Abstract
Human CD1a mediates foreign antigen recognition by a T cell clone, but the nature of possible T cell receptor interactions with CD1a-lipid are unknown. After incubating CD1a with a mycobacterial lipopeptide antigen, dideoxymycobactin (DDM), we identified and measured binding to a recombinant TCR (TRAV3/ TRBV3-1, KD ≈ 100µM). Detection of ternary CD1a-lipid-TCR interactions enabled development of CD1a tetramers and CD1a multimers with carbohydrate backbones (dextramers), which specifically stained T cells using a mechanism that was dependent on the precise stereochemistry of the peptide backbone and was blocked with a soluble TCR. Further, sorting of human T cells from unrelated tuberculosis patients for bright DDM-dextramer staining allowed recovery of T cells that were activated by CD1a and DDM. These studies demonstrate that the mechanism of T cell activation by lipopeptides occurs via ternary interactions of CD1a-antigen-TCR. Further, these studies demonstrate the existence of lipopeptide specific T cells in humans ex vivo.
Keywords: human, lipopeptide, antigens, T cell receptors, CD1
Introduction
Humans express four CD1 antigen presenting molecules, CD1a, CD1b, CD1c and CD1d [1]. Whereas the basic biology of CD1d and NKT cells has been extensively studied in mice lacking either CD1d or invariant NKT TCRs [2], CD1a proteins have been deleted from the murine genome [3], effectively limiting analysis to the study of a few human T cell clones propagated long-term in vitro [4, 5]. The mycobacterial lipopeptide, dideoxymycobactin (DDM), is the only well characterized foreign antigen in the CD1a system and has served as a model to understand the specificity and mechanism of action of CD1a-restricted T cells in vitro [4, 6, 7]. Building on the success of human MHC class I tetramers to quantitatively track fresh human T cells ex vivo [8], CD1 tetramers [9–16] capture lipid reactive T cells as populations for study ex vivo. Dextramers rely on the same principle, but use higher order multimers which allow detection of rare, lower affinity T cells [17]. In this study, we investigated whether human TCRs directly bind to CD1a-lipopeptide in order to develop tetramers and dextramers that identify polyclonal T cell populations, which recognize foreign antigen bound to CD1a.
Materials and Methods
Generation of soluble CD1a proteins
We produced soluble biotinylated CD1a monomers in lentivirus-transduced HEK293 T cells at the NIH Tetramer Core Facility (Emory University, Atlanta, GA) [14, 16].
Loading CD1a monomers with dideoxymycobactin
Dideoxymycobactin with defined stereochemistry (DDM 1S3R) or (DDM 1R3S) [7] was incubated with CD1a monomers and complexed with streptavidin coupled to allophycocyanin. Staining was optimized by analyzing T cells with CD1a tetramers generated after loading under varied conditions (Supplemental Figure 1). Optimal staining was seen with DDM solubilized in DMSO, sonicated into 30-fold excess 50 mM sodium citrate plus 1% CHAPS at pH 6 for 2 minutes, incubated at 42°C for one hour then added at 40-fold molar excess to CD1a monomers and incubated in a 37°C water bath for 1 hour prior to neutralization to pH 7.4 with 2ul TRIS pH 9.
CD1a tetramer and dextramer staining of clones
CD1a tetramers were generated and validated by staining the DDM-reactive αβ T cell clone CD8-2 [18] and using methods similar to those reported for CD1b tetramers [14]. We generated dextramers by diluting CD1a complexes to 0.1mg/ml in phosphate-buffered saline prior to incubation with 2µl of allophycocyanin-labelled dextramer backbone for 30 minutes at room temperature in the dark, followed by staining of T cells as previously reported [17]. For dual staining with TCR antibodies, TRBV3-1 (Beckman Coulter) was added after tetramer staining for the last 15 minutes of incubation. Cells were acquired on a FACSCanto II flow cytometer (Beckton Dickinson) and analyzed using Flowjo (Tree Star) software in the presence or absence of monoclonal antibodies or recombinant TCRs.
