Abstract
Physician orders, the concrete manifestation of clinical decision making, are enhanced by the distribution of clinical expertise in the form of order sets and corollary orders. Conventional order sets are top-down distributed by committees of experts, limited by the cost of manual development, maintenance, and limited end-user awareness. An alternative explored here applies statistical data-mining to physician order data (>330K order instances from >1.4K inpatient encounters) to extract clinical expertise from the bottom-up. This powers a corollary order suggestion engine using techniques analogous to commercial product recommendation systems (e.g., Amazon.com’s “Customers who bought this…” feature). Compared to a simple benchmark, the item-based association method illustrated here improves order prediction precision from 13% to 18% and further to 28% by incorporating information on the temporal relationship between orders. Incorporating statistics on conditional order frequency ratios further refines recommendations beyond just “common” orders to those relevant to a specific clinical context.
Introduction
In the course of clinical care, a physician may consider a broad differential diagnosis for a patient’s complaints and weigh the relative risks and benefits of many interventions. Ultimately however, physician orders (e.g., labs, imaging, medications) are the concrete manifestation of clinical decision making. Clinical orders can be enhanced with order sets and corollary orders that link commonly co-occurring orders. These may revolve around processes, such as the steps to completing a blood transfusion or heparin protocol, or clinical scenarios, such as standard diagnostics and therapeutics to approach a patient with chest pain or pneumonia.
Implementation of order sets and similar clinical decision support systems (CDSS) in an electronic medical record (EMR) with computerized physician order entry (CPOE) helps reinforce consistency among practitioners and support compliance with best-practice guidelines[ 1 ] [ 2 ]. While order sets and corollary orders can greatly benefit clinical practice, a top-down distribution model limits their benefit. The high cost of manually developing and maintaining them by a committee of subject matter experts ensures that only a finite number are ever produced and maintained[ 3 ]. Furthermore, even when useful order sets are produced, if they do not naturally integrate into the practicing community’s workflow, physicians will never look for them, let alone use them.
An alternative approach to developing clinical decision support content is to instead extract hidden or otherwise undocumented clinical knowledge and expertise from the bottom-up, by data-mining for patterns in large corpuses of electronic medical record data[ 4 ].
Background
Prior work in automated order set and corollary order development has focused primarily on identifying correlations among orders[ 5 ] as well as work to associate orders and diagnoses[ 4 ]. More recent work has refined the association algorithms and attempted some validation by human experts[ 6 ]. Thus far however, there has been limited effort to utilize the temporal relationships between elements, analyze the results with internal validation measures, and in general to actually translate the results into actionable tools for clinical use.
For comparison, similar techniques have been successfully applied for closely analogous problems in information retrieval with recommender systems[ 7 ], collaborative filtering, market basket analysis[ 5 ] and even natural language processing (NLP)[ 8 ]. By analogy to Amazon.com’s “Customers who bought A also bought B” recommender system[ 9 ], a physician order suggestion system could interact in real-time with a physician’s order entry workflow to infer their patient’s clinical context and advise “Based on prior usage patterns of physicians caring for similar patients who ordered A, X% also ordered B.”
Our approach here is to apply statistical data-mining methods to historical physician orders to identify associations that can power such an order suggestion system. This approach need not depend on the labor and expense of a top-down authoring effort, instead more fluidly crowd-sourcing the expertise of practicing physicians and readily evolving with practice patterns as further clinical usage data is integrated. Additionally, end-user physicians need not explicitly seek out relevant “order sets” to utilize the distributed expertise. Instead, the system would more organically observe ordering behavior to infer patient clinical context and make suggestions a natural part of the clinical workflow.
The method employed by Amazon.com’s core recommendation system, and the one largely employed in this work, is an item-based association method that assesses similarity between items based on the number of customers or patients that demonstrate co-occurrence of items. This approach is particularly useful for its scalability against large user history records and catalogs of items to choose from. After initial investment with an intensive pre-computation, item-based associations enable real-time suggestions with time complexity only O(m) where m is the number of prior items purchased / ordered for the current customer / patient, independent of the number of historical records.
