Abstract
Background
Treatment resistance resulting from the presence of cancer stem cells (CSCs) remains a challenge in cancer treatment. Little is known about possible markers of CSCs in treatment-resistant non-small cell lung cancer (NSCLC). We explored the coxsackie- and adenovirus receptor (CAR) as one such marker of CSCs in models of treatment-resistant NSCLC.
Materials and methods
Resistant H460 and A549 cell lines were established by repeated exposure to paclitaxel or fractionated radiation. CSC markers were measured by western blotting and flow cytometry. We also established stable CAR-overexpressing and stable shRNA-CAR-knockdown cell lines and assessed their survival, invasiveness, and tumorigenic capabilities with clonogenic, telomerase, Matrigel, and tumor formation assays.
Results
CAR expression was associated with CSC phenotype both in vitro and in vivo. CAR-overexpressing cells were more treatment-resistant, self-renewing, and tumorigenic than were parental cells, and shRNA-mediated knockdown of CAR expression was sufficient to inhibit these functions. CAR expression also correlated with the epithelial-mesenchymal transition.
Conclusions
We showed for the first time that CAR is a marker of CSCs and may affect the activities of CSCs in treatment-resistant NSCLC. CAR may prove to be a target for CSC treatment and a predictor of treatment response in patients with NSCLC.
Keywords: Coxsackie-adenovirus receptor, treatment resistance, NSCLC, cancer stem cells, molecular markers
INTRODUCTION
Tumor resistance to radiation or chemotherapy may be caused by the presence of cancer stem cells (CSCs) [1-4], and thus identification of biomarkers of CSCs and translation to CSC-based targeted therapy may represent a viable strategy for overcoming treatment resistance in patients with cancer. CSCs have been identified in and enriched from both clinical samples and cell lines of brain, breast, and colon cancers by techniques such as patterns of cell-surface marker expression [5-10], exclusion of the DNA-binding dye Hoechst 33342 (the so-called “side population” [SP]) [11-13], and increased aldehyde dehydrogenase (ALDH) activity [14,15]. Despite the relatively rapid pace of CSC research in these types of tumors, progress in lung cancer remains slower, in part because of the lack of reliable markers of human lung stem cells [15-17]. The membrane antigen CD133 has been described as a putative biomarker of CSC populations in brain, colon, and lung tumors [9, 10, 18-20], but conflicting results on the detection, abundance, and tumorigenicity of CD133+ cells and on the usefulness of CD133 as a marker of prognosis indicate the need for additional markers to interrogate lung CSCs [21-23]. Moreover, ALDH, which depends on the Notch signaling pathway, was recently found to identify a different CSC subpopulation than CD133 expression [15].
Coxsackie- and adenovirus receptor (CAR) has been well characterized as a cell surface attachment receptor for coxsackie and group C adenoviruses [24]. However, CAR is a multifaceted molecule expressed by many types of cells and is thought to have other cellular functions as well. CAR has been shown to be both a regulator of Notch signaling [25,26], with roles in embryonic development, and an epithelial intercellular adhesion molecule, with reported contributions to mucosal integrity and barrier functions [27,28]. CAR is essential for life; mice because of abnormal myocardial with embryonic stem cell-CAR knockout are non-viable development [29-31].
The contribution of CAR to the malignant phenotype is also under study. CAR is variably expressed in diverse tumors of both epithelial and mesenchymal origin [32-34]. Several reports have sought to link CAR expression with cell differentiation, tumorigenicity, or tumor progression. For example, one group postulated from cell culture models that CAR is a global tumor suppressor in prostate and bladder cancers [32, 33]. Another group found that upregulated CAR expression correlated with tumor progression [35], and another discovered that CAR expression is enhanced in the transition from preneoplastic lesions to invasive adenocarcinoma in a breast cancer model [36]. We previously published our findings of elevated CAR expression in radioresistant stem-like esophageal cancer cells [13]. Others have shown that downmodulation of CAR expression in a non-small cell lung cancer (NSCLC) cell line resulted in diminished colony formation in vitro and diminished tumorigenicity in vivo [37] and that CAR mediates the efficient tumor engraftment of mesenchymal-type lung cancer cells and has a putative role in the epithelial-mesenchymal transition (EMT) [38]. Therefore, CAR may have cell type specific functions that explain the discrepancies between these study findings.
To explore the potential role of CAR in CSCs in treatment-resistant lung cancer, we established paclitaxel-resistant (CR) or radiation-resistant (RR) NSCLC cell lines and found that these cell lines were enriched in CSCs, with significant increases in expression of stem cell markers. More importantly, expression of CAR also was increased in the treatment-resistant cells. We therefore hypothesized that CAR could be a new CSC marker and may have an important role in CSCs in treatment-resistant NSCLC. To test this hypothesis, we determined the effects of overexpressing CAR and inhibiting CAR expression by RNA interference in treatment-resistant NSCLC in vitro and on the tumorigenicity of SP- and CAR-positive (SP/CAR+) RR cells in vivo. This is the first report of the novel role of CAR in CSCs of treatment-resistant NSCLC.
MATERIALS AND METHODS
Cell lines and reagents
Human cell lines A549 and H460 were obtained from the American Type Culture Collection (ATCC) and routinely maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum, 10,000 U/mL of penicillin-streptomycin, and 2 mmol/L-glutamine. Normal human bronchial epithelial cells and normal human fibroblasts were purchased from Clonetics (San Diego, CA) and cultured as recommended by the manufacturer. Mesenchymal stem cells (used as a control in the assessment of inhibitory IC50 doses ) were isolated from bone marrow samples from healthy donors undergoing bone marrow harvest for use in allogeneic bone marrow transplantation (generous gift from Dr. F.C. Marini) and were cultured in α-MEM containing 20% fetal bovine serum, L-glutamine, and penicillin-streptomycin [39].
The identities of these cell lines were validated by short tandem repeat profiling (done by the institutional Characterized Cell Line Core) using the AmpFlSTR Identifiler PCR amplification kit (Applied Biosystems) according to the manufacturer’s instructions. The short tandem repeat profiles for these cell lines matched their known ATCC fingerprints. Paclitaxel was purchased from Bristol-Myers Squibb Co. (Stamford, CT). Hoechst 33342, insulin, and basic fibroblast growth factor were purchased from Sigma-Aldrich (St. Louis, MO), and methylcellulose-based medium from Stem Cell Technologies (Vancouver, BC, Canada). Antibodies against CD133 were purchased from Abcam, Inc. (Cambridge, MA), and epidermal β-catenin antibody from BD Biosciences, Inc. (Franklin Lakes, growth factor and anti-human NJ).
