Abstract
The cytochrome P450 (CYP) 2 gene family is the largest and most diverse CYP gene family in vertebrates. In zebrafish, we have identified 10 genes in a new subfamily CYP2AA, which does not show orthology to any human or other mammalian CYP genes. Here we report evolutionary and structural relationships of the 10 CYP2AA genes and expression of the first two genes, CYP2AA1 and CYP2AA2. Parsimony reconstruction of the tandem duplication pattern for the CYP2AA cluster suggests that CYP2AA1, CYP2AA2 and CYP2AA3 likely arose in the earlier duplication events and thus are most diverged in function from the other CYP2AAs. On the other hand, CYP2AA8 and CYP2AA9 are genes that arose in the latest duplication event, implying functional similarity between these two CYPs. A molecular model of CYP2AA1 showing the sequence conservation across the CYP2AA cluster reveals that the regions with the highest variability within the cluster map into CYP2AA1 near the substrate access channels, suggesting differing substrate specificity. Zebrafish CYP2AA1 transcript was expressed predominantly in intestine, while CYP2AA2 was most highly expressed in kidney, suggesting differing roles in physiology. In liver CYP2AA2 expression but not that of CYP2AA1, was increased by 1,4-bis [2-(3,5-dichloropyridyloxy)] benzene (TCPOBOP) and, to a lesser extent, by phenobarbital (PB). In contrast, pregnenolone 16α-carbonitrile (PCN) increased CYP2AA1, but not CYP2AA2 in liver. The results identify a CYP2 subfamily in zebrafish that includes genes apparently induced by PB-type chemicals and PXR agonists, the first concrete in vivo evidence for a PB-type response in fish.
Keywords: Cytochrome P450, CYP2, Zebrafish, Molecular evolution, Homology modeling, Phenobarbital
Introduction
Cytochrome P450 (CYP) enzymes play key roles in the synthesis and degradation of physiologically important endogenous substrates such as steroids, cytokines and fatty acids, and in the biotransformation of xenobiotics. The CYP2 gene family is the largest and most complex of the 18 CYP gene families in vertebrates. Currently there are over 40 CYP2 subfamilies and hundreds of genes, with differing numbers of genes even between closely related species, arising from differences in deletion or expansion in the number of genes (Kirischian et al., 2011; Kubota et al., 2011a; Nelson et al., 2013). Understanding the regulation and function of the CYP2s is important because of the broad range of pharmacological, toxicological, and endogenous substrates transformed by these proteins.
An intriguing question involving the CYP2s in fish concerns the long known yet not understood difference between mammals and fish in their response to phenobarbital (PB) type inducers. In mammals, PB and a number of structurally diverse xenobiotics, including the highly potent 1,4-bis [2-(3,5-dichloropyridyloxy)] benzene (TCPOBOP) and ortho-substituted polychlorinated biphenyl (PCB) congeners, strongly induce the expression of members of the CYP2B subfamily (Kopec et al., 2010; Nims et al., 1993; Smith et al., 1993a, 1993b; Vezina et al., 2004; Waxman, 1999). There is a long history of efforts to identify and demonstrate a CYP2B-like induction by PB or related compounds in fish (Bainy et al., 1999; Buhler and Wang-Buhler, 1998; Celander et al., 1996; Elskus and Stegeman, 1989; Förlin, 1980; Goksøyr, 1985; Iwata et al., 2002; Kleinow et al., 1990; Klotz et al., 1986; Stegeman et al., 1990). However, studies seeking PB-type induction involving analysis of microsomal enzymes or of RNA expression for CYPs related to CYP2B have been inconclusive. Some studies have shown increased expression of CYP1A in trout treated by PB, but this does not appear to involve mechanisms like those involved in CYP2B induction in mammals (Sadar et al., 1996).
Failure to detect a PB-type induction of CYP genes in fish as compared to mammals could reflect a difference in regulatory mechanisms, lack of CYP genes that are responsive to PB, or the use of the wrong endpoints. In mammals the constitutive androstane receptor (CAR) and pregnane X receptor (PXR) are involved in the induction of CYP genes by PB (Waxman, 1999). Teleost fish possess a PXR but apparently lack a CAR gene (Handschin et al., 2004; Maglich et al., 2003; Reschly and Krasowski, 2006). CAR (NR1I3) either arose from PXR(NR1I2) after the divergence of ray-finned fish and lobe-finned fish (the line leading to mammals), or was lost in teleosts (Mathas et al., 2012). In vitro reporter gene assays with a PXR ligand-binding domain have shown that a zebrafish PXR can be activated by structurally diverse chemicals, with differing potencies among chemicals and among species (Ekins et al., 2008; Milnes et al., 2008; Moore et al., 2002). Some in vivo studies with fish including zebrafish also revealed that known agonists for human PXR, including pregnenolone 16α-carbonitrile (PCN), increased expression levels of PXR itself and CYP3A (Bresolin et al., 2005). However, whether fish CYP2s are responsive to potential agonists for the PXR remains to be understood.
Our initial approach to find fish homologues of CYP2 genes that are responsive to PB and PB-type inducers was to amplify sequences with primers deigned from conserved regions in mammalian CYP2B genes. The first efforts resulted in identification of the CYP2Ns and CYP2Ps (Oleksiak et al., 2000, 2003). Subsequent efforts to find fish CYP2B-like genes and study their regulation have employed zebrafish and resulted in cloning of sequences defining a novel subfamily, 2AA. More recently we identified and annotated the full suite of CYP genes in zebrafish (Goldstone et al., 2010), including 10 CYP2AA genes. The CYP2AA genes are excellent examples of the evolution of sequence diversity within tandemly-duplicated gene clusters, and provide an important opportunity for understanding the evolution of functional diversity in toxicologically and pharmacologically relevant gene subfamilies. Here we report on cloning of the full-length CYP2AA1 and CYP2AA2, evolutionary and structural relationships of the 10 CYP2AAs, and the expression of the first two genes in vivo, including response to PB, TCPOBOP and PCN.
Materials and methods
Fish Husbandry
Sexually mature zebrafish (Danio rerio) of the Tupfel/long (TL) fin wild-type strain were used in all experiments. Fish were maintained under standard light and temperature conditions as described previously (Jönsson et al., 2007). The experimental procedures were approved by the Animal Care and Use Committee of the Woods Hole Oceanographic Institution.
Cloning strategy
Prior to the genomic sequence of zebrafish being available, a BLAST search using human CYP2B against the zebrafish ESTs database was carried out and matching regions were used for designing primers (in 2001). Adult specimens (n = 8) killed by cervical scission were used for cloning of a CYP2 subfamily in zebrafish. Messenger RNA (mRNA) was prepared from the liver of individual fish using the MicroPoly(A)Pure™ kit (Ambion). Oligo d(T)-primed cDNA synthesis from mRNA was carried out by RT using the SMART™ PCR cDNA synthesis kit (Clontech). Using the cDNA synthesized, an initial fragment was yielded with the forward primer 5′-CATTATCAAGCATTTCCCTGGTCC-3′ and the reverse primer 5′-CGCTGAATCTCGTGAACAGTGG-3′, after which the resulting sequences suggested two distinct CYP2 genes. Forward, reverse and nested primers distinct to each of these genes were designed to obtain PCR products that were used as templates for rapid amplification of cDNA ends (RACE). Primers to clone the full-length coding sequence of CYP genes were designed to the untranslated region of 5′ and 3′ ends.
