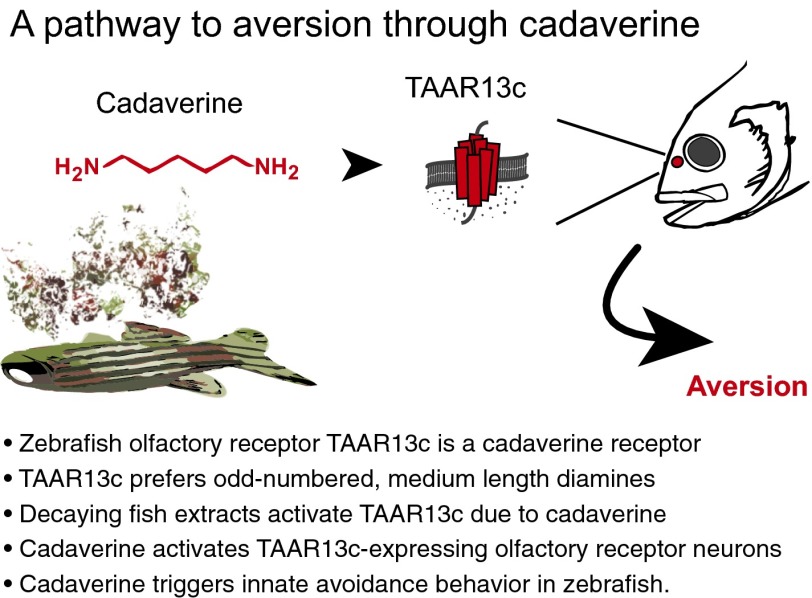
Significance
Cadaverine and putrescine, two diamines emanating from decaying flesh, are strongly repulsive odors to humans but serve as innate attractive or social cues in other species. Here we show that zebrafish, a vertebrate model system, exhibit powerful and innate avoidance behavior to both diamines, and identify a high-affinity olfactory receptor for cadaverine.
Keywords: Danio rerio, aversion, heterologous expression, polyamines
Abstract
Carrion smell is strongly repugnant to humans and triggers distinct innate behaviors in many other species. This smell is mainly carried by two small aliphatic diamines, putrescine and cadaverine, which are generated by bacterial decarboxylation of the basic amino acids ornithine and lysine. Depending on the species, these diamines may also serve as feeding attractants, oviposition attractants, or social cues. Behavioral responses to diamines have not been investigated in zebrafish, a powerful model system for studying vertebrate olfaction. Furthermore, olfactory receptors that detect cadaverine and putrescine have not been identified in any species so far. Here, we show robust olfactory-mediated avoidance behavior of zebrafish to cadaverine and related diamines, and concomitant activation of sparse olfactory sensory neurons by these diamines. The large majority of neurons activated by low concentrations of cadaverine expresses a particular olfactory receptor, trace amine-associated receptor 13c (TAAR13c). Structure-activity analysis indicates TAAR13c to be a general diamine sensor, with pronounced selectivity for odd chains of medium length. This receptor can also be activated by decaying fish extracts, a physiologically relevant source of diamines. The identification of a sensitive zebrafish olfactory receptor for these diamines provides a molecular basis for studying neural circuits connecting sensation, perception, and innate behavior.
Cadaverine, putrescine, and other biogenic diamines are strongly repulsive odors to humans, for whom these odors presumably signal bacterial contamination. It may be expected that animal species feeding on carcasses attribute a more positive valence to diamines, and indeed both putrescine and cadaverine have been reported to be feeding attractants for rats (1) as well as goldfish (2). Similarly, insects depositing their eggs in carcasses or other proteineacous materials are attracted by these diamines (3). Beyond signaling danger or food, putrescine and cadaverine also serve as social cues in several vertebrate species, both for marking of territories—for example, in feline species (4)—and for burial of conspecifics (5).
Very little is known about the molecular and cellular basis of cadaverine-driven behaviors. Cadaverine and putrescine evoke electrophysiological responses in the olfactory epithelium of two fish species (2, 6) and cadaverine-responsive olfactory sensory neurons and glomeruli have been identified in the mouse (7, 8). However, chemosensory receptors that detect cadaverine or related diamines are unknown in any species, and could provide valuable tools to study how the olfactory system mediates innate aversion or attraction.
Here, we show that cadaverine is a major product of zebrafish tissue decay, activates a zebrafish olfactory receptor (trace amine-associated receptor 13c, TAAR13c) with high affinity, and elicits a strong, low-threshold, and olfactory-mediated avoidance response in zebrafish. In vivo measurements indicate that high affinity cadaverine responses occur primarily in TAAR13c-expressing olfactory sensory neurons. These findings provide an important foundation for understanding the molecular basis of a powerful odor-driven behavior.
Results
Zebrafish Avoid a Cadaverine Odor Source.
The zebrafish has in recent years emerged as an important model system for understanding olfaction in vertebrates because of a remarkable similarity in the basic principles of olfactory representation (9) and some technical advantages over the mammalian system (10). However, behavioral responses of zebrafish to diamines have not been described. We report here that zebrafish, like humans, show pronounced innate aversion behavior for cadaverine (Fig. 1).