Generation of resistant cell lines
To produce chemoresistant cell lines, H460 and A549 cells were treated initially with 5 nM paclitaxel, and the surviving cells were then treated with increasing doses, up to 100 nM. Radioresistant subpopulations were established by exposing H460 and A549 cells to 2-Gy fractions once a week to a cumulative dose of up to 60 Gy [13].
Construction of the CAR retroviral vector
The cDNA for wild-type CAR was derived by polymerase chain reaction (PCR) using Vent polymerase and then cloned into pCR 2.1 via BamHI and EcoRI restriction sites and confirmed by DNA sequence analysis. The BamHI-EcoRI fragment was then inserted into pBabe-puro to produce pBabe-CAR as follows. pBabe-puro, an MuLV-based retroviral vector, was used to transduce the CAR gene. A consensus Kozak sequence, GCCACC, was incorporated immediately 5 to the translational initiation codon of CAR for optimal protein expression. The CAR oligonucleotide sequences for the upstream and downstream PCR primers were 5-ggatccgcagccaccatggcgctcctgctg-3 and 5-gaattcgctctatactatagacccatc-3. The control vector pGFP, which produces green fluorescent protein (GFP), was constructed in our lab by inserting pGFP and pCMV into pBabe-puro. The pBabe-CAR was introduced into 293PA cells by retroviral vector-mediated transduction; the pGFP control vector was also introduced into 293PA cells by retroviral vector-mediated transduction.
Establishment of stable CAR-overexpressing or shRNA-CAR-knockdown cell lines
H460 cells were infected with a pBabe-CAR retroviral vector or pGFP retroviral vector and selected in a medium containing puromycin for approximately 14 days. During this time, cells not transduced with the vector were eliminated by drug selection. Initially, puromycin-resistant colonies were individually picked, cloned, and expanded in culture. Three weeks after puromycin selection, abundant CAR or GFP expression could be detected and the stable CAR-expressing cell line H460/CAR established. To establish H460/RR/shCAR cells, H460/RR cells were transduced with SMART vector shRNA lentiviral particles targeted against the CAR gene (catalog number SK0036500025; Thermo Fisher Scientific, Lafayette, CO) with 8 μg/mL of polybrene (Sigma-Aldrich) to inhibit CAR gene expression. An empty SMART vector expressing TurboGFP (catalog number S00400001; Thermo Fisher Scientific) was used as a transduction control. The stable shRNACAR-cell line H460/RRshCAR was generated after 2 weeks of culture in puromycin.
Flow cytometry and side population analysis
Expression of β-catenin, Oct3/4, CD133, and CAR was assessed by flow cytometry and SP analysis as described elsewhere [13]. Apoptosis was quantified by flow cytometry [40, 41].
Matrigel invasion assay
The invasive potential of H460/CR and A549/CR cells was assessed with the CytoSelect 24-well cell migration and invasion assay, colorimetric format with fluorescence dye according to the manufacturer’s instructions (Cell Biolabs Inc., San Diego, CA). Each assay was repeated 3 times.
In vivo tumor formation assays
H460/RR cells were used to establish subcutaneous tumors in nu/nu mice as described in the Supplementary Methods. Mice were maintained and experiments done in accordance with NIH and institutional guidelines established for the Animal Core Facility at The University of Texas MD Anderson Cancer Center. Mice (4 per group) were injected subcutaneously with limiting dilutions of 105, 104, or 103 sorted SP or non-SP H460/RR cells and tumor formation was monitored weekly for the ensuing 16 weeks. These cells typically begin forming tumors by 1-2 weeks after inoculation.
Lung cancer sphere formation assay
Cells were suspended in 0.8% methylcellulose-based serum-free medium supplemented with 20 ng/mL epidermal growth factor, basic fibroblast growth factor, and 4 μg/mL insulin and plated at 2,000 cells/mL in ultralow-attachment 24-well plates (Corning Life Sciences, Lowell, MA). Sphere formation was assessed as described in Supplementary Methods.
Statistical analysis
Data were expressed as means with 95% confidence intervals (CIs) or as means ± standard deviations as appropriate. Analysis of variance (ANOVA) and two-tailed Student’s t tests were used to identify significant differences in growth of sorted cells in vivo and in vitro. Differences were considered statistically significant when the P value was 0.05 or less.
RESULTS
Confirmation of resistance and CSCs inchemo- and radiation-resistant NSCLC cell lines
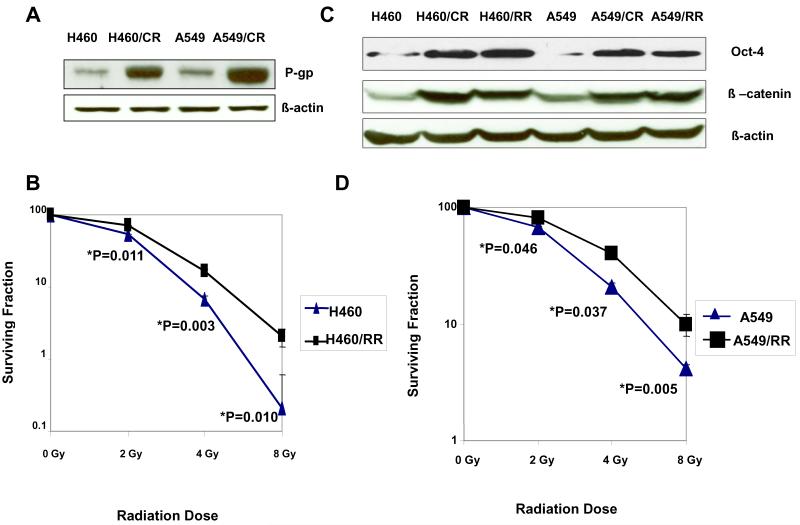
We established two paclitaxel-resistant lung cancer cell lines, H460/CR and A549/CR, by repeatedly treating parental H460 and A549 cells with paclitaxel. The resistance indices (the IC50 for resistant cells/the IC50 for parental cells) were 80 for the H460/CR cells and 12 for the A549/CR cells (Supplementary Table S1). Drug resistance was confirmed by P-glycoprotein expression, which was higher in the resistant variants than in the corresponding parental cells (Fig. 1A) We also established two radiation-resistant cell lines (H460/RR and A549/RR) by irradiating the parental cells in 2-Gy fractions to cumulative doses of up to 60 Gy. Clonogenic assays confirmed that both H460/RR cells (Fig. 1B) and A549/RR cells (Fig. 1D) were significantly more radiation-resistant than their parental counterparts (P<0.05); more H460/RR cells were in G2-M arrest at 48 and 72 hours after irradiation to 4 Gy (24% and 27%) than their irradiated parental counterparts (7% and 6%) (P<0.05); and fewer H460/RR cells were apoptotic (23%) than their parental counterparts (48%) (Supplementary Table S2). Cell-cycle distributions were no different for these two cell lines either before or after irradiation (Table S2).