Sequence and structural analyses of CYP2AA
CYP2AA1 and 2AA2 were cloned in 2002 prior to the sequencing of the zebrafish genome. Subsequently, additional CYP2AA genes were identified in a cluster in the zebrafish genome (Goldstone et al., 2010). Phylogenetic trees were constructed by analyzing inferred or confirmed amino acid sequences using maximum likelihood (RAxML 7.0.4) (Stamatakis, 2006) using the WAG model of amino acid substitution (Whelan and Goldman, 2001). Synteny (gene order) was examined using the Ensembl browser. Parsimony tandem duplication analysis was performed using DILTAG (Lajoie et al., 2010). Molecular models were constructed using Modeller [v9.8; (Eswar et al., 2007)]. Multiple models were built for each CYP2AA isoform based on structures of mammalian CYP2s (CYP2C5: 1DT6, CYP2B4:1PO5, CYP2A6:3EBS, CYP2C19: 4GQS). The best model was chosen on the basis of the discrete optimized potential energy (DOPE) score. Alignment scoring was determined using Scorecons (Valdar, 2002). Substrate access channels were determined using the Caver 3.0.1 plugin for PyMol (Chovancova et al., 2012).
Tissue-specific gene expression
Adult zebrafish were anesthetized by MS-222 and were euthanized by decapitation. The organs including intestine, liver, testes, ovary, heart, kidney, brain, and eye were collected by dissection. Three replicates were collected for both males and females, resulting from four individuals pooled per replicate for each organ. The dissected organs were flash frozen in liquid nitrogen and were stored at −70 °C until RNA isolation.
Chemical exposure
To study response of zebrafish to archetypal inducers of CYP2B genes, male specimens (n = 8) were injected i.p. with TCPOBOP (1.5 μg/g) or PB (70 μg/g) dissolved in a solution of 15% dimethylsulphoxide (DMSO) and 85% saline. Fish from the vehicle control group was injected with the DMSO/saline solution. As a separate experiment, male specimens (n = 6) were injected i.p. with PCN (20 μg/g) dissolved in DMSO. Fish from the vehicle control group was injected with DMSO. Stock solutions were prepared in order to inject 2 μl/g fish for each chemical. The fish were kept in system water (Jönsson et al., 2007) in 3 l tanks with constant aeration, and were fed with live brine shrimp (Artemia salina) twice a day for the first two days. Three days after the injection, fish were killed by cervical section and liver was excised immediately and frozen in liquid nitrogen. Total RNA was isolated from the liver of individual fish using RNA STAT-60 (Tel-Test B, Inc., Friendswood, TX). The quantity of RNA was determined spectrophotometrically (NanoDrop ND-1000; NanoDrop Technologies, Wilmington, DE, USA). cDNA was synthesized from 1 μg total RNA using iScript kit (Bio-Rad).
Quantitative real time PCR
Quantitative real time PCR was performed using the iQ SYBR Green Supermix (Bio-Rad) in a MyiQ Single-Color Real-Time PCR Detection System (Bio-Rad), according to the manufacture's instruction. Gene-specific primers for real time PCR were synthesized by Eurofins MWG Operon (Huntsville, AL, USA), and are shown in Supplemental Table S1. In each sample, the genes were analyzed in duplicate with the following protocol: 95 °C for 3 min and 95 °C for 15 s/62 °C for 1 min (45 cycles). A melt curve analysis was performed at the end of each PCR run to ensure that a single product was amplified. The preliminary study confirmed that the right product was amplified by qPCR. Relative mRNA expression of each target gene was normalized to that of ARNT2 and EF1α (E−ΔCt; where ΔCt = [Ct(target genes)−Ct(geometric mean of ARNT2 and EF1α)]). Relative changes due to treatment (TCPOBOP, PB, or PCN) were determined by E−ΔΔCt (E-ΔCt[sample]/mean E−Δ Ct[control]). PCR efficiencies (E) for within-experiment amplicon groups were determined by the LinRegPCR program (Ramakers et al., 2003; Ruijter et al., 2009).
Statistics
Messenger RNA expression levels of CYP2AA genes are presented as mean ± SD for tissue distribution analysis and as range with 25th to 75th percentile for chemical effect analysis. Significant differences in the mRNA levels among 7 tissues were determined for each gene by one-way ANOVA which if significant was followed by Tukey-Kramer test. Significance of differences in the mRNA expression levels between control and treatment groups (i.e., TCPOBOP or PB) was determined for each gene by one-way ANOVA which if significant was followed by Dunnett test. In a separate experiment, significance of differences in the mRNA expression levels between control and PCN-treated group was determined by Student t-test. Spearman's rank correlation test was conducted for relationships of expression levels of CYP2AA genes with PXR gene in each treatment group. The significant level was set at 0.05. All statistical analyses were performed with GraphPad Prism 5.0 (GraphPad Software, San Diego, CA).
Results
Molecular cloning of CYP2AAs
The initial identification of zebrafish CYP2AA genes was accomplished prior to sequencing of the genome and thus before it was possible to search the genome to annotate all CYPs. Initially, a fragment of 458 bp was amplified and the sequence analysis indicated the presence of consistent nucleotide differences in different clones, suggesting that there were at least two different sequences with 89% identity, indicating the presence of two genes or possible allelic variation. RACE analysis with primers designed to amplify the 5′ and 3′ cDNA ends of both genes resulted in full-length sequences of the coding regions. Alignment and comparison with known CYP2 family sequences resulted in a new subfamily designated CYP2AA, with the first sequences termed CYP2AA1 (AF497969) and CYP2AA2 (#######; see Supplemental Fig. S1). Open reading frames for CYP2AA1 and CYP2AA2 are 1494 bp each, coding for proteins of 498 amino acids.
The CYP2AA1 and CYP2AA2 nucleotide sequences share 88% identity, while the deduced amino acid sequences share 84 % identity. The amino acid sequence of the cloned CYP2AA2 is identical to the most recent Ensembl CYP2AA2 sequence entry (ENSDARP00000021614), while the cloned CYP2AA1 showed a 14 amino acid difference from its Ensembl entry (ENSDARP00000093164), suggesting an allelic variation of CYP2AA1. The CYP2AA1 and CYP2AA2 predicted protein sequences were 38-41% identical to mammalian CYP2Bs and killifish CYP2Ns and CYP2Ps. Several functional motifs (i.e., a putative heme-binding motif, conserved threonine residue, proline rich region, etc.) between zebrafish CYP2AAs and other vertebrate CYP2s were well conserved (Supplemental Fig. S1).