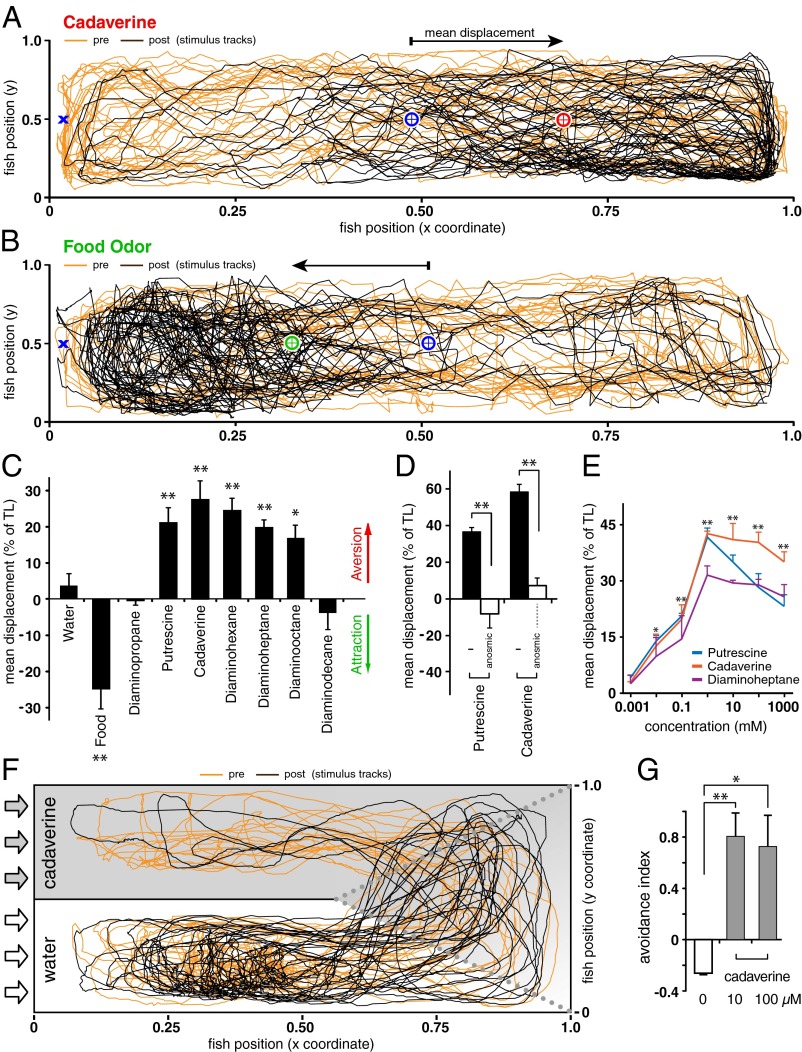
Fig. 1.
Aversive behavioral response of zebrafish to diamines. (A and B) Cadaverine-evoked aversion and food odor-evoked attraction of individual zebrafish as visualized by movement patterns before (orange tracks) and after (brown tracks) stimulus addition (1 mM, 180 µL). The position of stimulus deposition (blue X), as well as the mean location before (blue circle) and after (red, green circles) stimulus addition are indicated. The axes represent tank dimensions. (C) Mean displacement was expressed as a percentage of tank length (n = 6 ± SEM). Addition of tank water alone had no significant effect, whereas food odor elicited strong attraction. Similar avoidance behavior was observed in male and female fish. Significance was evaluated by Student t test, *P < 0.05, **P < 0.01. (D) Aversion behavior requires olfactory input. Nostril closure of zebrafish eliminates aversion to 1 mM cadaverine or putrescine. Black bars, no treatment; white bars, nostril closure; **P < 0.01, n = 3 ± SEM. (E) Avoidance behavior to diamines was dose-dependent. Responses to cadaverine and diaminoheptane were measured over a broad concentration range (1 µM to 1 M, n = 3 ± SEM). The behavior is clearly saturable at 1-mM stimulus concentration. Significance was evaluated by Student t test, asterisks refer to cadaverine, *P < 0.05, **P < 0.01. The slight decrease at higher concentrations is not significant except for putrescine (P < 0.05). (F) Cadaverine-evoked aversion of an individual zebrafish in a flow-through two-channel set-up (11) as visualized by movement patterns before (orange tracks) and after (brown tracks) change of the upper channel to a chronic concentration of 10 µM cadaverine. The arrows indicate direction of flow (cadaverine, gray; water, white). Dotted gray lines enclose the mixing zone not included in analysis of preference. The axes represent tank dimensions. (G) Aversion behavior of zebrafish in the two-channel preference test is maximal at 10 µM cadaverine. The avoidance index shows similar avoidance of the cadaverine channel for 10 and 100 µM cadaverine (gray bars). Significance in comparison with water was evaluated by Student t test, *P < 0.05, **P < 0.01.
We developed a valence assay to measure behavioral responses of zebrafish to olfactory cues. Zebrafish were habituated to a rectangular tank, and the position of each fish was recorded before and after odor delivery. Shifts in average position toward or away from the odor source were recorded as attraction and avoidance, respectively. Food odor, an attractant for zebrafish, caused a mean displacement of 0.25 tank lengths (TLs, P < 0.01) toward the odor source (Fig. 1B), but tank water alone had no effect (Fig. 1C and Table S1). In contrast, cadaverine caused a mean displacement of 0.28 TLs (P < 0.01) away from the odor source, and was thus aversive (Fig. 1 A and C, and Table S1). Furthermore, the fish spent several-fold less time in close approach (distances < 0.05 TL) to the stimulus application site (P < 0.0001), although this area was not completely avoided (P < 0.03) (Fig. 1A and Table S1), suggesting that the zebrafish did for short periods of time investigate the area, where stimulus was given. Mean velocity or total distance traveled was not altered during avoidance behavior (Table S1). Thus, the displacement observed is not caused by changes in motility but may result from an assessment of odor valence by the fish.
Next, we analyzed whether related diamines were similarly aversive. We tested diamines with different carbon chain lengths, ranging from C3 (diaminopropane) to C10 (diaminodecane). Avoidance behavior was observed (Fig. 1C) to putrescine (C4), cadaverine (C5), diaminohexane (C6), diaminoheptane (C7), and diaminooctane (C8), but not to diaminopropane (C3) or diaminodecane (C10). Time spent in close approach to the stimulus application site did not differ significantly from prestimulus values for diaminopropane and diaminodecane, but was reduced to one-third or less for the other diamines (P < 0.0001) (Table S1). Only cadaverine and putrescine are naturally abundant in carrion (see below); other diamines not found ecologically may be aversive because they function as agonists for cadaverine and putrescine-activated receptors.