TCR affinity measurements
The cDNAs of the α and β-chains of the CD1b-restricted TCR LDN5 and the CD1a-restricted, dideoxymycobactin-specific TCR CD8-2 were produced as previously described [14, 19]. Loaded CD1a-DDM was coupled to research-grade streptavidin-coated chips. Increasing concentrations of the CD8-2 TCR (0–533 μM) were injected over all flow cells. BIAevaluation version 3.1 software (Biacore AB) was used to fit the data to the 1:1 Langmuir binding model and the equilibrium data were analyzed with the Prism program (GraphPad).
Dextramer staining of human peripheral blood mononuclear cells (PBMC)
Work with human subjects was overseen by the institutional review boards of the Lemuel Shattuck Hospital (00000786), Partners Healthcare (2002-P-000061) and the Harvard Committee on Microbiologic Safety (08–184). PBMC were treated with dextramers at a 1 to 100 dilution for 15 minutes at room temperature followed by 15 minutes at 4°C, after which cells were washed and incubated with violet viability dye, CD3-FITC, CD14-PerCP-Cy5.5 and CD19 PerCP-Cy5.5 for 15 minutes at 4°C. Unfixed dextramer-positive cells were sorted using a FACSAria flow cytometer and expanded by stimulation with anti-CD3 (30ng/ml) in the presence of irradiated feeder cells and IL-2 (2nM). After 3 weeks, clones were analyzed for binding CD1a dextramers and were tested for DDM recognition in an ELISPOT assay [20].
Results and Discussion
Generation of antigenic CD1a-lipopeptide complexes
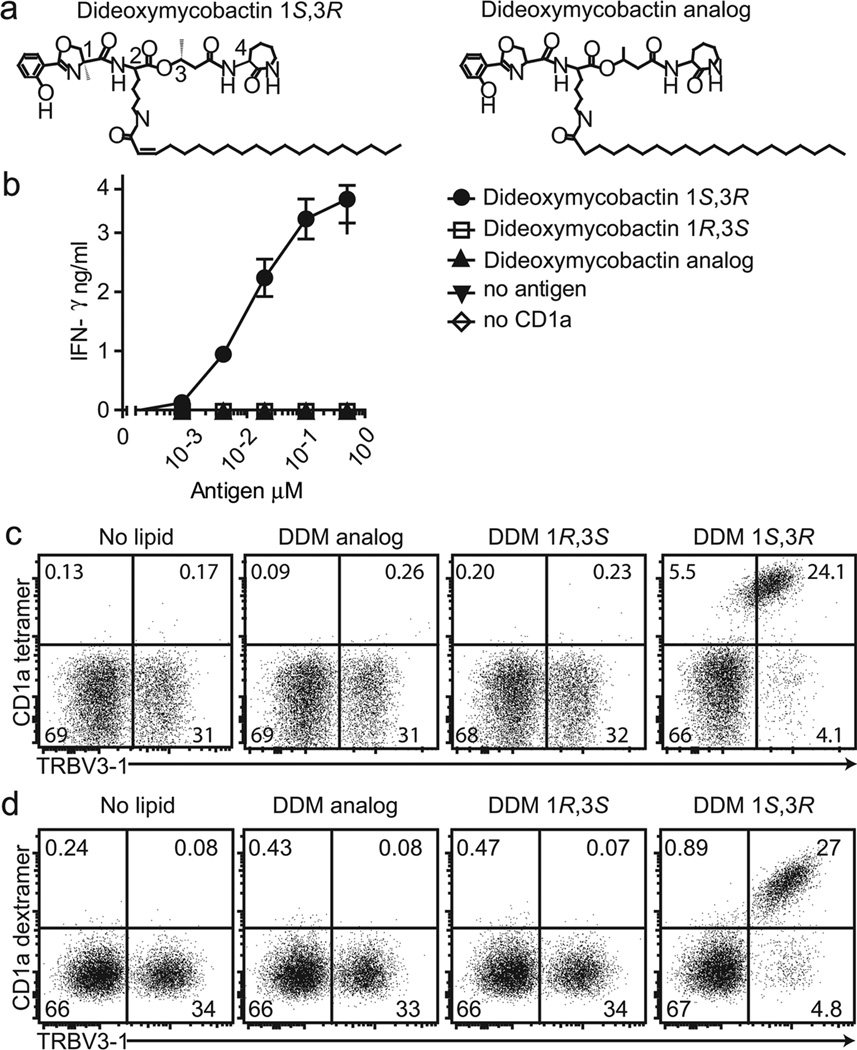
We generated transmembrane-truncated soluble biotinylated CD1a proteins based on methods previously developed for MHC, CD1d, CD1b and CD1c [8, 12, 14, 16]. To test the folding and antigen presenting function of this new CD1a construct, it was bound to streptavidin plates, treated with a synthetic dideoxymycobactin and used to activate T cells [7]. The synthetic antigen recapitulates the naturally occurring S or R stereochemistry among the 4 stereocenters in the natural deoxymycobactin peptide, including S and R stereochemistry at positions 1 and 3 present in M. tuberculosis DDM (DDM 1S3R) (Figure 1a) [7]. This lipopeptide activated the CD1a-restricted human T cell line CD8-2 in a dose-dependent manner. No activation was seen in response to dideoxymycobactin with the opposite stereo-configuration at positions 1 and 3 (DDM 1R3S) or a synthetic analog that deviated from the optimal natural DDM based on a fully saturated acyl chain and serine substituting for α-methyl serine (DDM analog, Figure 1b). Thus, soluble CD1a monomers were properly folded and were sufficient to present a lipopeptide antigen to human T cells.
