Abstract
Sarcopenia describes an age-related decline in skeletal muscle mass, strength, and function that ultimately impairs metabolism, leads to poor balance, frequent falling, limited mobility, and a reduction in quality of life. Here we investigate the pathogenesis of sarcopenia through a proteomic shotgun approach. Briefly, we employed tandem mass tags (TMT) to quantitate and compare the protein profiles obtained from young versus old rat slow-twitch type of muscle (soleus) and a fast-twitch type of muscle (extensor digitorum longus, EDL). Our results disclose 3452 and 1848 proteins identified from soleus and EDL muscles samples of which 78 and 174 were found to be differentially expressed, respectively. In general, most of the proteins were structural related, involved in energy metabolism, oxidative stress, detoxification, or transport. Aging affected soleus and EDL muscles differently and several proteins were regulated in opposite ways. For example, pyruvate kinase had its expression and activity different in both soleus and EDL muscles. We were able to verify with existing literature many of our differentially expressed proteins as candidate aging biomarkers, and most importantly, disclose several new candidate biomarkers such as the glioblastoma amplified sequence (GAS), zero beta-globin, and prolargin.
Keywords: Aging, EDL, proteome, rat, sarcopenia, soleus, TMT
INTRODUCTION
Sarcopenia describes an age-related decline in skeletal muscle mass, strength and function1,2,3,4 that ultimately results in impaired metabolism, poor balance, frequent falling, limited mobility and reduction in quality of life5’6. Therefore, the identification of new disease markers will aid in understanding the biology of this disease, and therefore pave the way to more effective diagnostic methods and treatments to stop or at least delay age-related muscle wasting.
Sarcopenia is a multi-factorial disease that is highly correlated with aging. Alterations in cells have been found to occur such as increased oxidative stress, increased apoptosis, metabolic abnormalities, alterations in hormone response, decreased protein synthesis of myofibrillar components, and impaired signal transduction3,7,8,9,10. Although contractile proteins are lost during atrophy, alterations can occur in non-contractile proteins, increasing the complexity of such alterations.
Aging affects the metabolic capacity of skeletal muscle and changes the activity of specific enzymes. In general, there is a decline in the oxidative capacity of skeletal muscles of aged rats as well as a decline in energy-rich molecules such as ATP and creatine phosphate (CrP)11,12. However, the effect on enzyme activity and expression cannot be generalized since data suggests that distinct muscles may respond differently to aging.
During aging there is a progressive loss of skeletal muscle proteins. Fractional synthesis rates of myofibrillar proteins are reduced in the elderly indicating that the old muscles have reduced capacity to synthesize new proteins. Furthermore, decline in mitochondrial proteins has also been reported (reviewed by CARMELI et al.13).
Oxidative stress appears to be a key component of sarcopenia, and it has been suggested that the deleterious effects of reactive oxygen species (ROS) are at least in part responsible for the aging process14. Free radicals are also responsible for dysfunctional mitochondria, which play a major role in muscle function decline. Alterations in mitochondrial volume, increased oxidative stress, reduced oxidative capacity, and an increase in mitochondrially mediated apoptosis have been described (reviewed by PETERSON et al.15).
Thirty-month old wistar rats represent an established animal model of skeletal muscle aging and a variety of studies have been performed using this animal strain16,17,18. Over the last decade several muscle proteomic studies, following such biological model, have been completed using different techniques such as two-dimensional gel electrophoresis (2DE)19,20,21 and shotgun based proteomics18,22,23,24 to catalogue muscle skeletal proteins. And within the 2DE universe, we note that several staining methods have been evaluated; such as Coomassie blue22, fluorescent Deep Purple labeling23 and fluorescent difference in-gel electrophoresis (DIGE)24. Taken together, these results describe various differentially expressed proteins related to structure, metabolic enzymes, or involved in ion homeostasis and stress response25. Previous reports have also investigated post-translational modifications such as phosphorylation26,27, glycosylation28,29,30, carbonylation31, and nytrosylation17.
Because soleus and extensor digitorum longus (EDL) muscles have different fiber composition, function and metabolic features, we argue that an in-depth proteomic analysis of these tissues could unveil complementary aspects in the biology of aging. Skeletal muscle is composed of two major types of muscle fibers, the slow- and the fast-twitch fibers, which can be distinguished by the enzymatic characteristics of myosin ATPase and species of myosin heavy chain isoforms. Slow twitch fibers express type I while type II predominate in the fast-twitch fibres32. Among skeletal muscles in the rat hind limb, the soleus muscle is a typical muscle composed mainly of slow-twitch fibers (more than 90% of type I fibers) which allow them prolonged, steady contractions. The EDL muscle is composed mainly of fast-twitch fibers (more than 90% of type II fibers), which ensures rapid and delicate movements33.
Therefore, the aim of our study was to evaluate the effect of aging in the protein expression patterns of soleus and EDL muscles. To accomplish this, we employed state of the art proteomic techniques such as multi-dimensional protein identification technology (MudPIT)34, high resolution mass spectrometry with an Orbitrap Velos (Thermo, San José), and an isobaric labeling quantitation approach using the tandem mass tags (TMT)35.
EXPERIMENTAL PROCEDURES
Animals
This research was approved by the Local Ethical Committee and the experiments were conducted in accordance with the National Research Council's Guidelines for the Care and Use of Laboratory Animals. Male Wistar rats were housed under controlled environmental conditions (temperature, 22°C; 12h dark period starting at 6:00 p.m.). Young rats had an average weight of 0.370kg ± 0.01 g (3 months of age) and old rats had an average weight of 0.500kg ± 0.08 g (24 months of age).
Serum and muscle oxidative stress
Serum oxidative stress was determined using ferrous oxidation-xylenol orange (FOX) assay as previously described36.
Creatine kinase and pyruvate kinase enzyme activity
Samples were homogenized in extraction buffer (0.05 M Imidazole-HCl, 0.12 M KCl, and 0.062 M MgSO4; pH 7.6) at 1:10 dilution (w/v). Homogenates were centrifuged at 7000 g for 10 minutes, at 4°C, and the supernatants used for the further assays. An aliquot of each homogenate was used for determining total protein content, using BSA a standard (Bradford et al., 1979), which was used for normalization of the results. Evaluation of CK and PK activities was performed by the methodologies described by Ainsworth and MacFarlane37 and Dinovo et al.38, respectively. Results are shown as µmol/min/mg protein.
Histological analysis
Muscles were embedded in tissue tek, cooled in isopentane, frozen in liquid nitrogen, and sectioned with a cryostat. The resulting 10-µm transverse sections were stained with hematoxylin and eosin (HE) for morphological analysis and NADH-TR. Fibers from soleus muscle were classified as slow-oxidative (SO) and fast-oxidative-glycolytic (FOG); fibers from EDL muscle were classified as SO, FOG, and fast-glycolytic (FG). Cross-sectional areas (CSAs) of ~400 muscle fibers of a muscle from each rat were measured using the Image Pro-Plus software.
Trypsin digestion
One hundred micrograms of protein were precipitated using the methanol/chloroform precipitation method. Briefly, 4 volumes of methanol and 1 volume of chloroform was added, the samples were mixed, centrifuged and then washed once with 4 volumes of methanol. The samples were then dissolved in 45 µL of TEAB 200 mM pH 8.0, 5 µL of 2% SDS and ultra-pure water was added to complete 100 µL. Reduction was performed with 10 µL TCEP 200mM for 60 min at 55 °C followed by 5 µL of of 375 mM iodoacetamide for 30 min in the dark at RT. The sample was precipitated in cold acetone (1:6), at −20 °C overnight and dissolved in 45 µL of TEAB 200 mM pH 8.0; 2.5 µL of SDS 2% and ultra-pure water. Trypsin digestion was carried out overnight at 37 °C with 2.5 µg of enzyme.
TMT labeling
Six-plex TMT labeling (Thermo Scientific) was performed according to the manufacturer’s instructions to isobarically label primary amino groups and thus allow to simultaneously comparison six samples. Briefly, each tube (0.8 mg) was reconstituted with 41 µL of anhydrous acetonitrile at RT and the reagents were dissolved by vortexing for 5 min. Each sample was labeled by adding 10 µL of tag, followed by incubation for 1 h at RT. The reaction was quenched with the addition of 5% hydroxylamine for 15 min incubation at RT. Samples were then pooled and stored at −80 °C until further analysis. Young muscle samples were labeled with 126, 127 and 128 while old muscle samples were labeled with 129, 130 and 131.
Multidimensional Protein Identification Technology Analysis (MudPIT)
Analysis was performed on an LTQ Orbitrap Velos (Thermo Scientific, San Jose, CA, USA) interfaced with a quaternary HP 1100 series HPLC pump (Agilent Technology, Santa Clara, CA, USA). The analytical column was a 100 µm diameter fused-silica capillary (J/W Scientific, Agilent Technology, Santa Clara, CA, USA), pulled with a P-2000 laser (Sutter Instrument Co., Novato, CA, USA) and packed with 12 cm of 5 µm C18 resin (Aqua, Phenomenex, Torrence, CA, USA). The biphasic microcapillary trapping column (5 cm of 250 µm diameter) consisted of a fritted capillary with Kasil 1624, packed with 2.5 cm reversed-phase C18 (Aqua, Phenomenex, Torrence, CA, USA) and 2.5 cm of strong cation exchange (5 µm Partisphere, Whatman, Maidstone, Kent, UK) packing material. The biphasic column was loaded offline with sample using a pressure pump at approximately 800 psi.
An automated 11-step chromatographic run was performed on each sample using three mobile phases consisting of buffer A (5% acetonitrile (ACN); 0.1% acid formic (FA), buffer B (80% ACN, 0.1% FA), and buffer C (500 mM ammonium acetate, 5% ACN, 0.1% FA). Step one consisted of a linear gradient from 0 to 100% buffer B (120min). Steps 2–9 had the following profile: 1 min 100% buffer A; 4 min (100 - X)% buffer A, X% buffer C; 80 min from 100% buffer A to 50% buffer A and 50% buffer B; 10 min from 50% to 100% buffer B; 1 min from 100% buffer B to 100% A; 10 min at 100% buffer A. For buffer C, X% was, respectively, 10%, 20%, 30%, 40%, 50%, 60%, 70%, and 100%. Steps 10 and 11 had the following profile: 2 min 100% buffer A; 4 min 10% buffer B, 90% buffer C; 45 min from 100% buffer A to 50% buffer A and 50% buffer B; 10 min from 50% to 100% buffer B; 1 min from 100% buffer B to 100% A; 10 min at 100% buffer A.
The application of the distal voltage of 2.5 kV electrospayed the eluted peptides directly into LTQ Orbitrap Velos (Thermo Scientific, San Jose, CA, USA). A cycle of one full scan was applied (300–2000 m/z, resolution 30 000) followed by 10 data dependent HCD dual MS/MS and repeated continuously through each MudPIT step. Full scans and higher energy collisional activated dissociation (HCD) scans were in Orbitrap, at resolutions of 30 000 and 7500, respectively. The normalized collision energy in the HCD was of 45% in HCD. The following parameters for dynamic exclusion were applied: 1 repeat count, 30 s repeat duration, 180 exclusion list size, 60 s exclusion duration, and a dynamic exclusion list of 60 seconds and an HCD fragmentation energy of 45. The number of microscans for ms1 and ms2 was 1 and a 2m/z isolation window. The mass spectrometer and HPLC were controlled by the Xcalibur data system (Thermo, San Jose, CA).
Database searching
Tandem mass spectra were extracted from the raw files using RawExtract 1.9.339. Database searching for protein identification was performed using the ProLuCID algorithm v.1.3.140, with the following parameters: carbamidomethylation of cysteines (57.021464 C) and TMT-labeled in the primary amino group (229.1629K) as static modifications; trypsin as enzyme (KR) allowing for semi-specific peptides; tolerance of 50 ppm and the ProLuCID isotopic peaks parameter was set to 341. The search was performed against the all Rattus sequences available for download from Uniprot on April 1st, 2013; these comprised 41,772 sequences plus other 127 sequences that we added as they are known to be common mass spectrometry contaminants. For each sequence, a reversed decoy sequence was included, doubling the number of sequences in the final database.
Assessment of PSMs
The validity of the Peptide Sequence Matches (PSMs) was assessed using the Search Engine Processor (SEPro). Briefly, identifications were grouped by charge state (+2 and +3) and then by tryptic status (fully tryptic, semi-tryptic), resulting in four distinct subgroups. For each group, the ProLuCID XCorr, DeltaCN, delta ppm, number of peaks matched, and ZScore values were used to generate a Bayesian discriminator. The identifications were sorted in a non-decreasing order according to the discriminator score. A cutoff score was established to accept a false-discovery rate (FDR) of 1% based on the number of decoys. This procedure was independently performed on each data subset, resulting in a false-positive rate that was independent of tryptic status or charge state. Additionally, a minimum sequence length of six amino acid residues was required. Results were post-processed to only accept PSMs with less than 8 ppm.
