Abstract
Few studies have addressed the phylogeography of species of the Cerrado, the largest savanna biome of South America. Here we aimed to investigate the phylogeographical structure of Dalbergia miscolobium, a widespread tree from the Cerrado, and to verify its concordance with plant phylogeographical and biogeographical patterns so far described. A total of 287 individuals from 32 populations were analyzed by sequencing the trnL intron of the chloroplast DNA and the internal transcribed spacer of the nuclear ribosomal DNA. Analysis of population structure and tests of population expansion were performed and the time of divergence of haplotypes was estimated. Twelve and 27 haplotypes were identified in the cpDNA and nrDNA data, respectively. The star-like network configuration and the mismatch distributions indicated a recent spatial and demographic expansion of the species. Consistent with previous tree phylogeographical studies of Cerrado trees, the cpDNA also suggested a recent expansion towards the southern Cerrado. The diversity of D. miscolobium was widespread but high levels of genetic diversity were found in the Central Eastern and in the southern portion of Central Western Cerrado. The combined analysis of cpDNA and nrDNA supported a phylogeographic structure into seven groups. The phylogeographical pattern showed many concordances with biogeographical and phylogeographical studies in the Cerrado, mainly with the Cerrado phytogeographic provinces superimposed to our sampling area. The data reinforced the uniqueness of Northeastern and Southeastern Cerrados and the differentiation between Eastern and Western Central Cerrados. The recent diversification of the species (estimated between the Pliocene and the Pleistocene) and the ‘genealogical concordances’ suggest that a shared and persistent pattern of species diversification might have been present in the Cerrado over time. This is the first time that an extensive ‘genealogical concordance’ between phylogeographic and phytogeographic patterns is shown for the Cerrado biome.
Introduction
Considerable advances have been made in disentangling biogeographical patterns across the globe, but the diversification of many biodiversity rich areas of the world remain poorly studied [1]. One such region is the Cerrado biome, a savanna vegetation that, together with the Seasonally Dry Tropical Forests (SDTF), covers a huge diagonal seasonally dry area in the interior of Eastern Tropical South America (ETSA) [1]–[3]. The Cerrado is typically found in dystrophic soils, often latosols, under a climate characterized by a marked dry season, whereas the SDTFs typically occur in areas with more fertile soils and longer dry seasons [2]–[4]. The Cerrado is the most diverse savanna in the world, particularly in terms of the number of plant species present [5], and it has high levels of endemisms and species adapted to stressed environments [3], [5], [6]. Its almost 1.8 million km2 of original area has been reduced by more than 50% in the last years [7], making the Cerrado one of the main global hotspots for biodiversity conservation [5]. Despite this relevance, many details about the historical biogeography of the Cerrado biome are unresolved [4], [8].
The Cerrado has a highly heterogeneous biota and many studies have tried to subdivide it into biogeographic provinces (see [4], [8] for reviews). Three broad phytogeographical studies have found similar patterns [9]–[11]. In studies analyzing 145 Cerrado areas, Castro [9] and Castro and Martins [10] recognized three main phytogeographic groups, a Southern Cerrados group (named ‘São Paulo’), a Central Cerrados group (named ‘Planalto Central’) and the Northeastern Cerrados. Largely concordant with it, Ratter et al. [11], based on the floristic similarity of a broader sampling effort involving 376 Cerrado areas, identified six phytogeographic provinces (see names and delimitation of them in Figure 1). Their results were largely concordant and they suggested that climate and soil, as well as past climatic events, might be among the main factors responsible for the patterns found. With regard to the impact of past climates on the Cerrado biota, many controversies exist, and expansion [12], stability [13], and reduction [14], [15] of its extension during the Last Glacial Maximum (LGM) have been proposed. Phylogeographical approaches would help to evaluate the persistence of these patterns at the intra-specific level and to help unravel the causes for them.
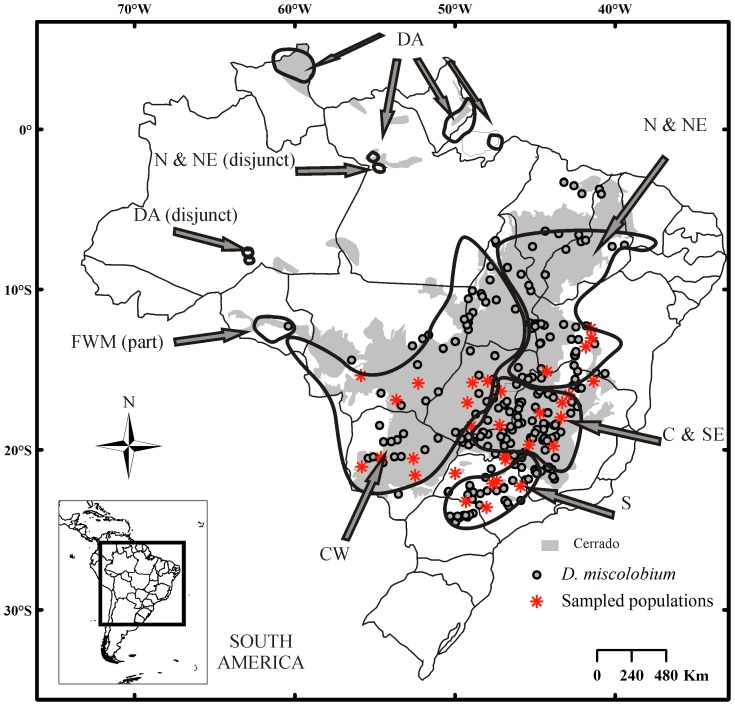
Figure 1. Geographic distribution of Dalbergia miscolobium (dots) across the Brazilian Cerrado biome (grey) in Eastern Tropical South America.
Each dot represents a record of D. miscolobium and each red asterisk a sampled population. The geographic distribution of D. miscolobium was estimated through field trips, herbarium records and databases of floristic checklists. The six phytogeographical provinces proposed by Ratter et al. [11] have been delimited: ‘S’, Southern, ‘C & SE’, Central and southeastern, ‘N & NE’, North and northeastern, ‘CW’, Central-western, ‘FWM’, Far western mesotrophic sites and ‘DA’, Disjunct Amazonian. The codes of the provinces are the same of the original publication.
Phylogeographical studies have proved to be valuable independent sources of evidence in comprehending biogeographical patterns and in reconstructing past vegetation dynamics [16]. Concordances between phylogeographical and biogeographical patterns and paleodata have been found in numerous instances (e. g. [17], [18]), thus helping understand the evolution and shared history of many species. Nevertheless, only a few studies have addressed the phylogeography of species from the Cerrado [1], [8] and phylogeographical studies that have focused on Cerrado trees are scarce [19]–[22]. Essentially, these studies found phylogeographic structuring and high levels of differentiation among populations in the respective species analyzed. Three of them observed major groups of populations subdivided longitudinally [20]–[22], and three [19], [21], [22] observed also patterns of recent range expansion of northern resources towards the southern Cerrado. These expansions have been hypothesized to have occurred after the return of wetter and warmer climatic conditions after the last glacial period. A better understanding of the evolutionary patterns of the Cerrado could be achieved with more phylogeographical studies of species with distinct ecological requirements together with wider samplings across the biome, including the peripheral regions. In the present study, we focused on a widespread tree species adapted to the coldest regions of the biome, and encompassing a large portion of the biome, including the southwest portion.
The species chosen for this study, Dalbergia miscolobium Benth. (Papilionoideae, Fabaceae), is endemic and widespread in the Cerrado (Figure 1). It is among the 121 most representative tree species of the biome and is the 9th and 12th most common tree species in the southeastern and the southern Cerrado, respectively [23]. Unlike other Cerrado trees subjects of phylogeographical studies so far, D. miscolobium seems to be more tolerant to the cold. It is common in the coldest (subject to eventual frosts during winter) and southernmost portions of the Cerrado, and is widely found at higher altitudes (>1,000 m) in the biome [24]. In the work of Bridgewater et al. [23], D. miscolobium was found in 82% of the 22 floristic surveys conducted in the southern Cerrado and in 75% of the 73 sites studied in the south-eastern Cerrado. On the other hand, the species was encountered in only 37% and 28% of the north-eastern and central-western Cerrado provinces, respectively. It is a tree pollinated by bees and its seeds are wind-dispersed [25]. It is an important timber species, and is a rapid colonizer of suitable environments.