Evolutionary and structural relationships of the CYP2AAs
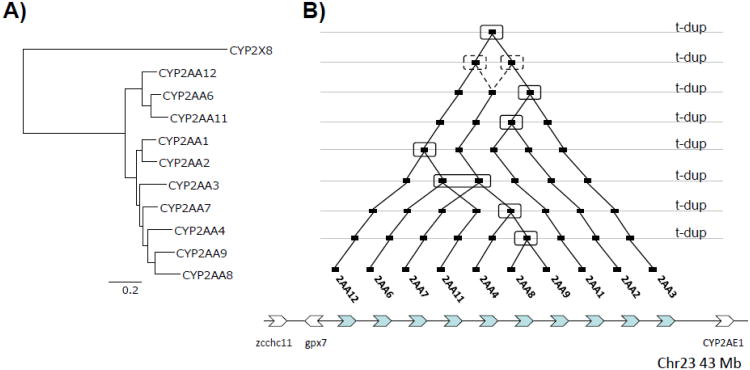
The zebrafish genome has 10 CYP2AA genes located on chromosome 23 in a duplicated array (Goldstone et al., 2010). The CYP2AAs are most closely related to the CYP2X subfamily, found in zebrafish and other fishes, and somewhat less closely related to the fish CYP2K, and mammalian CYP2A/B/F/G/S/T clusters (Goldstone et al., 2010; Kirischian et al., 2011). In order to understand evolutionary relationships within the CYP2AA gene cluster, we constructed a maximum likelihood phylogenetic tree (Fig. 1A) and conducted a parsimony reconstruction of the CYP2AA gene duplication pattern (Fig. 1B). There are likely eight independent tandem duplication events in this cluster (Fig. 1B). Multiple duplication patterns exhibit the same lowest parsimony score (Supplemental Fig. S2). Four alternative patterns with identical parsimony scores are possible in the second duplication. However, in all cases the last three rounds of duplication are identical. Parsimony reconstruction suggests that CYP2AA1, CYP2AA2, and CYP2AA3 are genes that likely arose in the earlier duplication events and thus are most diverged from the other CYP2AAs. In contrast, CYP2AA8 and CYP2AA9 seem to occur in the latest duplication event. The genomic arrangement of the CYP2AA cluster genes (Fig. 1B) shows that CYP2AA8 and CYP2AA9 are located next to each other, while CYP2AA1, CYP2AA2, and CYP2AA3 are present sequentially in the cluster.
Fig. 1.
Maximum likelihood phylogeny, parsimony reconstruction of tandem duplication, and syntenic relationships for the CYP2AA cluster. The maximum likelihood phylogeny (A) was constructed using RA×ML, with zebrafish CYP2X8 as the outgroup. The tandem duplication (t-dup) pattern (B) for the CYP2AA cluster located on Zv9 Chromosome 23 was constructed using DILTAG. Duplication events are noted with a box. Note that four alternative patterns with identical parsimony scores are possible, illustrated for two cases by the dashed lines in the second duplication. In all cases the last three rounds of duplication are identical.
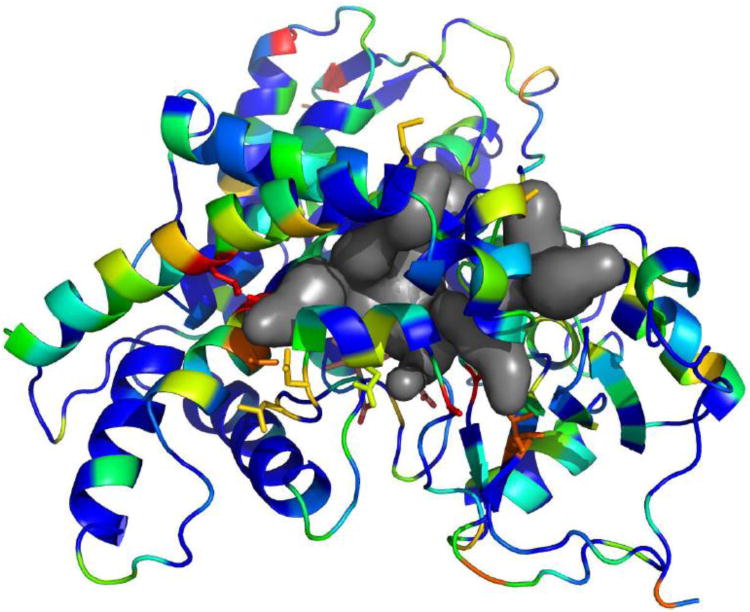
Although the CYP2AAs are all closely related, the existence of such a large tandemly duplicated array of genes implies some functional differences between the expressed proteins (Bergthorsson et al., 2007). The amino acid sequence identity between the duplicated genes ranges from 65-85%. The regions of highest amino acid variability were mapped onto a molecular model of CYP2AA1 based on the crystal structures of closely related mammalian CYP2s (37-44% identity over the length of available crystal structures). This mapping (Fig. 2) reveals that these variable regions occur primarily at the entrances to the substrate access channels, as well as a few residues at the N and C-termini of the structure. These substrate access channels are created by tertiary structure, which brings together amino acids that are distant in the primary sequence (e.g., the highly variable residues 105 and 372, both red in Fig 2). Notably, much of the variability is located with the substrate recognition sites (SRSs), identified from the analysis of primary sequence but which form the substrate access channels and active site (Gotoh, 1992). Separate model construction and analysis of other CYP2AAs showed that the position of these computed substrate access channels does not appear to be significantly altered in any of the CYP2AA models (data not shown).
Fig. 2.
Homology model of CYP2AA1 showing the sequence conservation across the CYP2AA cluster and computed substrate access channels. The model was constructed using Modeller 9v8 based on four mammalian CYP2 crystal structures. Colors indicate the range of BLOSUM80-based sequence conservation scores across the CYP2AA cluster, with red indicating the regions of highest variability and dark blue the regions without any substitutions. Also shown are the top substrate access channels computed with Caver 3.0. Note that the two primary regions with low conservation are located at the entrance to the channels with the highest computed throughput.