Avoidance behavior to cadaverine and putrescine was abolished (P < 0.01) in fish, whose nostrils were closed by tissue glue, showing the avoidance to be mediated by olfaction and not other sensory modalities (Fig. 1D). Avoidance responses to cadaverine, putrescine, and diaminoheptane exhibited similar dose-dependence (Fig. 1E), with complete saturation at 1 mM diamine in the stimulus, half-maximal values at 100 µM, and about one-third of maximal values at 10 µM (Fig. 1E). The actual concentrations encountered by the zebrafish at the time of decision-making are expected to be much lower than these values because of dilution of a small stimulus volume (180 µL) into a large tank volume (9 L). To obtain a more stringent estimate of behavioral sensitivity, we have therefore used a two-channel preference assay (11), with a constant cadaverine concentration in one of the two streams during the stimulus period. Cadaverine evoked robust avoidance responses in this paradigm, with similar levels of avoidance observed at 10 µM (avoidance index 0.81 ± 0.18 SEM, n = 4, P < 0.01) and 100 µM (avoidance index 0.73 ± 0.24 SEM, n = 4, P < 0.05). Such sensitive detection suggests the existence of specialized olfactory receptors for diamines.
Cadaverine and Other Diamines Activate Sparse Olfactory Sensory Neurons.
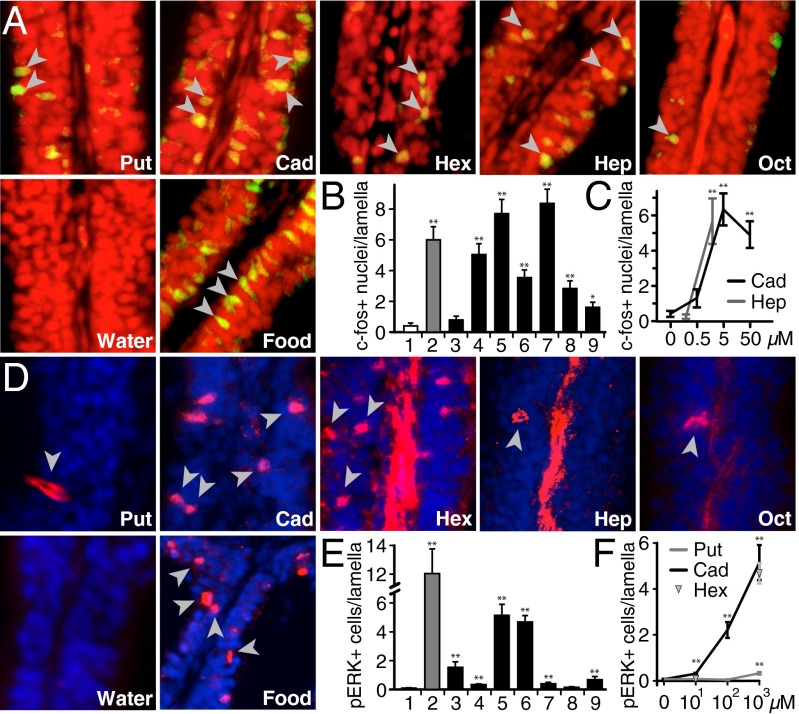
We observed diamine-evoked increases in c-Fos expression and pERK levels in sparse olfactory sensory neurons. ERK phosphorylation and c-Fos expression are induced by neuronal activity, and are widely used reporters for neuron responsiveness, including in the olfactory system (12, 13). Olfactory tissue was obtained from zebrafish (n = 102) exposed to diamines or control stimuli and stained using standard immunohistochemical (IHC) techniques. c-Fos expression was similarly and robustly induced by food odor, cadaverine, and other diamines (∼6.0 cells per lamella) but not tank water alone (<0.5 cells per lamella), with low background levels likely resulting from residual odors in tank water (Fig. 2 A and B, and Fig. S1). The frequency of pERK-containing cells was similar to that of c-Fos–expressing cells for food odor, cadaverine, and diaminohexane. Some differences in c-Fos induction and ERK phosphorylation were observed for other diamines, and could reflect differences in reporter sensitivity for lower affinity ligands or the much longer exposure time required for c-Fos expression. Dose-dependent analysis indicated threshold c-Fos responses to cadaverine and diaminoheptane at 2 and 5 µM, respectively (Fig. 2C). Low concentrations of cadaverine and putrescine resulted in very low frequencies of pERK-labeled cells (Fig. 2F), consistent with detection by a single olfactory receptor (cf. refs. 14 and 15). The range of diamine chain lengths (C3 to C10) that stimulate either c-Fos expression or ERK phosphorylation in olfactory tissue includes all diamines that promote aversive behavior (C4 to C8), consistent with this behavior being mediated by olfaction. However, olfactory receptors that detect cadaverine in zebrafish or any species have not been identified so far.
Fig. 2.
Diamines elicit c-Fos and pERK increase in olfactory sensory neurons. (A) Zebrafish (n = 21) were exposed to stimuli indicated (2 or 5 mM). c-Fos IHC (green) and nuclear staining (propidium iodide, red), enabled visualization of c-Fos+ nuclei (yellow), some emphasized by gray arrow heads. (B and C) Quantification of c-Fos+ nuclei/lamella as a function of diamine chain length (B) or concentration (C). Results from one representative experiment each are shown. Counting was done on randomized micrographs, values given represent mean ± SEM. Significance in comparison with water was evaluated by Student t test, *P < 0.05, **P < 0.01. (B) 1, water; 2, food extract; 3–8, numbers reflect carbon chain length of diamines; 9, diaminodecane. (D) Zebrafish (n = 14) were exposed to stimuli indicated (1 mM). Some pERK-labeled cells (red) are emphasized by gray arrowheads; nuclear counterstain (DAPI, blue). Red central stripes in some panels, unspecific label in the basal lamina outside the sensory region. (E and F) Quantification of pERK+ cells/lamella as function of chain length (E) or concentration (F). Values given represent mean ± SEM. Significance in comparison with water was evaluated by Student t test, **P < 0.01. (E) Results from two experiments are shown; 1, water; 2, food extract; 3–8, numbers reflect carbon chain length of diamines; 9, diaminodecane. (F) Evaluation was partly on randomized data, no difference was seen between randomized and nonrandomized evaluation.