Here, we undertook an extensive sampling of D. miscolobium populations encompassing a large portion of the Cerrado biome in order to investigate the species' phylogeographical structure and to verify its congruence with phylogeographical and biogeographical patterns reported to the Cerrado biome so far. More specifically, we aimed to answer the following questions: 1) is the phylogeographical structure of this wide range species congruent with the Cerrado biogeographical provinces proposed by Ratter et al. [11]? Considering that the historical casual factors that contributed to determine the phytogeographical provinces of Cerrado may also have contributed to the phylogeographical structure of D miscolobium we predicted some congruency between them; 2) in what extension is the phylogeographical structure of D. miscolobium similar to other Cerrado tree species studied so far? Based on previous studies [20]–[22] we predicted that the southern populations present lower genetic diversity than populations from central Cerrado, which in turn would present high diversity levels, consistent with a recent colonization of southernmost populations from more stable northernmost areas.
Materials and Methods
Ethics Statement
Dalbergia miscolobium is not included in the Brazilian Official List of Threatened Plants. The collection of samples of leaves in conservation units was realized with the legal authorization of the ‘Instituto Brasileiro do Meio Ambiente e dos Recursos Naturais Renováveis’ (IBAMA; populations DFE, MUC and PER - licenses 192/2003, 110/2005, 26.627.1) and of the ‘Instituto Estadual de Florestas – MG’ (population RPE – license 018/05). The collection of the samples in localities out of conservation units had authorization for collection and transport of IBAMA (licenses 099/2004 and 016/2006, 14035-2, 293770) and of the owners of the lands. Also we had authorization for the access to sampling of component of genetic patrimony of the Brazilian ‘Conselho de Gestão do Patrimônio Genético’ (CGEN/MMA) number 03/2004.
Population sampling and DNA amplification
The geographic distribution of D. miscolobium was estimated through field trips, databases of floristic checklists (personal communications by José Felipe Ribeiro and Ary T. Oliveira-Filho) and herbarium records (Figure 1). Expeditions were carried out from 2004 to 2010 to sample D. miscolobium across the Brazilian Cerrado. Leaf samples were collected from 287 adult trees in 32 populations covering most of its geographic distribution range, encompassing seven Brazilian states (Table 1 and Figure 1). Our sampling covered all the four Cerrado phytogeographic provinces proposed by Ratter et al. [11] in which the species is found: Southern (S), Central and southeastern (C & SE), North and northeastern (N & NE) and Central-western (CW) (Figure 1). We sampled a lower number of populations in the N & NE province, where the species is scarcely found (see Introduction). Three to 14 individuals were sampled in each population. Populations were sampled in natural reserves and remnant fragments of the Cerrado (Table 1). Genomic DNA was isolated by a modified cetyl trimethylammonium bromide (CTAB) extraction method [26] and quantification was carried out by visual inspection of agarose gels.
Table 1. Locations of Dalbergia miscolobium populations in the Brazilian Cerrado biome, genetic diversity indices and the distribution of the chloroplast DNA (cpDNA) and nuclear ribosomal DNA (nrDNA) haplotypes.
| cpDNA data | nrDNA data | |||||||
| Locality, State a (population code) | Latitude (S) - Longitude (W) | Sample size b | Haplotype diversity | Nucleotide diversity (X10−2) | Haplotypes | Haplotype diversity | Nucleotide diversity (X10−2) | Haplotypes |
| Água Clara, MS (AGU) | 20°32′36″– 52°34′26″ | 14 | 0.473 | 0.083 | C1, C2, C3 | 0.712 | 0.155 | N1, N2, N3, N4, N5 |
| Bataguassu, MS (BAT) | 21°36′05″– 52°28′19″ | 07 | 0.000 | 0.000 | C1 | 0.495 | 0.080 | N1, N2 |
| Nioque, MS (NIO) | 21°04′04″– 55°47′38″ | 11 | 0.327 | 0.054 | C1, C3 | 0.710 | 0.175 | N1, N2, N3, N4 |
| Anhanduí, MS (NHA) | 20°28′15″– 54°37′26″ | 06 | 0.000 | 0.000 | C4 | 0.712 | 0.181 | N1, N2, N3, N4 |
| Alto das Garças, MT (GAR) | 16°53′58″– 53°38′59″ | 10 | 0.000 | 0.000 | C5 | 0.747 | 0.171 | N1, N3, N4, N6 |
| Barra das Garças, MT (BGA) | 15°51′19″– 52°16′09″ | 10 | 0.000 | 0.000 | C6 | 0.000 | 0.000 | N4 |
| Salgadeira, MT (CGU) | 15°22′02″– 55°50′25″ | 10 | 0.000 | 0.000 | C1 | 0.468 | 0.080 | N3, N4, N6 |
| Professor Jamil, GO (PJA) | 17°04′03″– 49°13′24″ | 11 | 0.000 | 0.000 | C6 | 0.000 | 0.000 | N4 |
| Pirenópolis, GO (PNO) | 15°48′44″– 48°53′28″ | 10 (7) | 0.000 | 0.000 | C6 | 0.200 | 0.032 | N4, N6 |
| Brasília, DF (DFE) | 15°43′39″– 47°56′44″ | 10 | 0.000 | 0.000 | C1 | 0.440 | 0.071 | N3, N7 |
| Avanhandava, SP (AVA) | 21°28′45″– 49°58′13″ | 06 | 0.000 | 0.000 | C2 | 0.621 | 0.115 | N1, N2, N6 |
| Analândia, SP (ANA) | 22°07′40″– 47°38′48″ | 10 | 0.000 | 0.000 | C7 | 0.647 | 0.210 | N8, N9, N10, N11 |
| Emas, SP (EMA) | 21°56′01″– 47°22′23″ | 10 | 0.000 | 0.000 | C7 | 0.614 | 0.200 | N8, N9, N10, N12, N13 |
| Itapetininga, SP (IPE) | 23°35′49″– 48°01′42″ | 10 | 0.000 | 0.000 | C7 | 0.100 | 0.016 | N9, N12 |
| Piraju, SP (PIR) | 23°15′12″– 49°18′16″ | 10 | 0.000 | 0.000 | C7 | 0.358 | 0.116 | N9, N10, N12 |
| Bom Despacho, MG (BDE) | 19°44′00″– 45°15′00″ | 07 | 0.571 | 0.094 | C1, C8 | 0.485 | 0.078 | N3, N7 |
| Coromandel, MG (COR) | 18°29′03″–47°12′10″ | 10 | 0.533 | 0.088 | C7, C9 | 0.574 | 0.152 | N3, N7, N14, N15, N16 |
| Divisa Alegre, MG (DAL) | 15°43′32″– 41°20′42″ | 10 (9) | 0.000 | 0.000 | C7 | 0.835 | 0.236 | N3, N7, N14, N17, N18, N19 |
| Delfinopólis, MG (DEL) | 20°20′45″– 46°50′15″ | 03 | 0.667 | 0.110 | C1, C10 | 0.333 | 0.054 | N3, N7 |
| Grão Mogol, MG (GMO) | 16°34′00″– 42°53′00″ | 06 | 0.533 | 0.088 | C1, C7 | 0.682 | 0.132 | N3, N14, N17, N19 |
| Itacambira, MG (ICA) | 15°06′00″– 44°06′00″ | 06 (5) | 0.000 | 0.000 | C7 | 0.750 | 0.156 | N3, N17, N18 |
| Pouso Alegre, MG (PAL) | 22°16′26″– 45°53′27″ | 10 | 0.000 | 0.000 | C7 | 0.774 | 0.353 | N9, N10, N12, N20, N21, N22, N23 |
| Januária - Peruaçu, MG (PER) | 15°07′20″– 44°14′53″ | 10 | 0.000 | 0.000 | C1 | 0.495 | 0.080 | N3, N17 |
| São Gonçalo do Rio Preto, MG (RPE) | 18°00′00″– 43°23′00″ | 10 | 0.200 | 0.033 | C1, C7 | 0.673 | 0.154 | N3, N7, N14, N17, N18 |
| Santa Luzia, MG (SAL) | 19°46′00″– 43°51′00″ | 06 | 0.533 | 0.088 | C1, C8 | 0.409 | 0.066 | N3, N7 |
| São João Batista da Glória, MG (SJO) | 20°37′21″– 46°31′13″ | 03 | 0.000 | 0.000 | C5 | 0.600 | 0.097 | N3, N7 |
| Tupaciguara, MG (TUP) | 18°31′32″– 48°59′29″ | 12 (11) | 0.000 | 0.000 | C6 | 0.000 | 0.000 | N4 |
| Unaí, MG (UNA) | 16°22′14″– 47°07′21″ | 10 (8) | 0.467 | 0.154 | C7, C11 | 0.791 | 0.199 | N3, N7, N17, N18 |
| Várzea da Palma, MG (VPA) | 17°42′37″– 44°41′24″ | 10 | 0.533 | 0.088 | C1, C7 | 0.739 | 0.200 | N3, N14, N17, N18 |
| Mucugê, BA (MUC) | 13°03′13″– 41°28′30″ | 10 | 0.000 | 0.000 | C12 | 0.667 | 0.224 | N3, N7, N24, N25, N26 |
| Palmeiras, BA (PRA) | 12°26′36″– 41°29′49″ | 10 (9) | 0.000 | 0.000 | C12 | 0.933 | 0.345 | N3, N7, N24, N25, N27 |
| Rio de Contas, BA (RCO) | 13°35′21″– 41°49′07″ | 09 | 0.000 | 0.000 | C12 | 0.837 | 0.272 | N3, N7, N14, N24, N25, N26 |
Abbreviation of Brazilian States: MS, Mato Grosso do Sul; MT, Mato Grosso; GO, Goiás; DF, Distrito Federal; SP, São Paulo; MG, Minas Gerais; BA, Bahia.