Figure 1.
Establishing treatment-resistant NSCLC cell lines that express CSC markers. (A) Western blotting of P-glycoprotein (P-gp) expression in parental (H460 and A549) and paclitaxel chemotherapy-resistant cell lines (H460/CR, and A549/CR) show upregulation of P-gp in the resistant cells. (B) Clonogenic survival assays of H460 cells and radiation-resistant H460/RR cells confirm the resistance of the H460/RR variant. To determine surviving fractions, counts were normalized according to the plating efficiency of the unirradiated control. Each point is the mean from three experiments, each done in triplicate (n = 9). (C) Western blots indicates elevated expression of the stem cell survival markers β-catenin and Oct3/4 in the resistant cells. β-Actin was used as a loading control. These experiments were repeated 3 times with similar results. (D) Clonogenic survival assays of A549 cells and radiation-resistant A549/RR cells confirm the resistance of the A549/RR variant.
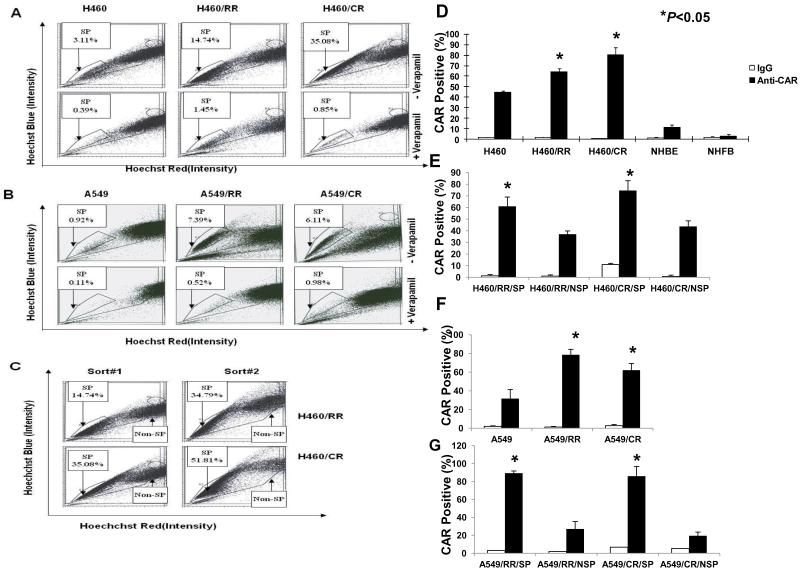
We further confirmed that both the chemosensitive and radiosensitive variants contained CSCs in a series of experiments as follows. Nuclear accumulation of β-catenin (a key event in stem cell activation) and expression of Oct3/4 (a putative embryonic marker) were increased in both the chemoresistant and radioresistant variants compared with the parental cell lines, suggesting that these treatment-resistant subpopulations contained CSCs (Fig. 1C). We also found that the proportions of SP cells (those that could exclude Hoechst 33342 dye) were higher in both the chemoresistant and radioresistant subpopulations than in the parental cells (P<0.05 for both H460 and A549 cells; Figs. 2A and B). In each case, the SP population was greatly decreased by treatment with verapamil (Figs. 2A and B), confirming that the populations were indeed SPs. Serial sorting and reanalysis of SP cells from cultured SP cells demonstrated enrichment of SP cells and production of non-SP cells in both the H460/CR and H460/RR variants (Fig. 2C), indicating the capacity for self-renewal. However, the non-SP cells produced mainly other non-SP cells (data not shown), suggesting asymmetric division.
Figure 2.
Enrichment of side population (SP) cells in established radioresistant (RR) and paclitaxel-resistant (CR) lung cancer cell lines. (A) and (B), H460, H460/RR, and H460/CR cells (A) or A549, A549/RR, and A549/CR cells (B) were labeled with Hoechst 33342 dye and the percentage of SP cells determined by flow cytometry before and after treatment with verapamil; all resistant cell lines demonstrated enrichment of SP cells. (C) Flow cytometric analysis of H460/RR and H460/CR SP cells after 7-10 days of culture followed by re-sorting (by flow cytometry) and a second 7- to 10-day culture revealed progressive enrichment of SP cells as well as the presence of non-SP cells. Data shown represent one of three independent experiments. (D) CAR expression measured by flow cytometry and specific fluorescein isothiocyanate conjugated anti-CAR antibodies revealed increased proportions of CAR-positive cells among the resistant variants but not in normal human bronchial epithelial cells (NHBE) or normal human fibroblasts (NHFB). IgG was used as a control. (E) CAR expression was increased in treatment-resistant H460 cells in the side population (SP) subset but not in the non-SP subset. (F) CAR expression was also increased in the resistant variants of A549 cells and in the SP subset Relative to the non-SP cells (G). Columns in (D) through (G) represent means of three independent experiments; bars are 95% confidence intervals.