TAAR13c Is an Olfactory Receptor for Cadaverine and Other Diamines.
As in mammals (16), zebrafish TAARs function as olfactory receptors (17). We reasoned that a zebrafish TAAR could mediate the cadaverine avoidance behavior because several rodent TAARs detect biogenic amines, including some highly aversive odors (16, 18–20). As a result of numerous gene-duplication events, the zebrafish TAAR family is large, with 112 receptors encoded by the zebrafish genome (17). So far, ligands have not been identified for any zebrafish TAAR, and identification of such ligands would be a key step toward understanding their physiological roles.
The dynamic evolution of the teleost Taar gene family led to widespread loss (17) of an amine-binding motif found in biogenic amine receptors (21). However, most class I and class II teleost TAARs retain this amine-binding motif (17), making them good candidates for contributing to amine perception by the fish olfactory system.
We initiated a chemical screen to identify agonists for zebrafish TAARs and selected representatives from each of the five TAAR subfamilies retaining the amine-binding motif and three TAARs without it (see Fig. 3D for phylogenetic position of genes analyzed). We previously developed a reporter gene system to measure ligand-induced TAAR activation (16, 19); here, we used this system to identify zebrafish TAAR agonists among 93 test odorants, including a large number of amines, diamines, polyamines, and amino acids.
Fig. 3.
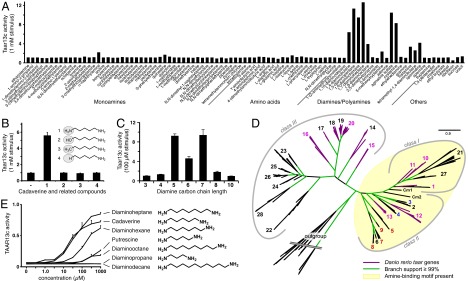
TAAR13c is a sensitive diamine detector. (A) HEK293 cells were transfected with TAAR13c plasmid and a reporter gene, incubated with 93 different test chemicals (1 mM), and assayed for reporter gene activity. TAAR13c activity is reported after normalization to responses from control stimuli (media alone). Only aliphatic and mixed aliphatic/aromatic diamines and polyamines activate TAAR13c. No responses to diamines were observed in cells transfected with reporter gene alone. (B) TAAR13c activation requires a divalent ligand with two amino groups. TAAR13c responses (n = 3 ± SEM, 1-mM stimuli) were measured for: 1, cadaverine; 2, 5-amino-1-pentanol; 3, hexylamine; 4, pentylamine; or “–” no ligand. (C) TAAR13c prefers odd-chained diamines. Responses of TAAR13c (n = 3 ± SEM) were measured to diamines of carbon chain length 3–10 (100 µM). (D) Phylogenetic tree for taar genes, gene set as described previously (17), but only three mammalian species included (mouse, rat, human). Class I and II TAARs retain the amine-binding motif (yellow shade). Numbers indicate TAAR subfamilies, including mammalian TAARs that detect primary (blue) or tertiary (red) amines, as well as zebrafish TAARs analyzed here (purple). TAAR13c terminal branch, light purple; Cm1, Cm2, elephant shark taar genes. (E) Dose-dependent activation of TAAR13c by aliphatic diamines; values are in relative units. A representative experiment is shown (n = 3 ± SEM). No responses to diamines were observed in cells transfected with reporter gene alone. Higher affinity is seen for odd-chained diamines.
One receptor tested, zebrafish TAAR13c, gave robust responses to cadaverine (1,5-diaminopentane) and related aliphatic diamines (Fig. 3 A and C). Cadaverine activated HEK293 cells expressing TAAR13c, but not control cells lacking TAAR13c, with a half-maximal response (EC50) occurring at 23 ± 3 µM (mean ± SD) and a threshold response occurring at 3 µM (Fig. 3E). Cadaverine variants, in which one amino group is replaced with a hydroxyl group, methyl group, or hydrogen, did not activate TAAR13c (Fig. 3B). Furthermore, 47 different aliphatic and aromatic monoamines with varying chain lengths, degrees of substitution, and functional groups, did not activate TAAR13c (Fig. 3A). This structure–activity analysis suggests that TAAR13c contains two remote cation recognition sites, both of which require occupancy for receptor activation.
TAAR13c Preferentially Detects Odd-Chained Diamines.
We next measured TAAR13c responses to diamines ranging from C3 to C10 across several concentrations (Fig. 3E) and found cadaverine (C5) and diaminoheptane (C7) to activate TAAR13c with highest affinity (EC50 = 23 ± 3 µM and 30 ± 2 µM, respectively). Diaminohexane (C6) had ∼fivefold reduced affinity, whereas putrescine (C4) and diaminooctane (C8) had >10-fold reduced affinity. Diaminopropane (C3) and diaminodecane (C10) did not activate TAAR13c at any concentration tested. Furthermore, other dibasic ligands, including cystamine, agmatine, and histamine, activated TAAR13c with reduced affinity (Fig. S1). Although TAAR13c detected numerous primary amines, it showed reduced activity for the tertiary amine derived from putrescine, tetramethyl-1,4-diaminobutane (Fig. 3A). Indeed, TAAR13c is phylogenetically closer to those mammalian TAARs that detect primary amines than to those preferring tertiary amines (Fig. 3D) (cf. ref. 18).
Interestingly, both fish avoidance behavior and sensory neuron responses showed no distinct preference for odd-chained diamines, suggesting the existence of other receptors tuned to even-chained diamines. This finding is consistent with electrophysiological studies indicating limited cross-adaptation of olfactory responses to cadaverine and putrescine (2). Here, we identified TAAR13c as a highly sensitive detector of odd-chained diamines that include the repulsive odor cadaverine.
A Physiological Source of Cadaverine Activates TAAR13c.