Quantitation and pinpointing differentially expressed proteins
The quantitation of confident identifications was assessed using SEPro’s quantitation module SEProQ41 which is another module from PatternLab for proteomics suite. Briefly, this module reads the intensity corresponding to each of the six TMT reporter ions (i.e., 126, 127, 128, 129, 130, and 131) of which the first three and the last three correspond to samples from different biological conditions. The data treatment options selected for this analysis were signal normalization by channel sum and impurity correction. For the later, we included the impurity and isotopic overlap information provided together with the TMT kit. SEProQ uses a peptide centric approach to pinpoint differentially expressed proteins. For this, it relies on the paired Student’s t-test to pinpoint differentially expressed proteins by considering, for each mass spectrum associated to a given protein, the differences in average of the reporter ions associated to each biological state. Finally, after associating a differential expression p-value to each protein, SEProQ applies the Benjamini-Hochberg43 to control the theoretical FDR to q < 0.05.
RESULTS
Animal characteristics
The body weight, soleus muscle wet weight, EDL muscle wet weight, and the muscle wet weight to body ratio (sarcopenia index) is shown in Table 1. The mean body weight for the young animals was 0.369kg ± 0.00574 and for the old group, 0.502kg ± 0.0250 (p < 0.0001). Despite differences in body weight, soleus wet weight (0.196g ± 0.00599 and 0.210g ± 0.0158; p = 0.415 for young and old respectively) and EDL wet weight (0.168g ± 0.00409 and 0.193g ± 0.0187; p = 0.2243 for young and old respectively) were not statistically different between the groups.
Table 1.
Animal characteristics
| Rat | Body weight (g) |
Soleus weight (g) |
EDL weight (g) |
SI (soleus)a | SI (EDL)b | |
|---|---|---|---|---|---|---|
| young | 1 | 0.37 | 0.23 | 0.17 | 0.62 | 0.45 |
| 2 | 0.36 | 0.20 | 0.17 | 0.55 | 0.47 | |
| 3 | 0.37 | 0.20 | 0.17 | 0.54 | 0.45 | |
| 4 | 0.39 | 0.21 | 0.18 | 0.53 | 0.45 | |
| 5 | 0.38 | 0.20 | 0.16 | 0.52 | 0.42 | |
| 6 | 0.33 | 0.17 | 0.16 | 0.51 | 0.48 | |
| 7 | 0.37 | 0.18 | 0.16 | 0.51 | 0.45 | |
| 8 | 0.36 | 0.18 | 0.15 | 0.51 | 0.42 | |
| 9 | 0.36 | 0.18 | 0.19 | 0.50 | 0.53 | |
| MEAN ± SEM | 0.369 ± 0.00574 | 0.196 ± 0.00599 | 0.168 ± 0.00409 | 0.532 ± 0.0117 | 0.458 ± 0.0117 | |
| old | 1 | 0.52 | 0.16 | 0.19 | 0.31 | 0.37 |
| 2 | 0.41 | 0.13 | 0.10 | 0.33 | 0.26 | |
| 3 | 0.55 | 0.25 | 0.23 | 0.46 | 0.43 | |
| 4 | 0.52 | 0.22 | 0.15 | 0.43 | 0.30 | |
| 5 | 0.54 | 0.23 | 0.31 | 0.44 | 0.57 | |
| 6 | 0.61 | 0.29 | 0.19 | 0.48 | 0.32 | |
| 7 | 0.35 | 0.20 | 0.16 | 0.42 | 0.46 | |
| 8 | 0.49 | 0.18 | 0.18 | 0.38 | 0.38 | |
| 9 | 0.51 | 0.21 | 0.19 | 0.41 | 0.37 | |
| MEAN ± SEM | 0.502 ± 0.0250 | 0.210 ± 0.0158 | 0.193 ± 0.0187 | 0.404 ± 0.0189 | 0.384 ± 0.0313 | |
SI = sarcopenia índex = soleus weight/body weight;
SI = EDL weight/body weight
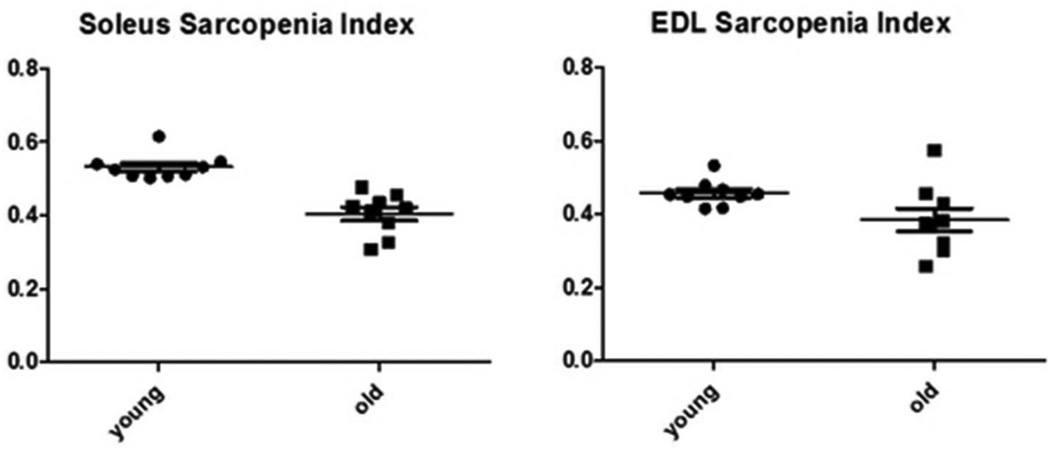
The sarcopenia index (SI) is defined as the relation between body weight and muscle weight25 and was calculated for both soleus and EDL muscles. For young and old soleus, the SI’s were 0.532 ± 0.0117 and 0.4044 ± 0.0189, respectively, with p < 0.0001. For EDL, the young and old SI’s were 0.458 ± 0.00117 and 0.384 ± 0.031, respectively with p < 0.04 (Figure 1). A drop in the sarcopenia index, as observed for both muscles, but more significantly in soleus, indicates muscle wasting.
Figure 1.
The relation between body weight and soleus and EDL muscles is shown as the sarcopenia index (SI, muscle wet weight over whole body weight). In aged animals there is a loss of muscle weight, which results in a drop in the sarcopenia index. A: young soleus SI (0.5323 ± 0.01169) versus old (0.4044 ± 0.01892) p < 0.0001; B: young EDL SI (0.4576 ± 0.001169) versus old (0.3845 ± 0.03129) p < 0.04.
Histological analysis
In soleus muscles, both slow-oxidative (SO) and fast oxidative glycolytic (FOG) fiber areas were significantly reduced in the old group when compared with the young group (Supplementary figure 1A; 1418 ± 334.7 vs. 115.2 ± 7.4 mm2 of SO fibers and 1719 ± 310.2 vs. 136.0 ± 2.7 mm2 of FOG fiber in young and old groups, respectively; p = 0.0065 and p = 0.0013). Furthermore, soleus cross sectional area (CSA) was significantly decreased in the old group when compared with the young group (Supplementary figure 1C; 3137 ± 640.2 vs. 251.2 ± 9.0 mm2 in young and old groups, respectively; p = 0.0028). Additionally, soleus muscles presented an increased number of centrally located nuclei, extensive variability in diameter and increased connective tissue (Supplementary figure 2B).
In contrast with soleus, EDL muscles presented no significant differences in fiber areas (Supplementary figure 1B; 890.4 ± 150.9 vs. 642.2 ± 185.3 mm2 of SO fibers, 1595 ± 281.1 vs. 1015 ± 303.3 mm2 of FOG fibers, and 2605 ± 533.9 vs. 1489 ± 458.3 mm2 of FG fibers in young and old groups, respectively; p > 0.05) and in CSA area (Supplementary figure 1C; 5091 ± 959.9 vs. 3146 ± 932.7 mm2 in young and old groups, respectively; p > 0.05). Likewise, EDL muscles presented an increased number of centrally located nuclei, extensive variability in diameter and increased connective tissue (Supplementary Figure 2A).
Differentially expressed proteins
Our soleus muscle shotgun proteomic analysis identified 3452 proteins (FDR at protein level = 0.98%) and a total of 6756 peptides (FDR at peptide level = 0.33%). PatternLab for proteomics41,44 provides an additional report indicating that these 3452 proteins can be reduced to 2027 when applying the bi-partite maximum parsimony algorithm to converge to a maximum parsimony protein list45 (i.e., a reduced list of proteins that explains all identified peptides). We note that 1006 of these proteins had at least one unique peptide. In our EDL muscle analysis, we identified 1848 proteins (FDR =0.97%, 1075 with maximum parsimony of which 523 had at least one unique peptide) and a total of 4255 peptides (FDR at peptide level = 0.19%). To determine which proteins were differentially regulated we used a fold change cutoff of 1.3 and a q-value of 0.05 (viz., for the experiment at hand, a corrected p-value cutoff of 0.03. Overall, 78 proteins had a change in abundance in soleus muscles, of which 57 were down regulated in old muscles and 21 up-regulated (considering maximum parsimony) (Table 2). Accordingly, for EDL muscles, the number of up and down-regulated proteins in the old muscles were 145 and 29, respectively, totaling 174 (Table 3).
Table 2.
Differentially expressed proteins in soleus muscles
| Protein accession no. |
P value |
−Log2 fold changea |
DESCRIPTION |
|---|---|---|---|
| Energy metabolism | |||
| Q6IMX3 | 0.017 | −0.8 | Acetyl-Coenzyme A dehydrogenase, short chain, isoform CRA_a |
| P13221 | < 0.001 | −0.8 | Aspartate aminotransferase, cytoplasmic |
| P13221 | 0.001 | −1.1 | Aspartate aminotransferase, mitochondrial |
| F1LP05 | < 0.001 | −0.9 | ATP synthase subunit alpha |
| P15429 | < 0.001 | −0.6 | Beta-enolase |
| Q68FY0 | 0.003 | −0.7 | Cytochrome b-c1 complex subunit 1, mitochondrial |
| P32551 | < 0.001 | −0.7 | Cytochrome b-c1 complex subunit 2, mitochondrial |
| P10888 | < 0.001 | −1.3 | Cytochrome c oxidase subunit 4 isoform 1, mitochondrial |
| Q68FU3 | 0.005 | −1.0 | Electron transfer flavoprotein subunit beta |
| Q66HF3 | 0.019 | −0.4 | Electron transfer flavoprotein-ubiquinone oxidoreductase, mitochondrial |
| P42123 | < 0.001 | 0.6 | L-lactate dehydrogenase B chain |
| G3V6H5 | 0.001 | 0.4 | Mitochondrial 2-oxoglutarate/malate carrier protein |
| P04636 | < 0.001 | −1.2 | Mitochondrial 2-oxoglutarate/malate carrier protein |
| P16617 | < 0.001 | −0.9 | Phosphoglycerate kinase 1 |
| P25113 | < 0.001 | −1.4 | Phosphoglycerate mutase 1 |
| F1LPA6 | 0.001 | 3.7 | Protein-arginine deiminase type-2 (Fragment) |
| Q6P7S0 | 0.001 | −0.9 | Pyruvate kinase |
| P15651 | 0.017 | −0.8 | Short-chain specific acyl-CoA dehydrogenase, mitochondrial |
| Q5RK08 | 0.010 | −22.1 | Glioblastoma amplified sequence |
| Q64428 | 0.005 | −0.7 | Trifunctional enzyme subunit alpha, mitochondrial |
| Structure and cell motility | |||
| F1LYK7 | 0.001 | 0.4 | Protein Cfl2 (cofilin 2) |
| P85972 | 0.002 | −0.5 | Vinculin |
| F1LMC6 | 0.016 | 17.8 | Troponin I, slow skeletal muscle (Fragment) |
| G3V885 | < 0.001 | 1.0 | Myosin-6 (Myh6) |
| G3V8B0 | < 0.001 | 1.0 | Myosin-7 (Myh7) |
| Q9EQP5 | 0.018 | 1.0 | Prolargin |
| F1LRV9 | 0.006 | 0.9 | Myosin-4 (Myh4) |
| F1M789 | < 0.001 | 0.7 | Protein Myh13 (Fragment) |
| G3V6E1 | 0.002 | 0.6 | Myosin-4 (Myh4) |
| Q62920 | 0.015 | −0.4 | PDZ and LIM domain protein 5 |
| F1LNH3 | 0.001 | −0.5 | Protein Col6a2 (Fragment) |
| D3ZCV0 | < 0.001 | −0.7 | Protein Actn2 |
| D3ZHA0 | < 0.001 | −0.8 | Protein Flnc |
| F2Z3T2 | 0.003 | −0.9 | Tropomyosin alpha-3 chain |
| P08733 | < 0.001 | −0.9 | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform |
| P68136 | 0.004 | −0.9 | Actin, alpha skeletal muscle |
| Q63781 | 0.002 | −1.0 | Myosin regulatory light chain |
| Q8K551 | 0.003 | −1.1 | Truncated alpha-actinin |
| P04466 | 0.009 | −1.5 | Myosin regulatory light chain 2, skeletal muscle isoform |
| P04692 | < 0.001 | −2.0 | Tropomyosin alpha-1 chain |
| P58775 | < 0.001 | −18.2 | Tropomyosin beta chain |
| P09495 | < 0.001 | −18.4 | Tropomyosin alpha-4 chain |
| Oxidative stress, detoxification and degradation | |||
| Q2TA66 | 0.018 | 2.6 | Ferritin (Fragment) GN=Fth1 PE=2 SV=1 |
| F1LVC6 | 0.002 | 1.2 | Glutathione S-transferase (Fragment) |
| B6DYQ2 | 0.011 | 1.1 | Glutathione S-transferase mu 2 |
| P63018 | < 0.001 | −0.4 | Heat shock cognate 71 kDa protein |
| Q66HD0 | 0.003 | −1.5 | Endoplasmin |
| P97541 | < 0.001 | −2.8 | Heat shock protein beta-6 |
| P35565 | 0.010 | −0.5 | Calnexin |
| D4ACB8 | 0.013 | −0.7 | Chaperonin subunit 8 (Theta) (Predicted), isoform CRA_a |
| P11598 | 0.003 | −0.7 | Protein disulfide-isomerase A3 GN=Pdia3 PE=1 SV=2 |
| G3V8T4 | 0.011 | −1.1 | DNA damage-binding protein 1 GN=Ddb1 PE=4 SV=1 |
| P0CC09 | 0.018 | −1.2 | Histone H2A type 2-A |
| Transport and catabolism | |||
| P02770 | < 0.001 | −1.0 | Serum albumin |
| Q63011 | 0.006 | −16.9 | Zero beta-globin (Fragment) |
| Q6MG90 | 0.013 | −0.4 | Complement component 4, gene 2 |
| P11442 | 0.004 | −0.4 | Clathrin heavy chain 1 |
| Q4V8H8 | 0.014 | −0.9 | EH domain-containing protein 2 |
| Additional differentially expressed proteins | |||
| Q5M7V3 | 0.013 | 1.8 | LOC367586 protein |
| P08932 | 0.002 | 1.4 | T-kininogen 2 |
| F1LPQ6 | < 0.001 | 0.9 | Uncharacterized protein (Fragment) |
| B2RZB2 | 0.005 | 0.7 | Putative uncharacterized protein |
| Q63041 | < 0.001 | 0.6 | Alpha-1-macroglobulin |
| B0BNM1 | 0.013 | 0.6 | NAD(P)H-hydrate epimerase |
| A0JPJ7 | 0.001 | 0.4 | Obg-like ATPase 1 |
| O08557 | 0.002 | −0.4 | N(G),N(G)-dimethylarginine dimethylaminohydrolase 1 |
| Q8R490 | 0.015 | −0.5 | Cadherin 13 |
| D4ABR6 | 0.002 | −0.5 | Annexin |
| P14046 | 0.017 | −0.9 | Alpha-1-inhibitor 3 |
| Q9WTT7 | 0.015 | −1.1 | Basic leucine zipper and W2 domain-containing protein 2 |
| D3ZHA7 | 0.001 | −1.2 | Protein LOC100359980 |
| D3ZCI9 | < 0.001 | −1.5 | Uncharacterized protein |
| D3ZH98 | < 0.001 | −21.1 | Uncharacterized protein |
| P05197 | < 0.001 | −0.9 | Elongation factor 2 |
| P11762 | < 0.001 | −1.1 | Galectin-1 GN=Lgals1 PE=1 SV=2 |
| Q62881 | 0.003 | −1.2 | Nucleolar protein 3 |
| Q7TP38 | 0.016 | −1.3 | Ab2-371 |
| O35814 | 0.003 | −1.0 | Stress-induced-phosphoprotein 1 |
fold change = young/old
Table 3.