Tissue distribution of CYP2AA1 and CYP2AA2 transcripts
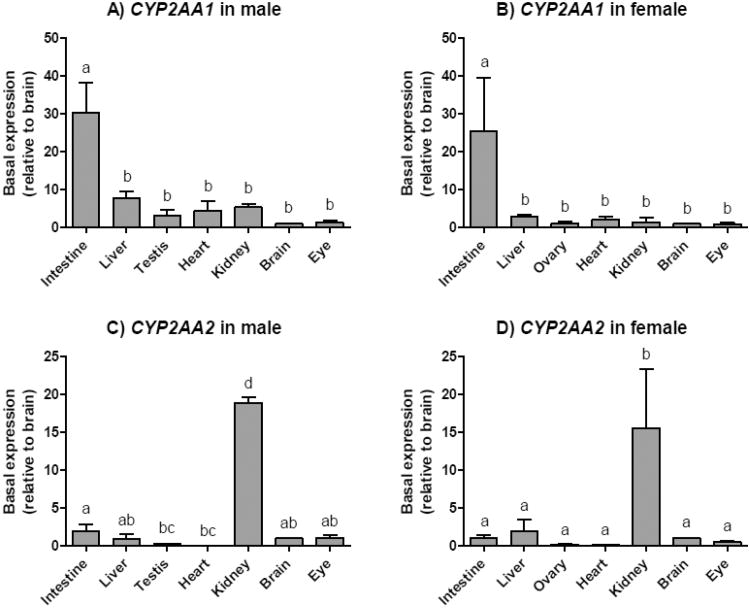
To determine the tissue distribution of the CYP2AA1 and CYP2AA2 expression, total RNA isolated from various tissues was reverse transcribed and internal primers designed to specific regions from both genes were used for quantitative real time PCR analysis. CYP2AA1 transcripts were more highly expressed in intestine than the other tissues analyzed (Fig. 3A, 3B), in which CYP2AA1 was evenly expressed. In contrast, CYP2AA2 transcripts were particularly abundant in kidney, while they were detected at very low levels in gonad and heart (Fig. 3C, 3D). No clear sex-difference in the tissue distribution profile was observed for either CYP2AA1 or CYP2AA2.
Fig. 3.
Tissue distribution of CYP2AA1 (A, B) and CYP2AA2 (C, D) in adult zebrafish. Transcript levels of CYP2AA1 and CYP2AA2 were determined by qPCR in seven tissues from males and females. Data were normalized by geometric mean of two separate reference genes, EF1α and ARNT2. Results are shown as values in each tissue relative to the average value in brain (mean + SD; n = 3 for each sex), as this organ showed one of the least inter-individual variability and sex-differences in the CYP2AA expression levels. Statistical differences in transcript levels among tissues were determined by one-way ANOVA followed by Tukey's multiple comparisons test and are shown by different letters (p < 0.05). Outliers were excluded based on the Grubbs test.
Response to select xenobiotics in adults
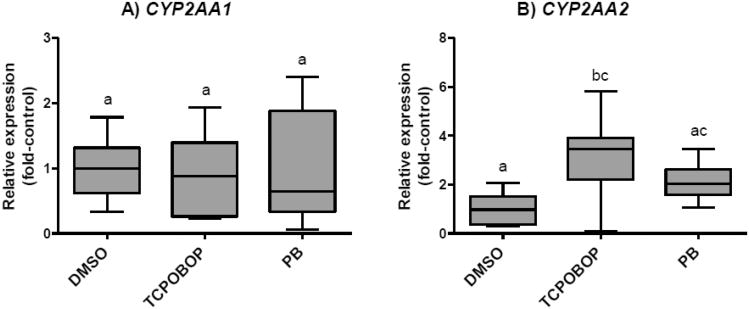
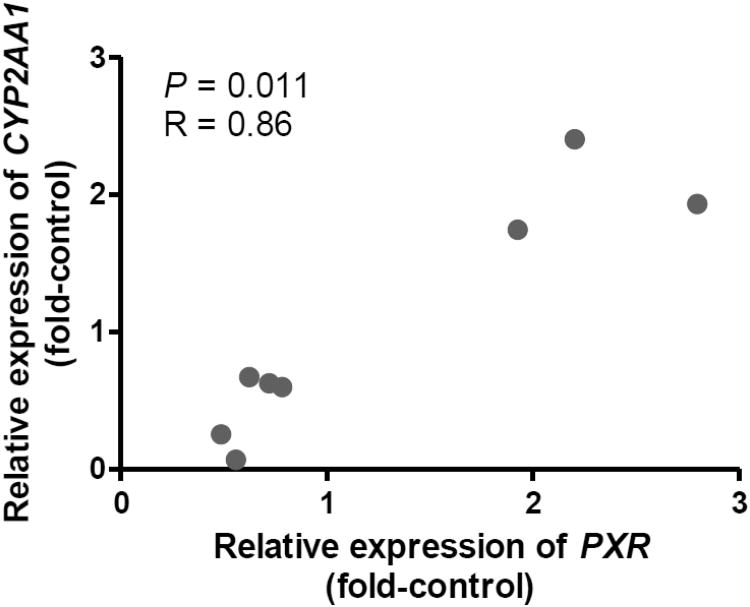
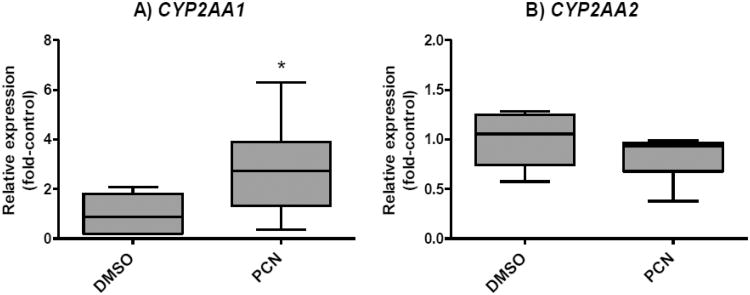
We first examined responses of CYP2AA1 and CYP2AA2 to PB and TCPOBOP in liver of adult zebrafish (Fig. 4). No significant differences in CYP2AA1 transcript levels were observed among treatments. In contrast, treatment with TCPOBOP caused significant up-regulation of CYP2AA2, with a 3.2-fold increase in the transcript levels relative to the vehicle control group. A smaller increase (2.1-fold) in CYP2AA2 transcript levels was also observed for PB-treated fish, although this was not statistically significant. In order to examine possible mechanisms by which these chemicals showed inter-individual variability of CYP2AA expression in each treatment group and by which TCPOBOP and, to a lesser extent, PB increased transcript levels of CYP2AA2, we performed correlation analyses between PXR and each of CYP2AA1 and CYP2AA2. Spearman's rank correlation tests showed a significant positive correlation between PXR and CYP2AA1 in the PB-treated group (Fig. 5). None of the other correlations examined between PXR and CYP2AA1 or CYP2AA2 was statistically significant (Supplemental Fig. S3). We also tested the response of the CYP2AA genes to PCN, another potential agonist for zebrafish PXR. Injection of adult zebrafish with PCN significantly increased transcript levels of CYP2AA1 in liver, but not CYP2AA2 (Fig. 6A, 6B). Yet, neither CYP2AA1 nor CYP2AA2 showed significant correlation with PXR in these samples (data not shown), although the number of samples was small for this analysis (n = 6).
Fig.4.
Effects of TCPOBOP and PB on mRNA expression levels of CYP2AA1 (A) and CYP2AA2 (B). CYP2AA transcript levels were determined by qPCR in liver of adult male zebrafish treated with vehicle DMSO, TCPOBOP (1.5 μg/g) or PB (70 μg/g). Data were normalized by geometric mean of two separate reference genes, EF1α and ARNT2. Results are shown as values in each treatment relative to the average value in DMSO control (fold-control). The whiskers show data range, while the box extends from the 25th to 75th percentile. Statistical differences in transcript levels among treatments were determined by one-way ANOVA followed by Tukey's multiple comparisons test and are shown by different letters (p < 0.05). N = 8.