A physiologically relevant source of diamine odors is decomposing carcasses, whose presence may signal danger. In these circumstances, cadaverine will be present in a complex mixture together with other potentially odorous chemicals that also result from tissue decay. Thus far we have examined TAAR13c responses to pure chemicals; next we asked whether TAAR13c could detect cadaverine produced during the natural process of tissue decomposition.
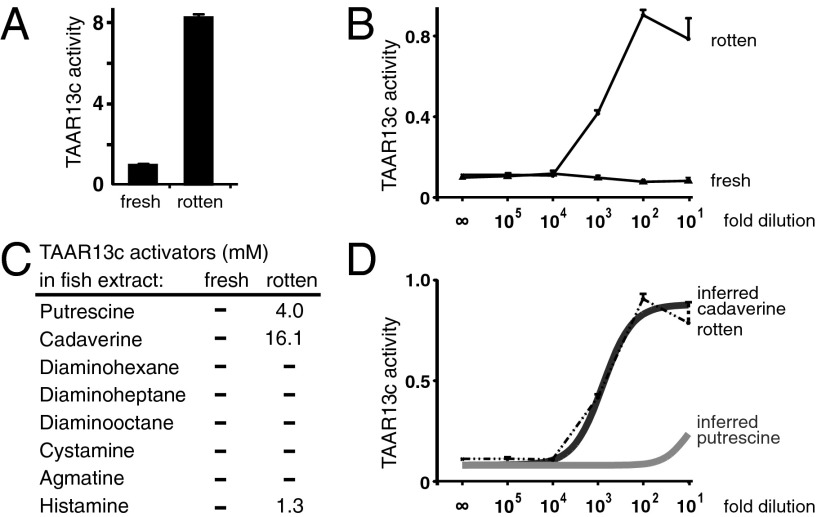
We prepared fish extracts by placing a killed zebrafish in PBS for 0 min (“fresh”) or 1 wk (“rotten”). PBS solutions were homogenized and centrifuged to remove debris, and resulting supernatants used as “extracts.” We found that rotten fish extracts provided a potent stimulus for TAAR13c, but that fresh extracts had no activity (Fig. 4A). Next, we analyzed the concentration dependence of fresh and rotten fish extracts. We observed a half-maximal response to rotten fish extracts diluted ∼1,000-fold from the initial preparation (Fig. 4B). In contrast, fresh fish extracts did not activate TAAR13c at any concentration tested, up to a 10-fold dilution (Fig. 4B). These results indicate that TAAR13c is able to detect diamines in a complex and physiologically relevant mixture.
Fig. 4.
TAAR13c is activated by a biological source of diamines. (A) TAAR13c detects rotten but not fresh fish extracts diluted 100-fold. Values are given as signal-to-blank ratios. (B) Rotten fish extract activates TAAR13c in a dose-dependent manner, whereas fresh extract shows no activity at any concentration. (C) LC/MS analysis showed that rotten but not fresh fish extracts contain cadaverine, putrescine, and histamine at concentrations indicated. No other TAAR13c activators were detected (“–”, below detection limit of 0.5 mM). (D) TAAR13c activation by rotten fish extract can be explained by cadaverine content. Gray solid curves indicate inferred TAAR13c activation by cadaverine and putrescine present in the various dilutions of rotten fish extract and are superimposed on a curve (dashed lines) reporting measured dose-dependent TAAR13c activation by rotten fish extract (see B).
Cadaverine Is the Principal TAAR13c Activator in Decayed Fish.
To determine the most relevant TAAR13c ligands in rotten fish extracts, we quantified the levels of diamines using liquid chromatography and tandem mass spectrometry (LC/MS). The number of ion counts with the mass-charge ratio (m/z) corresponding to nine different diamines (putrescine, cadaverine, diaminohexane, diaminoheptane, diaminooctane, agmatine, cystamine, histamine, and cysteamine) were separately graphed over time. The retention time and integrated area of observed peaks were compared with standards for chemical assignment and quantification. None of these nine amines were detected in fresh fish extracts, and only cadaverine, putrescine, and histamine were detected in rotten fish extract (Fig. 4C). Cadaverine was the most abundant diamine detected, and levels of cadaverine, but not putrescine or histamine, were sufficient to explain the striking sensitivity of TAAR13c for decomposed tissue (Fig. 4D).
High-Affinity Cadaverine Responses Occur Primarily in TAAR13c-Expressing Neurons.
We next sought to determine whether high-affinity cadaverine responses occurred primarily in TAAR13+ or TAAR13c− olfactory sensory neurons. We generated a polyclonal antibody that recognizes a highly divergent region of the TAAR13c sequence that is not conserved in closely related TAAR13 family members (Fig. S1). This antibody labeled a 55-kDa protein in olfactory epithelium, but not other tissues by Western blot analysis, and an extremely sparse population of olfactory sensory neurons (0.3 cells per lamella) (Fig. S1) by IHC analysis. Two-color analysis indicated colabeling of olfactory sensory neurons with TAAR13c antibody and Taar13c cRNA riboprobe, with Taar13c riboprobe labeling three- to sixfold more cells, likely because of cross-hybridization to the four other Taar13c family members. Neurons labeled by TAAR13c antibody showed a ciliary morphology (Fig. S1) and occurred at a frequency similar to that predicted for expression of individual olfactory receptor genes (15).