In parentheses sample size analyzed through nrDNA when this was different from that of cpDNA.
The following regions were chosen for DNA sequencing: the trnL intron of the chloroplast DNA (cpDNA) and the internal transcribed spacer (ITS) region of the 18S–26S nuclear ribosomal DNA (nrDNA) genes (which includes the ITS1 and ITS2 intergenic spacers and the 5.8S gene). The trnL intron and ITS region were amplified and sequenced using primers as described by Taberlet et al. [27] and Delgado-Salinas et al. [28], respectively. The cpDNA region was chosen, because it presented specific amplification and the highest variability among 15 non-coding cpDNA regions evaluated (information is available on request). Special care was taken with regard to the specificities of the ITS region and the main guidelines proposed by Feliner and Rosselló [29] were followed in the amplification and analysis of this region. The amplification of both regions was performed in 25 µl total volume containing 10–20 ng of DNA, 1X PCR buffer with 2.0 mM MgCl2, 200 µM of each dNTP, 0.2 ng bovine serum albumin (BSA), 0.5 µM of each primer, 1 U Taq DNA polymerase (Phoneutria, Belo Horizonte, Brazil), and autoclaved deionized water. The nrDNA region reaction mixture also contained 2% dimethyl sulfoxide (DMSO) and 1 M Betaine (N3-trimethyglycine). Thermal conditions of the reaction were as follows: initial denaturation at 94°C for 2 min, followed by 35 cycles at 94°C for 1 min, 56°C (cpDNA) or 60°C (nrDNA) for 1 min, and 72°C for 1 min, and a final elongation at 72°C for 7 min. Each PCR product was then double-strand sequenced using the DYEnamic ET dye terminator sequencing Kit (GE Healthcare, UK). Sequencing reactions were analyzed on a MegaBACE 1000 automated sequencer (GE Healthcare) following the manufacturer's instructions.
Sequence analysis
Sequences were assembled using the package PHRED/PHRAP/CONSED to generate consensus sequences. The cpDNA and nrDNA sequences were deposited in GenBank under accession numbers JQ612719 to JQ612730 and JQ582850 to JQ582876, respectively. The consensus sequences were aligned using CLUSTAL-W [30] implemented in the MEGA 5 program [31], followed by careful manual adjustment. The ends of the sequences were pruned to eliminate fragments that could not be obtained for all individuals and to preserve only high confidence bases.
For nrDNA sequences, all variable sites were carefully checked in the original electropherograms and, when heterozygous nucleotide positions identified by double peaks were present, samples were re-sequenced independently in order to confirm the genotype. To reconstruct the nrDNA haplotypes, we used PHASE 2.1 software [32], which uses a Bayesian statistical method to infer haplotypes of individuals that have more than one polymorphic site based on genotypic data. It was run under default conditions, allowing for multiallelic loci (-d option) and for 10,000 iterations. In the first run, one multiallelic site was shown to be highly homoplasious in our data and, based on this, we removed this site prior to further analysis. Only haplotypes recovered with >0.90 posterior probability were used in subsequent analyses, a precaution adopted by other studies that have used PHASE [33]. To perform subsequent genetic analyses, the output of PHASE was transformed through the pipeline described by Machado et al. [34].
Population genetic and phylogeographical analyses
Genetic diversity indices were estimated in ARLEQUIN 3.5 [35] and in DNAsp 5.10 [36]. The hypothesis of population expansion was tested by pairwise mismatch distributions [37] and the neutrality tests Tajima's D [38] and Fu's FS [39]. These analyses were performed with the ARLEQUIN and DNAsp programs. Allelic richness was computed in Contrib [40] and index of differentiation among populations was estimated by D [41] in SMOGD [42]. Both the indices and the tests were performed for the whole species as well as for the phylogeographic groups of populations delimited in the analyses (see Results and Table 2 for details about their composition). Analyses of molecular variance (AMOVA; [43]) using pairwise differences were conducted in ARLEQUIN to assess population genetic structure in D. miscolobium based on cpDNA and nrDNA data.
Table 2. Genetic diversity indices for the phylogeographic groups of Dalbergia miscolobium based on chloroplast DNA (cpDNA) and nuclear ribosomal DNA (nrDNA) data.
| Phylogeographic groupsa | Data | Sample size | Total haplotypes | Shared haplotypes | Exclusive haplotypes | Haplotype diversity (SD) | Nucleotide diversity (SD) (X10−2) | Allelic richnessc |
| All populations | cpDNA | 287 | 12 | - | - | 0.794 (0.013) | 0.289 (0.190) | 6.542 |
| nrDNA | 251b | 27 | - | - | 0.893 (0.006) | 0.510 (0.290) | 8.905 | |
| Central Eastern 1 (CE1) | cpDNA | 62 | 3 | C1,C7 | C11 | 0.516 (0.042) | 0.095 (0.086) | 1.696 |
| nrDNA | 49 | 6 | N3,N7,N14 | N17,N18,N19 | 0.758 (0.024) | 0.185 (0.134) | 3.954 | |
| Central Eastern 2 (CE2) | cpDNA | 39 | 6 | C1,C5,C7 | C8,C9,C10 | 0.739 (0.056) | 0.193 (0.141) | 4.599 |
| nrDNA | 35 | 5 | N3,N7,N14 | N15,N16 | 0.521 (0.047) | 0.104 (0.091) | 2.101 | |
| Central Western 1 (CW1) | cpDNA | 20 | 2 | C1,C5 | - | 0.526 (0.036) | 0.087 (0.084) | 1.000 |
| nrDNA | 17 | 4 | N1,N3,N4,N6 | - | 0.658 (0.064) | 0.144 (0.114) | 2.874 | |
| Central Western 2 (CW2) | cpDNA | 44 | 4 | C1 | C2,C3,C4 | 0.599 (0.066) | 0.315 (0.203) | 2.826 |
| nrDNA | 44 | 6 | N1,N3,N4,N6 | N2,N5 | 0.705 (0.021) | 0.164 (0.123) | 2.963 | |
| Central Cerrados (CC) | cpDNA | 43 | 1 | - | C6 | 0.00 | 0.00 | 0.00 |
| nrDNA | 37 | 2 | N4,N6 | - | 0.027 (0.026) | 0.044 (0.016) | 0.230 | |
| Southern Cerrados (SC) | cpDNA | 50 | 1 | C7 | - | 0.00 | 0.00 | 0.00 |
| nrDNA | 49 | 10 | - | N8,N9,N10,N11, N12,N13,N20, N21,N22,N23 | 0.596 (0.049) | 0.235 (0.159) | 3.587 | |
| Northeastern Cerrados (NC) | cpDNA | 29 | 1 | - | C12 | 0.00 | 0.00 | 0.00 |
| nrDNA | 20 | 7 | N3,N7,N14 | N24,N25,N26, N27 | 0.815 (0.034) | 0.297 (0.193) | 5.023 | |
| Central Eastern (CE1+CE2) | cpDNA | 101 | 7 | C1,C5,C7 | C8,C9,C10,C11 | 0.675 (0.029) | 0.146 (0.114) | 4.532 |
| nrDNA | 84 | 8 | N3,N7,N14 | N15,N16,N17, N18,N19 | 0.751 (0.017) | 0.193 (0.137) | 5.026 | |
| Central Western (CW1+CW2) | cpDNA | 64 | 5 | C1,C5 | C2,C3,C4 | 0.638 (0.055) | 0.271 (0.179) | 3.817 |
| nrDNA | 61 | 6 | N3,N4,N6 | N1,N2,N5 | 0.780 (0.012) | 0.205 (0.144) | 4.291 |
The phylogeographic groups are composed by following populations: CE1 = DAL, GMO, ICA, PER, RPE,UNA,VPA; CE2 = BDE, COR, DEL, SAL, SJO, DFE; CW1 = GAR, CGU; CW2 = AGU, BAT, NIO, NHA, AVA; CC = BGA, PNO, PJO, TUP; SC = ANA, EMA, IPE, PIR, PAL; NC = MUC, PRA, RCO. See also the Figure 3D.
