Abstract
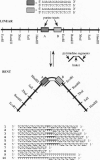
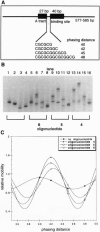
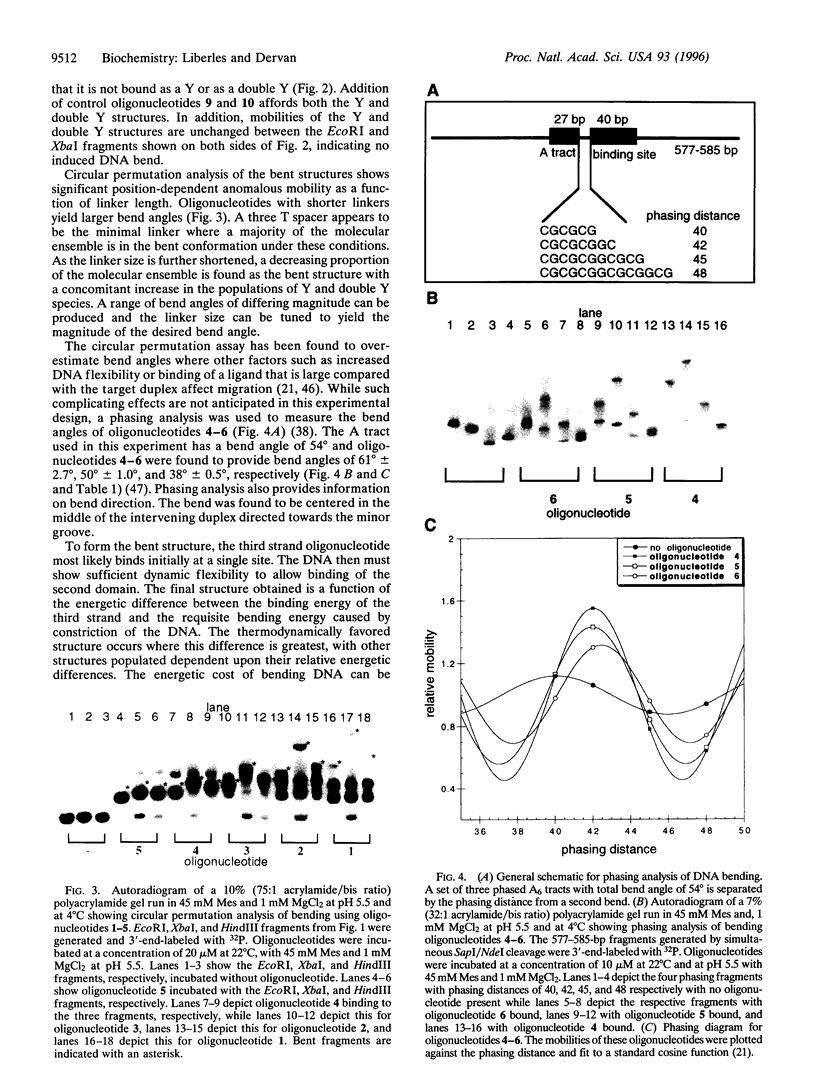
Proteins that bend DNA are important regulators of biological processes. Sequence-specific DNA bending ligands have been designed that bind two noncontiguous sites in the major groove and induce a bend in the DNA. An oligonucleotide containing pyrimidine segments separated by a central variable linker domain simultaneously binds by triple helix formation two 15-bp purine tracts separated by 10 bp. Bend angles of 61 degrees, 50 degrees, and 38 degrees directed towards the minor groove were quantitated by phasing analysis for linkers of four, five, and six T residues, respectively. The design and synthesis of nonnatural architectural factors may provide a new class of reagents for use in biology and human medicine.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beal P. A., Dervan P. B. Second structural motif for recognition of DNA by oligonucleotide-directed triple-helix formation. Science. 1991 Mar 15;251(4999):1360–1363. doi: 10.1126/science.2003222. [DOI] [PubMed] [Google Scholar]
- Beal P. A., Dervan P. B. The influence of single base triplet changes on the stability of a pur.pur.pyr triple helix determined by affinity cleaving. Nucleic Acids Res. 1992 Jun 11;20(11):2773–2776. doi: 10.1093/nar/20.11.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenowitz M., Senear D. F., Shea M. A., Ackers G. K. "Footprint" titrations yield valid thermodynamic isotherms. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8462–8466. doi: 10.1073/pnas.83.22.8462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney M., Czernuszewicz G., Postel E. H., Flint S. J., Hogan M. E. Site-specific oligonucleotide binding represses transcription of the human c-myc gene in vitro. Science. 1988 Jul 22;241(4864):456–459. doi: 10.1126/science.3293213. [DOI] [PubMed] [Google Scholar]
- Finzi L., Gelles J. Measurement of lactose repressor-mediated loop formation and breakdown in single DNA molecules. Science. 1995 Jan 20;267(5196):378–380. doi: 10.1126/science.7824935. [DOI] [PubMed] [Google Scholar]
- Fisher R. F., Long S. R. Interactions of NodD at the nod Box: NodD binds to two distinct sites on the same face of the helix and induces a bend in the DNA. J Mol Biol. 1993 Oct 5;233(3):336–348. doi: 10.1006/jmbi.1993.1515. [DOI] [PubMed] [Google Scholar]
- Fossella J. A., Kim Y. J., Shih H., Richards E. G., Fresco J. R. Relative specificities in binding of Watson-Crick base pairs by third strand residues in a DNA pyrimidine triplex motif. Nucleic Acids Res. 1993 Sep 25;21(19):4511–4515. doi: 10.1093/nar/21.19.4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman S. D., Nicholson S. C., Nash H. A. Deformation of DNA during site-specific recombination of bacteriophage lambda: replacement of IHF protein by HU protein or sequence-directed bends. Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):11910–11914. doi: 10.1073/pnas.89.24.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goryshin IYu, Kil Y. V., Reznikoff W. S. DNA length, bending, and twisting constraints on IS50 transposition. Proc Natl Acad Sci U S A. 1994 Nov 8;91(23):10834–10838. doi: 10.1073/pnas.91.23.10834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallet B., Rezsöhazy R., Mahillon J., Delcour J. IS231A insertion specificity: consensus sequence and DNA bending at the target site. Mol Microbiol. 1994 Oct;14(1):131–139. doi: 10.1111/j.1365-2958.1994.tb01273.x. [DOI] [PubMed] [Google Scholar]
- Haqq C. M., King C. Y., Ukiyama E., Falsafi S., Haqq T. N., Donahoe P. K., Weiss M. A. Molecular basis of mammalian sexual determination: activation of Müllerian inhibiting substance gene expression by SRY. Science. 1994 Dec 2;266(5190):1494–1500. doi: 10.1126/science.7985018. [DOI] [PubMed] [Google Scholar]