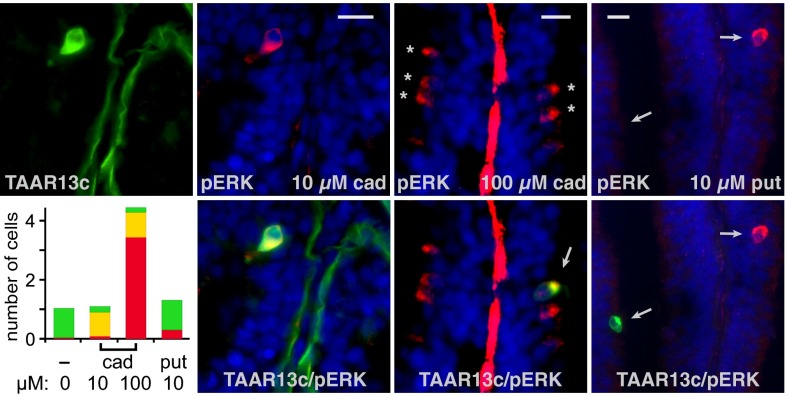
Next, we asked whether cells labeled by the TAAR13c antibody responded to diamines using two-color IHC analysis for pERK and TAAR13c. We found that cadaverine-responsive neurons could be classified as low or high affinity based on response sensitivity. High-affinity (10 µM) cadaverine responses occurred primarily (∼90%) in TAAR13c-expressing cells (Fig. 5), whereas increasing cadaverine concentration 10-fold resulted in increased numbers of responsive neurons, suggesting recruitment of additional low-affinity receptors (Fig. 5). TAAR13c cells were distinct from those activated by low concentrations (10 µM) of putrescine (Fig. 5), consistent with our findings that TAAR13c prefers odd-chained diamines. Taken together, these results are consistent with TAAR13c being a major component in high-affinity cadaverine recognition by zebrafish.
Fig. 5.
Cadaverine activates TAAR13c-expressing neurons. Zebrafish were exposed to cadaverine (cad) and putrescine (put) at concentrations indicated and processed for concomitant IHC of TAAR13c (green fluorescence) and pERK (red fluorescence). DAPI was used as nuclear counterstain (blue). Sometimes the basal lamina was stained unspecifically (green and red stripes in the center of some lamellae). Asterisks, pERK+ cells; arrows, colabeled cells (yellow) and pERK+/TAAR13c− cells (red). (Scale bars: 10 μm.) (Lower Left) Quantitative evaluation, values are given as normalized cell numbers (120–250 cells per condition were analyzed); green bar, TAAR13c+/pERK− cells; yellow bar, double label; red bar, TAAR13c−/pERK+ cells.
Discussion
Cadaverine and putrescine are death-associated odors produced by microbe-mediated decarboxylation of basic amino acids (22). Chemosensory receptors that detect these odors are unknown in any species and could provide valuable tools to study how the olfactory system mediates innate aversion or attraction (cf. ref. 23).
Here, we show that zebrafish TAAR13c detects cadaverine with high sensitivity and specificity. This study is unique in reporting a ligand for any of the 112 zebrafish TAARs, and following identification of two amino acid-activated receptors from the V2R-related receptor family (24, 25), constitutes the third deorphanization of any fish olfactory receptor. TAAR13c is strongly activated by primary amines and indeed is phylogenetically closer to those rodent TAARs that prefer primary over tertiary amines (18). Moreover, TAAR13c has distinct molecular recognition properties compared with many biogenic amine-activated G protein-coupled receptors in that it is selective for diamines compared with monoamines. Structure activity analysis indicates an unusual divalent ligand binding pocket requiring two remote positive charges for activation. A conserved aspartic acid in biogenic amine receptors that forms a salt bridge with the ligand amino group is retained in TAAR13c (Asp3.32), but residues important for recognition of the second amine are not known. Nevertheless, the existence of a second amine contact site raises the possibility for a unique inverted mode of monoamine recognition by G protein-coupled receptors that lose the conserved Asp3.32 but retain the second recognition site.
TAAR13c is a narrowly tuned receptor that prefers medium-length, odd-chained diamines. Although a pronounced tuning to chain length is commonly found for olfactory receptors (26, 27), TAAR13c is peculiar in its strong preference for odd-chained diamines. Odd- and even-chained diamines have significant differences in the relative orientation and positioning of the two amino groups, and their cognate olfactory receptors likely have negatively charged counterions in distinct locations of the agonist binding pocket. Interestingly, odd-chained and even-chained diamines did elicit comparable aversive behavior, which suggests the presence of additional zebrafish olfactory receptors activated by even-chained diamines, consistent with data from cross-adaptation studies (6).
Some estimates about the conceivable size of the cadaverine receptor repertoire can be derived from our quantitative analysis of sensory neuron responses. We find that many receptor neurons show increased pERK levels at high cadaverine concentrations, consistent with the presence of several olfactory receptors that can detect cadaverine. However, at low concentrations an extremely sparse population of receptor neurons (0.3 cells per lamella, about 100 cells per olfactory rosette) is activated, corresponding to the lower limit of cell numbers found for individual olfactory receptor genes (cf. ref. 15). Moreover, the large majority of these cells expresses TAAR13c, consistent with this receptor being a major component of high affinity cadaverine detection. Further investigation, in particular TAAR13c loss-of-function analysis, will be required to delineate the exact role of TAAR13c in generating avoidance behavior to diamines.
The dose-dependence of cadaverine-evoked avoidance behaviors, c-Fos and pERK induction, and TAAR13c activation are all understandably different, because stimulus application, signal detection threshold, and signal/noise ratio are specific to each method. Receptor affinities can be much lower in heterologous systems than in vivo (28–30), where expression of receptors and signaling components are presumably optimized. Nevertheless, the threshold of cadaverine detection by TAAR13c of 3 µM is very similar to in vivo thresholds observed for the intact olfactory system (6, 31) and to thresholds measured for isolated olfactory sensory neurons (2). Although it is difficult to estimate naturally occurring cadaverine concentrations close to dead fish, the cadaverine concentration we measured in rotten zebrafish extracts was several orders of magnitude higher than the TAAR13c activation threshold, and presumably high enough to allow detection of that odor source from some distance. Importantly, the behavioral response in the two-choice assay is maximal at the same low concentration of cadaverine, which elicits neuronal activity predominantly in TAAR13c-expressing cells. Thus, the available data are consistent with TAAR13c being a significant part of the receptor repertoire that detects cadaverine present in ecologically relevant sources.