Figure 1. CD1a tetramers and dextramers stain human T cells.
(a) Dideoxymycobactin is a biosynthetic precursor to mycobactin siderophores composed of a fatty acyl tail and a peptide backbone that contains four chiral centers (1,2,3,4), which are in the S or R configuration as indicated. Natural DDM occurs as the (1S,3R) diastereomer. The non-stimulatory dideoxymycobactin analog is presumed to be a mixture of 1S,3R and 1R,3S diastereomers [7]. (b) Tetramerizable CD1a monomers were bound to streptavidin coated plates, treated with antigen overnight at 37°C and used to activate IFN-γ release by the CD1a-restricted human T cell line CD8-2 (mean+/− SD). (c) The CD8-2 T cell line, which contains cells the DDM-specific TCR β chain, TRBV3-1, as well as cells with other TCRs, was stained with CD1a tetramers loaded with the indicated DDM isomer and or TRBV3-1 specific mAb. (d) CD8-2 T cells were stained by CD1a dextramers loaded with the indicated DDM isomer followed by T cell receptor anti Vβ chain antibody (TRBV3-1). Data (b-d) are representative of three or more experiments.
CD1a tetramers and dextramers stain T cells
After optimizing loading conditions based on pH, time, temperature and solvent variables (Supplemental Figure 1), we observed that DDM (1S3R) treated CD1a tetramers selectively stain T cells expressing the clonotypic TRBV3-1 TCR, but not other T cells (Figure 1c). T cells were not stained by CD1a tetramers that were not exposed to lipids or tetramers treated with the two non-antigenic DDMs that substantially mimic DDM (1S3R). Thus, tetramer staining was dependent on the structure of the added lipopeptide and was specific for the clonotypic TCR that defines CD8-2. These data established a working CD1a tetramer and strongly supported the model of direct binding of an αβ TCR to CD1a. To further increase the avidity of interaction we developed CD1a dextramers, which are composed of 10–14 CD1a monomers on a flexible, fluorescently labeled dextran backbone [17]. We observed bright, selective and highly reproducible staining of the TRBV3-1 subset of CD8-2 T cells (Fig. 1d) with DDM (1S3R) loaded dextramers. Thus, dextramers provided a second reagent for probing the interaction of CD1a with T cells.