We further confirmed the greater clonogenic efficiency of H460/CR cells (26.2% vs. 15.1% for H460 cells) and A549/CR cells (31.7% vs. 18.1% for A549 cells) and in H460/CR-SP cells (38.9%) and A549/CR-SP cells (59.7%) (Figs. S1A and B). A Matrigel assay revealed that H460/CR-SP and A549/CR-SP cells were significantly more invasive than their parental counterparts (P<0.05, Figs. S1C and D), another characteristic of stem cells. We further measured telomerase activity in the H460 variants and found that that activity was higher in the H460/RR and H460/CR than in the H460 cells (P<0.05) and was higher in the sorted H460/RR-SP and H460/CR-SP cells than in the H460/RR-non-SP and H460/CR-non SP cells (P<0.05; Fig. S1E). Notably, the telomerase activity of all of these cancer cell lines was considerably higher than that of normal cell lines (P<0.001).
Finally, we also tested the tumorigenicity of SP cells in vivo and found that the difference in tumor incidence was greatest for the lowest dilution of tumor cells tested: whereas 103 SP cells could generate tumors in mice, the same number of non-SP cells failed to generate any detectable tumors (Table 1), indicating that H460/RR-SP cells were enriched in tumor-initiating cells. Tumors generated from 105 H460/RR-SP cells were also larger and grew faster than tumors from their non-SP counterparts (P<0.05, data not shown). These results, in combination with those of the in vitro experiments, indicate that the treatment-resistant SP cells (H460/RR-SP, H460/CR-SP, A549/RR-SP, and A549/CR-SP) all have characteristics of CSCs.
Table 1.
Tumorigenicity of cell line variants in nude mice according to side population and expression of coxsackie- and adenovirus receptor
| No. of Cells Innoculated | |||
|---|---|---|---|
| Cell Variant | 105 | 104 | 103 |
| H460 | 1 /4 | 0/4 | 0/4 |
| H460/RR | 3/4 | 2/4 | 1 /4 |
| H460/RR/SP | 4/4 | 3/4 | 2/4 |
| H460/RR/NSP | 3/4 | 1 /4 | 0/4 |
| H460/RR/SP/CAR+ | 4/4 | 4/4 | 3/4 |
| H460/RR/SP/CAR− | 4/4 | 2/4 | 0/4 |
| H460/RR/NSP/CAR+ | 4/4 | 2/4 | 0/4 |
| H460/RR/NSP/CAR− | 2/4 | 0/4 | 0/4 |
RR, radioresistant; SP, side population; CAR, coxsackie- and adenovirus receptor.
Values are numbers of tumor produced /number of mice.
Overexpression of CAR and tumorigenicity in treatment-resistant NSCLC cells and CSCs
Our previous finding that CAR was overexpressed in radioresistant stem-like esophageal adenocarcinoma cells [13] led us to investigate whether CAR could be a novel marker of CSCs in treatment-resistant NSCLC. We tested this hypothesis by measuring the expression of CAR in parental and treatment-resistant H460 cells (adenocarcinoma) and 2 normal human lung cell lines (bronchial epithelial cells and lung fibroblasts). Expression of CAR was significantly higher in the H460/RR and H460/CR cells (P<0.05; Fig. 2D); CAR expression was also higher in H460/RR/SP and H460/CR/SP cells than in their non-SP counterparts (P<0.05; Fig. 2E). Similar results were observed for A549 cells (P<0.05; Figs. 2F and G).
Next, to determine whether expression of CAR affected tumorigenicity, we isolated H460/RR cells expressing or not expressing CAR from SP and non-SP populations by flow cytometry and implanted them subcutaneously at limiting dilutions of 105, 104, or 103 cells into nude mice. SP/CAR+ cells were significantly more tumorigenic than SP/CAR−, non-SP/CAR+, or non-SP/CAR-cells (Table 1). Notably, no tumors were observed with SP/CAR-cells unless at least 104 cells were injected (2 of 4 mice); all 4 mice injected with 104 SP/CAR+ cells developed tumors. Similar results were obtained with non-SP/CAR+ and non-SP/CAR-cells.
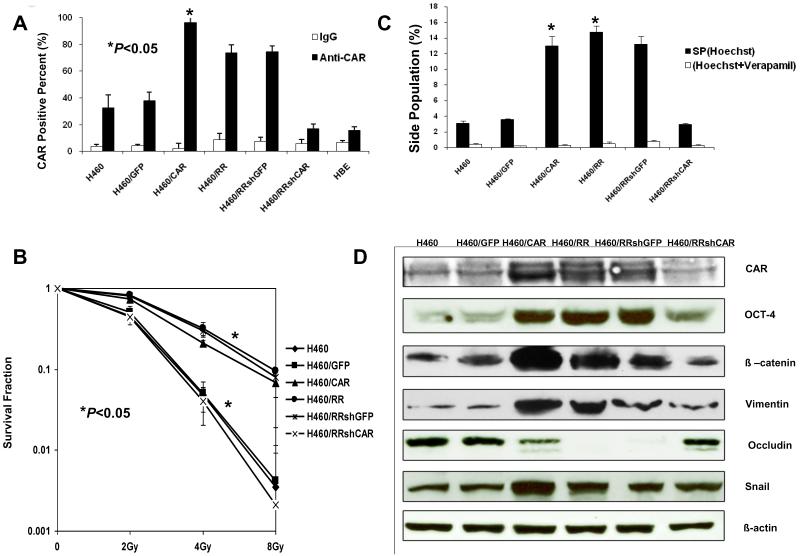
CSC properties in CAR-overexpressing stable cell lines
To explore the role of CAR in CSCs, we constructed a pBabe-CAR retroviral vector and transduced it into parental H460 cells to establish a CAR-overexpressing stable cell line (H460/CAR). Relative proportions of cell variants expressing CAR are shown in Figure 3A. The percentage of H460/RR cells positive for CAR (73.6 ± 4.1%) was significantly higher than that of H460 cells (32.5 ± 3.6%, P<0.05), and percentages in both cell lines were higher than that of normal human bronchial epithelial cells. Moreover, the percentage of cells positive for CAR in the H460/CAR stable cell line (94.4 ± 5.5%) was the highest of all cell lines tested in this experiment. Clonogenic survival curves showed that the H460/CAR cells were significantly more radioresistant than H460 cells or the H460/GFP vector controls (P<0.05; Fig. 3B). Moreover, significantly greater percentages of H460/CAR cells (11.99%) and H460/RR cells (13.38%) were SP cells than H460 (1.33%) or H460/GFP cells (0.97%, P<0.05; Fig. 3C), indicating enrichment of CSCs in H460/RR and H460/CAR. More importantly, H460/RR and H460/CAR cells overexpressed markers associated with CSCs, such as OCT-4 and β-catenin (Fig. 3D).