TAAR13c arose during teleost evolution and orthologs are not found in rodents and humans who also detect cadaverine. Thus, cadaverine-activated olfactory receptors in mammals may present a case of convergent evolution, either within the vertebrate TAAR family or between different olfactory receptor families (cf. ref. 27). In vitro studies did not identify a high-affinity cadaverine receptor among mouse, rat, or human TAARs (18), although cadaverine reportedly activates TAAR-containing glomeruli in mice at high concentrations (8). Other mammalian TAARs also detect aversive amines; for example, isoamylamine (TAAR3) and 2-phenylethylamine (TAAR4), both likewise produced by decarboxylation of amino acids (16, 18, 19, 32). Indeed, amines are an odor group that is chemically suited both to aquatic and airborne detection. Interestingly, trimethylamine, a TAAR5 agonist, is an aversive odor to humans and rats (20, 33) but attractive to mice (20). This finding is reminiscent of cadaverine, which is attractive to goldfish (2) but aversive to zebrafish (present results).
Taking these data together, we have shown that TAAR13c emerges as a sensitive olfactory receptor for the death-associated odor cadaverine, both in isolation and as part of a complex mixture. Cadaverine at low concentrations activates a sparse population of TAAR13c-expressing olfactory sensory neurons and elicits powerful and innate avoidance behavior in zebrafish, a vertebrate model system. See Fig. 6 for a graphical summary of key findings. Such an association of odors and cognate receptors with a powerful avoidance response provides a molecular basis for studying neural circuits connecting sensation with perception of odor valence.
Fig. 6.
Graphical summary of key findings.
Materials and Methods
TAAR Cloning.
TAAR13c cDNA (National Center for Biotechnology Information accession no. NM_001083040.1) was cloned from zebrafish genomic DNA using standard methods. For other clones, primer, and vector information, see SI Materials and Methods.
TAAR Phylogenetic Analysis.
The TAAR gene dataset was from ref. 17. For algorithms used, see SI Materials and Methods.
TAAR Functional Assays.
The reporter assay was performed as described previously (16). Details for assay and receptor sequences are provided in SI Materials and Methods.
Antibody Generation.
A unique peptide of 16 amino acids served as immunogen for TAAR13c. Details are provided in SI Materials and Methods.
Western Blot.
The Western blot was performed as described previously (34). Details are provided in SI Materials and Methods.
Immunohistochemistry.
Standard procedures were used. Protocols are provided in SI Materials and Methods.
Immunohistochemistry Combined with in Situ Hybridization.
In situ hybridizaiton-IHC was performed as described previously (35). Protocols and details for antibody and probe are supplied in SI Materials and Methods.
Behavioral Analysis.
Fish motion was tracked pre- and poststimulus addition. Two set-ups were used: single arena and two channel. Details of set-ups and analysis are provided in SI Materials and Methods.
Quantitative LC/MS Analysis of Diamines.
Protocols for sample preparation, processing, and analysis are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Walter Nadler for help with programming; Yuichiro Oka for critical review of the manuscript; Wayne Korzan, Matthias Gruhn, and Kim Korsching for technical advice; Jamie Lemon and Priyanka Maity for technical support; and Ansgar Bueschges for housing our behavioral set-up. This work was supported by Deutsche Forschungsgemeinschaft Award KO-1046/3 (to S.I.K.); an International Graduate School in Genetics and Functional Genomics stipend (to A.H. and L.R.S.); an International Graduate School in Development Health and Disease stipend (to G.A.); a Boehringer Ingelheim travel grant (to L.R.S.); and a grant (Award Number R01DC010155) from the National Institute on Deafness and Other Communicative Disorders (to S.D.L.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1318596110/-/DCSupplemental.
References
- 1.Heale VR, Petersen K, Vanderwolf CH. Effect of colchicine-induced cell loss in the dentate gyrus and Ammon’s horn on the olfactory control of feeding in rats. Brain Res. 1996;712(2):213–220. doi: 10.1016/0006-8993(95)01416-0. [DOI] [PubMed] [Google Scholar]
- 2.Rolen SH, Sorensen PW, Mattson D, Caprio J. Polyamines as olfactory stimuli in the goldfish Carassius auratus. J Exp Biol. 2003;206(Pt 10):1683–1696. doi: 10.1242/jeb.00338. [DOI] [PubMed] [Google Scholar]
- 3.Hamana K, Matsuzaki S. Unusual polyamines in slime molds Physarum polycephalum and Dictyostelium discoideum. J Biochem. 1984;95(4):1105–1110. doi: 10.1093/oxfordjournals.jbchem.a134698. [DOI] [PubMed] [Google Scholar]
- 4.Burger BV, et al. Chemical characterization of territorial marking fluid of male Bengal tiger, Panthera tigris. J Chem Ecol. 2008;34(5):659–671. doi: 10.1007/s10886-008-9462-y. [DOI] [PubMed] [Google Scholar]
- 5.Pinel JP, Gorzalka BB, Ladak F. Cadaverine and putrescine initiate the burial of dead conspecifics by rats. Physiol Behav. 1981;27(5):819–824. doi: 10.1016/0031-9384(81)90048-2. [DOI] [PubMed] [Google Scholar]
- 6.Michel WC, Sanderson MJ, Olson JK, Lipschitz DL. Evidence of a novel transduction pathway mediating detection of polyamines by the zebrafish olfactory system. J Exp Biol. 2003;206(Pt 10):1697–1706. doi: 10.1242/jeb.00339. [DOI] [PubMed] [Google Scholar]
- 7.Nara K, Saraiva LR, Ye X, Buck LB. A large-scale analysis of odor coding in the olfactory epithelium. J Neurosci. 2011;31(25):9179–9191. doi: 10.1523/JNEUROSCI.1282-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pacifico R, Dewan A, Cawley D, Guo C, Bozza T. An olfactory subsystem that mediates high-sensitivity detection of volatile amines. Cell Rep. 2012;2(1):76–88. doi: 10.1016/j.celrep.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshihara Y. Molecular genetic dissection of the zebrafish olfactory system. Results Probl Cell Differ. 2009;47:97–120. doi: 10.1007/400_2008_1. [DOI] [PubMed] [Google Scholar]