CD1a multimers bind to the αβ TCR
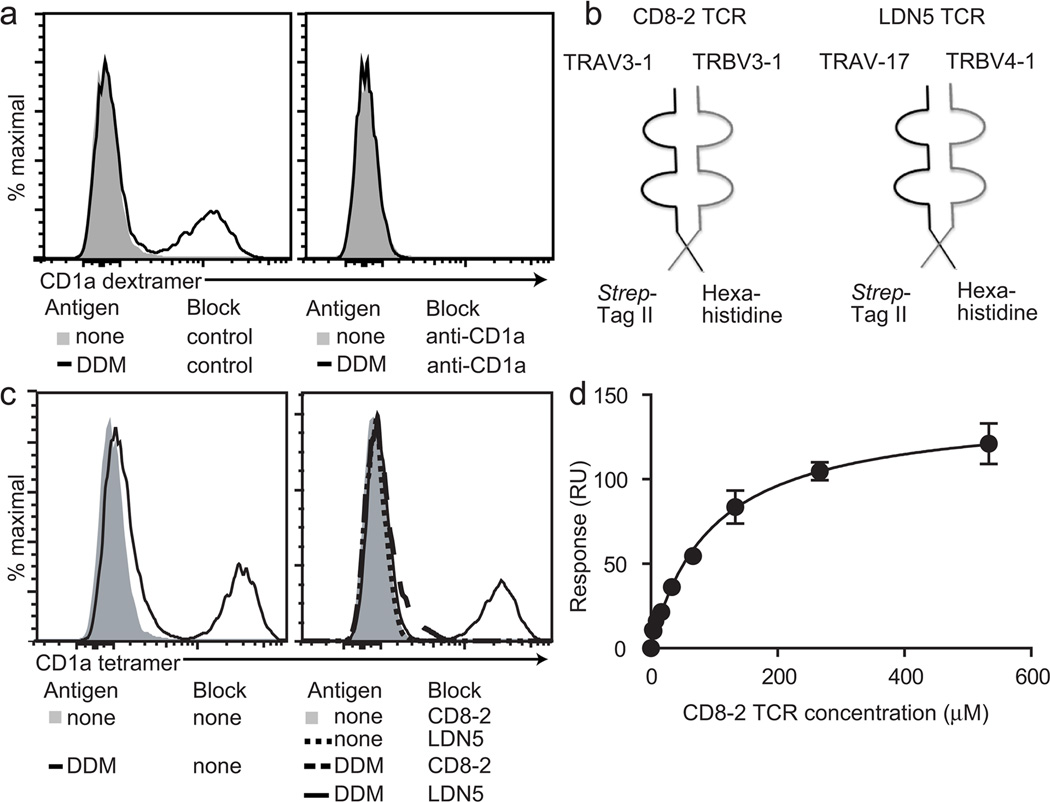
CD1a-DDM tetramer and dextramer binding to TRBV3-1+ T cells strongly implicated a cognate model in which a ternary interaction of CD1a-lipid binds to the TCR. We performed experiments to directly test binding between lipopeptide-CD1a complexes and the clonotypic human αβTCR versus all other surface receptors. Pre-incubation of dextramers with anti-CD1a antibody blocked staining to background (Figure 2a), confirming that staining was mediated by CD1a. We generated soluble TCRs containing the TCR α and β chains from CD8-2 (αβ heterodimers encoded by the TRAV3-1 and TRBV3-1 variable regions) or analogous constructs from the CD1b-restricted T cell LDN5 (composed of the TRAV17 and TRBV4-1 variable regions) (Figure 2b). Pre-incubation with soluble LDN5 TCR minimally impacted staining, whereas pre-incubation with soluble CD8-2 TCR blocked T cell staining to background levels (Figure 2c). Thus CD1a-DDM staining of cells is mediated by the αβ TCR.
Figure 2. Tetramer and dextramer staining demonstrates a trimolecular interaction among CD1a, dideoxymycobactin and the clonotypic TCR.