Differentially expressed proteins in EDL muscles
| Protein accession no. |
P value | −Log2 fold changea |
DESCRIPTION |
|---|---|---|---|
| Energy metabolism | |||
| Q52KS1 | <0.001 | 0.6 | 6-phosphofructokinase |
| G3V796 | 0.005 | −0.4 | Acetyl-Coenzyme A dehydrogenase, medium chain |
| Q9ER34 | 0.014 | 0.9 | Aconitate hydratase, mitochondrial |
| P10760 | 0.009 | 1.4 | Adenosylhomocysteinase |
| F1LN88 | < 0.001 | 1.6 | Aldehyde dehydrogenase, mitochondrial |
| Q63041 | < 0.001 | 0.8 | Alpha-1-macroglobulin |
| P04764 | < 0.001 | 1.1 | Alpha-enolase |
| D4AEH9 | < 0.001 | 0.6 | Amylo-1, 6-glucosidase, 4-alpha-glucanotransferase, isoform CRA_a |
| P13221 | < 0.001 | 1.3 | Aspartate aminotransferase, cytoplasmic |
| P00507 | < 0.001 | 1.1 | Aspartate aminotransferase, mitochondrial |
| P15999 | < 0.001 | 0.8 | ATP synthase subunit alpha, mitochondrial |
| P19511 | < 0.001 | 0.7 | ATP synthase subunit b, mitochondrial |
| P10719 | < 0.001 | 0.9 | ATP synthase subunit beta, mitochondrial |
| G3V7Y3 | 0.001 | 0.6 | ATP synthase subunit delta, mitochondrial |
| Q06647 | < 0.001 | −0.4 | ATP synthase subunit O, mitochondrial |
| Q7TP35 | 0.002 | 0.7 | ATPase family AAA domain-containing protein 1 |
| B4F7E5 | < 0.001 | −0.4 | ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| P15429 | < 0.001 | 1.2 | Beta-enolase |
| G3V936 | < 0.001 | 1.3 | Citrate synthase |
| B0BNC0 | < 0.001 | 0.7 | Ckmt2 protein (creatine kinase, mitochondrial) |
| P32551 | < 0.001 | 0.7 | Cytochrome b-c1 complex subunit 2, mitochondrial |
| P10888 | < 0.001 | 0.7 | Cytochrome c oxidase subunit 4 isoform 1, mitochondrial |
| P11951 | < 0.001 | −0.5 | Cytochrome c oxidase subunit 6C-2 |
| P62898 | 0.003 | 0.8 | Cytochrome c, somatic |
| D3ZFQ8 | 0.011 | −0.6 | Cytochrome c-1 (Predicted), isoform CRA_c |
| P11348 | 0.002 | −26.4 | Dihydropteridine reductase |
| Q68FU3 | < 0.001 | 0.4 | Electron transfer flavoprotein subunit beta |
| D3ZJT8 | 0.007 | 1.0 | Enolase |
| Q5BJ93 | < 0.001 | 1.1 | Enolase 1, (Alpha) |
| G3V900 | < 0.001 | 0.7 | Fructose-bisphosphate aldolase |
| P09117 | 0.004 | 1.0 | Fructose-bisphosphate aldolase C |
| Q5M964 | < 0.001 | 0.7 | Fumarate hydratase 1 |
| P07323 | < 0.001 | 1.0 | Gamma-enolase |
| Q6P6V0 | < 0.001 | 0.7 | Glucose-6-phosphate isomerase |
| D4A6J7 | < 0.001 | 0.8 | Glyceraldehyde-3-phosphate dehydrogenase |
| D4A3W5 | < 0.001 | 0.6 | Glyceraldehyde-3-phosphate dehydrogenase |
| P04797 | < 0.001 | 0.5 | Glyceraldehyde-3-phosphate dehydrogenase |
| O35077 | < 0.001 | 0.5 | Glycerol-3-phosphate dehydrogenase [NAD(+)], cytoplasmic |
| P41565 | < 0.001 | −0.4 | Isocitrate dehydrogenase [NAD] subunit gamma 1, mitochondrial |
| P56574 | < 0.001 | 0.5 | Isocitrate dehydrogenase [NADP], mitochondrial |
| P04642 | < 0.001 | 1.9 | L-lactate dehydrogenase A chain |
| P42123 | < 0.001 | 0.8 | L-lactate dehydrogenase B chain |
| P15650 | < 0.001 | 1.3 | Long-chain specific acyl-CoA dehydrogenase, mitochondrial |
| O88989 | < 0.001 | 1.5 | Malate dehydrogenase, cytoplasmic |
| P04636 | < 0.001 | 1.0 | Malate dehydrogenase, mitochondrial |
| P08503 | 0.005 | −0.4 | Medium-chain specific acyl-CoA dehydrogenase, mitochondrial |
| Q02253 | 0.007 | 0.5 | Methylmalonate-semialdehyde dehydrogenase [acylating], mitochondrial |
| G3V6H5 | < 0.001 | −0.6 | Mitochondrial 2-oxoglutarate/malate carrier protein |
| Q5RJN0 | < 0.001 | −0.4 | NADH dehydrogenase (Ubiquinone) Fe-S protein 7 |
| B0BNE6 | 0.002 | 0.7 | NADH dehydrogenase (Ubiquinone) Fe-S protein 8 (Predicted), isoform CRA_a |
| Q641Y2 | 0.006 | 1.6 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 2, mitochondrial |
| Q66HF1 | 0.013 | 1.2 | NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial |
| A1A5L2 | < 0.001 | 1.2 | Pgm1 protein (Fragment) |
| Q499Q4 | < 0.001 | 1.2 | Phosphoglucomutase 1 |
| P16617 | < 0.001 | 0.8 | Phosphoglycerate kinase 1 |
| P25113 | < 0.001 | 0.8 | Phosphoglycerate mutase 1 |
| P16290 | < 0.001 | 0.7 | Phosphoglycerate mutase 2 |
| D3ZAP9 | 0.004 | 0.7 | Protein Gpd1l (glycerol-3-phosphate dehydrogenase 1-like) |
| D4ADX5 | < 0.001 | −0.4 | Protein Ndufs7(NADH dehydrogenase (ubiquinone)Fe-S protein 7) |
| P49432 | 0.001 | 5.3 | Pyruvate dehydrogenase E1 component subunit beta, mitochondrial |
| Q6P7S0 | < 0.001 | 0.8 | Pyruvate kinase |
| P11980 | < 0.001 | 0.8 | Pyruvate kinase isozymes M1/M2 |
| F8WG21 | < 0.001 | 1.3 | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial |
| Q64428 | < 0.001 | 1.0 | Trifunctional enzyme subunit alpha, mitochondrial |
| Q60587 | 0.009 | 0.7 | Trifunctional enzyme subunit beta, mitochondrial |
| P48500 | 0.001 | 1.1 | Triosephosphate isomerase |
| Structure and cell motility | |||
| P51886 | < 0.001 | 1.2 | Lumican |
| D4A6M0 | 0.003 | 1.1 | Mitsugumin 29 (Predicted) |
| D4A111 | < 0.001 | 1.5 | Procollagen, type VI, alpha 3 (Predicted), isoform CRA_a |
| P45592 | 0.014 | 1.2 | Cofilin-1 |
| P50609 | < 0.001 | 0.8 | Fibromodulin |
| D3ZVD7 | < 0.001 | −0.9 | Keratocan (Predicted) |
| D3ZVB7 | < 0.001 | 1.2 | Osteoglycin (Predicted) |
| Q9EQP5 | < 0.001 | 23.5 | Prolargin |
| F1LYK7 | < 0.001 | 1.2 | Protein Cfl2 (Fragment) |
| F1M7Q6 | < 0.001 | −1.9 | Protein Myh13 (Fragment) (myosin heavy chain 13) |
| F2Z3S8 | < 0.001 | 1.2 | Protein Tnnc2 (Fragment) (troponin C type 2) |
| F1LNH3 | 0.003 | 23.1 | Protein Col6a2 (Fragment) |
| F2Z3T2 | 0.019 | 1.6 | Tropomyosin alpha-3 chain |
| D4A115 | < 0.001 | 1.6 | Protein Col6a3 |
| D3ZHA0 | 0.015 | 1.4 | Protein Flnc |
| D4A2S4 | < 0.001 | 1.3 | Myosin-binding protein C, slow-type |
| Q9Z1P2 | 0.006 | 1.2 | Alpha-actinin-1 |
| P60711 | < 0.001 | 1.2 | Actin, cytoplasmic 1 |
| Q63518 | < 0.001 | 1.2 | Myosin-binding protein C, slow-type (Fragment) |
| P68136 | 0.013 | 1.2 | Actin, alpha skeletal muscle |
| D3ZCV0 | < 0.001 | 1.0 | Protein Actn2 |
| F1LP83 | < 0.001 | 0.6 | Tropomyosin beta chain |
| P27768 | < 0.001 | 0.6 | Troponin I, fast skeletal muscle |
| P17209 | < 0.001 | 0.5 | Myosin light chain 4 |
| P04466 | < 0.001 | 0.5 | Myosin regulatory light chain 2, skeletal muscle isoform |
| Q63781 | 0.001 | 0.5 | Myosin regulatory light chain |
| Q5FVG5 | < 0.001 | 0.4 | Similar to tropomyosin 1, embryonic fibroblast-rat, isoform CRA_c |
| G3V7K1 | < 0.001 | 0.4 | Myomesin 2 |
| P02600 | < 0.001 | 0.4 | Myosin light chain 1/3, skeletal muscle isoform |
| G3V8B0 | < 0.001 | −0.5 | Myosin-7 (Myh7) |
| G3V6E1 | < 0.001 | −0.7 | Myosin-4 (Myh4) |
| F1M8F6 | < 0.001 | −0.7 | Myosin-8 (Myh8) |
| F1LRV9 | < 0.001 | −0.8 | Myosin-4 (Myh4) |
| Q4FZU2 | 0.010 | −0.9 | Keratin, type II cytoskeletal 6A |
| Q6P6Q2 | 0.014 | −1.0 | Keratin, type II cytoskeletal 5 |
| P02454 | < 0.001 | −1.1 | Collagen alpha-1(I) chain |
| P31000 | 0.006 | −1.2 | Vimentin |
| F1LMU0 | < 0.001 | −1.3 | Myosin-4 (Myh4) |
| Oxidative stress, detoxification and degradation | |||
| P15865 | < 0.001 | 1.2 | Histone H1.4 |
| D3Z8U0 | < 0.001 | 1.7 | Histone H2B |
| Q6PDW8 | 0.028 | 26.8 | Glutathione peroxidase |
| P04785 | < 0.001 | 0.7 | Protein disulfide-isomerase |
| P08009 | < 0.001 | 4.2 | Glutathione S-transferase Yb-3 |
| P08010 | 0.001 | 3.3 | Glutathione S-transferase Mu 2 |
| P07895 | < 0.001 | 1.5 | Superoxide dismutase [Mn], mitochondrial (SOD 2) |
| B6DYQ7 | < 0.001 | 1.5 | Glutathione S-transferase pi |
| P63018 | < 0.001 | 1.2 | Heat shock cognate 71 kDa protein |
| P07632 | 0.002 | 0.9 | Superoxide dismutase [Cu-Zn] (SOD 1) |
| P34058 | < 0.001 | 0.7 | Heat shock protein HSP 90-beta |
| Transport and catabolism | |||
| P02770 | 0.016 | 2.8 | Serum albumin |
| Q7TP52 | 0.026 | 2.7 | Carboxymethylenebutenolidase homolog |
| Q62669 | < 0.001 | 1.1 | Protein Hbb-b1 |
| Q05962 | < 0.001 | 1.0 | ADP/ATP translocase 1 |
| P12346 | < 0.001 | 0.9 | Serotransferrin |
| Q9ER30 | < 0.001 | 0.9 | Kelch repeat and BTB domain-containing protein 10 |
| P02767 | < 0.001 | −0.5 | Transthyretin |
| Additional differentially expressed proteins | |||
| P35213 | < 0.001 | 0.4 | 14-3-3 protein beta/alpha |
| P62260 | < 0.001 | 1.0 | 14-3-3 protein epsilon |
| P68511 | 0.018 | −1.5 | 14-3-3 protein ETA |
| P63102 | 0.015 | 22.8 | 14-3-3 protein zeta/delta |
| P39069 | < 0.001 | 1.0 | Adenylate kinase isoenzyme 1 |
| P14046 | < 0.001 | 1.2 | Alpha-1-inhibitor 3 |
| P08461 | 0.002 | 2.1 | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial |
| P06399 | 0.002 | 0.8 | Fibrinogen alpha chain |
| P06866 | < 0.001 | 0.7 | Haptoglobin |
| Q5I0J0 | 0.004 | 1.8 | Immunoglobulin heavy chain (Gamma polypeptide) |
| F1MAN9 | < 0.001 | 1.2 | Myelin protein P0 |
| P19804 | < 0.001 | 1.2 | Nucleoside diphosphate kinase B |