Twenty-seven individuals were excluded in PHASE analysis due to unresolved genotypes.
Allelic richness after rarefaction to 29 and 20 for cpDNA and nrDNA, respectively.
The phylogenetic relationships among haplotypes were inferred using the Median-Joining (MJ) network method [44] and Statistical Parsimony [45] as implemented in NETWORK 4.6 (fluxus-engineering.com) and TCS 1.21 [46], respectively. Insertion/deletions (indels) were considered as fifth character states and were coded as single mutations, regardless of their size.
The phylogeographic structure of populations was evaluated through a Bayesian Analysis of genetic Population Structure in BAPS 5.3 [47]. Despite the availability of some other software for Bayesian spatial analyses [48], we opted to use BAPS because of its efficiency in unveiling population structures [49] and its extensive usage for definition of clustering of genetic variation using geographical information (e.g. [50]–[53]). The analysis was performed at the group level, considering each population as a group, and using their respective geographical coordinates. The BAPS analysis was performed for the cpDNA and nrDNA data both separately and concatenated. The best number of clusters (k) was determined after 15 runs under a maximum of 20 clusters. We also made some analysis with prior numbers of clusters based on the number of Cerrado phytogeographic provinces superimposed to our sampled area (k = 4 equivalent to Ratter et al.'s consensus analysis and k = 5 to Ratter et al.'s Twinspan analysis; see Results for details). The location of potential geographic barriers was explored through the Monmonier's algorithm in BARRIER 2.2 [54]. As genetic data inputs, we used Nei unbiased distance matrices [55] generated through GenALEx 6.5 [56]. We used the matrices of cpDNA and nrDNA together to compute one to ten barriers. To confirm the phylogeographical structure and to test the hypothesis of concordance between phylogeography and biogeography, hierarchical AMOVAs [43] were conducted in ARLEQUIN, following Brante et al. [57]. The AMOVAs were run with the same clusters identified by BAPS with cpDNA, nrDNA and concatenated data, as well as with the population clusters established by BAPS in the analysis with prior of 4 and 5 clusters (see above). The fixation indices FCT, FSC and FST were computed considering the pairwise distances between haplotypes. The level of significance was computed by 1000 nonparametric permutations.
The divergence times among the cpDNA and nrDNA haplotypes identified in D. miscolobium were estimated through a Bayesian coalescent approach implemented in BEAST 1.6 [58]. We assumed a Bayesian skyline plot tree prior with two groups and a piecewise-linear model. Independent runs were carried out for the cpDNA and nrDNA datasets, both under a HKY substitution model and a strict clock, and each composed of three runs in a total of 2.2×108 generations. Substitution rates for the trnL-trnF and ITS regions of Inga species, estimated to be 1.3×10−9 and 2.34×10−9 substitutions per site per year, respectively [59], were used to estimate divergence times. The genus Inga shares many features with Dalbergia. In particular, both species belong to the Leguminosae family, are composed of trees, and occur in the neotropics. Thus, the estimated rates for sequence evolution of Inga are suitable for use in our study.
Results
Genetic diversity
Consensus sequences were obtained from 287 and 278 individuals of D. miscolobium for the cpDNA and nrDNA data, respectively. For cpDNA, the alignment produced a total of 606 bp with 13 detected polymorphic sites, which included 11 base substitutions and two indels (Table S1). Twelve cpDNA haplotypes were shared across D. miscolobium populations. For the nrDNA region, in a total alignment of 618 bp, we found 15 polymorphic sites (all represented by base substitutions), resulting in 27 haplotypes (Table S2). After the PHASE analysis, 27 sampled individuals (54 allele copies) were excluded from the nrDNA analysis due to unresolved genotypes (p<0.90), thus leaving 251 individuals in the analysis totaling 502 allele copies. Because of the nature of nrDNA region, some heterozygotes we observed could be in fact paralogous copies that escaped concerted evolution [29], and because of that we did not perform any analysis concerning the heterozygosity of sampled individuals.
The molecular diversity indices and haplotypes distributions for each D. miscolobium population are described in Table 1. The majority of the populations had no genetic variability in the cpDNA data (22 populations of the 32 analyzed), whereas only three populations (BGA, PJA, and TUP) were monomorphic in the nrDNA data (Table 1). For the cpDNA and nrDNA data, the total haplotype diversity was 0.794 and 0.893, and the total nucleotide diversity was equal to 0.00289 and 0.00510, respectively (Table 2). The FST values were estimated in 0.952 and 0.740 for cpDNA and nrDNA respectively, showing high differentiation among populations [60], which were corroborated by the Jost's D values of 0.799 and 0.805 for cpDNA and nrDNA data, respectively.
Phylogeographical structure
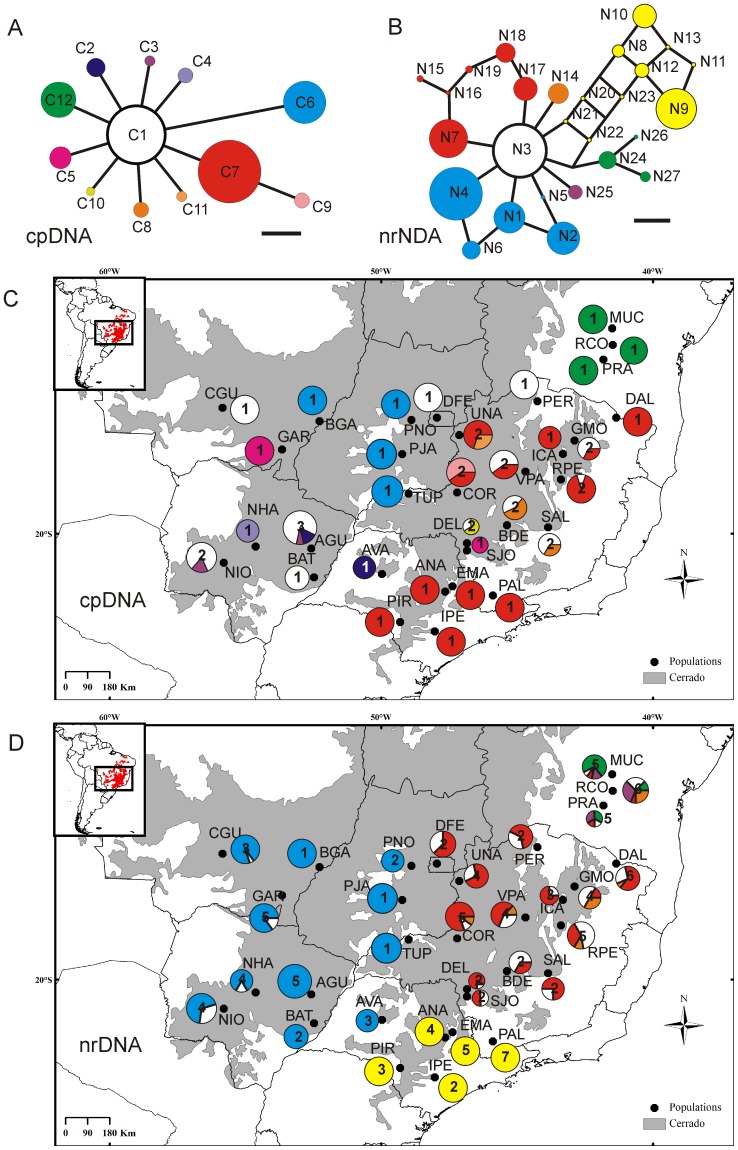
The phylogenetic relationships among haplotypes inferred through both network methods had very similar topologies; therefore, only the MJ networks are shown (Figure 2A, B). For both cpDNA and nrDNA data, the networks exhibited a central haplotype that was common and the most widespread, and from which many other haplotypes diverged in a star-like fashion. For cpDNA data, all haplotypes (except C9) were directly linked to central haplotype (C1), and only C6 was separated from it by more than one mutation event (three base substitutions; Figure 2A and Table S1). For nrDNA, the pattern was similar but the number and level of divergence of haplotypes were greater (Figure 2B and Table S2). Many reticulations connected the groups of haplotypes that derived from the central haplotype (Figure 2B), indicating equally probable evolutionary pathways. These reticulations at the nrDNA level have been observed in other plant species (e.g.: [61], [62]) and we believe that it does not affect our main conclusions.