Figure 3.
Confirmation of CSC properties in CAR-overexpressing stable cell lines and shRNA-mediated CAR knockdown stable cell lines. (A) Transduction of H460/RR cells with P-Babe-CAR led to overexpression of CAR and transduction with shRNA suppressed CAR expression in resistant variants of H460 cells. GFP was used as a transduction control. Columns indicate means of three independed experiments, repeated three times; and error bars are SDs (B) Clonogenic survival assay shows increased survival of the resistant variants H460/RR and H460/CAR and correspondingly lower survival of the H460/RR/shCAR cells and H460. Error bars are SDs. (C) Flow cytometric analysis indicates enriched SP cells in the resistant variant H460/RR and the H460/RR/CAR cells, but not in the shCAR cells. Asterisks indicate significant differences; error bars are SDs. (D) Western blots of the H460 variants with antibodies to markers of stem cells (CAR, OCT-4, β-catenin) and the epithelial-mesenchymal transition pathway (vimentin, occludin, SNAIL) show that H460/RR and H460/CAR cells overexpressed markers of CSCs and mesenchymal proteins (β-catenin, vimentin, SNAIL) but not the epithelial marker occludin, but shCAR did not. β-Actin was used as a loading control.
Effects of inhibiting CAR expression in CSCs of treatment-resistant NSCLC
To further confirm that the characteristics of CSCs described in the previous paragraph were a specific consequence of CAR expression, we transduced H460/RR cells with SMART vector shRNA lentiviral particles targeted against CAR to inhibit expression of the gene. This shRNA-mediated knockdown of CAR resulted in significantly lower CAR protein levels (Fig. 3A) and lesser expression of the CSC marker proteins OCT-4 and β-catenin (Fig. 3D) in H460/RR/shCAR stable cells compared with H460/RR/shGFP cells. Radioresistance and proportions of SP cells were both significantly lower in H460/RR/shCAR than in H460RR and H460/RR/shGFP cells (P<0.05, Figs. 3B and C).
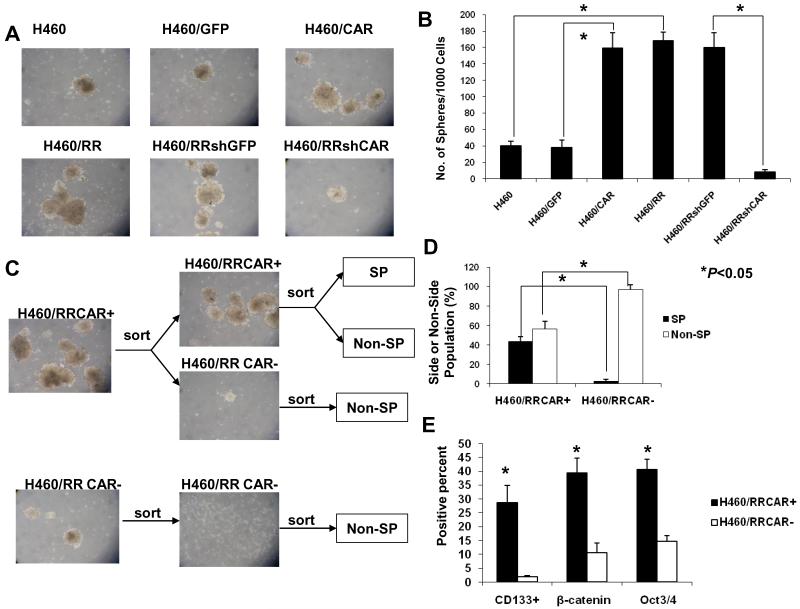
We further tested whether H460/CAR and H460/RR/shCAR cells could form spheres when grown in suspension cultures under stem cell selective conditions, an in vitro measure of CSC activity. The H460/CAR cells formed 4 times more spheres than the H460/GFP or H460 cells and produced nearly the same numbers of spheres as the H460/RR cells (Figs. 4A and B). These findings, plus the observation that shCAR almost completely blocked sphere formation, indicate that CAR could be an important marker of CSCs. This conclusion was supported by results of the self-renewal assessment, in which equal numbers of H460/RR/CAR+ and H460/RR/CAR-cells were sorted and cultured separately under the stem cell selective conditions (Fig. 4C). As expected, cultures initiated with H460/ RR/CAR+ cells reproduced the parental distribution of CAR+ and CAR-cells, whereas the cultures derived from CAR-cells generated primarily CAR-cells. Furthermore, H460/RRCAR+ cells generated both SP and non-SP cells, whereas H460RR/CAR-cells generated only non-SP cells (Fig. 4D). Finally, more of the H460/RR/CAR+ cells expressed CD133+ (28.8%), β-catenin (39.4%), and Oct3/4 (40.7%) than did H460/RR/CAR-cells (1.9%, 10.6%, and 14.7%, respectively (P<0.05; Fig. 4E).
Figure 4.
CAR and growth of lung CSCs. (A) Phase-contrast microscopy images of tumor spheres grown from the indicated cell lines under stem cell selective conditions for 10 days reveal increased sphere formation among the resistant cells and the CAR-overexpressing cells; shCAR, in contrast, blocked sphere formation. (B) Quantitiave comparison of numbers of spheres formed by the indicated cell line variants under stem cell selective conditions for 2 weeks. (C) Phase-contrast microscopy images of spheres formed by CAR+ and CAR-H460/RR cells. CAR+ cells were more tumorigenic and had greater self-renewal capability than CAR-cells. (D) CAR+ and CAR-cells (isolated by fluorescence-activated cell sorting from H460/RR) were grown in culture for 2 weeks and reanalyzed for proportions of SP cells and non-SP cells. The CAR+ cells produced more SP cells (42.5%) than the CAR-cells (3.1%); the CAR-cells produced predominantly non-SP cells, indicating asymmetric division. (E) Quantitative comparison of CSC marker expression confirmed that the H460/RR/CAR+ cells expressed these markers but the H460/RR/CAR-cells did not. Columns are the means of three independent experiments; error bars are 95% confidence intervals.