(a) The CD8-2 T cell line was stained with unloaded CD1a dextramers that were pre-incubated with isotype control antibody or anti-CD1a antibody (10µg/ml). (b) Soluble TCR α–chains with hexahistidine tags and β–chains with Strep-tag II were formed into soluble TCR dimers. (c) Loaded and unloaded tetramers were pre-incubated with 50-fold molar excess of soluble TCR. Data are representative of three or more experiments. (d) CD1a monomers were affixed to streptavidin-coated chips, loaded with dideoxymycobactin and then treated with soluble CD8-2 T cell receptors at the indicated concentrations to measure a dissociation constant of 95.78 +/− 13.51 µM. Data are representative of two experiments; error bars are shown for each data point (mean ± SEM).
We then measured the affinity of interaction between soluble-transmembrane truncated CD8-2 TCR and CD1a proteins alone, or pretreated with DDM using surface plasmon resonance (Figure 2d). No binding was seen to CD1a alone; the determined dissociation constant (KD) of 95.78+/−13.51 µM approximates that of weak self-antigens, (500µM). This affinity is significantly lower than that of TCRs recognizing α-galactosyl ceramide-CD1d (<1µM) and glucose monomycolate-CD1b (~1µM)[15]) but approximates the affinity of NKT TCRs recognizing β-linked glycolipids [21].
CD1a dextramers detect human lipopeptide-specific T cells ex vivo
To determine if DDM-reactive T cells exist as cell populations ex vivo, PBMC from subjects with active tuberculosis or positive tuberculin skin tests were stained with dextramers treated with DDM (1S3R) and then gated on CD3+CD14−CD19− live lymphocytes (Supplemental Figure 2a). Rare CD3+ cells were identified with the DDM-loaded dextramer among four subjects with mycobacterial exposure (Figure 3 and Supplemental Fig. 2b). This result suggested that DDM-reactive T cells are present in the blood of unrelated human donors.
Figure 3. CD1a dextramers stain polyclonal cells ex vivo.
PBMC from one subject with active tuberculosis (subject A24) as well as 3 tuberculin skin test positive subjects (subjects A22, A32 and C58) were stained with CD1a dextramers in addition to CD3 FITC, CD14 PercP-Cy5.5, CD19 PerCP-Cy5.5 and violet viability dye after which they were gated on live lymphocytes.
To determine whether the rare dextramer+ cells recognize CD1a and DDM, T cell yields were increased through leukapheresis (subject C58) or ex vivo expansion using anti-CD3 antibody and IL-2 (subject A32). We then used DDM treated dextramers to sort cells (Supplemental Figure 2c), cloned them using limiting dilution and tested them for reactivity in ELISPOT assays. Dextramer-based sorting generated many T cell clones that were brightly stained using DDM-loaded tetramers and these clones secreted TNF-α (clones 3, 6, 9, 21) or interferon-γ (clones P1, P5, P7, P9) in response to CD1a-expressing antigen presenting cells treated with DDM (Figure 4a and 4b). Three of the clones express CD8 and the other 5 express CD4. We therefore conclude that CD1a and DDM reactive T cells are present as populations among genetically diverse donors.
Figure 4. CD1a dextramers detect human lipopeptide-specific T cells.
T cell clones derived from tuberculin skin test positive subject A32 (a) or subject C58 (b) were originally obtained by sorting based on CD1a-DDM dextramer binding as shown in Supplementary Fig. S2b. Clones were harvested and stained with CD1a dextramers as in Figure 1. Positively staining clones were incubated with K562 antigen presenting cells transfected with empty vector or the noted CD1 isoform, with or without 1uM DDM (mean +/−SD) in an ELISPOT assay.
Overall, these results distinguish between indirect and cognate TCR interaction models, illustrating that direct CD1a-DDM-TCR interactions are the mechanism of T cell activation by CD1a and lipopeptide. These studies extend prior work describing polyclonal responses to lipoprotein mixtures [18] and lipopeptide-specific T cell clones [22] to prove that lipopeptide reactive T cells exist in the in vivo state. More generally, generation of working CD1a multimers completes the set of tools needed to study human CD1-restricted T cells, allowing determination of the phenotype and function of T cells recognizing CD1a bound to self and foreign antigens.