Methods
The particular data analyzed for this work was extracted from the STRIDE project[ 10 ], collecting 2 weeks of inpatient admissions to the Stanford University Hospital representing >1,4K patient encounters and >330K instances of physician orders, starting from their initial (emergency room) encounter and ending with their hospital discharge. These physician order instances, analogous to words in a document, derive from >4.3K unique orders, analogous to the vocabulary of words that are available. These unique orders include >1.3K medication, >550 laboratory, >350 imaging, and >500 nursing orders. The medication order data was further normalized with RxNorm mappings[ 11 ] to emphasize only the qualitative information relevant for clinical decision making, including the active ingredients and routes of medications, while resolving out mixtures and ignoring dosages.
With the above data, a pre-computation step collects statistics on physician order patterns, based on the definitions in Table 1 . The order suggestion system uses these ordering frequencies to approximate ordering probabilities that guide the suggestions based on Bayesian conditional probabilities[ 12 ]. Approximations used are outlined in Table 2 .
Table 1:
Pre-computed statistics from analysis of physician order data. Repeats allowed in counting.
| Notation | Definition |
|---|---|
| n A | Number of occurrences of order A |
| n ABt | Number of occurrences of order B following an order A within time t |
| N | Total number of patients / encounters |
Table 2:
Bayesian probability estimates based on order frequency statistics.
| Probability | Estimate | Notation / Notes |
|---|---|---|
| P(A) | nA / N | baselineFreq(A) |
| P(AB) | nAB / N | “Support.” Note that this is not quite the joint probability because nAB only counts the directed association where order A occurs before B. |
| P(B|A) = P(AB) / P(A) | nAB / nA |
conditionalFreq(B|A)
“Confidence.” Interpret as percentage of patients with order B, given order A done. Because repeat orders counted, may have values >1. In such cases, interpret as average number of times B is ordered after A. |
| P(B|A) / P(B) = P(AB) / P(A)*P(B) | (nAB/nA) / (nB/N) |
freqRatio(B|A)
Estimates likelihood ratio. Expect = 1, if A and B occur independently |
Order suggestions are based on a patient’s initial query set of A orders, from which a ranked list of suggested B orders is produced, sorted by scores derived from the above probability estimates. Specifically, given a query with n q items (number of query A orders), query for a separate suggested list of B orders for each query A i order (scored by conditionalFreq(B|A i )), then aggregate the suggested lists of B orders based on a weighted average of the individual list scores. Aggregation weighting is based on the inverse frequency of each query item A i , (w i = 1/baselineFreq(A i )). This weighting method favors less common query items, which are expected to provide more specific suggestions
With any recommendation approach, a validation metric is important to help assess the relative quality of different approaches. Short of end-user observations and surveys however, there is no commonly accepted notion of recommendation quality[ 7 ]. Different methods are favored for different applications, with prediction accuracy being the most commonly employed off-line, internal validation metric. In this scenario, a test patient’s initial orders are used to query for a set of recommended orders that are compared against the patient’s set of actual subsequent orders. Several benchmark recommender methods are described in Table 3 .
Table 3:
For 100 test patients, their first 20 orders are used as query items to get 10 recommended orders from each respective method. These top 10 recommendations are compared against the actual next set of up to 10 orders for each patient. Recall, precision, and F1-score is calculated for each method and averaged across all test patients.