CAR expression and EMT
Our observation that CAR-expressing subpopulations (H460/RR/CAR+ and H460/CAR) resemble mesenchymal cells, and that low-CAR-expressing cells (H460/RR/CAR- and H460/RR/shCAR) are more representative of epithelioid cells (Fig. 2S), prompted us to consider whether CAR is implicated in the EMT. To determine whether expression of CAR was associated with molecular characteristics of the EMT, we used western blorring to assess markers of EMT in H460/CAR and H460/RR/shCAR cells. We found that the CAR-overexpressing cells expressed primarily mesenchymal proteins (e.g., β-catenin, vimentin, SNAIL) and little to no epithelial cytokeratins (e.g., occludin) (Figs. 3D and 4E). In contrast, none of these mesenchymal proteins were upregulated in H460/RR/shCAR cells (Figs. 3D and 4E). Hence we conclude that CAR expression correlates with the EMT.
DISCUSSION
We showed in this study that CAR can be used to identify and enrich a subpopulation of treatment-resistant NSCLC cells having many of the properties ascribed to CSCs [37,42], suggesting that CAR can be considered a novel marker of CSCs and may be important in the activity of CSCs associated with treatment-resistant NSCLC. We also found that CAR expression was associated with the EMT.
Our experimental findings may have relevance with regard to the clinical treatment of NSCLC, which typically involves combined chemotherapy and radiation. Chemotherapy is typically administered in cycles that are separated by 3-week intervals to allow restoration of hematopoietic and other normal cells damaged by the drugs. However, some reports indicate that tumor cells can aggressively repopulate during these intervals and restore the tumor to its pretreatment size [43]. Our experimental findings lead us to suspect that conventional chemotherapy may increase the proportion of CSCs by eliminating drug-sensitive, differentiated tumor cells, which could also be restored during the 3-week intervals between chemotherapy cycles. Development of radioresistance after standard-fractionation radiotherapy parallels this clinical situation [13]. We showed here that treatment with paclitaxel or fractioned radiation can actually enrich CSCs. Another report that the enzymatic activity of the putative stem cell marker ALDH1 correlates with radiation-induced radioresistance in cancer cell lines [4] suggests that, in cases where chemotherapy or radiation treatment fails to completely eradicate the disease, the residual cancer cells could promote recurrence because of persistent CSCs. This notion is supported by clinical observations that, after conventional chemotherapy, breast tumors have an increased proportion of cells with a CD44hi/CD24lo marker profile and show increased sphere-forming ability [44]. Collectively, these findings suggest that further characterization of CSCs and their markers, as well as investigations into the mechanisms by which they mediate treatment resistance, may provide key information on relevant signaling pathways to be targeted to increase the therapeutic response.
Several lines of evidence support CAR as a novel CSC marker that could be used to predict treatment-resistant NSCLC. First, CAR expression was significantly greater in treatment-resistant NSCLC cells than in their parental counterparts and in 2 normal lung cell lines (Figs. 1A and 3A), particularly for the SP+ population (Figs. 2B and C). These results are consistent with our previous report that radiation-resistant SP cells expressed higher CAR levels than did non-SP cells from an esophageal adenocarcinoma line [13]. Moreover, downregulation of CAR expression in an NSCLC cell line has been shown to lead to diminished in vivo tumorigenicity and in vitro colony formation [37] and CAR expression was shown to mediate efficient tumor engraftment (which correlated with invasive mesenchymal phenotypes) in lung cancer cell lines [38]. More importantly, compared with their CAR-counterparts, isolated CAR+ treatment-resistant NSCLC cells were more tumorigenic (Table 1) and were able to regenerate both CAR+ and CAR-cells. Our observation that CAR-H460/RR cells formed much smaller and slower-growing tumors in mice than CAR+ H460/RR cells could indicate residual CSCs in CAR-cells or contamination with some CAR+ cells. We further verified the importance of CAR in CSCs in treatment-resistant NSCLC by overexpressing CAR (in CAR-transduced stable cells) and by inhibiting CAR expression (by shRNA). We analyzed the expression of CAR and molecular features involving targetable signaling pathways known to be involved in stem cell physiology and found that expression of CAR was associated with β-catenin and EMT signaling pathways, which we confirmed by overexpressing and silencing CAR in stable cell lines (Fig. 3D). β-Catenin is an essential component of both intercellular junctions and the canonical Wnt signaling pathway that has been implicated in stem cell survival. Overexpression of β-catenin leads to self-renewal of stem cells and tumorigenesis [45]. Cell-surface staining of CAR has been shown to overlap and coimmunoprecipitate with the adherent junction protein β-catenin in A549 cells [28], and CAR expression has been associated with increased expression of Zeb1, SNAIL, and vimentin in mesenchymal-type lung cancer cells [38].
Overexpression of vimentin and SNAIL is traditionally used to identify cells that have undergone the EMT [46], a key developmental program that is often activated during cancer invasion and metastasis [47] and promotes the generation of CSCs [48]. Here we provided functional validation that increasing CAR expression in a lung cancer adenocarcinoma cell (H460/CAR and H460/RR) induced EMT in two ways: first, by showing that expression of mesenchymal proteins (β-catenin, vimentin, SNAIL) was markedly upregulated by CAR overexpression, whereas expression of epithelial cytokeratins (e.g., occludin) was downregulated; and second, by showing that inhibiting CAR expression in the same lung cancer cell line (H460/RR/shCAR) induced MET: expression of epithelial cytokeratins was markedly upregulated, whereas expression of mesenchymal proteins was downregulated (Figs. 3D and 4E). Collectively, our findings implicate CAR during the EMT, although the mechanisms underlying this role remain elusive.
At this time, we cannot say if our results will prove to be applicable in the clinical treatment of patients with NSCLC. We are conducting further studies to determine whether CAR+ cells from samples derived from patients with NSCLC treated with chemotherapy or radiotherapy can be expanded or generate long-term cultures in vitro. Those findings will allow us to take the next step to validate CAR as a potential target for CSC treatment and as a predictor of treatment response in the clinical setting of NSCLC. In the meantime, the findings from the current study deepen our understanding of the tumor biology and tumorigenesis of lung cancer, and they may allow development of a novel approach to predicting response to treatment and guiding personalized CSC targeted therapy for treatment-resistant NSCLC.