Supplementary Material
Acknowledgments
This work was supported by grants from the Howard Hughes Medical Institute KwaZulu-Natal Research Institute for Tuberculosis and HIV (AGK and DBM), the Harvard University Global Health Initiative (AGK), the Burroughs Wellcome Fund program in Translational Research (DBM), the NIH (T-32 AI 007306-22, T-32 AR 007530-23, K08 AI089858 (AGK), R01 AI49313, R01 AR048632 (DBM), R01 A1042266 (IAW)) and the Nederlands Wetenschappelijk Onderzoek (Meervoud 836.08.001, IVR). SG is supported by an Australian Research Council Future Fellowship; JR is supported by a National Health and Medical Research Council Australia Fellowship.
Footnotes
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Yu CY, Milstein C. A physical map linking the five CD1 human thymocyte differentiation antigen genes. Embo J. 1989;8:3727–3732. doi: 10.1002/j.1460-2075.1989.tb08548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bedel R, Matsuda JL, Brigl M, White J, Kappler J, Marrack P, Gapin L. Lower TCR repertoire diversity in Traj18-deficient mice. Nat Immunol. 2012;13:705–706. doi: 10.1038/ni.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller MM, Wang C, Parisini E, Coletta RD, Goto RM, Lee SY, Barral DC, Townes M, Roura-Mir C, Ford HL, Brenner MB, Dascher CC. Characterization of two avian MHC-like genes reveals an ancient origin of the CD1 family. Proc Natl Acad Sci U S A. 2005;102:8674–8679. doi: 10.1073/pnas.0500105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moody DB, Young D, Cheng T, Porcelli S, Brenner M. T cell activation by lipopeptide antigens. Science. 2004;303:527–531. doi: 10.1126/science.1089353. [DOI] [PubMed] [Google Scholar]
- 5.Manolova V, Kistowska M, Paoletti S, Baltariu GM, Bausinger H, Hanau D, Mori L, De Libero G. Functional CD1a is stabilized by exogenous lipids. Eur J Immunol. 2006;36:1083–1092. doi: 10.1002/eji.200535544. [DOI] [PubMed] [Google Scholar]
- 6.Zajonc DM, Crispin MDM, Bowden TA, Young T-Y, Cheng DC, Hu J, Costello CE, Rudd PM, Dwek RA, Miller MJ, Brenner MB, Moody DB, Wilson IA. Molecular mechanism of lipopeptide presentation by CD1a. Immunity. 2005;22:209–219. doi: 10.1016/j.immuni.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 7.Young DC, Kasmar A, Moraski G, Cheng TY, Walz AJ, Hu J, Xu Y, Endres GW, Uzieblo A, Zajonc D, Costello CE, Miller MJ, Moody DB. Synthesis of dideoxymycobactin antigens presented by CD1a reveals T cell fine specificity for natural lipopeptide structures. J Biol Chem. 2009;284:25087–25096. doi: 10.1074/jbc.M109.000802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altman JD, Moss P, Goulder P, Barouch D, Bell M, McMichael A, Davis M. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 9.Benlagha K, Weiss A, Beavis A, Teyton L, Bendelac A. In vivo identification of glycolipid antigen-specific T cells using fluorescent CD1d tetramers. J Exp Med. 2000;191:1895–1903. doi: 10.1084/jem.191.11.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuda JL, Naidenko OV, Gapin L, Nakayama T, Taniguchi M, Wang CR, Koezuka Y, Kronenberg M. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J Exp Med. 2000;192:741–754. doi: 10.1084/jem.192.