Figure 2. Median-joining networks depicting the relationships among haplotypes of Dalbergia miscolobium based on cpDNA (A) and nrDNA (B) data.
Geographic distribution of the cpDNA (C) and nrDNA (D) haplotypes across sampled populations. Population and haplotype codes correspond to those in Table 1 and Table 2. The network colours are equivalent to those at the maps for the respective genomes. The full circles are proportional to the number of individuals. The numbers inside the circles correspond to number of haplotypes found in that population.
Similar phylogeographical structure was observed in the cpDNA and nrDNA data (Figure 2C, D). In both data sets the central haplotype was very common and was widespread in the central, western, and southeastern regions of the Cerrado. The ‘peripheral haplotypes’ in the nrDNA network were, in general, more restricted to certain geographic regions and most of them also less frequent. Furthermore, the haplotypes were not randomly distributed, and a clear structure could be observed (Figure 2).
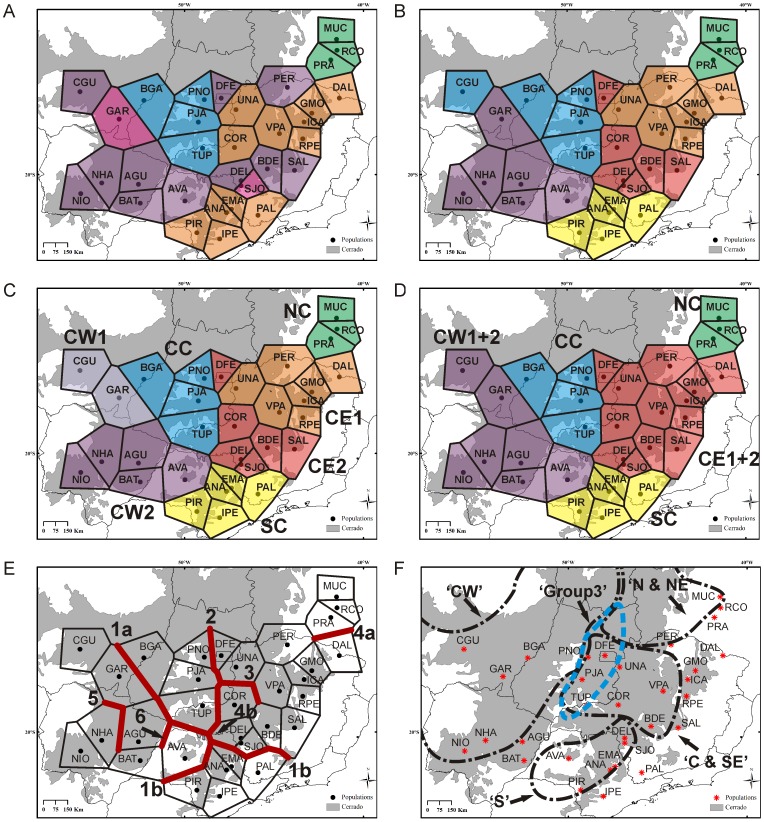
The Bayesian Analysis of Population Structure (BAPS) pointed to the most probable existence of five and six clusters of populations for the cpDNA and the nrDNA datasets, respectively (Figure 3A, B). With the data concatenated, the analysis suggested seven clusters (Figure 3C). Two clusters, ‘Northeastern Cerrados’ and the ‘Central Cerrados’, remained the same in the three types of datasets used (cpDNA, nrDNA and concatenated; Figure 3A–C). The divisions between ‘Central-Eastern’ and ‘Central-Western’ as well as between ‘Central-Eastern’ and ‘Southern Cerrados’ were weaker for the cpDNA data, showing signs of previous connections and relationships between those regions. The nrDNA and the concatenated results were very similar to each other (Figure 3B, C). As the concatenated clusterization takes into account both genomes and as there was a considerably concordance among the analyses for the three datasets (cpDNA, nrDNA and concatenated), we selected the concatenated results (Figure 3C) as the best representation of D. miscolobium phylogeographical structure. The AMOVA showed that the most of genetic variation was distributed among phylogeographical groups for both cpDNA (83.4% and 75.6% for five and seven clusters, respectively) and nrDNA (71.7% and 71.2% for six and seven clusters, respectively) (Table 3), confirming the consistency of the clusters determined by BAPS.
Figure 3. Results from population structure analyses of Dalbegia miscolobium populations (A to E) and the phytogeographical provinces proposed by Ratter et al.
[11] that are superimposed to our sampling area (F). In each panel is represented a result of a different analysis: A) Bayesian Analysis of Population Structure (BAPS) with cpDNA only; B) BAPS with nrDNA only; C) BAPS with cpDNA and nrDNA concatenated; D) BAPS with cpDNA and nrDNA concatenated with a prior of k = 5; E) Analysis with the Monmonier's algorithm implemented in Barrier 2.2. In panels A to E, the polygons are the result of Voronoi tessellation and each one is correspondent to a population whose code is the same as in Table 1. Polygons with the same color belong to the same BAPS cluster. In panel E, the red lines correspond to the consensus barriers among the cpDNA and nrDNA distance matrices. The barriers are numbered in order of their appearance which is related to their probability of existence. The larger abbreviations in panels C and D are for the phylogeographic groups as detailed in Results and Table 2. In panel F, the black traces are for the phytogeographical provinces of Ratter et al. [11] and the blue trace is correspondent to Group 3 from their Twinspan analysis. The codes of the provinces are the same of the original publication. For names of provinces see Figure 1 and for further details about them, refer to the text.
Table 3. Analyses of molecular variance (AMOVA) of cpDNA and nrDNA data among clusters identified by Bayesian Analysis of Population Structure (BAPS) with cpDNA, nrDNA and concatenated data, as well as with the population clusters established by BAPS in the analysis with prior of 4 and 5 clusters.
| BAPS clusters | Source of variation | Variation % | F-statisticsa |
| cpDNA data | |||
| 5 clusters | Among groups | 83.4 | FCT = 0.83 |
| Among populations within groups | 12.2 | FSC = 0.73 | |
| Within populations | 4.4 | FST = 0.95 | |
| 7 clusters | Among groups | 75.6 | F CT = 0.75 |
| Among populations within groups | 19.3 | FSC = 0.78 | |
| Within populations | 5.1 | FST = 0.94 | |
| 4 clusters a priori | Among groups | 45.8 | FCT = 0.45 |
| Among populations within groups | 49.2 | FSC = 0.90 | |
| Within populations | 5.0 | FST = 0.95 | |
| 5 clusters a priori | Among groups | 76.2 | FCT = 0.76 |
| Among populations within groups | 19.0 | FSC = 0.79 | |
| Within populations | 4.8 | FST = 0.95 | |
| nrDNA data | |||
| 6 clusters | Among groups | 71.7 | FCT = 0.71 |
| Among populations within groups | 5.1 | FSC = 0.17 | |
| Within populations | 23.2 | FST = 0.76 | |
| 7 clusters | Among groups | 71.2 | FCT = 0.71 |
| Among populations within groups | 5.2 | FSC = 0.18 | |
| Within populations | 23.6 | FST = 0.76 | |
| 4 clusters a priori | Among groups | 67.4 | FCT = 0.67 |
| Among populations within groups | 11.2 | FSC = 0.34 | |
| Within populations | 21.4 | FST = 0.78 | |
| 5 clusters a priori | Among groups | 68.2 | FCT = 0.68 |
| Among populations within groups | 9.5 | FSC = 0.29 | |
| Within populations | 22.3 | FST = 0.77 |
P-value<0.0001 for all analyses.
See methods and results for more details about the clusters.
