Panda D. SepF increases the assembly and bundling of FtsZ polymers and stabilizes FtsZ protofilaments by binding along its length. J Biol Chem. 2008;283(45):31116–31124. doi: 10.1074/jbc.M805910200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fadda D, et al. Characterization of divIVA and other genes located in the chromosomal region downstream of the dcw cluster in Streptococcus pneumoniae. J Bacteriol. 2003;185(20):6209–6214. doi: 10.1128/JB.185.20.6209-6214.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamoen LW, Meile JC, de Jong W, Noirot P, Errington J. SepF, a novel FtsZ-interacting protein required for a late step in cell division. Mol Microbiol. 2006;59(3):989–999. doi: 10.1111/j.1365-2958.2005.04987.x. [DOI] [PubMed] [Google Scholar]
- 9.Miyagishima SY, Wolk CP, Osteryoung KW. Identification of cyanobacterial cell division genes by comparative and mutational analyses. Mol Microbiol. 2005;56(1):126–143. doi: 10.1111/j.1365-2958.2005.04548.x. [DOI] [PubMed] [Google Scholar]
- 10.Jensen SO, Thompson LS, Harry EJ. Cell division in Bacillus subtilis: FtsZ and FtsA association is Z-ring independent, and FtsA is required for efficient midcell Z-ring assembly. J Bacteriol. 2005;187(18):6536–6544. doi: 10.1128/JB.187.18.6536-6544.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pichoff S, Lutkenhaus J. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol Microbiol. 2005;55(6):1722–1734. doi: 10.1111/j.1365-2958.2005.04522.x. [DOI] [PubMed] [Google Scholar]
- 12.Strahl H, Hamoen LW. Membrane potential is important for bacterial cell division. Proc Natl Acad Sci USA. 2010;107(27):12281–12286. doi: 10.1073/pnas.1005485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hale CA, de Boer PA. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell. 1997;88(2):175–185. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- 14.Geissler B, Elraheb D, Margolin W. A gain-of-function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc Natl Acad Sci USA. 2003;100(7):4197–4202. doi: 10.1073/pnas.0635003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh JK, Makde RD, Kumar V, Panda D. A membrane protein, EzrA, regulates assembly dynamics of FtsZ by interacting with the C-terminal tail of FtsZ. Biochemistry. 2007;46(38):11013–11022. doi: 10.1021/bi700710j. [DOI] [PubMed] [Google Scholar]
- 16.Król E, et al. Bacillus subtilis SepF binds to the C-terminus of FtsZ. PLoS ONE. 2012;7(8):e43293. doi: 10.1371/journal.pone.0043293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishikawa S, Kawai Y, Hiramatsu K, Kuwano M, Ogasawara N. A new FtsZ-interacting protein, YlmF, complements the activity of FtsA during progression of cell division in Bacillus subtilis. Mol Microbiol. 2006;60(6):1364–1380. doi: 10.1111/j.1365-2958.2006.05184.x. [DOI] [PubMed] [Google Scholar]
- 18.Mosyak L, et al. The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. EMBO J. 2000;19(13):3179–3191. doi: 10.1093/emboj/19.13.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szwedziak P, Wang Q, Freund SM, Löwe J. FtsA forms actin-like protofilaments. EMBO J. 2012;31(10):2249–2260. doi: 10.1038/emboj.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Holm L, Rosenstrom P (2010) Dali server: Conservation mapping in 3D. Nucleic Acids Res 38(Web Server issue):W545–W549. [DOI] [PMC free article] [PubMed]
- 21.Katoh E, et al. High precision NMR structure of YhhP, a novel Escherichia coli protein implicated in cell division. J Mol Biol. 2000;304(2):219–229. doi: 10.1006/jmbi.2000.4170. [DOI] [PubMed] [Google Scholar]
- 22.Ikeuchi Y, Shigi N, Kato J, Nishimura A, Suzuki T. Mechanistic insights into sulfur relay by multiple sulfur mediators involved in thiouridine biosynthesis at tRNA wobble positions. Mol Cell. 2006;21(1):97–108. doi: 10.1016/j.molcel.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 23.Levin PA, Kurtser IG, Grossman AD. Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. Proc Natl Acad Sci USA. 1999;96(17):9642–9647. doi: 10.1073/pnas.96.17.9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beall B, Lutkenhaus J. Impaired cell division and sporulation of a Bacillus subtilis strain with the ftsA gene deleted. J Bacteriol. 1992;174(7):2398–2403. doi: 10.1128/jb.174.7.2398-2403.