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karadimitris A, Gadola S, Altamirano M, Brown D, Woolfson A, Klenerman P, Chen JL, Koezuka Y, Roberts IA, Price DA, Dusheiko G, Milstein C, Fersht A, Luzzatto L, Cerundolo V. Human CD1d-glycolipid tetramers generated by in vitro oxidative refolding chromatography. Proc Natl Acad Sci U S A. 2001;98:3294–3298. doi: 10.1073/pnas.051604498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gumperz JE, Miyake S, Yamamura T, Brenner MB. Functionally distinct subsets of CD1d-restricted natural Killer T cells revealed by CD1d tetramer staining. J Exp Med. 2002;195:625–636. doi: 10.1084/jem.20011786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee PT, Benlagha K, Teyton L, Bendelac A. Distinct functional lineages of human V(alpha)24 natural killer T cells. J Exp Med. 2002;195:637–41. doi: 10.1084/jem.20011908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kasmar AG, van Rhijn T-Y, Cheng I, Turner M, Seshadri C, Schiefner A, Kalathur RC, Annand JW, de Jong A, Shires J, Leon L, Brenner M, Wilson IA, Altman JD, Moody DB. CD1b tetramers bind αβ T cell receptors to identify a mycobacterial glycolipid-reactive T cell repertoire in humans. J Exp Med. 2011;208:1741–1747. doi: 10.1084/jem.20110665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Rhijn I, Kasmar A, de Jong A, Gras S, Bhati M, Doorenspleet ME, de Vries N, Godfrey DI, Altman JD, de Jager W, Rossjohn J, Moody DB. A conserved human T cell population targets mycobacterial antigens presented by CD1b. Nat Immunol. 2013;14:706–13. doi: 10.1038/ni.2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ly D, Kasmar T-Y, Cheng AG, de Jong A, Huang S, Roy S, Bhatt A, van Summeren RP, Altman JD, Jacobs WR, Adams EJ, Minnaard AJ, Porcelli SA, Moody DB. CD1c tetramers detect ex vivo T cell responses to processed phosphomycoketide antigens. The Journal of Experimental Medicine. 2013;210:729–741. doi: 10.1084/jem.20120624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Batard P, Peterson DA, Devêvre E, Guillaume J-C, Cerottini P, Rimoldi D, Speiser DE, Winther L, Romero P. Dextramers: New generation of fluorescent MHC class I/peptide multimers for visualization of antigen-specific CD8+ T cells. J Immunol Methods. 2006;310:136–148. doi: 10.1016/j.jim.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 18.Rosat J-P, Grant EP, Beckman EM, Dascher CC, Sieling PA, Frederique D, Modlin RL, Porcelli SA, Furlong ST, Brenner MB. CD1-restricted microbial lipid antigen-specific recognition found in the CD8+ ab T cell pool. J Immunol. 1999;162:366–371. [PubMed] [Google Scholar]
- 19.Gras S, Burrows SR, Kjer-Nielsen L, Clements CS, Liu YC, Sullivan LC, Bell MJ, Brooks AG, Purcell AW, McCluskey J, Rossjohn J. The shaping of T cell receptor recognition by self-tolerance. Immunity. 2009;30:193–203. doi: 10.1016/j.immuni.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 20.de Jong A, Pena-Cruz T-Y, Cheng V, Clark RA, Van Rhijn I, Moody DB. CD1a-autoreactive T cells are a normal component of the human ab T cell repertoire. Nat Immunol. 2010;11:1102–1109. doi: 10.1038/ni.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pellicci DG, Clarke AJ, Patel O, Mallevaey T, Beddoe T, Le Nours J, Uldrich AP, McCluskey J, Besra GS, Porcelli SA, Gapin L, Godfrey DI, Rossjohn J. Recognition of [beta]-linked self glycolipids mediated by natural killer T cell antigen receptors. Nat Immunol. 2011;12:827–833. doi: 10.1038/ni.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Rhijn I, Young DC, De Jong A, Vazquez T-Y, Cheng J, Talekar R, Barral DC, León L, Brenner MB, Katz JT, Riese R, Ruprecht RM, O'Connor PB, Costello CE, Porcelli SA, Briken V, Moody DB. CD1c bypasses lysosomes to present a lipopeptide antigen with 12 amino acids. J Exp Med. 2009;206:1409–1422. doi: 10.1084/jem.20082480. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.