| Method | Recall | Precision | F1-Score | Method Description |
|---|---|---|---|---|
| Random | 0.3% | 0.3% | 0.3% | Items randomly recommended from available catalog |
| BaselineFreq | 14.4% | 13.2% | 13.5% | General “best seller” list, recommending overall most common orders |
| ItemAssociation | 19.8% | 18.2% | 18.7% | Items ranked based on conditionalFreq(B|A) ∼ P(B|A) |
| NextDay | 30.1% | 27.8% | 28.4% | Same as above, but uses nABday (only counts co-occurrences <1 day) |
| NextHour | 23.8% | 21.7% | 22.2% | Same as above, but uses nABhour |
Results
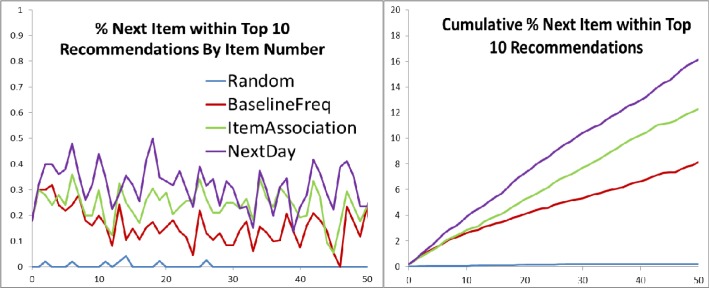
Table 3 compares the benchmark recommender methods based on recommendation accuracy for the initial orders from a randomly selected set of test patients. Figure 1 illustrates a similar analysis, charting the recommendation accuracy for all values of n q (number of query orders) up to 50. Table 4 provides example recommended orders for given a query order (C. diff toxin assay) based on the NextDay method (ranking by conditionalFreq(B|A) day ), with the respective scores for conditionalFreq(B|A) day , baselineFreq(B), and freqRatio(B|A) day .
Figure 1:
Recommendation accuracy for all values of n q (number of query orders, x-axis) up to 50, averaged across 50 test patients. Data points calculated by recommending 10 orders based on the first n q orders for a patient, and counting whether the (n q +1) th order was recommended. Graphs represent the average across all test patients in point probability and cumulative distribution forms.
Table 4:
Top suggestions when query by C. diff stool assay with the NextDay method, scoring and ranking by conditionalFreq(B|A) day . This common query order yields non-specific suggestions, reflecting orders that are simply common overall. Example interpretations: The first score column reflects conditionalFreq(B|A) day , indicating that, among patients for whom a C. diff stool assay is ordered, ∼67% subsequently have an order for IV Sodium Chloride (saline). The second score column reflects baselineFreq(B), indicating that, among all patients (baseline population), Basic Metabolic Panels are ordered an average of 2.1 times for each patient. Note that these values may be >1 as repeat orders are each counted.
| Rank | Order Description | Frequency / Likelihood | ||
|---|---|---|---|---|
| Conditional | Baseline | Ratio | ||
| 1 | Sodium Chloride (Intravenous) | 0.67 | 3.44 | 0.19 |
| 2 | ISTAT G3+, ARTERIAL | 0.38 | 1.84 | 0.20 |
| 3 | MAGNESIUM, SERUM/PLASMA | 0.33 | 1.73 | 0.19 |
| 4 | Potassium Chloride (Intravenous) | 0.33 | 1.54 | 0.22 |
| 5 | METABOLIC PANEL, BASIC | 0.31 | 2.11 | 0.14 |
Discussion
This recommender system approach demonstrates how meaningful and practical clinical knowledge, similar to authored clinical order sets, can be extracted by statistical data-mining of computerized physician order entry data. It does so by offering scored suggestions for related orders given initial query orders that can be observed naturally from a physician’s normal workflow. Furthermore, it does this through automated analysis of large bodies of historical usage data from the bottom-up, enabling discovery and dissemination of expertise without the labor and expense of top-down authoring.
Internal validation metrics from Table 3 demonstrate that the item-association method employed here can significantly improve the accuracy of predicting subsequent physician orders compared to simple baseline benchmarks. Furthermore, these results confirm that incorporating the temporal relationship between orders can further improve suggestion accuracy. This is reflected in the item-association recommender providing the best prediction results when limiting co-occurrence counts to only those order pairs that occurred within 1 day of each other. This is sensible as orders co-occurring across a wider expanse of time are less likely to have a direct clinical relationship. Of note, narrowing the time threshold further to only counting order pairs occurring within 1 hour of each other actually reduces prediction accuracy, indicating that too narrow a time frame will fail to capture the normal distribution of inter-order time variation. The choice of n q (number of query orders) and n r (number of recommended orders) for the metrics in Table 3 are somewhat arbitrary, but Figure 1 provides reassurance that the trends remain consistent over different values of n q , and a value of n r = 10 seems a reasonable upper limit on the number of recommendations a human user would be willing to process in a live setting.