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zimmerberg J, Kozlov MM. How proteins produce cellular membrane curvature. Nat Rev Mol Cell Biol. 2006;7(1):9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- 26.Sapay N, Guermeur Y, Deléage G. Prediction of amphipathic in-plane membrane anchors in monotopic proteins using a SVM classifier. BMC Bioinformatics. 2006;7:255. doi: 10.1186/1471-2105-7-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szeto TH, Rowland SL, Rothfield LI, King GF. Membrane localization of MinD is mediated by a C-terminal motif that is conserved across eubacteria, archaea, and chloroplasts. Proc Natl Acad Sci USA. 2002;99(24):15693–15698. doi: 10.1073/pnas.232590599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu Z, Lutkenhaus J. A conserved sequence at the C-terminus of MinD is required for binding to the membrane and targeting MinC to the septum. Mol Microbiol. 2003;47(2):345–355. doi: 10.1046/j.1365-2958.2003.03321.x. [DOI] [PubMed] [Google Scholar]
- 29.Salje J, van den Ent F, de Boer P, Löwe J. Direct membrane binding by bacterial actin MreB. Mol Cell. 2011;43(3):478–487. doi: 10.1016/j.molcel.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelly SM, Jess TJ, Price NC. How to study proteins by circular dichroism. Biochim Biophys Acta. 2005;1751(2):119–139. doi: 10.1016/j.bbapap.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 31.Drin G, Antonny B. Amphipathic helices and membrane curvature. FEBS Lett. 2010;584(9):1840–1847. doi: 10.1016/j.febslet.2009.10.022. [DOI] [PubMed] [Google Scholar]
- 32.Szeto TH, Rowland SL, Habrukowich CL, King GF. The MinD membrane targeting sequence is a transplantable lipid-binding helix. J Biol Chem. 2003;278(41):40050–40056. doi: 10.1074/jbc.M306876200. [DOI] [PubMed] [Google Scholar]
- 33.Kuchibhatla A, Bellare J, Panda D. Cationic lipid enhances assembly of bacterial cell division protein FtsZ: A possible role of bacterial membrane in FtsZ assembly dynamics. Int J Biol Macromol. 2011;49(4):737–741. doi: 10.1016/j.ijbiomac.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 34.Erickson HP, Taylor DW, Taylor KA, Bramhill D. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc Natl Acad Sci USA. 1996;93(1):519–523. doi: 10.1073/pnas.93.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Venema G, Pritchard RH, Venema-Schroeder T. Fate of transforming deoxyribonucleic acid in Bacillus Subtilis. J Bacteriol. 1965;89:1250–1255. doi: 10.1128/jb.89.5.1250-1255.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anagnostopoulos C, Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81(5):741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamoen LW, Smits WK, de Jong A, Holsappel S, Kuipers OP. Improving the predictive value of the competence transcription factor (ComK) binding site in Bacillus subtilis using a genomic approach. Nucleic Acids Res. 2002;30(24):5517–5528. doi: 10.1093/nar/gkf698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morimoto T, et al. Six GTP-binding proteins of the Era/Obg family are essential for cell growth in Bacillus subtilis. Microbiology. 2002;148(Pt 11):3539–3552. doi: 10.1099/00221287-148-11-3539. [DOI] [PubMed] [Google Scholar]
- 39.Cho E, Ogasawara N, Ishikawa S. The functional analysis of YabA, which interacts with DnaA and regulates initiation of chromosome replication in Bacillus subtilis. Genes Genet Syst. 2008;83(2):111–125. doi: 10.1266/ggs.83.111. [DOI] [PubMed] [Google Scholar]
- 40.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55(Pt 4):849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Laue TM, Shah BD, Ridgeway TM, Pelletier SL (1992) Analytical Ultracentrifugation in Biochemistry and Polymer Science (Royal Society of Chemistry, Cambridge, UK)
- 46.Scheffers DJ. The effect of MinC on FtsZ polymerization is pH dependent and can be counteracted by ZapA. FEBS Lett. 2008;582(17):2601–2608. doi: 10.1016/j.febslet.2008.06.038. [DOI] [PubMed] [Google Scholar]
- 47.Wang X, Lutkenhaus J. The FtsZ protein of Bacillus subtilis is localized at the division site and has GTPase activity that is dependent upon FtsZ concentration. Mol Microbiol. 1993;9(3):435–442. doi: 10.1111/j.1365-2958.1993.tb01705.x. [DOI] [PubMed] [Google Scholar]
- 48.Heitkamp T, et al. K+-translocating KdpFABC P-type ATPase from Escherichia coli acts as a functional and structural dimer. Biochemistry. 2008;47(11):3564–3575. doi: 10.1021/bi702038e. [DOI] [PubMed] [Google Scholar]
- 49.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37(8):911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.