- 10.Lieschke GJ, Currie PD. Animal models of human disease: Zebrafish swim into view. Nat Rev Genet. 2007;8(5):353–367. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- 11.Gerlach G, Atema J, Kingsford MJ, Black KP, Miller-Sims V. Smelling home can prevent dispersal of reef fish larvae. Proc Natl Acad Sci USA. 2007;104(3):858–863. doi: 10.1073/pnas.0606777104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guthrie KM, Anderson AJ, Leon M, Gall C. Odor-induced increases in c-fos mRNA expression reveal an anatomical “unit” for odor processing in olfactory bulb. Proc Natl Acad Sci USA. 1993;90(8):3329–3333. doi: 10.1073/pnas.90.8.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mirich JM, Illig KR, Brunjes PC. Experience-dependent activation of extracellular signal-related kinase (ERK) in the olfactory bulb. J Comp Neurol. 2004;479(2):234–241. doi: 10.1002/cne.20325. [DOI] [PubMed] [Google Scholar]
- 14.Weth F, Nadler W, Korsching S. Nested expression domains for odorant receptors in zebrafish olfactory epithelium. Proc Natl Acad Sci USA. 1996;93(23):13321–13326. doi: 10.1073/pnas.93.23.13321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato Y, Miyasaka N, Yoshihara Y. Hierarchical regulation of odorant receptor gene choice and subsequent axonal projection of olfactory sensory neurons in zebrafish. J Neurosci. 2007;27(7):1606–1615. doi: 10.1523/JNEUROSCI.4218-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liberles SD, Buck LB. A second class of chemosensory receptors in the olfactory epithelium. Nature. 2006;442(7103):645–650. doi: 10.1038/nature05066. [DOI] [PubMed] [Google Scholar]
- 17.Hussain A, Saraiva LR, Korsching SI. Positive Darwinian selection and the birth of an olfactory receptor clade in teleosts. Proc Natl Acad Sci USA. 2009;106(11):4313–4318. doi: 10.1073/pnas.0803229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrero DM, et al. Agonists for 13 trace amine-associated receptors provide insight into the molecular basis of odor selectivity. ACS Chem Biol. 2012;7(7):1184–1189. doi: 10.1021/cb300111e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferrero DM, et al. Detection and avoidance of a carnivore odor by prey. Proc Natl Acad Sci USA. 2011;108(27):11235–11240. doi: 10.1073/pnas.1103317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Q, et al. Synchronous evolution of an odor biosynthesis pathway and behavioral response. Curr Biol. 2013;23(1):11–20. doi: 10.1016/j.cub.2012.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang ES. Construction of a sequence motif characteristic of aminergic G protein-coupled receptors. Protein Sci. 2003;12(7):1360–1367. doi: 10.1110/ps.0305603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mietz JL, Karmas E. Polyamine and histamine content of rockfish, salmon, lobster, and shrimp as an indicator of decomposition. J Assoc Off Anal Chem. 1978;61(1):139–145. [Google Scholar]
- 23.Mori K, Sakano H. How is the olfactory map formed and interpreted in the mammalian brain? Annu Rev Neurosci. 2011;34:467–499. doi: 10.1146/annurev-neuro-112210-112917. [DOI] [PubMed] [Google Scholar]
- 24.Speca DJ, et al. Functional identification of a goldfish odorant receptor. Neuron. 1999;23(3):487–498. doi: 10.1016/s0896-6273(00)80802-8. [DOI] [PubMed] [Google Scholar]
- 25.Demaria S, et al. Role of a ubiquitously expressed receptor in the vertebrate olfactory system. J Neurosci. 2013;33(38):15235–15247. doi: 10.1523/JNEUROSCI.2339-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fuss SH, Korsching SI. Odorant feature detection: Activity mapping of structure response relationships in the zebrafish olfactory bulb. J Neurosci. 2001;21(21):8396–8407. doi: 10.1523/JNEUROSCI.21-21-08396.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saito H, Chi Q, Zhuang H, Matsunami H, Mainland JD. Odor coding by a mammalian receptor repertoire. Sci Signal. 2009;2(60):ra9. doi: 10.1126/scisignal.2000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mombaerts P. Genes and ligands for odorant, vomeronasal and taste receptors. Nat Rev Neurosci. 2004;5(4):263–278. doi: 10.1038/nrn1365. [DOI] [PubMed] [Google Scholar]
- 29.Oka Y, et al. Odorant receptor map in the mouse olfactory bulb: In vivo sensitivity and specificity of receptor-defined glomeruli. Neuron. 2006;52(5):857–869. doi: 10.1016/j.neuron.2006.10.019. [DOI] [PubMed] [Google Scholar]
- 30.Krautwurst D, Yau KW, Reed RR. Identification of ligands for olfactory receptors by functional expression of a receptor library. Cell. 1998;95(7):917–926. doi: 10.1016/s0092-8674(00)81716-x. [DOI] [PubMed] [Google Scholar]
- 31.Friedrich RW, Korsching SI. Combinatorial and chemotopic odorant coding in the zebrafish olfactory bulb visualized by optical imaging. Neuron. 1997;18(5):737–752. doi: 10.1016/s0896-6273(00)80314-1. [DOI] [PubMed] [Google Scholar]
- 32.Kobayakawa K, et al. Innate versus learned odour processing in the mouse olfactory bulb. Nature. 2007;450(7169):503–508. doi: 10.1038/nature06281. [DOI] [PubMed] [Google Scholar]
- 33.Mitchell SC, Smith RL. Trimethylaminuria: The fish malodor syndrome. Drug Metab Dispos. 2001;29(4 Pt 2):517–521. [PubMed] [Google Scholar]
- 34.Ahuja G, et al. Zebrafish crypt neurons project to a single, identified mediodorsal glomerulus. Sci Rep. 2013;3:2063. doi: 10.1038/srep02063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oka Y, Saraiva LR, Korsching SI. Crypt neurons express a single V1R-related ora gene. Chem Senses. 2012;37(3):219–227. doi: 10.1093/chemse/bjr095. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.