| P02625 | < 0.001 | 0.7 | Parvalbumin alpha |
| G3V6L9 | 0.017 | −0.4 | Peptidyl-prolyl cis-trans isomerase |
| P49744 | 0.008 | 1.1 | Thrombospondin-4 |
| Q63581 | < 0.001 | 3.9 | Rat T-kininogen (T-KG) |
| Q63654 | < 0.001 | 2.8 | Polyubiquitin (Fragment) |
| P14141 | < 0.001 | 2.5 | Carbonic anhydrase 3 |
| F1LPQ6 | 0.004 | 1.7 | Uncharacterized protein (Fragment) |
| P08932 | < 0.001 | 1.6 | T-kininogen 2 |
| P31044 | < 0.001 | 1.4 | Phosphatidylethanolamine-binding protein 1 |
| Q5BJZ2 | < 0.001 | 1.4 | LOC367586 protein |
| P20717 | 0.005 | 1.3 | Protein-arginine deiminase type-2 |
| P07943 | 0.001 | 1.3 | Aldose reductase |
| F1LNB5 | < 0.001 | 1.2 | Murinoglobulin-2 |
| Q01129 | < 0.001 | 1.2 | Decorin |
| F1LX07 | 0.002 | 1.2 | Protein LOC100360985 (Fragment) |
| P11517 | < 0.001 | 1.3 | Hemoglobin subunit beta-2 |
| P02650 | < 0.001 | 0.6 | Apolipoprotein E |
| P62632 | < 0.001 | 1.0 | Elongation factor 1-alpha 2 |
| P05197 | 0.004 | 1.0 | Elongation factor 2 |
| F8WG42 | 0.002 | −0.4 | Eukaryotic translation initiation factor 3 subunit J (Fragment) |
| Q3T1J1 | < 0.001 | 0.7 | Eukaryotic translation initiation factor 5A-1 |
| B1H216 | < 0.001 | 1.3 | Hemoglobin alpha, adult chain 2 |
| P02091 | < 0.001 | 1.9 | Hemoglobin subunit beta-1 |
| P20059 | < 0.001 | 1.0 | Hemopexin |
| F1LNF1 | < 0.001 | 0.7 | Heterogeneous nuclear ribonucleoproteins A2/B1 (Fragment) |
| P85125 | 0.004 | 1.3 | Polymerase I and transcript release factor |
| F1LWG8 | < 0.001 | 1.1 | Uncharacterized protein (Fragment) |
| P19633 | < 0.001 | 1.0 | Calsequestrin-1 |
| D3ZYG6 | 0.009 | 0.9 | Protein Rtn2 (reticulon 2) |
| Q6WN19 | 0.009 | 0.9 | RTN2-B |
| Q6IMZ3 | < 0.001 | 0.9 | Annexin |
| Q07936 | 0.004 | 0.8 | Annexin A2 |
| D3ZQT0 | < 0.001 | 0.8 | Uncharacterized protein |
| Q9ERS3 | 0.030 | 0.8 | Voltage-dependent calcium channel subunit alpha-2/delta-1 |
| P04276 | 0.004 | 0.6 | Vitamin D-binding protein |
| D4AAC3 | < 0.001 | 0.6 | Protein Sh3bgr |
| D4A3D2 | < 0.001 | 0.4 | Protein Smyd1 |
| Q64649 | 0.024 | −0.5 | Phosphorylase b kinase regulatory subunit alpha, skeletal muscle isoform |
| Q6IRH6 | < 0.001 | −0.6 | Phosphate carrier protein, mitochondrial |
| D3Z8G0 | 0.019 | −0.6 | Phosphorylase b kinase regulatory subunit alpha, skeletal muscle isoform |
fold change = young/old
Most of the differentially expressed proteins are involved in metabolism (glycolysis, oxidative metabolism and cellular processes), contractile apparatus, cellular stress response and detoxification.
Energy metabolism
Most of the differentially expressed proteins in soleus muscles were down-regulated in the old tissue (Table 2). Glycolitic enzymes such as beta-enolase, phosphoglycerate kinase and pyruvate kinase (PK) were down-regulated during aging. PK mediates one of the rate-limiting steps catalyzing the final stage of the glycolytic pathway. Decreased expression of PK during aging were previously described23,46; however, some authors have found increased levels of this protein with aging47,48. Proteins involved in oxidative metabolism, such as ATP synthase (subunit alpha), electron transfer flavoprotein and mitochondrial cytochrome complex subunits 1 and 2 were also down-regulated.
Among the up-regulated proteins were protein-arginine deiminase type-2, L-lactate dehydrogenase β chain (LDH)), and mitochondrial 2-oxoglutarate/malate carrier protein.
The glioblastoma amplified sequence (GBAS), also known as NIPSNAP2, is most abundant in brain, heart and skeletal muscle49,50. Interestingly, this protein was one of the most down-regulated proteins in our study (−Log2 fold change = −22.10) and it may be at least partly responsible for the reduction in oxidative capacity that is found in sedentary elderly adults51. To the best of our knowledge, this is the first report of altered levels of this protein with aging and its specific role in the process requires further studies.
Most of the differentially regulated proteins identified in EDL muscles are metabolic enzymes and in contrast to what was observed in soleus muscles, the majority of the proteins (56) were up-regulated in the aged muscle, while only 10 proteins were down-regulated in this condition (Table 3). Glycolitic enzymes such as beta-enolase, alpha enolase, gamma-enolase, pyruvate kinase, glyceraldehyde-3-phosphate dehydrogenase, fructose-biphosphate aldolase, phosphoglycerate kinase and phosphoglycerate mutase were up-regulated during aging.Several proteins involved in oxidative metabolism were also up-regulated in the old EDL muscles. Among them were subunits of ATP synthase (subunit alpha, b, beta and delta), NADH-ubiquinone oxidoreductase, malate dehydrogenase, isocitrate dehydrogenase and citrate synthase. Increased levels of the mitochondrial enzymes such as ATP synthase, malate dehydrogenase and isocitrate dehydrogenase have also been found during the aging of rat gastrocnemius muscles46,48,52,53.
Structural proteins
Several structural proteins were up-regulated in old soleus muscles (Table 2). Among them were troponin I, several isoforms of myosin heavy chain (4, 6, 7, and 13), and prolargin. We hypothesize that Troponin may be strongly correlated with scarponia. Recently, ZHANG et al.54 have demonstrated a role for troponin in nuclear imbalance and muscle aging and overexpression of TnT fragments resulted in defects in nuclear shape and caused high levels of apoptosis.
Among the down-regulated proteins were actin, myosin regulatory light chain, Actn2, Col6a2, Fln C, and different chains of tropomyosin (alpha-1, alpha-3 and alpha-4). Other proteins that are involved in myofibrils assembly such as PDZ and LIM domain protein 5 were also down-regulated.
In EDL muscles, among the up-regulated proteins were two alpha chains of type VI collagen (Col6a2 and Col6a3), FlnC (filamin C), myosin-binding protein C, alpha-actinin 1, tropomyosin beta chain, Actn2 (actinin, alpha 2), and several chains of myosin regulatory light chain (light chain 1/3, light chain 2 and light chain 4) (Table 3).
Collagen proteins play a role in maintaining the integrity of various tissues. Age-related studies of collagen indicate that collagen content varies with age and type of muscle and a 40% increase in total collagen has been found in fast-twich type of muscles55. Col6a2 along with prolargin were the two proteins with the greatest increase in expression (−Log2 fold change = +23.10 and + 23.5, respectively).
The down-regulated group of proteins contained myosin heavy chain (chains 4, 7, and 8), cytoskeletal type II keratin, collagen alpha, and vimentin. Vimentin is a constituent of cytoskeletal filaments and in gastrocnemius rat muscle it is increased with age47.
Oxidative stress, detoxification and degradation
In the up-regulated proteins of the soleus muscle, we identified two isoforms of glutathione S-transferase and ferritin (Table 2). Increased levels of glutathione S-transferase suggests increased detoxification of cytotoxic products, while higher levels of ferritin indicate an altered iron metabolism.
On the other hand, some heat shock proteins such as Hsp90, heat shock 71kDa protein, Hspb6, endoplasmin and chaperonin subunit 8 were down regulated. Deficits in chaperone function have been reported in several age-related diseases and literature also supports that synthesis of heat shock proteins is impaired in aging57, 58. The DNA damage-binding protein 1 is responsible for repair of UV-damaged DNA59; therefore, reduced levels of this protein may leave the DNA more susceptible to damage.
In EDL muscles, proteins involved in scavenging of reactive oxygen species, as well as enzymes involved in the detoxification of cytotoxic products were found to be up-regulated in old muscles (Table 3). Among them were several isoforms of glutathione-S-transferase (GSTs), glutathione peroxidase (GPx), heat shock proteins (71kDa protein and HSP 90-beta) and the antioxidant enzymes superoxide dismutase [Cu/Zn] and superoxide dismutase [Mn]. The up-regulation of glutathione transferase suggests increased detoxification of cytotoxic products while the up-regulation of glutathione peroxidase may be a counterbalance for increased levels of oxidative stress. Higher levels of chaperones may be needed to fold an increased number of misfolded proteins in aged muscles.
Transport and catabolism
In the transport and catabolism class, all of the identified proteins were down-regulated in aged soleus muscles (Table 2). These proteins are clathrin heavy chain 1, complement component 4, the EH domain-containing protein 2, serum albumin and zero beta-globin.