Supplementary Material
ACKNOWLEDGMENTS
Financial support for this work was provided by the Radiological Society of North America Research Scholar Award (J.Y.C.), a Career Development Award from The University of Texas Lung Cancer Specialized Programs of Research Excellence grant from the National Cancer Institute (J.Y.C.), National Cancer Institute grants R01 CA 092487 and R01 CA 098582 (B.F.) and Cancer Center Core Grant CA016672, and Lockton Grant-Matching Funds (B.F.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest with regard to this work.
REFERENCES
- [1].Baumann M, Krause M, Hill R. Exploring the role of cancer stem cells in radioresistance. Nat Rev Cancer. 2008;8:545–54. doi: 10.1038/nrc2419. [DOI] [PubMed] [Google Scholar]
- [2].Brunner TB, Kunz-Schughart LA, Grosse-Gehling P, Baumann M. Cancer stem cells as a predictive factor in radiotherapy. Semin Radiat Oncol. 2012;22:151–74. doi: 10.1016/j.semradonc.2011.12.003. [DOI] [PubMed] [Google Scholar]
- [3].Krause M, Yaromina A, Eicheler W, Koch U, Baumann M. Cancer stem cells: targets and potential biomarkers for radiotherapy. Clin Cancer Res. 2011;17:7224–9. doi: 10.1158/1078-0432.CCR-10-2639. [DOI] [PubMed] [Google Scholar]
- [4].Mihatsch J, Toulany M, Bareiss PM, et al. Selection of radioresistant tumor cells and presence of ALDH1 activity in vitro. Radiother Oncol. 2011;99:300–6. doi: 10.1016/j.radonc.2011.06.003. [DOI] [PubMed] [Google Scholar]
- [5].Al-Hajj M, Wicha M, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–88. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65:10946–51. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- [7].Hosen N, Park C, Tatsumi N, et al. CD96 is a leukemic stem cell-specific marker in human acute myeloid leukemia. Proc Natl Acad Sci U S A. 2007;104:11008–13. doi: 10.1073/pnas.0704271104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Li C, Heidt DG, Dalerba P, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–37. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- [9].Ricci-Vitiani L, Lombardi DG, Pilozzi E, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–15. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- [10].Richardson GD, Robson CN, Lang SH, Neal DE, Maitland NJ, Collins AT. CD133, a novel marker for human prostatic epithelial stem cells. J Cell Sci. 2004;117:3539–45. doi: 10.1242/jcs.01222. [DOI] [PubMed] [Google Scholar]
- [11].Hadnagy A, Gaboury L, Beaulieu R, Balicki D. SP analysis may be used to identify cancer stem cell populations. Exp Cell Res. 2006;312:3701–10. doi: 10.1016/j.yexcr.2006.08.030. [DOI] [PubMed] [Google Scholar]
- [12].Ho MM, Ng AV, Lam S, Hung JY. Side population in human lung cancer cell lines and tumors is enriched with stem-like cancer cells. Cancer Res. 2007;67:4827–33. doi: 10.1158/0008-5472.CAN-06-3557. [DOI] [PubMed] [Google Scholar]
- [13].Zhang X, Komaki R, Wang L, Fang B, Chang JY. Treatment of radioresistant stem-like esophageal cancer cells by an apoptotic gene-armed, telomerase-specific oncolytic adenovirus. Clin Cancer Res. 2008;14:2813–23. doi: 10.1158/1078-0432.CCR-07-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Christ O, Lucke K, Imren S, et al. Improved purification of hematopoietic stem cells based on their elevated aldehyde dehydrogenase activity. Haematologica. 2007;92:1165–72. doi: 10.3324/haematol.11366. [DOI] [PubMed] [Google Scholar]
- [15].Sullivan JP, Spinola M, Dodge M, et al. Aldehyde dehydrogenase activity selects for Notch signaling dependent non-small cell lung cancer stem cells. Cancer Res. 2010;70:9937–48. doi: 10.1158/0008-5472.CAN-10-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kim CF, Jackson EL, Woolfenden AE, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–35. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- [17].Giangreco A, Groot KR, Janes SM. Lung cancer and lung stem cells Strange bedfellows? Am J Resp Crit Care Med. 2007;175:547–53. doi: 10.1164/rccm.200607-984PP. [DOI] [PubMed] [Google Scholar]
- [18].Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- [19].Eramo A, Lotti F, Sette G, et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008;15:504–14. doi: 10.1038/sj.cdd.4402283. [DOI] [PubMed] [Google Scholar]
- [20].Bertolini G, Roz L, Perego P, et al. Highly tumorigenic lung cancer CD133+ cells display stem like features and are spared by cisplatin treatment. Proc Natl Acad Sci U S A. 2009;106:16281–86. doi: 10.1073/pnas.0905653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang J, Sakariassen PØ, Tsinkalovsky O, et al. CD133 negative glioma cells form tumors in nude rats and give rise to CD133 positive cells. Int J Cancer. 2008;122:761–68. doi: 10.1002/ijc.23130. [DOI] [PubMed] [Google Scholar]
- [22].Shmelkov SV, Butler JM, Hooper AT, et al. CD133 expression is not restricted to stem cells, and both CD133+ and CD133-metastatic colon cancer cells initiate tumors. J Clin Invest. 2008;118:2111–20. doi: 10.1172/JCI34401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sullivan JP, Minna JD, Shay JW. Evidence for self-renewing lung cancer stem cells and their implications in tumor initiation, progression, and targeted therapy. Cancer Metastasis Rev. 2010;29:61–72. doi: 10.1007/s10555-010-9216-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bergelson JM, Cunningham JA, Droguett G, et al. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2. Science. 1997;275:1320–23. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- [25].Sollerbrant K, Raschperger E, Mirza M, et al. The coxsackievirus adenovirus receptor (CAR) forms a complex with the PDZ domain-containing protein Ligand-of-Numb protein-X (LNX) J Biol Chem. 2003;278:7439–44. doi: 10.1074/jbc.M205927200. [DOI] [PubMed] [Google Scholar]