- Henriquez V., Milisavljevic V., Kahn J. D., Gennaro M. L. Sequence and structure of cmp, the replication enhancer of the Staphylococcus aureus plasmid pT181. Gene. 1993 Nov 30;134(1):93–98. doi: 10.1016/0378-1119(93)90179-7. [DOI] [PubMed] [Google Scholar]
- Kerppola T. K., Curran T. DNA bending by Fos and Jun: the flexible hinge model. Science. 1991 Nov 22;254(5035):1210–1214. doi: 10.1126/science.1957173. [DOI] [PubMed] [Google Scholar]
- Kerppola T. K., Curran T. Selective DNA bending by a variety of bZIP proteins. Mol Cell Biol. 1993 Sep;13(9):5479–5489. doi: 10.1128/mcb.13.9.5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler D. J., Pettitt B. M., Cheng Y. K., Smith S. R., Jayaraman K., Vu H. M., Hogan M. E. Triple helix formation at distant sites: hybrid oligonucleotides containing a polymeric linker. Nucleic Acids Res. 1993 Oct 11;21(20):4810–4815. doi: 10.1093/nar/21.20.4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. L., Nikolov D. B., Burley S. K. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993 Oct 7;365(6446):520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- Kim J., Klooster S., Shapiro D. J. Intrinsically bent DNA in a eukaryotic transcription factor recognition sequence potentiates transcription activation. J Biol Chem. 1995 Jan 20;270(3):1282–1288. doi: 10.1074/jbc.270.3.1282. [DOI] [PubMed] [Google Scholar]
- Kim Y., Geiger J. H., Hahn S., Sigler P. B. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993 Oct 7;365(6446):512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- Kiyama R., Oishi M. Protection of DNA sequences by triplex-bridge formation. Nucleic Acids Res. 1995 Feb 11;23(3):452–458. doi: 10.1093/nar/23.3.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H. S., Drak J., Rice J. A., Crothers D. M. Determination of the extent of DNA bending by an adenine-thymine tract. Biochemistry. 1990 May 1;29(17):4227–4234. doi: 10.1021/bi00469a027. [DOI] [PubMed] [Google Scholar]
- Kuprash D. V., Rice N. R., Nedospasov S. A. Homodimer of p50 (NF kappa B1) does not introduce a substantial directed bend into DNA according to three different experimental assays. Nucleic Acids Res. 1995 Feb 11;23(3):427–433. doi: 10.1093/nar/23.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Doan T., Perrouault L., Praseuth D., Habhoub N., Decout J. L., Thuong N. T., Lhomme J., Hélène C. Sequence-specific recognition, photocrosslinking and cleavage of the DNA double helix by an oligo-[alpha]-thymidylate covalently linked to an azidoproflavine derivative. Nucleic Acids Res. 1987 Oct 12;15(19):7749–7760. doi: 10.1093/nar/15.19.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., van deSande H. Chain length determination of small double- and single-stranded DNA molecules by polyacrylamide gel electrophoresis. Biochemistry. 1975 Aug 26;14(17):3787–3794. doi: 10.1021/bi00688a010. [DOI] [PubMed] [Google Scholar]
- Manning G. S., Ebralidse K. K., Mirzabekov A. D., Rich A. An estimate of the extent of folding of nucleosomal DNA by laterally asymmetric neutralization of phosphate groups. J Biomol Struct Dyn. 1989 Apr;6(5):877–889. doi: 10.1080/07391102.1989.10506519. [DOI] [PubMed] [Google Scholar]
- Meacock S., Pescini-Gobert R., DeLamarter J. F., Hooft van Huijsduijnen R. Transcription factor-induced, phased bending of the E-selectin promoter. J Biol Chem. 1994 Dec 16;269(50):31756–31762. [PubMed] [Google Scholar]
- Milot E., Belmaaza A., Rassart E., Chartrand P. Association of a host DNA structure with retroviral integration sites in chromosomal DNA. Virology. 1994 Jun;201(2):408–412. doi: 10.1006/viro.1994.1310. [DOI] [PubMed] [Google Scholar]