The complement component 4 (C4) is involved in classical complement activation and there is evidence in the literature suggesting that the immune system deteriorates with age, rendering old animals less able to mount an immune response61. C4 prevents early stage autoimmune diseases62 and mice with a disrupted C4 locus show impaired immune response63.
In EDL muscles an ADP/ATP translocase 1, BSA, carboxymethylenebutenolidase, kelch repear and BTB domain-containing protein, protein Bgg-b1, serum albumin and serotransferrin were up-regulated with aging while transthyretin was down-regulated (Table 3).
Serum albumin was found to be increased, suggesting a shift to more aerobic-oxidative metabolism in aged fibres23. Serum albumin plays a crucial role in maintaining the osmotic blood pressure and apparently also plays a role as radical and heme scavenger64. Serotransferrin was also increased and it is involved in the control of stress and iron levels by increasing iron uptake.
Additional differentially expressed proteins
In soleus muscles (Table 2), among the up-regulated proteins present were putative uncharacterized proteins, T-kininogen 2, alpha-1-macroglobulin, NAD(P)H-hydrate epimerase and Obg-like ATPase 1. T-kininogen´s (T-KG) expression has been shown to be considerably increased during aging in the liver of Sprague-Dawley rats65 and serum of Fisher rats. Some authors66,67 have suggested that T-KG may be a reliable biomarker for senescence in rats66,67. This protein may be involved in the deterioration of the immune system that occurs with aging. On the other hand, annexin, cadherin 13, alpha-1-inhibitor 3, N(G),N(G)-dimethylarginine dimethylaminohydrolase 1, basic leucine zipper, W2 domain-containing protein 2, glioblastoma amplified sequence, and two uncharacterized proteins were present at reduced levels.
Several proteins were up-regulated in old EDL muscles (Table 3). Among them, T-kininogen was also up-regulated in soleus muscles, presented a greater increase in EDL (−Log2 fold change = 1.8 and 3.9 for soleus and EDL, respectively). Calsequestrin and parvalbumin (PV) are Ca2+ binding proteins that were up-regulated in old EDL muscles. Ca2+ translocation from the myofibril to the sarcoplasmic reticulum (SR) is facilitated by parvalbumin in the fast-twitch skeletal muscle. The energy-dependent Ca2+ uptake into the SR is mediated by the SR ATPase, that is regulated by both Ca2+ - and CaM-dependent phosphorylation. The Ca2+ cycle is completed by binding of Ca2+ to the high-capacity, low-affinity Ca2+ -binding protein calsequestrin. Therefore, altered levels of both parvalbumin and calsequestrin may be related to the age-related impairment of intrinsic SR function and influence the speed of contraction in old fast-twitch motor units68.
A voltage-dependent calcium channel (Q9ERS3) was also up-regulated in old muscles. Up-regulation of anion-selective channel proteins in aging has already been reported47 and it may be a way to facilitate the access of kinases to ATP, therefore bypassing the restriction exerted by the mitochondrial outer membrane on the permeability for metabolites69.
Carbonic anhydrase III (CAIII) is a cytosolic zinc-containing enzyme which facilitates the transport of CO2 in skeletal muscle by catalyzing the reversible hydration of CO2. This activity is probably involved in the maintenance of ionic balance and acid-base homoeostasis within the muscle tissue. This enzyme is detected in large amounts in red slow-twitch muscles such as soleus70,71, but is virtually absent from adult white fast-twitch muscle, such as rodent anterior tibialis (AT) and EDL70. The age-dependent expression of this protein is still controversial in the literature. While some authors have demonstrated down regulation of CA3 in slow-type muscles such as gastrocnemius24,46,60, others have shown up-regulation47,72 in the same muscle type.
Oxidative stress in plasma and muscle
Taking in consideration the increased levels of antioxidant and enzymes involved in detoxification such as glutathione S-transferase, superoxide dismutase [Mn], and superoxide dismutase [Cu/Zn] that were up-regulated in both soleus and EDL muscles we decided to verify whether there were increased levels of oxidative stress in plasma and muscle of the old animals.
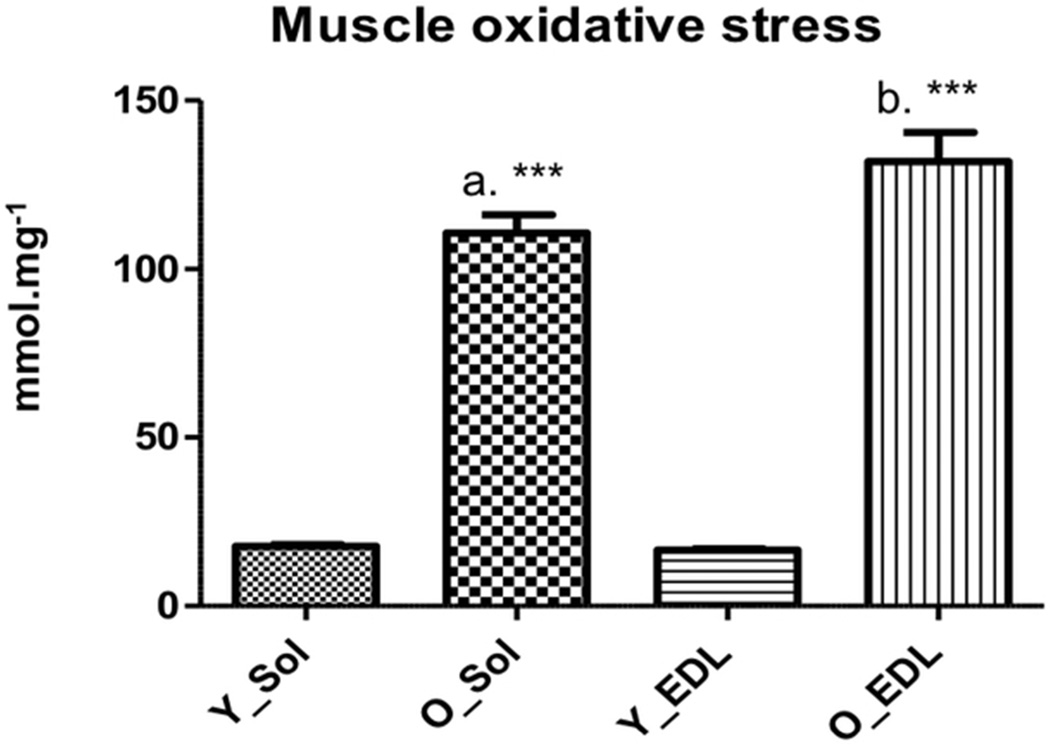
We determined oxidative stress levels in plasma and muscle of young and old animals using the FOX assay36. For the experiment at hand, we were unable to show statistical differences in the plasma of young versus old animals (1.08 ± 0.10mmol.mg−1 and 1.12 ± 0.07 mmol.mg−1, p = 0.33, respectively) (). However muscle oxidative stress was higher in old animals for both soleus and EDL muscles (17.71 ± 1.42 mmol.mg−1 and 110.69 ± 16.21 mmol.mg−1, p < 0.0001, for young and old soleus, respectively, and 16.54 ± 1.13 mmol.mg−1 and 131.89 ± 26.06 mmol.mg−1, p < 0.0001 for young and old EDL, respectively) (Figure 2).
Figure 2.
Soleus and EDL FOX values in the young and old group. Y_Sol = young group, soleus; O_Sol = old group, soleus; Y_EDL = young group, EDL; O_EDL = old group, EDL. a = p < 0,0001 vs. Y_Sol;; b = p < 0,0001 vs. Y_EDL; c = p < 0,0001 vs. YH. No statistical differences were found between O_Sol × O_EDL and Y_Sol × Y_EDL.
Creatine kinase and pyruvate kinase activity
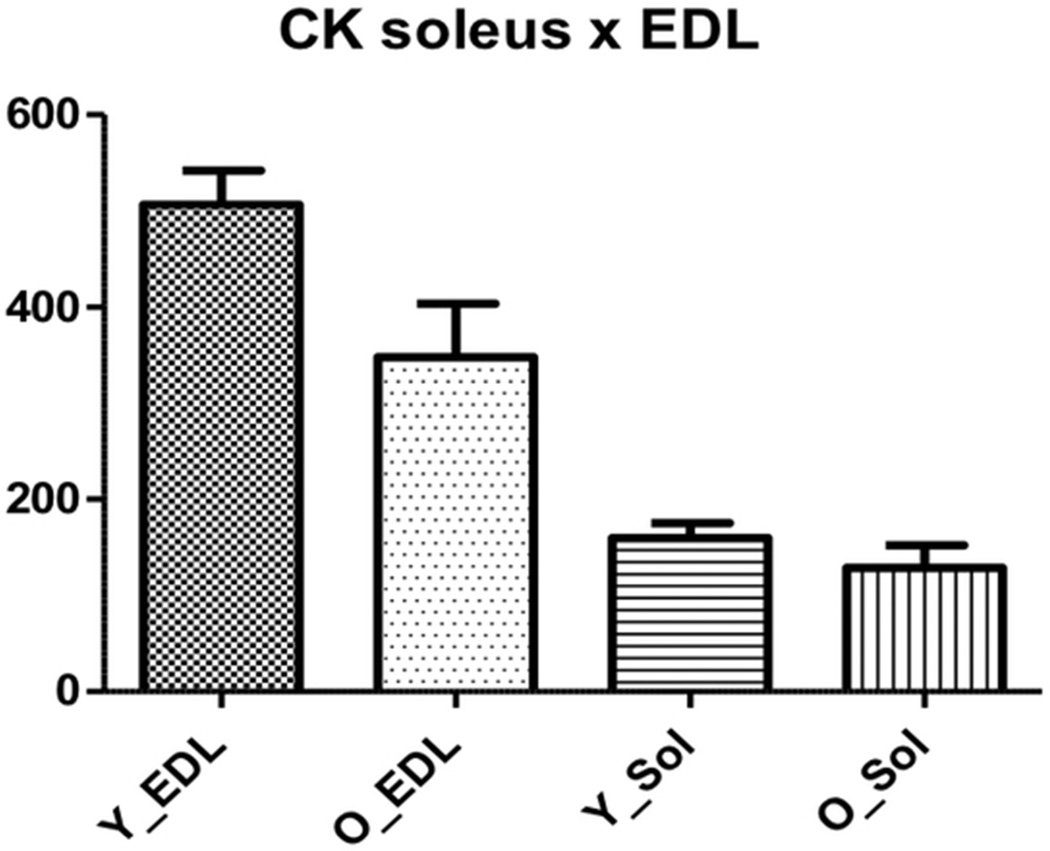
Creatine kinase catalyzes the reversible transfer of phosphate between ATP and creatine. The effect of aging on the levels of muscle CK has been controversial. We found that creatine kinase was up-regulated in old EDL muscles while there were no changes in expression for soleus muscles. Capitanio et al.47 and O’Connel et al.23 found decreased levels of this enzyme in gastrocnemius muscles while Doran et al.46 found a two-fold increase in the same muscle type and Donogue et al.48 reported differential effects of CK in 2D-gels of gastrocnemius muscles.
Therefore we evaluated CK activity to investigate whether an altered expression also resulted in different activities. When comparing the different muscles we found that CK activity was higher in EDL muscles, for both young and old (506.88 ± 145.62 and 159.74 ± 63.22, p < 0.0001 for young EDL and young soleus respectively and 347.52 ± 194.31 and 128.52 ± 78.52, p = 0.0022 for old EDL and old soleus muscles respectively) (Figure 3).
Figure 3.
Soleus and EDL creatine kinase (CK) activity in the young and old group. Y_Sol = young group, soleus; O_Sol = old group, soleus; Y_EDL = young group, EDL; O_EDL = old group, EDL. a. Young EDL × young soleus, p < 0.0001; b. old EDL × old soleus, p = 0.0022; c: young EDL × old EDL, p = 0.01.
When we compared young versus old soleus muscles we found that CK activity was not different (159.74 ± 63.22 and 128.52 ± 78.52, p = 0.25 for young old muscles respectively) (Figure 3). On the other hand, CK´s activity was lower in old EDL muscles (506.88 ± 145.62 and 347.52 ± 194.31, p = 0.01 for young and old muscles respectively) (Figure 3). Therefore, increased levels of CK may be a physiological response to counterbalance the reduced activity of this enzyme with aging. An age-related decline in this enzyme´s activity has been shown to occur in humans and rodents73,74.