Fig. 5.
Spearman's rank correlation of gene expression between PXR and CYP2AA1 in liver of adult zebrafish treated with PB (n = 8). A significant positive correlation was only seen in this relationship and none of the other correlations examined between PXR and CYP2AA1 or CYP2AA2 was statistically significant (see also Supplemental Fig. S3).
Fig. 6.
Effects of PCN on mRNA expression levels of CYP2AA1 (A) and CYP2AA2 (B). CYP2AA transcript levels were determined by qPCR in liver of adult zebrafish treated with vehicle DMSO or PCN (μg/g). Data were normalized by geometric mean of two separate reference genes, EF1α and ARNT2. Results are shown as values in each treatment relative to the average value in DMSO control (fold-control). The whiskers show data range, while the box extends from the 25th to 75th percentile. Statistical differences in transcript levels between vehicle control and PCN-treated group were determined by Student t-test and are shown by an asterisk (p < 0.05). N = 6.
Discussion
The CYP2 gene family is the largest CYP gene family in vertebrates. The diversity of CYP2 genes in teleost fishes is as great as that in mammals, but the function and regulation of CYP2s in fish is, with some exceptions, unknown. Establishing similarities and differences between CYP2s in different taxa is made more difficult by evolutionary distance, which can obscure homologous relationships and result in distinct classification for orthologs. For example, based on sequence, the CYP2N and CYP2P subfamilies found in fish do not occur in mammals, yet molecular phylogeny and syntenic arrangement of the genes indicate that the CYP2Ns and CYP2Ps all are co-orthologs of mammalian CYP2Js (Goldstone et al., 2010; Kirischian et al., 2011). Moreover, nearly identical regio- and enantio-selectivity for oxidation of arachidonic acid indicates that at least one CYP2P (CYP2P3) and human CYP2J2 are closely related functionally (Oleksiak et al., 2000; Oleksiak et al., 2003).
Understanding the roles of CYPs in zebrafish is important to the use of this premier non-mammalian species in toxicological, pharmacological and carcinogenesis research (e.g., (Kubota et al., 2011b; Teraoka et al., 2009). In zebrafish there are 44 CYP2 genes in 11 subfamilies (Goldstone et al., 2010), most of which do not show obvious orthology to human or other mammalian genes. These include genes in the new CYP2AA subfamily. The CYP2AA subfamily consists of 10 genes that are tandemly duplicated in a locus on chromosome 23 but that locus does not share synteny with any mammalian CYP2 genes. This lack of shared synteny appears to reflect lineage-specific gene subfamily expansion and translocation, occurring as well with CYPs in other vertebrate species (Kubota et al., 2011a; Nelson et al., 2013; Nelson et al., 2004). The CYP2AA subfamily is phylogenetically closely related to the fish CYP2X subfamily, another CYP2 gene subfamily that does not appear to share synteny with any mammalian CYP2s.
While the biological significance of these tandemly duplicated CYP2AA genes is not known, we suggest here that analysis of the pattern of tandem duplication as well as the genomic arrangement may help to predict functional and evolutionary diversity within a particular cluster of CYP genes (Fig. 1). We examined evolutionary relationships within the set of CYP2AA genes, with the objective of inferring functional similarity and difference within the subfamily. Parsimony reconstruction of the tandem duplication pattern for the CYP2AA cluster suggests that CYP2AA8 and CYP2AA9 likely are similar in original function, while CYP2AA1, CYP2AA2 and CYP2AA3 are likely to be most divergent in function from the other CYP2AAs. Although CYP2AA1 and CYP2AA9 are located next to each other, these two genes appear to have arisen from an earlier duplication event, implying that functional diversity could arise despite their proximity to one another.
Sub- and neo-functionalization within the CYP2AA cluster may proceed in both expression patterns (as observed in CYP19A1/CYP19A2 expression in ovary versus brain) and in protein function [as in the CYP17A1/CYP17A2 functional divergence (Zhou et al., 2007)]. Detailed expression patterns of the complete CYP2AA set will provide further insight into possible spatiotemporal divergence. Regarding functional divergence, effort was made to express CYP2AA1 and CYP2AA2 proteins in COS-7 cells, yet there was no detectable activity with potential substrates, including alkoxyresorufins. Although the analysis of substrate differences requires detailed functional studies, interesting details can be gleaned from a closer examination of the primary protein sequence and molecular modeling. Mapping of the observed sequence variability within the CYP2AA cluster onto a three dimensional protein model provides insight into the spatial location of the protein regions that determine substrate specificity (Fig. 2). As noted above, the regions with the highest variability within the CYP2AA cluster occur near the substrate access channels, suggesting that there are indeed differences in substrate specificity, and that these differences could be controlled by the (relatively few) variable residues near the entrances to the substrate channels.
In a recent study of the complement of CYPs in Atlantic cod (Gadus morhua) neither EST- nor genome-assemblies contained CYP genes that clustered in the CYP2AA clade (Karlsen et al., 2012). Our survey of fish genomes of seven other species currently available on Ensembl (i.e., coelacanth, fugu, medaka, platyfish, stickleback, tetraodon, and tilapia) also failed to identify any CYP genes that are readily annotated as members of the CYP2AA subfamily. Whether the genes in CYP2AA subfamily are specific to Cypriniformes to which zebrafish belong, or common to a broader lineage of fish requires further investigation as more fish genomes being available.
We, and others, have long sought some fish CYP genes that, like mammalian CYP2Bs, are responsive to PB and PB-type chemicals (Bainy et al., 1999; Buhler and Wang-Buhler, 1998; Celander et al., 1996; Elskus and Stegeman, 1989; Förlin, 1980; Goksøyr, 1985; Iwata et al., 2002; Kleinow et al., 1990; Klotz et al., 1986; Stegeman et al., 1990). In mammals, PB-type induction of CYP2B genes occurs via both CAR and PXR (Waxman, 1999). While zebrafish lack CAR, they do possess PXR (Maglich et al., 2003; Reschly and Krasowski, 2006), and several lines of evidence indicate that a variety of structurally diverse chemicals can activate PXR of zebrafish (Ekins et al., 2008; Milnes et al., 2008; Moore et al., 2002). These prior studies led us to hypothesize that fish respond to PB and related chemicals through PXR.