- Mondragón A., Harrison S. C. The phage 434 Cro/OR1 complex at 2.5 A resolution. J Mol Biol. 1991 May 20;219(2):321–334. doi: 10.1016/0022-2836(91)90568-q. [DOI] [PubMed] [Google Scholar]
- Moser H. E., Dervan P. B. Sequence-specific cleavage of double helical DNA by triple helix formation. Science. 1987 Oct 30;238(4827):645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- Müller H. P., Varmus H. E. DNA bending creates favored sites for retroviral integration: an explanation for preferred insertion sites in nucleosomes. EMBO J. 1994 Oct 3;13(19):4704–4714. doi: 10.1002/j.1460-2075.1994.tb06794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M., Sheikh Q. I., Yamaoka K., Yui Y., Kajiwara S., Shishido K. Bending of DNA segments with Saccharomyces cerevisiae autonomously replicating sequence activity, isolated from basidiomycete mitochondrial linear plasmids. Mol Gen Genet. 1993 Feb;237(1-2):1–9. doi: 10.1007/BF00282777. [DOI] [PubMed] [Google Scholar]
- Natesan S., Gilman M. Z. DNA bending and orientation-dependent function of YY1 in the c-fos promoter. Genes Dev. 1993 Dec;7(12B):2497–2509. doi: 10.1101/gad.7.12b.2497. [DOI] [PubMed] [Google Scholar]
- Pérez-Martín J., Rojo F., de Lorenzo V. Promoters responsive to DNA bending: a common theme in prokaryotic gene expression. Microbiol Rev. 1994 Jun;58(2):268–290. doi: 10.1128/mr.58.2.268-290.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. W., Crothers D. M. Specificity and stringency in DNA triplex formation. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9397–9401. doi: 10.1073/pnas.88.21.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougée M., Faucon B., Mergny J. L., Barcelo F., Giovannangeli C., Garestier T., Hélène C. Kinetics and thermodynamics of triple-helix formation: effects of ionic strength and mismatches. Biochemistry. 1992 Sep 29;31(38):9269–9278. doi: 10.1021/bi00153a021. [DOI] [PubMed] [Google Scholar]
- Singleton S. F., Dervan P. B. Equilibrium association constants for oligonucleotide-directed triple helix formation at single DNA sites: linkage to cation valence and concentration. Biochemistry. 1993 Dec 7;32(48):13171–13179. doi: 10.1021/bi00211a028. [DOI] [PubMed] [Google Scholar]
- Singleton S. F., Dervan P. B. Influence of pH on the equilibrium association constants for oligodeoxyribonucleotide-directed triple helix formation at single DNA sites. Biochemistry. 1992 Nov 17;31(45):10995–11003. doi: 10.1021/bi00160a008. [DOI] [PubMed] [Google Scholar]
- Strauss J. K., Maher L. J., 3rd DNA bending by asymmetric phosphate neutralization. Science. 1994 Dec 16;266(5192):1829–1834. doi: 10.1126/science.7997878. [DOI] [PubMed] [Google Scholar]
- Thompson J. F., Landy A. Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res. 1988 Oct 25;16(20):9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Xodo L. E., Manzini G., Quadrifoglio F., van der Marel G. A., van Boom J. H. Effect of 5-methylcytosine on the stability of triple-stranded DNA--a thermodynamic study. Nucleic Acids Res. 1991 Oct 25;19(20):5625–5631. doi: 10.1093/nar/19.20.5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimm B. H., Levene S. D. Problems and prospects in the theory of gel electrophoresis of DNA. Q Rev Biophys. 1992 May;25(2):171–204. doi: 10.1017/s0033583500004662. [DOI] [PubMed] [Google Scholar]
- Zinkel S. S., Crothers D. M. Comparative gel electrophoresis measurement of the DNA bend angle induced by the catabolite activator protein. Biopolymers. 1990 Jan;29(1):29–38. doi: 10.1002/bip.360290106. [DOI] [PubMed] [Google Scholar]
- Zinkel S. S., Crothers D. M. DNA bend direction by phase sensitive detection. Nature. 1987 Jul 9;328(6126):178–181. doi: 10.1038/328178a0. [DOI] [PubMed] [Google Scholar]