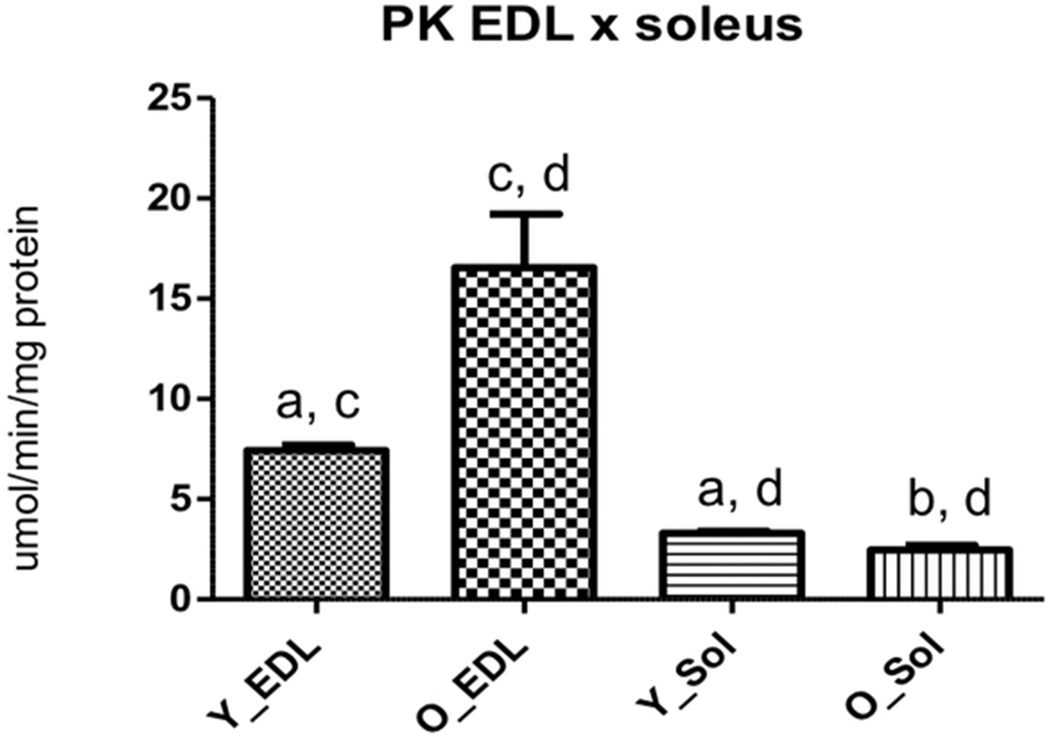
Pyruvate kinase (PK), a key enzyme in the glycolytic pathway, was differentially regulated in both soleus and EDL muscles. In EDL, it was up-regulated in the old muscles while in soleus muscles the result was the opposite. We evaluated PK activity and found that it was higher for EDL when compared to soleus (7.43 ± 1.09 and 3.30 ± 0.42, p < 0.0001 for young EDL and young soleus muscles respectively and 16.55 ± 9.25 and 2.47 ± 0.79, p < 0.0001 for old EDL and old soleus respectively) (Figure4). Furthermore, in EDL old muscles there was an increased activity for this enzyme (7.43 ± 1.09 and 16.55 ± 9.25, p < 0.0001 for young and old, respectively) (Figure4), while for soleus muscles, PK activity decreased with age (3.30 ± 0.42 and 2.47 ± 0.79, p = 0.0018 for young and old, respectively) (Figure4).
Figure 4.
Soleus and EDL pyruvate kinase (PK) activity in the young and old group. Y_Sol = young group, soleus; O_Sol = old group, soleus; Y_EDL = young group, EDL; O_EDL = old group, EDL. a. Young EDL × young soleus, p < 0.0001; b. old EDL × old soleus, p < 0.0001; c. young EDL × old EDL, p < 0.0001; d. young soleus × old soleus, p = 0.0018.
DISCUSSION
Over the last few years several proteomic studies have investigated changes in the protein complement of aging skeletal muscles in humans and animal considered as aging models (reviewed by DORAN et al.24). Most of these studies used two-dimensional electrophoresis and mass spectrometry as the main tool to identify differentially regulated proteins (reviewed by DORAN et al. 24).
As far as we know, the use of TMT coupled with LC/LC/MS/MS mass spectrometry based proteomics allowed us to identify and quantify a larger number of proteins from both soleus and EDL muscles than any preceding report (total of 5300 proteins, out of which 252 were differentially regulated in aging).
We chose to analyze two different muscles, one in which fast-twich type II fibers predominate (EDL) and the other in which the type I, slow-twich fibers are more abundant (soleus) in order to have a comprehensive overview of the effects of aging on different types of muscles. Our investigation revealed many differences in the protein expression pattern of young versus old muscles, and there were considerable differences between soleus and EDL muscles as well.
Histological analysis of soleus and EDL showed striking features of aged muscles which include extensive variability in fiber diameter, a higher frequency of longitudinal splitting, an increased number of centrally located nuclei and thickening of the endomysium25, 75 which indicate that the old rats used in this study (24 months old) did show signs of sarcopenia.
Furthermore, the sarcopenia index, which indicates the degree of muscle wasting was decreased for both muscles, and the decrease was more pronounced in the soleus than in the EDL muscle. Although sarcopenia is widely considered to preferentially impact fast twitch muscles76 this notion may not be applied at more advanced ages. Hagen et al.77 have shown that the relative protection of the slow twitch soleus muscle from age-related atrophy is present only until middle age with a great degree of atrophy present thereafter.
Our results are in agreement with those of Carter et al.78 which have shown that slow twitch soleus muscles undergo large phenotypic alterations in very old age and also that, for several measures, it is of greater magnitude than fast twitch muscle. Furthermore, there are several reports of atrophy, force decline, altered MHC expression and myofiber loss in aged rat soleus muscles77,79,80.
The identification of several metabolic enzymes among the differentially regulated proteins suggests perturbations in the energy metabolism of old skeletal muscles. Only one enzyme, L-lactate dehydrogenase (LDH) was up-regulated in both soleus and EDL muscles. The effect of aging in the expression levels of this enzyme is still controversial in the literature. While Doran et al.46 and Donogue et al.48 found LDH to be one of the most drastically up-regulated proteins in old gastrocnemius muscle, Capitanio et al.47 showed decreased levels of this enzyme in the same muscle type.
The mitochondria is the major cellular site for ATP production. During aging, the respiratory chain function (RCF) falls considerably in humans between ages 17 and 90 accompanied by an impaired function of cytochrome c oxidase in old muscles81.
Down-regulation of several components of the respiratory chain was observed for soleus muscles, while this trend was not true for EDL muscles, in which some components were up while others were down-regulated. Soleus muscles may be more susceptible to oxidative stress that plays a role in mitochondrial dysfunction and may therefore contribute to decreased expression of mitochondrial enzymes.
Interestingly, 11 metabolic enzymes were regulated in opposite ways in both muscles (down-regulated in soleus and up-regulated in EDL) (Table 4). These enzymes are involved in amino acid metabolism (aspartate aminotransferase, both the cytoplasmic and mitochondrial isoforms), glycolysis/gluconeogenesis (beta-enolase, phosphoglycerate kinase, phosphoglycerate mutase and pyruvate kinase) and oxidative metabolism (ATP synthase subunit beta, cytochrome c oxidase and electron transfer flavoprotein).
Table 4.
Metabolic enzymes regulated in opposite ways in soleus and EDL muscles
| SOLEUS | EDL | ||||
|---|---|---|---|---|---|
| Protein accession no. |
DESCRIPTION | P value | −LOG2 fold change |
P value | −LOG2 fold change |
| P13221 | Aspartate aminotransferase, cytoplasmic | < 0.001 | −0.8 | < 0.001 | 1.3 |
| P13221 | Aspartate aminotransferase, mitochondrial | 0.001 | −1.1 | < 0.001 | 1.1 |
| F1LP05 | ATP synthase subunit alpha | < 0.001 | −0.9 | < 0.001 | 0.8 |
| P15429 | Beta-enolase | < 0.001 | −0.6 | < 0.001 | 1.2 |
| P10888 | Cytochrome c oxidase subunit 4 isoform 1, mitochondrial |
< 0.001 | −1.3 | < 0.001 | 0.7 |
| Q68FU3 | Electron transfer flavoprotein subunit beta | 0.005 | −1.0 | < 0.001 | 0.4 |
| P42123 | L-lactate dehydrogenase B chain | < 0.001 | 0.6 | < 0.001 | 0.8 |
| P16617 | Phosphoglycerate kinase 1 | < 0.001 | −0.9 | < 0.001 | 0.8 |
| P25113 | Phosphoglycerate mutase 1 | < 0.001 | −1.4 | < 0.001 | 0.8 |
| Q6P7S0 | Pyruvate kinase | 0.001 | −0.9 | < 0.001 | 0.8 |
| Q64428 | Trifunctional enzyme subunit alpha, mitochondrial |
0.005 | −0.7 | < 0.001 | 1.0 |
The glioblastoma amplified sequence (GAS), according to our quantitative strategy, showed to be the most down-regulated protein (Log2 fold change = −22.1), along with a yet uncharacterized protein (D3ZH98, Log2 fold change = −21.1). Zero beta-globin, a protein that participates in oxygen transport, was also drastically decreased (Log2 fold change = −16.9). Therefore, the down-regulation of both GAS and zero beta-globin may contribute for the reduction of oxidative capacity in the elderly.
In muscle, the interaction between myosin and actin filaments is responsible for force production and several of the differentially expressed proteins belong to the myosin-actin complex, suggesting that force production may be compromised in old rats due to a more disorganized structure (some of the components were down and other components were up-regulated). In soleus muscles, the structural proteins tropomyosin (alpha and beta chains) were among the most down-regulated proteins (Log2 fold change = −18.4 and −18.2, respectively) while troponin I was among the most up-regulated (Log2 fold change = 17.8).
The thin filament regulatory proteins troponin (Tn) and tropomyosin (Tm) are essential for contraction of striated muscle (skeletal and cardiac), which is regulated by the concentration of intracellular calcium82,83. Such a drastic decrease in tropomyosin proteins may be responsible for a lower force contraction and muscle velocity in the elderly. The up-regulated of troponin confirms previous studies which have described this protein as a potential new biomarker for sarcopenia45.
During aging there is an increase in the production of reactive oxygen species (ROS) due to the functional deterioration of mitochondria, and the increase in free radicals generation may contribute to several age-related pathologies84,85,86. Enhanced ROS production may contribute to an accumulation of mtDNA damage and increased apoptosis of muscles fibers, which may represent a key mechanism underlying sarcopenia87. Glutathione S-transferase (GSTs) was up-regulated in both muscles while two of the main antioxidant enzymes, glutathione peroxidase (GPx), which controls the levels of hydrogen peroxide and superoxide dismutase (SOD) were up-regulated only in EDL muscles. In EDL muscles, GPx was among the most up-regulated (Log2 fold change = 26.8). Up-regulation of GPx has been reported in Duchenne muscular distrohpy (DMD), a progressive muscle wasting disorder that impairs muscle function and ultimately results in muscle degeneration and death88. Therefore, an increase in GPx expression is most likely a compensatory attempt to counterbalance increased levels of oxidative stress.
We also performed an assay (FOX) to determine the levels of oxidative stress in plasma and muscles. We confirmed that old muscles had significantly higher oxidative stress than young muscles, which suggests that a higher level of antioxidant enzymes may be an attempt to counterbalance oxidative stress. However, we were unable to find a convincing statistical difference between soleus and EDL. Nevertheless, we encourage future studies to consider complementary features such as the activity of specific antioxidant enzymes, or the content of reduced glutathione to further investigate the difference between these muscles. The soleus muscle is known to be more susceptible to oxidative stress than EDL due to its higher oxidative potential when compared to a fast-twitch muscle89.
There is still controversy in the literature to whether aging increases or decreases the enzymatic antioxidant defenses in the cell. Studies conducted in both humans and rats have indicated that although the antioxidant system undergoes significant changes, both up, down-regulation or no change of the antioxidant enzymes may occur, suggesting that the results may be dependent of muscle fiber composition and other factors such as age and sex48,60,87,90.
Our results show that aging affects slow-twich and fast-twitch muscles in different ways and that the most significant differences were found for metabolic enzymes. Furthermore, EDL muscles had a higher number of differentially expressed proteins (16.2% of the total identified proteins had a change in abundance against 3.8% of the total proteins for soleus muscles), suggesting that aging affects protein expression more drastically in a fast-twitch type of muscle.
Compared to previous age-related muscle proteome studies24 we identified novel muscle proteins with a change in abundance during aging. Among these were the glioblastoma amplified sequence (GAS), with a marked down-regulated in old soleus muscles, the uncharacterized protein D3ZH98, zero beta-globin and prolargin.
Therefore, the confirmation of many already identified proteins involved with aging and the identification of new candidate biomarkers will help in the establishment of a more comprehensive database for the study of sarcopenia and aid in the development of new diagnostic methods and treatment for age-associated muscular disorders.
Supplementary Material
Acknowledgments
This work was funded by FAPESP, grants 2009/52022-2 and 2010/10852-6 J.J.M. and J.R.Y. were supported by the National Center for Research Resources (P41RR011823), National Institute of General Medical Sciences (P41GM103533), and National Institute on Aging (AG027463). PCC was supported by Fiocruz – PDTIS. DBL was supported by IBMP and CAPES.