The present study showed evidence for the induction of transcripts of CYP2AA1 and CYP2AA2 in liver of zebrafish treated with the mammalian PXR agonist PCN and with a PB-type inducer TCPOBOP. An earlier study also reported that PCN induces the mRNA expression level of PXR in liver of zebrafish, suggesting a positive self-regulation (Bresolin et al., 2005). Indeed, we also found a significant up-regulation of the PXR mRNA levels in liver of zebrafish treated with PCN (Kubota et al., unpublished results). However, we failed to find coordinate induction of PXR and CYP2AA1, as no significant correlation was found between levels of transcripts for these two genes in the PCN-treated group, although with a limited number of samples (n = 6). There also was no significant up-regulation of PXR mRNA expression in the TCPOBOP-treated group (n = 8) (data not shown), nor significant correlation between expression levels of PXR and CYP2AA2. TCPOBOP is known to activate human PXR, but not zebrafish PXR in vitro, although under differing conditions of the reporter assay (e.g., the expression plasmids differed, consisting of a full-length for human PXR and a ligand binding domain for zebrafish PXR) (Ekins et al., 2008; Milnes et al., 2008; Moore et al., 2002). Overall, there still is an unresolved question about mechanisms underlying up-regulation of CYP2AA genes in response to PCN and TCPOBOP. Whether the PCN- and TCPOBOP-induced transcript expression of CYP2AA1 and CYP2AA2 observed in the present study depends upon the PXR signaling pathway requires further investigation.
There was no significant difference in the transcript level of CYP2AA1 between the vehicle control group and the PB-treated group. Nevertheless, the significant positive correlation between PXR and CYP2AA1 in fish treated with PB is pronounced and presumably represents a shared mechanism of the transcript regulation of these two genes in response to PB. In vitro studies have shown that PB has a potency to activate zebrafish PXR (Moore et al., 2002; Reschly et al., 2007), yet the efficiency of PB for the activation was the lowest among the chemicals tested, including androstanol, 5α-cyprinol 27-sulfate and, and TCDD (Reschly et al., 2007). It is likely that since PB has a weak potency to activate zebrafish PXR in vitro, the induction of its potential target genes, perhaps including CYP2AA1, may be weak in vivo. Alternatively, there is a possibility that PXR alleles identified in zebrafish (Bainy et al., unpublished results) contribute to the inter-individual differences in the PXR and CYP2AA1 responses, as some fish showed no or very weak response to PB but others showed substantial response in the present study.
While the results here show a CYP2 transcript induction by PB-type compounds, there are still important questions about this observation, and about PB response(s) in fish in general. Is the magnitude of the response biologically significant, and are protein levels induced as well? Does the mechanism of induction involve ligand activation of PXR or some other transcription factor? How might cellular context influence the response of the CYP2AAs? Are increased rates of metabolism of substrates, such as observed with PCBs in vivo in fish exposed to congeners that are mammalian CYP2B-like inducers (Buckman et al., 2007), linked to CYP2AAs or to other CYPs yet to be described but that also respond to PB-type inducers? Inference from substrate metabolism alone is weak, without linkage of the activity to a specific catalyst, but can suggest directions for research. The CYP2AA genes offer subjects for further addressing the question of PB responsiveness in fish. However, the indications of CYP2AA response do not preclude the possibility of other CYPs that respond to PB-type inducers, or involvement of CYPs that are not PB responsive in catalytic functions with substrates that in mammals are metabolized by CYP2Bs. Identification of responsive genes may be forthcoming, as full transcriptome analyses are completed.
At present, as described above, there still is uncertainty about the breadth of occurrence of the CYP2AA subfamily in the fishes. Whether the PB-type induction of CYP2AA genes observed in zebrafish is conserved in other fish awaits the complete annotation of more fish genomes and subsequent studies on the chemical effect on the gene expression. It also will be interesting to determine whether fish have CYP2 genes other than CYP2AAs that are responsive to PB and related chemicals. For example, zebrafish CYP2Y3 and CYP2Y4 are syntenic and therefore orthologous to genes in human CYP2A/B/F/G/S/T cluster, which includes PB responsive genes (Zhang et al., 2010)
In summary, on zebrafish chromosome 23 there are 10 CYP2AA genes that presumably arose from eight independent tandem duplication events. We cloned the cDNAs of the first two genes, CYP2AA1 and CYP2AA2, and found that they are expressed in extrahepatic tissues. Our finding of the prominent expression of zebrafish CYP2AA1 in intestine and CYP2AA2 in kidney opens the possibility that these enzymes have distinct physiological roles in intestinal and renal functions. We provided evidence showing a PB-type response of CYP2 genes, including CYP2AA1 and CYP2AA2, with differing responses between the two genes. However, further studies are necessary to address regulatory mechanisms of these CYP2 genes upon exposure to PB-type inducers, PXR agonists, and other xenobiotic inducers, including AHR ligands. Mapping of the regulatory elements upstream of each of these tandemly duplicated genes could indicate shared or divergent regulation. Knocking down of PXR expression in zebrafish embryos using morpholino antisense oligonucleotides and subsequent exposure of the embryos to PXR agonists also is one possible approach currently underway to address this question.
Supplementary Material
Highlights.
A tandemly duplicated cluster of ten CYP2AA genes was described in zebrafish.
Parsimony and duplication analyses suggest pathways to CYP2AA diversity.
Homology models reveal amino acid positions possibly related to functional diversity.
The CYP2AA locus does not share synteny with any CYP2 subfamily in mammals.
Induction of CYP2AA1 and CYP2AA2 indicates a phenobarbital-type response in fish.
Acknowledgments
The authors thank Dr. Benjamin Lemaire and Mr. Matthew Takata for collection of zebrafish tissue samples. This study was supported by National Institute of Health Grant (R01ES015912) to J.J.S., Superfund Research Program (5P42ES007381) to J.J.S., and Grant-in-Aid for JSPS Fellows to A.K. from Japan Society for the Promotion of Science. JSPS Postdoctoral Fellowship and of Postdoctoral Fellowships for Research Abroad to A. K. are acknowledged (nos. 4313 and 820, respectively). A.C.D.B. was recipient of Postdoctoral Fellowship from CAPES, Ministry of Education, Brazil. A.C.D.B. is recipient of CNPq Productivity Fellowship. The sponsors had no involvement in performing or in the decision to publish this study. The U.S. and Japanese Governments are authorized to produce and distribute reprints for governmental purposes notwithstanding any copyright notation that may appear hereon.