We named each one of the seven clusters using as a basis the biogeographic units described by Ratter et al. [11], as they have been used as a reference to some subsequent biogeographic studies (e. g. [63]), and we will use that nomenclature from now on to refer to them (Table 2 and Figure 3C). The groups Central Cerrados (CC), Southern Cerrados (SC) and Northeastern Cerrados (NC) are the first ones to be separated from the rest of the populations in the BAPS analyses with lower values of k (k<7), what highlights their consistency as distinct phylogeographic units. Moreover each one has very peculiar characteristics with regard to levels and patterns of genetic diversity in cpDNA and nrDNA, relationships with the other provinces and haplotypes composition and uniqueness (Figures 2 and 3 and Table 2). The other four groups, CW1, CW2, CE1 and CE2, on the other hand, appear to be more connected to each other, as they share the most common and widespread haplotypes of both cpDNA and nrDNA genomes (Figure 2A, B). These groups do not have a genetic uniformity and they do have some neighboring populations with completely different genetic compositions. Probably because of that, in BAPS analyses, they are not separated with cpDNA data, and the subdivisions between CW1 and CW2 groups as well as between CE1 and CE2 groups do not appear in BAPS analysis with concatenated data with lower k values (k = 5 and k = 4).
In order to test for the fitness of our data to the Cerrado phytogeographical provinces proposed by Ratter et al. [11] we also ran BAPS using as priors four and five clusters for the concatenated data. By doing this we aimed to verify if our seven groups could be partitioned into Ratter et al's phytogeographical Cerrado provinces. Those priors correspond to the number of Cerrado's provinces that are superimposed to our sampling area in two ways: Ratter et al's Consensus results (k = 4; the four provinces ‘CW’, ‘S’, ‘C & SE’ and ‘N & NE’) and Twinspan results (k = 5; province ‘C & SE’ is subdivided; Figure 3F). Compared to the partition without priors (seven groups; Figure 3C), the prior of four clusters (k = 4) resulted in the merging of the groups NC, CE1 and CE2 into one and the groups CW1 and CW2 into another (Figure S1). With the prior of five clusters (k = 5), group NC is detached from CE1+CE2 (Figure 3D). Both configurations, but mainly the five clusters partition, considerably match the provinces of Ratter et al. [11] as analyzed by the Twinspan method (Figures 3D, F and S1). The AMOVAs with the five clusters partition showed high FCT values for cpDNA (FCT = 0.76) and nrDNA (FCT = 0.68), which was higher than with the four clusters division (FCT = 0.45 and 0.67, respectively; Table 3), reinforcing greater concordance of the phylogeographical groups with the provinces proposed by Ratter et al. [11], analyzed by the Twinspan method.
To further investigate the support of those subdivisions we explored the potential place of genetics barriers through the Monmonier's algorithm implemented in BARRIER. We added barriers from one to ten, using multiple matrices (cpDNA and nrDNA), and we show only the consensus barriers among the matrices (Figure 3E). With the adding of 10 barriers, six are consensual among cpDNA and nrDNA and the subdivisions determined by them are almost the same as those observed in BAPS concatenated seven clusters. Altogether, the four most probable barriers are placed almost exactly at the limits among BAPS five clusters (Barriers 1 to 4 in Figure 3E).
Exclusive haplotypes could be found in six of the seven groups (Table 2) showing a widespread distribution of diversity. But only two centres of genetic diversity were common for cpDNA and nrDNA data. The first centre was composed by the CE1 and CE2 groups, comprised mainly of populations from the state of Minas Gerais, and the second was the CW2 group. These centers have many exclusive haplotypes in addition to the central one, higher level of intrapopulation variation and high levels of genetic diversity (Table 2, Figure 2C, D). Two other groups, SC and NC were completely monomorphic for the cpDNA but medium (SC) to highly (NC) diverse for nrDNA (Table 2).
Population expansion and time of divergence
Population expansion, both demographic and spatial, of the species was corroborated by the mismatch distributions for the cpDNA and nrDNA data, where the observed values did not differ from the expected values under a sudden-expansion model. For the phylogeographic groups, only the CW2 and CE1 for nrDNA and CW1 for cpDNA did not exhibit population expansion pattern by mismatch distributions, as well as the groups of CE1+CE2 and CW+CW2 for both cpDNA and nrDNA (Table 4). The neutrality tests (Tajima's D and Fu's FS), on the other hand, did not have significant values for all populations together or for any of the seven groups (data not shown), and thus do not support any population expansions. The neutrality tests can be more sensitive to sequence variability [64] and the low level of sequence diversity observed in our data could be responsible for the absence of significance for them.
Table 4. Mismatch distribution analysis (parameters of demographic and spatial expansion) for phylogeographic groups of Dalbergia miscolobium based on chloroplast DNA (cpDNA) and nuclear ribosomal DNA (nrDNA) data.
| Demographic expansion | Spatial expansion | ||||
| Phylogeographic groups a | Data | SSD (p-value) | Raggedness (p-value) | SSD (p-value) | Raggedness (p-value) |
| All populations | cpDNA | 0.016 (0.130b) | 0.070 (0.285b) | 0.013 (0.057b) | 0.070 (0.150b) |
| nrDNA | 0.168 (0.000) | 0.043 (0.993b) | 0.020 (0.106b) | 0.043 (0.188b) | |
| Central Eastern 1 (CE1) | cpDNA | 0.015 (0.051b) | 0.160 (0.064b) | 0.015 (0.002) | 0.160 (0.072b) |
| nrDNA | 0.009 (0.056b) | 0.110 (0.008) | 0.009 (0.004) | 0.110 (0.006) | |
| Central Eastern 2 (CE2) | cpDNA | 0.003 (0.529b) | 0.074 (0.355b) | 0.003 (0.358b) | 0.074 (0.338b) |
| nrDNA | 0.007 (0.139b) | 0.121 (0.174b) | 0.007 (0.045) | 0.121 (0.183b) | |
| Central Western 1 (CW1) | cpDNA | 0.028 (0.046) | 0.280 (0.043) | 0.028 (0.011) | 0.280 (0.046) |
| nrDNA | 0.005 (0.355b) | 0.099 (0.268b) | 0.005 (0.215b) | 0.099 (0.258b) | |
| Central Western 2 (CW2) | cpDNA | 0.051 (0.049) | 0.137 (0.198b) | 0.047 (0.181b) | 0.137 (0.668b) |
| nrDNA | 0.008 (0.074b) | 0.099 (0.037) | 0.008 (0.022) | 0.099 (0.037) | |
| Central Cerrados (CC) | cpDNA | - | - | - | - |
| nrDNA | 0.000 (0.126b) | 0.895 (0.912b) | 0.000 (0.111b) | 0.895 (0.904b) | |
| Southern Cerrados (SC) | cpDNA | - | - | - | - |
| nrDNA | 0.459 (0.000) | 0.124 (1.000b) | 0.017 (0.593b) | 0.124 (0.670b) | |
| Northeastern Cerrados (NC) | cpDNA | - | - | - | - |
| nrDNA | 0.019 (0.109b) | 0.072 (0.239b) | 0.017 (0.165b) | 0.072 (0.357b) | |
| Central Eastern (CE1+CE2) | cpDNA | 0.017 (0.028) | 0.144 (0.000) | 0.017 (0.000) | 0.144 (0.000) |
| nrDNA | 0.005 (0.062b) | 0.083 (0.029) | 0.005 (0.017) | 0.083 (0.020) | |
| Central Western (CW1+CW2) | cpDNA | 0.029 (0.025) | 0.126 (0.031) | 0.029 (0.000) | 0.126 (0.050) |
| nrDNA | 0.005 (0.070b) | 0.088 (0.020) | 0.005 (0.022) | 0.088 (0.023) |
The times to the most recent common ancestor (TMRCA) for cpDNA and nrDNA haplotypes were estimated in 2.03×106 (0.49–3.88) and 2.94×106 (0.75–5.71) years before the present (YBP), respectively. These dates place the beginning of diversification of D. miscolobium lineages at the transition between Pliocene and Pleistocene, with confidence intervals spanning from Late Miocene to Middle Pleistocene. This estimated recent origin might be the cause of the low resolution of the networks. The results are very concordant between the cpDNA and nrDNA analyses, with the estimated dates being very close to each other and having overlapping confidence intervals.