Footnotes
Associated Content - Supporting Information: This material is available free of charge via the internet at http://publs.acs.org
Availability: The RAW data, .sqt files and SEPro files are available via internet at http://proteomics.fiocruz.br/daniela/jpr2013-1.
References
- 1.Melton LJ, III, Khosla S, Crowson CS, O'Connor MK, O'Fallon WM, Riggs BL. Epidemiology of sarcopenia. J Am Geriatr Soc. 2000;48(6):625–630. [PubMed] [Google Scholar]
- 2.Morley JE, Baumgartner RN, Roubenoff R, Mayer J, Nair KS. Sarcopenia. J Lab Clin Med. 2001;137(4):231–243. doi: 10.1067/mlc.2001.113504. [DOI] [PubMed] [Google Scholar]
- 3.Greenlund LJ, Nair KS. Sarcopenia--consequences, mechanisms, and potential therapies. Mech Ageing Dev. 2003;124(3):287–299. doi: 10.1016/s0047-6374(02)00196-3. [DOI] [PubMed] [Google Scholar]
- 4.Vandervoot AA, Symons TB. Functional and metabolic consequences of sarcopenia. Can J Appl Physiol. 2001;26(1):90–101. doi: 10.1139/h01-007. [DOI] [PubMed] [Google Scholar]
- 5.Janssen I, Heymsfield SB, Ross R. Low relative skeletal muscle mass (sarcopenia) in older persons is associated with functional impairment and physical disability. J Am Geriatr Soc. 2002;50(5):889–896. doi: 10.1046/j.1532-5415.2002.50216.x. [DOI] [PubMed] [Google Scholar]
- 6.Doherty TJ. Invited review: Aging and sarcopenia. J Appl Physiol. 2003;95(4):1717–1727. doi: 10.1152/japplphysiol.00347.2003. [DOI] [PubMed] [Google Scholar]
- 7.Dirks A, Leeuwenburgh C. Apoptosis in skeletal muscle with aging. Am J Physiol Regul Integr Comp Physiol. 2002;282(2):R519–R527. doi: 10.1152/ajpregu.00458.2001. [DOI] [PubMed] [Google Scholar]
- 8.Renault V, Thornell LE, Eriksson PO, Butler-Browne G, Mouly V. Regenerative potential of human skeletal muscle during aging. Aging Cell. 2002;1(2):132–139. doi: 10.1046/j.1474-9728.2002.00017.x. [DOI] [PubMed] [Google Scholar]
- 9.Vandervoort AA. Aging of the human neuromuscular system. Muscle Nerve. 2002;25(1):17–25. doi: 10.1002/mus.1215. [DOI] [PubMed] [Google Scholar]
- 10.Balagopal P, Rooyackers OE, Adey DB, Ades PA, Nair KS. Effects of aging on in vivo synthesis of skeletal muscle myosin heavy-chain and sarcoplasmic protein in humans. Am J Physiol. 1997;273(4 Pt 1):E790–E800. doi: 10.1152/ajpendo.1997.273.4.E790. [DOI] [PubMed] [Google Scholar]
- 11.Pastoris O, Boschi F, Verri M, Baiardi P, Felzani G, Vecchiet J, Dossena M, Catapano M. The effects of aging on enzyme activities and metabolite concentrations in skeletal muscle from sedentary male and female subjects. Exp Gerontol. 2000;35(1):95–104. doi: 10.1016/s0531-5565(99)00077-7. [DOI] [PubMed] [Google Scholar]
- 12.Taylor DJ, Kemp GJ, Thompson CH, Radda GK. Ageing: effects on oxidative function of skeletal muscle in vivo. Mol Cell Biochem. 1997;174(1–2):321–324. [PubMed] [Google Scholar]
- 13.Carmeli E, Coleman R, Reznick AZ. The biochemistry of aging muscle. Exp Gerontol. 2002;37(4):477–489. doi: 10.1016/s0531-5565(01)00220-0. [DOI] [PubMed] [Google Scholar]
- 14.Harman D. The aging process. Proc Natl Acad Sci U S A. 1981;78(11):7124–7128. doi: 10.1073/pnas.78.11.7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peterson CM, Johannsen DL, Ravussin E. SkeletalMuscleMitochondria and Aging: A Review. J Agin Res. 2012:1–20. doi: 10.1155/2012/194821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanski J, Alterman MA, Schoneich C. Proteomic identification of age-dependent protein nitration in rat skeletal muscle. Free Radic Biol Med. 2003;35(10):1229–1239. doi: 10.1016/s0891-5849(03)00500-8. [DOI] [PubMed] [Google Scholar]
- 17.Kanski J, Hong SJ, Schoneich C. Proteomic analysis of protein nitration in aging skeletal muscle and identification of nitrotyrosine-containing sequences in vivo by nanoelectrospray ionization tandem mass spectrometry. J Biol Chem. 2005;280(25):24261–24266. doi: 10.1074/jbc.M501773200. [DOI] [PubMed] [Google Scholar]
- 18.Piec I, Listrat A, Alliot J, Chambon C, Taylor RG, Bechet D. Differential proteome analysis of aging in rat skeletal muscle. FASEB J. 2005;19(9):1143–1145. doi: 10.1096/fj.04-3084fje. [DOI] [PubMed] [Google Scholar]
- 19.Isfort RJ. Proteomic analysis of striated muscle. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;771(1–2):155–165. doi: 10.1016/s1570-0232(02)00056-9. [DOI] [PubMed] [Google Scholar]
- 20.Doran P, Donoghue P, O'Connell K, Gannon J, Ohlendieck K. Proteomic profiling of pathological and aged skeletal muscle fibres by peptide mass fingerprinting (Review) Int J Mol Med. 2007;19(4):547–564. [PubMed] [Google Scholar]
- 21.Doran P, Gannon J, O'Connell K, Ohlendieck K. Proteomic profiling of animal models mimicking skeletal muscle disorders. Proteomics Clin Appl. 2007;1(9):1169–1184. doi: 10.1002/prca.200700042. [DOI] [PubMed] [Google Scholar]
- 22.Gelfi C, Vigano A, Ripamonti M, Pontoglio A, Begum S, Pellegrino MA, Grassi B, Bottinelli R, Wait R, Cerretelli P. The human muscle proteome in aging. J Proteome Res. 2006;5(6):1344–1353. doi: 10.1021/pr050414x. [DOI] [PubMed] [Google Scholar]
- 23.O'Connell K, Gannon J, Doran P, Ohlendieck K. Proteomic profiling reveals a severely perturbed protein expression pattern in aged skeletal muscle. Int J Mol Med. 2007;20(2):145–153. [PubMed] [Google Scholar]
- 24.Doran P, Donoghue P, O'Connell K, Gannon J, Ohlendieck K. Proteomics of skeletal muscle aging. Proteomics. 2009;9(4):989–1003. doi: 10.1002/pmic.200800365. [DOI] [PubMed] [Google Scholar]
- 25.Edstrom E, Altun M, Bergman E, Johnson H, Kullberg S, Ramirez-Leon V, Ulfhake B. Factors contributing to neuromuscular impairment and sarcopenia during aging. Physiol Behav. 2007;92(1–2):129–135. doi: 10.1016/j.physbeh.2007.05.040. [DOI] [PubMed] [Google Scholar]
- 26.Gannon J, Staunton L, O'Connell K, Doran P, Ohlendieck K. Phosphoproteomic analysis of aged skeletal muscle. Int J Mol Med. 2008;22(1):33–42. [PubMed] [Google Scholar]
- 27.Hojlund K, Bowen BP, Hwang H, Flynn CR, Madireddy L, Geetha T, Langlais P, Meyer C, Mandarino LJ, Yi Z. In vivo phosphoproteome of human skeletal muscle revealed by phosphopeptide enrichment and HPLC-ESI-MS/MS. J Proteome Res. 2009;8(11):4954–4965. doi: 10.1021/pr9007267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cieniewski-Bernard C, Bastide B, Lefebvre T, Lemoine J, Mounier Y, Michalski JC. Identification of O-linked N-acetylglucosamine proteins in rat skeletal muscle using two-dimensional gel electrophoresis and mass spectrometry. Mol Cell Proteomics. 2004;3(6):577–585. doi: 10.1074/mcp.M400024-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.O'Connell K, Doran P, Gannon J, Ohlendieck K. Lectin-based proteomic profiling of aged skeletal muscle: decreased pyruvate kinase isozyme M1 exhibits drastically increased levels of N-glycosylation. Eur J Cell Biol. 2008;87(10):793–805. doi: 10.1016/j.ejcb.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 30.Hedou J, Bastide B, Page A, Michalski JC, Morelle W. Mapping of O-linked beta-N-acetylglucosamine modification sites in key contractile proteins of rat skeletal muscle. Proteomics. 2009;9(8):2139–2148. doi: 10.1002/pmic.200800617. [DOI] [PubMed] [Google Scholar]
- 31.Feng J, Xie H, Meany DL, Thompson LV, Arriaga EA, Griffin TJ. Quantitative proteomic profiling of muscle type-dependent and age-dependent protein carbonylation in rat skeletal muscle mitochondria. J Gerontol A Biol Sci Med Sci. 2008;63(11):1137–1152. doi: 10.1093/gerona/63.11.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rivero JL, Talmadge RJ, Edgerton VR. Correlation between myofibrillar ATPase activity and myosin heavy chain composition in equine skeletal muscle and the influence of training. Anat Rec. 1996;246(2):195–207. doi: 10.1002/(SICI)1097-0185(199610)246:2<195::AID-AR6>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 33.Gupta RC, Misulis KE, Dettbarn WD. Activity dependent characteristics of fast and slow muscle: biochemical and histochemical considerations. Neurochem Res. 1989;14(7):647–655. doi: 10.1007/BF00964874. [DOI] [PubMed] [Google Scholar]
- 34.Washburn MP, Wolters D, Yates JR., III Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19(3):242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 35.Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Johnstone R, Mohammed AK, Hamon C. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem. 2003;75(8):1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 36.Nourooz-Zadeh J. Ferrous ion oxidation in presence of xylenol orange for detection of lipid hydroperoxides in plasma. Methods Enzymol. 1999;300:58–62. doi: 10.1016/s0076-6879(99)00113-5. [DOI] [PubMed] [Google Scholar]
- 37.Ainsworth S, MacFarlane N. A kinetic study of rabbit muscle pyruvate kinase. Biochem J. 1973;131(2):223–236. doi: 10.1042/bj1310223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dinovo EC, Miyada DS, Nakamura RM. Evaluation of direct and indirect coupled enzyme assay systems for measurement of creatine kinase activity. Clin Chem. 1973;19(9):994–997. [PubMed] [Google Scholar]
- 39.McDonald WH, Tabb DL, Sadygov RG, MacCoss MJ, Venable J, Graumann J, Johnson JR, Cociorva D, Yates JR., III MS1, MS2, and SQT-three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid Commun Mass Spectrom. 2004;18(18):2162–2168. doi: 10.1002/rcm.1603. [DOI] [PubMed] [Google Scholar]
- 40.Xu T, Venable JD, Park SK, Cociorva D, Liao L, Wohlschlegel J, Hewel J, Yates JR. ProLuCID, a fast and sensitive tandem mass spectra-based protein identification program. Mol.Cell.Proteomics. 2006;5(S174) [Google Scholar]
- 41.Carvalho PC, Fischer JS, Xu T, Yates JR, III, Barbosa VC. PatternLab: from mass spectra to label-free differential shotgun proteomics. Curr Protoc Bioinformatics. 2012 doi: 10.1002/0471250953.bi1319s40. Chapter 13:Unit13. [DOI] [PubMed] [Google Scholar]
- 42.Carvalho PC, Fischer JS, Xu T, Cociorva D, Balbuena TS, Valente RH, Perales J, Yates JR, III, Barbosa VC. Search engine processor: Filtering and organizing peptide spectrum matches. Proteomics. 2012;12(7):944–949. doi: 10.1002/pmic.201100529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. 1. J.R.Statist.Soc.B. 1995;57:289–300. [Google Scholar]
- 44.Carvalho PC, Fischer JS, Chen EI, Yates JR, III, Barbosa VC. PatternLab for proteomics: a tool for differential shotgun proteomics. BMC Bioinformatics. 2008;9:316. doi: 10.1186/1471-2105-9-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang B, Chambers MC, Tabb DL. Proteomic parsimony through bipartite graph analysis improves accuracy and transparency. J Proteome Res. 2007;6(9):3549–3557. doi: 10.1021/pr070230d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doran P, O'Connell K, Gannon J, Kavanagh M, Ohlendieck K. Opposite pathobiochemical fate of pyruvate kinase and adenylate kinase in aged rat skeletal muscle as revealed by proteomic DIGE analysis. Proteomics. 2008;8(2):364–377. doi: 10.1002/pmic.200700475. [DOI] [PubMed] [Google Scholar]