Footnotes
Conflict of Interest Statement: None of the authors has any conflict of interest regarding the research described in this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bainy ACD, Woodin BR, Stegeman JJ. Elevated levels of multiple cytochrome P450 forms in tilapia from Billings Reservoir-Saõ Paulo, Brazil. Aquat Toxicol. 1999;44:289–305. [Google Scholar]
- Bergthorsson U, Andersson DI, Roth JR. Ohno's dilemma: evolution of new genes under continuous selection. Proc Natl Acad Sci U S A. 2007;104:17004–17009. doi: 10.1073/pnas.0707158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresolin T, de Freitas Rebelo M, Bainy ACD. Expression of PXR, CYP3A and MDR1 genes in liver of zebrafish. Comp Biochem Physiol C. 2005;140:403–407. doi: 10.1016/j.cca.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Buckman AH, Brown SB, Small J, Muir DC, Parrott J, Solomon KR, Fisk AT. Role of temperature and enzyme induction in the biotransformation of polychlorinated biphenyls and bioformation of hydroxylated polychlorinated biphenyls by rainbow trout (Oncorhynchus mykiss) Environ Sci Technol. 2007;41:3856–3863. doi: 10.1021/es062437y. [DOI] [PubMed] [Google Scholar]
- Buhler DR, Wang-Buhler JL. Rainbow trout cytochrome P450s: purification, molecular aspects, metabolic activity, induction and role in environmental monitoring. Comp Biochem Physiol C. 1998;121:107–137. doi: 10.1016/s0742-8413(98)10033-6. [DOI] [PubMed] [Google Scholar]
- Celander M, Stegeman JJ, Förlin L. CYP1A1-, CYP2B- and CYP3A-like proteins in rainbow trout (Oncorhynchus mykiss) liver: CYP1A1-specific down-regulation after prolonged exposure to PCB. Mar Environ Res. 1996;42:283–286. [Google Scholar]
- Chovancova E, Pavelka A, Benes P, Strnad O, Brezovsky J, Kozlikova B, Gora A, Sustr V, Klvana M, Medek P, Biedermannova L, Sochor J, Damborsky J. CAVER 3.0: a tool for the analysis of transport pathways in dynamic protein structures. PLoS Comput Biol. 2012;8:e1002708. doi: 10.1371/journal.pcbi.1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins S, Reschly EJ, Hagey LR, Krasowski MD. Evolution of pharmacologic specificity in the pregnane X receptor. BMC Evol Biol. 2008;8:103. doi: 10.1186/1471-2148-8-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elskus AA, Stegeman JJ. Further consideration of phenobarbital effects on cytochrome P-450 activity in the killifish, Fundulus heteroclitus. Comp Biochem Physiol C. 1989;92:223–230. doi: 10.1016/0742-8413(89)90045-5. [DOI] [PubMed] [Google Scholar]
- Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. Comparative protein structure modeling using MODELLER. Curr Protoc Protein Sci. 2007;Chapter 2 doi: 10.1002/0471140864.ps0209s50. Unit 29. [DOI] [PubMed] [Google Scholar]
- Förlin L. Effects of clophen A50, 3-methylcholanthrene, pregnenolone-16 alpha-carbonitrile, and phenobarbital on the hepatic microsomal cytochrome P-450-dependent monooxygenase system in rainbow trout, Salmo gairdneri, of different age and sex. Toxicol Appl Pharmacol. 1980;54:420–430. doi: 10.1016/0041-008x(80)90169-6. [DOI] [PubMed] [Google Scholar]
- Goksøyr A. Purification of hepatic microsomal cytochromes P-450 from beta-naphthoflavone-treated Atlantic cod (Gadus morhua), a marine teleost fish. Biochim Biophys Acta. 1985;840:409–417. [PubMed] [Google Scholar]
- Goldstone JV, McArthur AG, Kubota A, Zanette J, Parente T, Jönsson ME, Nelson DR, Stegeman JJ. Identification and developmental expression of the full complement of Cytochrome P450 genes in Zebrafish. BMC Genomics. 2010;11:643. doi: 10.1186/1471-2164-11-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J Biol Chem. 1992;267:83–90. [PubMed] [Google Scholar]
- Handschin C, Blattler S, Roth A, Looser R, Oscarson M, Kaufmann MR, Podvinec M, Gnerre C, Meyer UA. The evolution of drug-activated nuclear receptors: one ancestral gene diverged into two xenosensor genes in mammals. Nucl Recept. 2004;2:7. doi: 10.1186/1478-1336-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata H, Yoshinari K, Negishi M, Stegeman JJ. Species-specific responses of constitutively active receptor (CAR)-CYP2B coupling: lack of CYP2B inducer-responsive nuclear translocation of CAR in marine teleost, scup (Stenotomus chrysops) Comp Biochem Physiol C. 2002;131:501–510. doi: 10.1016/s1532-0456(02)00038-8. [DOI] [PubMed] [Google Scholar]
- Jönsson ME, Orrego R, Woodin BR, Goldstone JV, Stegeman JJ. Basal and 3,3′,4,4′,5-pentachlorobiphenyl-induced expression of cytochrome P450 1A, 1B and 1C genes in zebrafish. Toxicol Appl Pharmacol. 2007;221:29–41. doi: 10.1016/j.taap.2007.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsen OA, Puntervoll P, Goksøyr A. Mass spectrometric analyses of microsomal cytochrome P450 isozymes isolated from beta-naphthoflavone-treated Atlantic cod (Gadus morhua) liver reveal insights into the cod CYPome. Aquat Toxicol. 2012;108:2–10. doi: 10.1016/j.aquatox.2011.08.018. [DOI] [PubMed] [Google Scholar]
- Kirischian N, McArthur AG, Jesuthasan C, Krattenmacher B, Wilson JY. Phylogenetic and functional analysis of the vertebrate cytochrome P450 2 family. J Mol Evol. 2011;72:56–71. doi: 10.1007/s00239-010-9402-7. [DOI] [PubMed] [Google Scholar]
- Kleinow KM, Haasch ML, Williams DE, Lech JJ. A comparison of hepatic P450 induction in rat and trout (Oncorhynchus mykiss): delineation of the site of resistance of fish to phenobarbital-type inducers. Comp Biochem Physiol C. 1990;96:259–270. doi: 10.1016/0742-8413(90)90006-u. [DOI] [PubMed] [Google Scholar]
- Klotz AV, Stegeman JJ, Woodin BR, Snowberger EA, Thomas PE, Walsh C. Cytochrome P-450 isozymes from the marine teleost Stenotomus chrysops: their roles in steroid hydroxylation and the influence of cytochrome b5. Arch Biochem Biophys. 1986;249:326–338. doi: 10.1016/0003-9861(86)90009-3. [DOI] [PubMed] [Google Scholar]
- Kopec AK, Burgoon LD, Ibrahim-Aibo D, Mets BD, Tashiro C, Potter D, Sharratt B, Harkema JR, Zacharewski TR. PCB153-elicited hepatic responses in the immature, ovariectomized C57BL/6 mice: comparative toxicogenomic effects of dioxin and non-dioxin-like ligands. Toxicol Appl Pharmacol. 2010;243:359–371. doi: 10.1016/j.taap.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota A, Stegeman JJ, Goldstone JV, Nelson DR, Kim EY, Tanabe S, Iwata H. Cytochrome P450 CYP2 genes in the common cormorant: Evolutionary relationships with 130 diapsid CYP2 clan sequences and chemical effects on their expression. Comp Biochem Physiol C. 2011a;153:280–289. doi: 10.1016/j.cbpc.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota A, Stegeman JJ, Woodin BR, Iwanaga T, Harano R, Peterson RE, Hiraga T, Teraoka H. Role of zebrafish cytochrome P450 CYP1C genes in the reduced mesencephalic vein blood flow caused by activation of AHR2. Toxicol Appl Pharmacol. 2011b;253:244–252. doi: 10.1016/j.taap.2011.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajoie M, Bertrand D, El-Mabrouk N. Inferring the evolutionary history of gene clusters from phylogenetic and gene order data. Mol Biol Evol. 2010;27:761–772. doi: 10.1093/molbev/msp271. [DOI] [PubMed] [Google Scholar]