Discussion
The combined analysis of a non-coding region of the cpDNA and the ITS region from nrDNA, performed by means of four different approaches, phylogenetic relationships estimation among haplotypes, Bayesian analyses of population genetic structure, a Monmonier's algorithm to infer genetic barriers and analyses of molecular variance (AMOVAs), revealed a phylogeographic structure in D. miscolobium (Figure 3 and Table 2) largely concordant with some of the main phytogeographical patterns reported for the Cerrado to date. Remarkably, the observed phylogeographic structure shows a clear similarity to the Cerrado phytogeographical provinces proposed by Ratter et al. [11] which have been delimited based on a comprehensive sampling effort in the biome. Based on Twinspan and consensus analyses, Ratter et al. proposed five and four provinces respectively [11], when considering only those that are superimposed to our sampling area (Figure 3) and our data strongly support them. The main difference between Ratter's four consensus provinces and five Twinspan provinces is the status of province ‘C & SE’ (Figure 1), which is a single province in the consensus analysis but is subdivided into two in the Twinspan analysis, named 2 and 3 (Figure 3D, F) groups. In this region our genetic data also allowed the identification of two phylogeographical groups, CC and CE1+2, which correspond to groups 3 and 2, respectively in the Twinspan analysis. Therefore, the observed structure of D. miscolobium is in accordance to the Cerrado phytogeographical patterns observed by Ratter et al. [11].
Some other longstanding phytogeographical hypotheses also are corroborated by our data. Several different authors have proposed Southern Cerrados and Northeastern Cerrados as completely different units from the remaining Cerrado [10], [11], [63], [65], [66], and this pattern was also observed in our data, although a wider sampling of Northeastern Cerrados would be desirable to confirm its characteristics found by our study. In some degree, these patterns were also observed by two other phylogeographical studies [21], [22]. With regard to the separation between Eastern and Western Central Cerrados, not only the phytogeographical provinces [9]–[11] but also phylogeographic studies have showed it [20], [22]. Finally, another striking phylogeographic pattern we observed is the consistent separation into two groups of the populations in the southern limits of the Cerrado (below 20° S). They can consistently be separated into an eastern group (our SC group) and a western group (part of the CW2 group; Figure 2C, D), a pattern that have been reported by Durigan et al. [67] based on floristic data.
Such ‘genealogical concordances’ [17], [68] between phylogeographic and phytogeographical patterns and provinces have been found in other regions of the globe (e.g. [18], [69], [70]), but this is the first time it is explicitly shown in the Cerrado biome. Given the relatively recent diversification of D. miscolobium, estimated at the Pliocene/Pleistocene (see results), these ‘genealogical concordances’ suggest that a shared and persistent pattern of species diversification might have been present on the Cerrado over time. Ecological as well as historical biogeographic factors have acted collectively in shaping species diversification and distribution in the biome and the relative importance of each of these factors has been the object of considerable debate [3], [4], [71]–[73]. Castro and Martins [10] and Ratter et al. [11] proposed that climate (mainly duration of the dry season and mean temperature), soil fertility and drainage are the main factors responsible for the subdivisions of the Cerrado in phytogeographical provinces observed, with variation in altitude and past climatic changes also playing a part in the observed diversification. In a recent study, Simon et al. [73], by analyzing time-calibrated phylogenies and plant adaptations, highlighted the importance of fire and the emergence of C4 flammable grasses to the diversification of the Cerrado flora, as has also been pointed out by others [6]. For Southern Cerrados, the duration of the dry season and the contact with other vegetation types, as the Atlantic Forest, were hypothesized as probable determinant factor in differentiating eastern and western [67]. Studies based on molecular phylogenies have pointed that the diversification on the neotropical region resulted from many evolutionary forces acting in different spatial and temporal scales, which include the climatic changes of Quaternary, Neogene tectonic events and paleogeographical reorganizations [3], [12], [74]–[76]. None of these hypotheses can be discarded, and, indeed, the complex interaction among these factors across the Cerrado appears to have determined the occurrence of the biome itself [72], as well as the phytogeographical and phylogeographical patterns that occur within it. But our results suggest that some of those driving forces have persisted over time, and lead to the recent intra-specific diversification of D. miscolobium in a similar way to the events of speciation that are behind the current phytogeographic patterns. Future study on genetic, morphological, and physiological diversity of widespread Cerrado species might provide the basis for better understanding the contribution of each of these factors to the observed patterns of diversification.
The pattern of cpDNA diversity of D. miscolobium in the southern Cerrado (SC group) is also very similar to that reported for three other tree species studied in the Cerrado, Caryocar brasiliense [19], Hymenaea stigonocarpa [22] and Plathymenia reticulata [21]. In this region, D. miscolobium presents populations with low genetic diversity, probably descent from northernmost sources, a star-like network and mismatch distributions not significantly different from the demographic and expansion model distribution. Together, these patterns point to a recent range expansion [77] of the species to southern Cerrados, from northernmost sources. The retraction of the Cerrado vegetation in the present-day southern portion of Cerrado during the colder and drier periods of the LGM has been proposed by paleoenvironmental studies [14], [78] and it was one of the least probable regions of the Cerrado to persist during the LGM, according to a modeling study [79]. The subtropical SDTF [2], [80] and/or grassland species [14], [81] from the southernmost regions may have expanded northwards during the LGM, replacing the Cerrado species. Subsequent climate amelioration [15] may have allowed the range expansion of the Cerrado species southwards again. Thus, even though, among the Cerrado species, D. miscolobium seems to better tolerate lower temperatures, it seems to have been similarly affected by the extremely arid and cold conditions of the LGM. It is suggestive that maybe, the entirely Cerrado biota, including its more cold-tolerant species, were unable to resist to the coldest conditions of LGM times in the southern portion of the Cerrado. This same pattern was not observed for nrDNA data, whose variation, in SC group, has contrasting patterns, that is, high genetic diversity and genetic uniqueness. Rapid expansion of those populations coupled with the high rate of mutation of nrDNA, nrDNA paralogue evolution, seed/pollen bias due to cpDNA gene flow only by seeds and even hybridization/introgression are among the factors that could have been responsible for those results [29], [82]. Analyses with more markers with different evolutionary rates could help to reveal the causes of these differences between cpDNA and nrDNA diversity levels in southern Cerrado. In this regard, next-generation sequencing to produce maps of model species of Leguminosae could help to find useful markers in D. miscolobium.
The heterogeneous distribution of genetic diversity of D. miscolobium is in accordance to the commonly reported heterogeneity of Cerrado biome in terms of species distribution [3], [8], [11], [23]. Also, the high level of differentiation among populations observed in D. miscolobium has been reported to three other species [19], [21], [22]. Together with the widespread occurrence of exclusive haplotypes, these results reinforce the importance to treat the different Cerrado provinces as different biodiversity assemblages. Among these, two centres of genetic diversity deserve some attention: the CW2 and the CE1+CE2 regions. The high diversity of CE1+CE2 regions have been reported in the plant phylogeographic studies of Hymenaea stigonocarpa [22] and Plathymenia reticulata [21] as well as in animal [3], [83], [84] and plant [85] endemism studies. CE1+CE2 regions are also relatively close to the recently proposed principal Cerrado refugium [79]. The spatial heterogeneity and/or climate stability of this region, especially in its northern portion, could have helped to give rise to and maintain this diversity. Together with the CW1+CW2 groups, the CE1+CE2 were the only groupings of populations that did not show mismatch distributions of sequences in accordance with a recent population expansion for both cpDNA and nrDNA, in a sign that they might have been more stable than the other groups (Table 3). Based on that, we reinforce the important role that the central region of the Cerrado could have in maintaining the genetic diversity of widespread Cerrado species. With regard to CW2 group, this is the first time a phylogeographic study has performed a wide sampling in the south-western portion of the Cerrado and we have found unexpected high levels of genetic diversity and also signs of population stability on it. This region is currently under considerable economic growth and therefore, research should focus on it in order to aid conservation efforts and its sustainable exploitation.
These many patterns found rise novel questions for future consideration. Do these biogeographic patterns persist in other widespread Cerrado species? What would have occurred to species endemic to these provinces, especially those found commonly in the southern Cerrado? How did the different ecological and historical factors lead to the diversification of the Cerrado biota? Phylogeography of the Cerrado is in its infancy, and many studies are still needed to improve our understanding of the historical biogeography of this rich and threatened biome to help in its conservation and sustainable use.
Supporting Information
Results from Bayesian Analysis of Population Structure (BAPS) of Dalbergia miscolobium populations with a prior of k = 4. See text and Figure 3 for details.
(TIF)
Variable sites of aligned cpDNA sequences.
(XLS)
Variable sites of aligned nrDNA sequences.