While prediction accuracy, primarily precision, correlates well with end-user satisfaction of recommendations[ 13 ], recommendations[ 13 ], the lack of a well-accepted notion for recommendation quality results in a difficult to resolve struggle between “common” vs. “interesting” suggestions. Table 4 demonstrates the top suggestions based on the NextDay method using conditional frequency (recommending what is most common) given a common, non-specific query order for a C. diff toxin stool assay. While the results are likely accurate in terms of predicting the most likely orders to follow, they mostly reflect orders that are very common overall, but which are not very interesting as suggestions. A potential solution comes from the TF*IDF (term frequency * inverse document frequency) concept from natural language processing[ 8 ]. The frequency ratio approximates the likelihood ratio for a suggested item, with values >1 indicating orders more specific to the given context. Simply using freqRatio(B|A) as a ranking method will yield non-useful results however, as it is overly sensitive to rare orders with baselineFreq approaching 0, pointing towards order associations that are not statistically significant.
Table 5 illustrates more intuitively meaningful and useful suggestions by combining metrics for commonality (conditionalFreq) and relevance (freqRatio). While this and many other possible composite metrics may be useful, they re-introduce the challenge of internal validation. In particular, once the scoring system recommends anything other than the most common expected items, prediction accuracy is no longer a valid metric as it will inherently perform worse in such cases.
Table 5:
Top suggestions by C. diff stool assay, scored and ranked by conditionalFreq(B|A) day , but filtered to only include those with freqRatio(B|A) day >= 1. Suggestions are more meaningfully associated with the query order, including diagnostics for diarrhea (stool culture, ova & parasites) as well as therapeutics and management for C. diff colitis (metronidazole, oral vancomycin, contact isolation, lactobacillus probiotics).
| Rank | Order Description | Frequency / Likelihood | ||
|---|---|---|---|---|
| Conditional | Baseline | Ratio | ||
| 1 | Metronidazole (Oral) | 0.14 | 0.05 | 2.56 |
| 2 | STOOL CULTURE | 0.13 | 0.03 | 4.24 |
| 3 | CONTACT ISOLATION | 0.12 | 0.06 | 1.97 |
| 4 | OVA AND PARASITES | 0.10 | 0.02 | 4.07 |
| 5 | Metronidazole (Intravenous) | 0.09 | 0.06 | 1.51 |
| 6 | Vancomycin (Oral) | 0.06 | 0.02 | 3.19 |
| 7 | FUNGAL CULTURE AND KOH | 0.05 | 0.04 | 1.36 |
| 8 | TRIGLYCERIDES, PLEURAL | 0.03 | 0.02 | 1.33 |
| 9 | CMV IGM | 0.03 | 0.02 | 1.70 |
| 10 | Lactobacillus Acidophilus (Oral) | 0.03 | 0.02 | 1.33 |
Limitations and Future Directions
Some natural concerns and limitations arise from using an automated recommender system for developing and distributing clinical expertise. For example, a recommender system in the context of clinical care may increase costs by encouraging additional orders that may not be necessary. This can be counter-balanced by using the same framework to intercept manual clinician orders with suggestions against orders established to be uncommon in such clinical contexts. Perhaps the most pressing concern is that an automated suggestion method tends to only reinforce the most common practice patterns. While this is useful for naturally adapting to local practice preferences with ease of integration into the source electronic medical record system, the concern is when the most common practice patterns are not actually the best . In this case, there is an ongoing role for top-down expertise to review guidelines and shift practice patterns. Even then, the statistics gleaned from this system’s analysis can be used to identify those scenarios where best practices are known, and yet the community practice patterns reflect divergent behavior, identifying opportunities for intervention.
Other methods can take advantage of the system’s framework to answer public health questions such as inverting the query to start from adverse events (e.g., inpatient deaths) and “suggest” which orders and events tend to occur before the query event. Future work will also enhance the quality of any suggestions by incorporating larger bodies of order data (several million order instances from a year’s worth of inpatient data) as well as incorporating non-order data to better define a patient’s specific clinical context, such as abnormal lab values, problem list / diagnosis codes, demographic information, and keywords extracted from clinical notes.
In closing, this work reflects another step towards mature clinical decision support systems that will not only propose content for expert review, but directly and fluidly interact with a clinical workflow to optimize efficiency and improve quality of patient care, while adapting itself automatically to evolving practice patterns by extracting clinical knowledge and expertise from the ever growing body of electronic medical record data.