- Maglich JM, Caravella JA, Lambert MH, Willson TM, Moore JT, Ramamurthy L. The first completed genome sequence from a teleost fish (Fugu rubripes) adds significant diversity to the nuclear receptor superfamily. Nucleic Acids Res. 2003;31:4051–4058. doi: 10.1093/nar/gkg444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathas M, Burk O, Qiu H, Nusshag C, Godtel-Armbrust U, Baranyai D, Deng S, Romer K, Nem D, Windshugel B, Wojnowski L. Evolutionary history and functional characterization of the amphibian xenosensor CAR. Mol Endocrinol. 2012;26:14–26. doi: 10.1210/me.2011-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milnes MR, Garcia A, Grossman E, Grun F, Shiotsugu J, Tabb MM, Kawashima Y, Katsu Y, Watanabe H, Iguchi T, Blumberg B. Activation of steroid and xenobiotic receptor (SXR, NR1I2) and its orthologs in laboratory, toxicologic, and genome model species. Environ Health Perspect. 2008;116:880–885. doi: 10.1289/ehp.10853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LB, Maglich JM, McKee DD, Wisely B, Willson TM, Kliewer SA, Lambert MH, Moore JT. Pregnane X receptor (PXR), constitutive androstane receptor (CAR), and benzoate X receptor (BXR) define three pharmacologically distinct classes of nuclear receptors. Mol Endocrinol. 2002;16:977–986. doi: 10.1210/mend.16.5.0828. [DOI] [PubMed] [Google Scholar]
- Nelson DR, Goldstone JV, Stegeman JJ. The cytochrome P450 genesis locus: the origin and evolution of animal cytochrome P450s. Phil Trans R Soc B. 2013;368:20120474. doi: 10.1098/rstb.2012.0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DR, Zeldin DC, Hoffman SM, Maltais LJ, Wain HM, Nebert DW. Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative-splice variants. Pharmacogenetics. 2004;14:1–18. doi: 10.1097/00008571-200401000-00001. [DOI] [PubMed] [Google Scholar]
- Nims RW, Sinclair PR, Sinclair JF, Dragnev KH, Jones CR, Mellini DW, Thomas PE, Lubet RA. Dose-response relationships for the induction of P450 2B by 1,4-bis[2-(3,5-dichloropyridyloxy)]benzne (TCPOBOP) in rat and cultured rat hepatocytes. Xenobiotica. 1993;23:1411–1426. doi: 10.3109/00498259309059450. [DOI] [PubMed] [Google Scholar]
- Oleksiak MF, Wu S, Parker C, Karchner SI, Stegeman JJ, Zeldin DC. Identification, functional characterization, and regulation of a new cytochrome P450 subfamily, the CYP2Ns. J Biol Chem. 2000;275:2312–2321. doi: 10.1074/jbc.275.4.2312. [DOI] [PubMed] [Google Scholar]
- Oleksiak MF, Wu S, Parker C, Qu W, Cox R, Zeldin DC, Stegeman JJ. Identification and regulation of a new vertebrate cytochrome P450 subfamily, the CYP2Ps, and functional characterization of CYP2P3, a conserved arachidonic acid epoxygenase/19-hydroxylase. Arch Biochem Biophys. 2003;411:223–234. doi: 10.1016/s0003-9861(02)00734-8. [DOI] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 2003;339:62–66. doi: 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- Reschly EJ, Bainy AC, Mattos JJ, Hagey LR, Bahary N, Mada SR, Ou J, Venkataramanan R, Krasowski MD. Functional evolution of the vitamin D and pregnane X receptors. BMC Evol Biol. 2007;7:222. doi: 10.1186/1471-2148-7-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reschly EJ, Krasowski MD. Evolution and function of the NR1I nuclear hormone receptor subfamily (VDR, PXR, and CAR) with respect to metabolism of xenobiotics and endogenous compounds. Curr Drug Metab. 2006;7:349–365. doi: 10.2174/138920006776873526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, Moorman AF. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009;37:e45. doi: 10.1093/nar/gkp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadar MD, Ash R, Sundqvist J, Olsson PE, Andersson TB. Phenobarbital induction of CYP1A1 gene expression in a primary culture of rainbow trout hepatocytes. J Biol Chem. 1996;271:17635–17643. doi: 10.1074/jbc.271.30.17635. [DOI] [PubMed] [Google Scholar]
- Smith G, Harrison DJ, East N, Rae F, Wolf H, Wolf CR. Regulation of cytochrome P450 gene expression in human colon and breast tumour xenografts. Br J Cancer. 1993a;68:57–63. doi: 10.1038/bjc.1993.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G, Henderson CJ, Parker MG, White R, Bars RG, Wolf CR. 1,4-Bis[2-(3,5-dichloropyridyloxy)]benzene, an extremely potent modulator of mouse hepatic cytochrome P-450 gene expression. Biochem J. 1993b;289:807–813. doi: 10.1042/bj2890807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stegeman JJ, Woodin BR, Waxman DJ. Structual relatedness of mammalian cytochromes P450 IIB and cytochrome P450 B from the marine fish scup (Stenotomus chrysops) FASEB J. 1990;4:A739. [Google Scholar]
- Teraoka H, Kubota A, Dong W, Kawai Y, Yamazaki K, Mori C, Harada Y, Peterson RE, Hiraga T. Role of the cyclooxygenase 2-thromboxane pathway in 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced decrease in mesencephalic vein blood flow in the zebrafish embryo. Toxicol Appl Pharmacol. 2009;234:33–40. doi: 10.1016/j.taap.2008.09.021. [DOI] [PubMed] [Google Scholar]
- Valdar WS. Scoring residue conservation. Proteins. 2002;48:227–241. doi: 10.1002/prot.10146. [DOI] [PubMed] [Google Scholar]
- Vezina CM, Walker NJ, Olson JR. Subchronic exposure to TCDD, PeCDF, PCB126, and PCB153: effect on hepatic gene expression. Environ Health Perspect. 2004;112:1636–1644. doi: 10.1289/txg.7253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman DJ. P450 gene induction by structurally diverse xenochemicals: central role of nuclear receptors CAR, PXR, and PPAR. Arch Biochem Biophys. 1999;369:11–23. doi: 10.1006/abbi.1999.1351. [DOI] [PubMed] [Google Scholar]
- Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- Zhang JG, Ho T, Callendrello AL, Crespi CL, Stresser DM. A multi-endpoint evaluation of cytochrome P450 1A2, 2B6 and 3A4 induction response in human hepatocyte cultures after treatment with beta-naphthoflavone, phenobarbital and rifampicin. Drug Metab Lett. 2010;4:185–194. doi: 10.2174/187231210792928224. [DOI] [PubMed] [Google Scholar]
- Zhou LY, Wang DS, Shibata Y, Paul-Prasanth B, Suzuki A, Nagahama Y. Characterization, expression and transcriptional regulation of P450c17-I and -II in the medaka, Oryzias latipes. Biochem Biophys Res Commun. 2007;362:619–625. doi: 10.1016/j.bbrc.2007.08.044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.