- 47.Capitanio D, Vasso M, Fania C, Moriggi M, Vigano A, Procacci P, Magnaghi V, Gelfi C. Comparative proteomic profile of rat sciatic nerve and gastrocnemius muscle tissues in ageing by 2-D DIGE. Proteomics. 2009;9(7):2004–2020. doi: 10.1002/pmic.200701162. [DOI] [PubMed] [Google Scholar]
- 48.Donoghue P, Staunton L, Mullen E, Manning G, Ohlendieck K. DIGE analysis of rat skeletal muscle proteins using nonionic detergent phase extraction of young adult versus aged gastrocnemius tissue. J Proteomics. 2010;73(8):1441–1453. doi: 10.1016/j.jprot.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 49.Seroussi E, Pan HQ, Kedra D, Roe BA, Dumanski JP. Characterization of the human NIPSNAP1 gene from 22q12: a member of a novel gene family. Gene. 1998;212(1):13–20. doi: 10.1016/s0378-1119(98)00098-5. [DOI] [PubMed] [Google Scholar]
- 50.Wang XY, Smith DI, Liu W, James CD. GBAS, a novel gene encoding a protein with tyrosine phosphorylation sites and a transmembrane domain, is co-amplified with EGFR. Genomics. 1998;49(3):448–451. doi: 10.1006/geno.1998.5239. [DOI] [PubMed] [Google Scholar]
- 51.Conley KE, Jubrias SA, Esselman PC. Oxidative capacity and ageing in human muscle. J Physiol. 2000;526(Pt 1):203–210. doi: 10.1111/j.1469-7793.2000.t01-1-00203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang J, Cornell JE, Van Remmen H, Hakala K, Ward WF, Richardson A. Effect of aging and caloric restriction on the mitochondrial proteome. J Gerontol A Biol Sci Med Sci. 2007;62(3):223–234. doi: 10.1093/gerona/62.3.223. [DOI] [PubMed] [Google Scholar]
- 53.O'Connell K, Ohlendieck K. Proteomic DIGE analysis of the mitochondria-enriched fraction from aged rat skeletal muscle. Proteomics. 2009;9(24):5509–5524. doi: 10.1002/pmic.200900472. [DOI] [PubMed] [Google Scholar]
- 54.Zhang T, Birbrair A, Wang ZM, Taylor J, Messi ML, Delbono O. Troponin T nuclear localization and its role in aging skeletal muscle. Age (Dordr) 2013;35(2):353–370. doi: 10.1007/s11357-011-9368-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mohan S, Radha E. Age-related changes in rat muscle collagen. Gerontology. 1980;26(2):61–67. doi: 10.1159/000212396. [DOI] [PubMed] [Google Scholar]
- 56.Sohal RS, Orr WC. Relationship between antioxidants, prooxidants, and the aging process. Ann N Y Acad Sci. 1992;663:74–84. doi: 10.1111/j.1749-6632.1992.tb38651.x. [DOI] [PubMed] [Google Scholar]
- 57.Hall DM, Xu L, Drake VJ, Oberley LW, Oberley TD, Moseley PL, Kregel KC. Aging reduces adaptive capacity and stress protein expression in the liver after heat stress. J Appl Physiol. 2000;89(2):749–759. doi: 10.1152/jappl.2000.89.2.749. [DOI] [PubMed] [Google Scholar]
- 58.Pahlavani MA, Harris MD, Moore SA, Weindruch R, Richardson A. The expression of heat shock protein 70 decreases with age in lymphocytes from rats and rhesus monkeys. Exp Cell Res. 1995;218(1):310–318. doi: 10.1006/excr.1995.1160. [DOI] [PubMed] [Google Scholar]
- 59.Dualan R, Brody T, Keeney S, Nichols AF, Admon A, Linn S. Chromosomal localization and cDNA cloning of the genes (DDB1 and DDB2) for the p127 and p48 subunits of a human damage-specific DNA binding protein. Genomics. 1995;29(1):62–69. doi: 10.1006/geno.1995.1215. [DOI] [PubMed] [Google Scholar]
- 60.Lombardi A, Silvestri E, Cioffi F, Senese R, Lanni A, Goglia F, de Lange P, Moreno M. Defining the transcriptomic and proteomic profiles of rat ageing skeletal muscle by the use of a cDNA array, 2D- and Blue native-PAGE approach. J Proteomics. 2009;72(4):708–721. doi: 10.1016/j.jprot.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 61.Linton PJ, Dorshkind K. Age-related changes in lymphocyte development and function. Nat Immunol. 2004;5(2):133–139. doi: 10.1038/ni1033. [DOI] [PubMed] [Google Scholar]
- 62.Paul E, Pozdnyakova OO, Mitchell E, Carroll MC. Anti-DNA autoreactivity in C4-deficient mice. Eur J Immunol. 2002;32(9):2672–2679. doi: 10.1002/1521-4141(200209)32:9<2672::AID-IMMU2672>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 63.Gadjeva M, Verschoor A, Brockman MA, Jezak H, Shen LM, Knipe DM, Carroll MC. Macrophage-derived complement component C4 can restore humoral immunity in C4-deficient mice. J Immunol. 2002;169(10):5489–5495. doi: 10.4049/jimmunol.169.10.5489. [DOI] [PubMed] [Google Scholar]
- 64.Ascenzi P, Fasano M. Serum heme-albumin: an allosteric protein. IUBMB Life. 2009;61(12):1118–1122. doi: 10.1002/iub.263. [DOI] [PubMed] [Google Scholar]
- 65.Sierra F, Coeytaux S, Juillerat M, Ruffieux C, Gauldie J, Guigoz Y. Serum T-kininogen levels increase two to four months before death. J Biol Chem. 1992;267(15):10665–10669. [PubMed] [Google Scholar]
- 66.Acuna-Castillo C, Leiva-Salcedo E, Gomez CR, Perez V, Li M, Torres C, Walter R, Murasko DM, Sierra F. T-kininogen: a biomarker of aging in Fisher 344 rats with possible implications for the immune response. J Gerontol A Biol Sci Med Sci. 2006;61(7):641–649. doi: 10.1093/gerona/61.7.641. [DOI] [PubMed] [Google Scholar]
- 67.Walter R, Murasko DM, Sierra F. T-kininogen is a biomarker of senescence in rats. Mech Ageing Dev. 1998;106(1–2):129–144. doi: 10.1016/s0047-6374(98)00107-9. [DOI] [PubMed] [Google Scholar]
- 68.Larsson L. The age-related motor disability: underlying mechanisms in skeletal muscle at the motor unit, cellular and molecular level. Acta Physiol Scand. 1998;163(3):S27–S29. doi: 10.1046/j.1365-201x.1998.00375.x. [DOI] [PubMed] [Google Scholar]
- 69.Rostovtseva TK, Bezrukov SM. VDAC regulation: role of cytosolic proteins and mitochondrial lipids. J Bioenerg Biomembr. 2008;40(3):163–170. doi: 10.1007/s10863-008-9145-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carter ND, Wistrand PJ, Isenberg H, Askmark H, Jeffery S, Hopkinson D, Edwards Y. Induction of carbonic anhydrase III mRNA and protein by denervation of rat muscle. Biochem J. 1988;256(1):147–152. doi: 10.1042/bj2560147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Larsson L, Salviati G. Effects of age on calcium transport activity of sarcoplasmic reticulum in fast- and slow-twitch rat muscle fibres. J Physiol. 1989;419:253–264. doi: 10.1113/jphysiol.1989.sp017872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Owens EL, Lynch CJ, McCall KM, Carter ND, Vary TC. Altered expression of skeletal muscle proteins during sepsis. Shock. 1994;2(3):171–178. doi: 10.1097/00024382-199409000-00003. [DOI] [PubMed] [Google Scholar]
- 73.Steinghagen-Thiessen E, Hilz H. The age-dependent decrease in creatine kinase and aldolase activities in human striated muscle is not caused by an accumulation of faulty proteins. Mech Ageing Dev. 1976;5(6):447–457. doi: 10.1016/0047-6374(76)90043-9. [DOI] [PubMed] [Google Scholar]
- 74.Nuss JE, Amaning JK, Bailey E, Doford JH, Dimayuga VL, Rabek JP, Papaconstantinou J. Oxidative modification and aggregation of creatine kinase from aged mouse skeletal muscle. Aging. 2009;1(6) doi: 10.18632/aging.100055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Marsh DR, Criswell DS, Hamilton MT, Booth FW. Association of insulin-like growth factor mRNA expressions with muscle regeneration in young, adult, and old rats. Am J Physiol. 1997;273(1 Pt 2):R353–R358. doi: 10.1152/ajpregu.1997.273.1.R353. [DOI] [PubMed] [Google Scholar]
- 76.Dirks AJ, Hofer T, Marzetti E, Pahor M, Leeuwenburgh C. Mitochondrial DNA mutations, energy metabolism and apoptosis in aging muscle. Ageing Res Rev. 2006;5(2):179–195. doi: 10.1016/j.arr.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 77.Hagen JL, Krause DJ, Baker DJ, Fu MH, Tarnopolsky MA, Hepple RT. Skeletal muscle aging in F344BN F1-hybrid rats: I. Mitochondrial dysfunction contributes to the age-associated reduction in VO2max. J Gerontol A Biol Sci Med Sci. 2004;59(11):1099–1110. doi: 10.1093/gerona/59.11.1099. [DOI] [PubMed] [Google Scholar]
- 78.Carter EE, Thomas MM, Murynka T, Rowan SL, Wright KJ, Huba E, Hepple RT. Slow twitch soleus muscle is not protected from sarcopenia in senescent rats. Exp Gerontol. 2010;45(9):662–670. doi: 10.1016/j.exger.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 79.Edstrom E, Ulfhake B. Sarcopenia is not due to lack of regenerative drive in senescent skeletal muscle. Aging Cell. 2005;4(2):65–77. doi: 10.1111/j.1474-9728.2005.00145.x. [DOI] [PubMed] [Google Scholar]
- 80.Snow LM, McLoon LK, Thompson LV. Adult and developmental myosin heavy chain isoforms in soleus muscle of aging Fischer Brown Norway rat. Anat Rec A Discov Mol Cell Evol Biol. 2005;286(1):866–873. doi: 10.1002/ar.a.20218. [DOI] [PubMed] [Google Scholar]
- 81.Boffoli D, Scacco SC, Vergari R, Solarino G, Santacroce G, Papa S. Decline with age of the respiratory chain activity in human skeletal muscle. Biochim Biophys Acta. 1994;1226(1):73–82. doi: 10.1016/0925-4439(94)90061-2. [DOI] [PubMed] [Google Scholar]
- 82.Tobacman LS. Thin filament-mediated regulation of cardiac contraction. Annu Rev Physiol. 1996;58:447–481. doi: 10.1146/annurev.ph.58.030196.002311. [DOI] [PubMed] [Google Scholar]
- 83.Szczesna D, Potter JD. The role of troponin in the Ca(2+)-regulation of skeletal muscle contraction. Results Probl Cell Differ. 2002;36:171–190. doi: 10.1007/978-3-540-46558-4_13. [DOI] [PubMed] [Google Scholar]
- 84.McArdle A, Vasilaki A, Jackson M. Exercise and skeletal muscle ageing: cellular and molecular mechanisms. Ageing Res Rev. 2002;1(1):79–93. doi: 10.1016/s0047-6374(01)00368-2. [DOI] [PubMed] [Google Scholar]
- 85.Sastre J, Pallardo FV, Vina J. The role of mitochondrial oxidative stress in aging. Free Radic Biol Med. 2003;35(1):1–8. doi: 10.1016/s0891-5849(03)00184-9. [DOI] [PubMed] [Google Scholar]
- 86.Messina S, Vita GL, Aguennouz M, Sframeli M, Romeo S, Rodolico C, Vita G. Activation of NF-kappaB pathway in Duchenne muscular dystrophy: relation to age. Acta Myol. 2011;30(1):16–23. [PMC free article] [PubMed] [Google Scholar]
- 87.Rossi P, Marzani B, Giardina S, Negro M, Marzatico F. Human skeletal muscle aging and the oxidative system: cellular events. Curr Aging Sci. 2008;1(3):182–191. doi: 10.2174/1874609810801030182. [DOI] [PubMed] [Google Scholar]
- 88.Emery AE. The muscular dystrophies. Lancet. 2002;359(9307):687–695. doi: 10.1016/S0140-6736(02)07815-7. [DOI] [PubMed] [Google Scholar]
- 89.Oh-Ishi S, Kizaki T, Yamashita H, Nagata N, Suzuki K, Taniguchi N, Ohno H. Alterations of superoxide dismutase iso-enzyme activity, content, and mRNA expression with aging in rat skeletal muscle. Mech Ageing Dev. 1995;84(1):65–76. doi: 10.1016/0047-6374(95)01637-f. [DOI] [PubMed] [Google Scholar]
- 90.Sohal RS, Wennberg-Kirch E, Jaiswal K, Kwong LK, Forster MJ. Effect of age and caloric restriction on bleomycin-chelatable and nonheme iron in different tissues of C57BL/6 mice. Free Radic Biol Med. 1999;27(3–4):287–293. doi: 10.1016/s0891-5849(99)00052-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.