Acknowledgments
Project supported by a pilot grant from the Stanford Translational Research and Applied Medicine (TRAM) program in the Department of Medicine (DOM). R.B.A. is supported by NIH/National Institute of General Medical Sciences PharmGKB resource, R24GM61374, as well as LM05652 and GM102365. Additional support is from the Stanford NIH/National Center for Research Resources CTSA award number UL1 RR025744.
Patient data extracted and de-identified by Tanya Podchiyska of the STRIDE (Stanford Translational Research Integrated Database Environment) project, a research and development project at Stanford University to create a standards-based informatics platform supporting clinical and translational research. The STRIDE project described was supported by the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through grant UL1 RR025744. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
References
- [1]. Kaushal R , Shojania KG , Bates DW . “Effects of computerized physician order entry and clinical decision support systems on medication safety: a systematic review.,” . Archives of Internal Medicine . 2003 ; 163 ( 12 ): 1409 – 1416 . doi: 10.1001/archinte.163.12.1409. [DOI] [PubMed] [Google Scholar]
- [2]. Overhage J , Tierney W . “A randomized trial of ‘corollary orders’ to prevent errors of omission,” . Journal of the American Medical Informatics Association . 1997 ; 4 ( 5 ): 364 – 75 . doi: 10.1136/jamia.1997.0040364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3]. Bates D , Kuperman G . “Ten commandments for effective clinical decision support: making the practice of evidence-based medicine a reality,” . Journal of the American Medical Informatics Association . 2003 ; 10 ( 6 ): 523 – 530 . doi: 10.1197/jamia.M1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4]. Wright A , Sittig DF . “Automated development of order sets and corollary orders by data mining in an ambulatory computerized physician order entry system.,” . AMIA Annual Symposium Proceedings . 2006 ; 2006 : 819 – 823 . [PMC free article] [PubMed] [Google Scholar]
- [5]. Doddi S , Marathe a , Ravi SS , Torney DC . “Discovery of association rules in medical data.,” . Medical informatics and the Internet in medicine . 2001 ; 26 ( 1 ): 25 – 33 . [PubMed] [Google Scholar]
- [6]. Klann J , Schadow G , McCoy JM . “A recommendation algorithm for automating corollary order generation.,” . AMIA Annual Symposium Proceedings . 2009 Jan. ; 2009 : 333 – 7 . [PMC free article] [PubMed] [Google Scholar]
- [7]. Shani G , Gunawardana A . “Evaluating Recommendation Systems,” . Recommender Systems Handbook . 2011 ; 12 ( 19 ): 1 – 41 . [Google Scholar]
- [8]. Manning CD , Schütze H . Foundations of Statistical Natural Language Processing . 2 . 26 . MIT Press ; 1999 . pp. 277 – 279 . [Google Scholar]
- [9]. Linden G , Smith B , York J . “Amazon.com recommendations: item-to-item collaborative filtering,” . IEEE Internet Computing . 2003 ; 7 ( 1 ): 76 – 80 . [Google Scholar]
- [10]. Lowe HJ , Ferris Ta , Hernandez PM , Weber SC . “STRIDE--An integrated standards-based translational research informatics platform.,” . AMIA Annual Symposium Proceedings . 2009 Jan. ; 2009 : 391 – 5 . [PMC free article] [PubMed] [Google Scholar]
- [11]. Hernandez P , Podchiyska T , Weber S , Ferris T , Lowe H . “Automated mapping of pharmacy orders from two electronic health record systems to RxNorm within the STRIDE clinical data warehouse.,” . AMIA Annual Symposium Proceedings . 2009 Jan. ; 2009 ( 2 ): 244 – 8 . [PMC free article] [PubMed] [Google Scholar]
- [12]. Bernardo JM , Smith AFM . “Bayesian Theory,” . Measurement Science and Technology . 2001 ; 12 ( 2 ): 221 . [Google Scholar]
- [13]. Wit JD . “Evaluating Recommender Systems,” . University of Twente ; 2008 . [Google Scholar]