- [26].Mirza M, Raschperger E, Philipson L, Pettersson RF, Sollerbrant K. The cell surface protein coxsackie-adenovirus receptor (CAR) directly associates with the Ligand-of-Numb Protein-X2 (LNX2) Exp Cell Res. 2005;309:110–20. doi: 10.1016/j.yexcr.2005.05.023. [DOI] [PubMed] [Google Scholar]
- [27].Cohen CJ, Shieh JT, Pickles RJ, Okegawa T, Hsieh JT, Bergelson JM. The coxsackievirus adenovirus receptor is a transmembrane component of the tight junction. Proc Natl Acad Sci USA. 2001;98:15191–96. doi: 10.1073/pnas.261452898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Walters RW, Freimuth P, Moninger TO, Ganske I, Zabner J, Welsh MJ. Adenovirus fiber disrupts CAR-mediated intercellular adhesion allowing virus escape. Cell. 2002;110:789–99. doi: 10.1016/s0092-8674(02)00912-1. [DOI] [PubMed] [Google Scholar]
- [29].Dorner AA, Wegmann F, Butz S, et al. Coxsackievirus-adenovirus receptor (CAR) is essential for early embryonic cardiac development. J Cell Sci. 2005;118:3509–21. doi: 10.1242/jcs.02476. [DOI] [PubMed] [Google Scholar]
- [30].Asher DR, Cerny AM, Weiler SR, et al. Coxsackievirus adenovirus receptor is essential for cardiomyocyte development. Genesis. 2005;42:77–85. doi: 10.1002/gene.20127. [DOI] [PubMed] [Google Scholar]
- [31].Chen JW, Zhou B, Yu QC, et al. Cardiomyocyte-specific deletion of the coxsackievirus adenovirus receptor results in hyperplasia of the embryonic left ventricle abnormalities of sinuatrial valves. Circ Res. 2006;98:923–30. doi: 10.1161/01.RES.0000218041.41932.e3. [DOI] [PubMed] [Google Scholar]
- [32].Okegawa T, Li Y, Pong RC, Bergelson JM, Zhou J, Hsieh JT. The dual impact of coxsackie adenovirus receptor expression on human prostate cancer gene therapy. Cancer Res. 2000;60:5031–36. [PubMed] [Google Scholar]
- [33].Okegawa T, Pong RC, Li Y, Bergelson JM, Sagalowsky AI, Hsieh JT. The mechanism of the growth-inhibitory effect of coxsackie adenovirus receptor (CAR) on human bladder cancer: a functional analysis of CAR protein structure. Cancer Res. 2001;61:6592–6600. [PubMed] [Google Scholar]
- [34].Cripe TP, Dunphy EJ, Holub AD, et al. Fiber knob modifications overcome low heterogeneous expression of the coxsackievirus-adenovirus receptor that limits adenovirus gene transfer oncolysis for human rhabdomyosarcoma cells. Cancer Res. 2001;61:2953–60. [PubMed] [Google Scholar]
- [35].Anders M, Hansen R, Ding RX, Rauen KA, Bissell MJ, Korn WM. Disruption of 3D tissue integrity facilitates adenovirus infection by deregulating the coxsackievirus adenovirus receptor. Proc Natl Acad Sci USA. 2003;100:1943–48. doi: 10.1073/pnas.0337599100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Bruning A, Stickeler E, Diederich D, et al. Coxsackie adenovirus receptor promotes adenocarcinoma cell survival is expressionally activated after transition from preneoplastic precursor lesions to invasive adenocarcinomas. Clin Cancer Res. 2005;11:4316–20. doi: 10.1158/1078-0432.CCR-04-2370. [DOI] [PubMed] [Google Scholar]
- [37].Qin M, Escuadro B, Dohadwala M, Sharma S, Batra RK. A novel role for the coxsackie adenovirus receptor in mediating tumor formation by lung cancer cells. Cancer Res. 2004;64:6377–80. doi: 10.1158/0008-5472.CAN-04-1490. [DOI] [PubMed] [Google Scholar]
- [38].Veena MS, Qin M, Andersson A, Sharma S, Batra RK. CAR mediates efficient tumor engraftment of mesenchymal type lung cancer cells. Lab Invest. 2009;89:875–86. doi: 10.1038/labinvest.2009.56. [DOI] [PubMed] [Google Scholar]
- [39].Studeny M, Marini FC, Dembinski JL, et al. Mesenchymal stem cells: Potential precursors for tumor stroma and targeted-delivery vehicles for anticancer agents. J Natl Cancer Inst. 2004;96:1593–1603. doi: 10.1093/jnci/djh299. [DOI] [PubMed] [Google Scholar]
- [40].Zhang X, Cheung RM, Komaki R, Fang B, Chang JY. Radiotherapy sensitization by tumor-specific TRAIL gene targeting improves survival of mice bearing human non-small cell lung cancer. Clin Cancer Res. 2005;11:6657–68. doi: 10.1158/1078-0432.CCR-04-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Chang JY, Zhang X, Komaki R, Cheung R, Fang B. Tumor-specific apoptotic gene targeting overcomes radiation resistance in esophageal NSCLC. Int J Radiat Oncol Biol Phys. 2006;64:1482–94. doi: 10.1016/j.ijrobp.2005.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Sullivan JP, Minna JD, Shay JW. Evidence for self-renewing lung cancer stem cells and their implications in tumor initiation, progression, and targeted therapy. Cancer Metastasis Rev. 2010;29:61–72. doi: 10.1007/s10555-010-9216-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kim JJ, Tannock IF. Repopulation of cancer cells during therapy: an important cause of treatment failure. Nat Rev Cancer. 2005;5:516–25. doi: 10.1038/nrc1650. [DOI] [PubMed] [Google Scholar]
- [44].Li X, Lewis MT, Huang J, et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst. 2008;100:672–79. doi: 10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]
- [45].Reya T, Duncan AW, Ailles L, et al. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003;423:409–14. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- [46].Roussos ET, Keckesova E, Haley JD, Epstein DM, Weinberg RA, Condeelis JS. AACR special conference on epithelial-mesenchymal transition and cancer progression and treatment. Cancer Res. 2010;70:7360–64. doi: 10.1158/0008-5472.CAN-10-1208. [DOI] [PubMed] [Google Scholar]
- [47].Gupta PB, Onder TT, Jiang G, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–59. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Mani SA, Guo W, Liao MJ, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.