(XLS)
Acknowledgments
We thank the Instituto Brasileiro do Meio Ambiente e dos Recursos Naturais Renováveis (IBAMA) and Instituto Estadual de Florestas (IEF) which provided research facilities for the fieldwork. We thank Fabrício R. Santos for providing facilities for DNA sequencing. We are grateful to Danielle Moura Santos for her help with laboratory procedures, Maíra F. Goulart for the collect of some leaf samples and to Moara Machado and Giordano B. S. Souza for assistance with the data analyses. We also thank José Felipe Ribeiro and James Ratter for providing maps of the Cerrado range and provinces and José Felipe Ribeiro and Ary T. Oliveira-Filho for providing Dalbergia miscolobium occurrence data from floristic checklists. Finally we thank two anonymous reviewers for their contributions on improving the manuscript.
Funding Statement
This research was funded by the Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG) and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). The Pro-Reitoria de Pesquisa of the Universidade Federal de Minas Gerais provided resources for editing the language of the manuscript. R.A.R., M.B.L., and J.P.L-F. received research fellowships from CNPq. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Beheregaray LB (2008) Twenty years of phylogeography: the state of the field and the challenges for the Southern Hemisphere. Molecular Ecology 17: 3754–3774. [DOI] [PubMed] [Google Scholar]
- 2.Pennington RT, Ratter JA, Lewis GP (2006) Neotropical savannas and seasonally dry forests: plant diversity, biogeography, and conservation. Boca RatonFL: CRC/Taylor & Francis. 484 p. [Google Scholar]
- 3. Werneck FP (2011) The diversification of eastern South American open vegetation biomes: Historical biogeography and perspectives. Quaternary Science Reviews 30: 1630–1648. [Google Scholar]
- 4.Scariot A, Sousa-Silva JC, Felfili JM, editors (2005) Cerrado: ecologia, biodiversidade e conservação. Brasília: Ministério do Meio Ambiente. 439 p. [Google Scholar]
- 5. Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J (2000) Biodiversity hotspots for conservation priorities. Nature 403: 853–858. [DOI] [PubMed] [Google Scholar]
- 6. Eiten G (1972) The Cerrado vegetation of Brazil. Botanical Review 38: 201–341. [Google Scholar]
- 7. Klink CA, Machado RB (2005) Conservation of the Brazilian cerrado. Conservation Biology 19: 707–713. [Google Scholar]
- 8. Fiaschi P, Pirani JR (2009) Review of plant biogeographic studies in Brazil. Journal of Systematics and Evolution 47: 477–496. [Google Scholar]
- 9.Castro AAJF (1994) Comparação florístico-geográfica (Brasil) e fitossociológica (Piauí-São Paulo) de amostras de Cerrado [Ph.D. Thesis]. SP, Brazil: Universidade Estadual de Campinas.
- 10. Castro AAJF, Martins FR (1999) Cerrados do Brasil e do Nordeste: caracterização, área de ocupação e considerações sobre a sua fitodiversidade. Pesq Foco, São Luís 7: 147. [Google Scholar]
- 11. Ratter JA, Bridgewater S, Ribeiro JF (2003) Analysis of the floristic composition of the Brazilian Cerrado vegetation III: comparison of the woody vegetation of 376 areas. Edinburgh Journal of Botany 60: 57–109. [Google Scholar]
- 12. Haffer J (1969) Speciation in amazonian forest birds. Science 165: 131–137. [DOI] [PubMed] [Google Scholar]
- 13. Mayle FE (2004) Assessment of the Neotropical dry forest refugia hypothesis in the light of palaeoecological data and vegetation model simulations. Journal of Quaternary Science 19: 713–720. [Google Scholar]
- 14. Behling H, Lichte M (1997) Evidence of dry and cold climatic conditions at glacial times in tropical southeastern Brazil. Quaternary Research 48: 348–358. [Google Scholar]
- 15.Ledru M-P (2002) Late quaternary history and evolution of the Cerrados as revealed by palynological records. In: Oliveira PS, Marquis RJ, editors. The cerrados of Brazil: ecology and natural history of a neotropical savanna. New York: Columbia University Press. pp. 33–68.
- 16. Hickerson MJ, Carstens BC, Cavender-Bares J, Crandall KA, Graham CH, et al. (2010) Phylogeography's past, present, and future: 10 years after Avise, 2000. Molecular Phylogenetics and Evolution 54: 291–301. [DOI] [PubMed] [Google Scholar]
- 17.Avise JC (2000) Phylogeography: the history and formation of species. Cambridge, Mass: Harvard University Press. viii, 447 p. p.
- 18.Weiss S, Ferrand N (2007) Phylogeography of southern European refugia: evolutionary perspectives on the origins and conservation of European biodiversity. Dordrecht: Springer. ix, 377 p. p.
- 19. Collevatti RG, Grattapaglia D, Hay JD (2003) Evidences for multiple maternal lineages of Caryocar brasiliense populations in the Brazilian Cerrado based on the analysis of chloroplast DNA sequences and microsatellite haplotype variation. Molecular Ecology 12: 105–115. [DOI] [PubMed] [Google Scholar]
- 20. Collevatti RG, Rabelo SG, Vieira RF (2009) Phylogeography and disjunct distribution in Lychnophora ericoides (Asteraceae), an endangered cerrado shrub species. Annals of Botany 104: 655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Novaes RM, Lemos-Filho JP, Ribeiro RA, Lovato MB (2010) Phylogeography of Plathymenia reticulata (Leguminosae) reveals patterns of recent range expansion towards northeastern Brazil and southern Cerrados in Eastern Tropical South America. Molecular Ecology 19: 985–998. [DOI] [PubMed] [Google Scholar]
- 22. Ramos ACS, Lemos-Filho JP, Ribeiro RA, Santos FR, Lovato MB (2007) Phylogeography of the tree Hymenaea stigonocarpa (Fabaceae: Caesalpinioideae) and the influence of Quaternary climate changes in the Brazilian cerrado. Annals of Botany 100: 1219–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bridgewater S, Ratter JA, Felipe Ribeiro J (2004) Biogeographic patterns, β-diversity and dominance in the cerrado biome of Brazil. Biodiversity and Conservation 13: 2295–2317. [Google Scholar]
- 24.Carvalho AM (1989) Systematic studies of the genus Dalbergia L.f. in Brazil [Ph.D. Thesis]. Reading, England: University of Reading.
- 25.Silva Júnior MC (2005) 100 árvores do Cerrado: guia de campo. Brasília, DF: Rede de Sementes do Cerrado.
- 26. Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantitie.s of fresh leaf tissue. Phytochemical Bulletin 19: 11–15. [Google Scholar]
- 27. Taberlet P, Gielly L, Pautou G, Bouvet J (1991) Universal primers for amplification of 3 noncoding regions of chloroplast DNA. Plant Molecular Biology 17: 1105–1109. [DOI] [PubMed] [Google Scholar]
- 28. Delgado-Salinas A, Turley T, Richman A, Lavin M (1999) Phylogenetic analysis of the cultivated and wild species of Phaseolus (Fabaceae). Systematic Botany 24: 438–460. [Google Scholar]
- 29. Feliner GN, Rosselló JA (2007) Better the devil you know? Guidelines for insightful utilization of nrDNA ITS in species-level evolutionary studies in plants. Molecular Phylogenetics and Evolution 44: 911–919. [DOI] [PubMed] [Google Scholar]
- 30. Thompson JD, Higgins DG, Gibson TJ (1994) Clustal-W - improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology and Evolution 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. The American Journal of Human Genetics 68: 978–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Garrick RC, Sunnucks P, Dyer RJ (2010) Nuclear gene phylogeography using PHASE: dealing with unresolved genotypes, lost alleles, and systematic bias in parameter estimation. BMC Evolutionary Biology 10: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Machado M, Magalhães WCS, Sene A, Araújo B, Faria-Campos AC, et al. (2011) Phred-Phrap package to analyses tools: a pipeline to facilitate population genetics re-sequencing studies. Investigative Genetics 2: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Excoffier L, Lischer HE (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources 10: 564–567. [DOI] [PubMed] [Google Scholar]
- 36. Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- 37. Rogers AR, Harpending H (1992) Population growth makes waves in the distribution of pairwise genetic differences. Molecular Biology and Evolution 9: 552–569. [DOI] [PubMed] [Google Scholar]
- 38. Tajima F (1989) Statistical-method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147: 915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Petit RJ, El Mousadik A, Pons O (1998) Identifying populations for conservation on the basis of genetic markers. Conservation Biology 12: 844–855. [Google Scholar]
- 41. Jost L (2008) G(ST) and its relatives do not measure differentiation. Molecular Ecology 17: 4015–4026. [DOI] [PubMed] [Google Scholar]
- 42. Crawford NG (2010) SMOGD: software for the measurement of genetic diversity. Molecular Ecology Resources 10: 556–557. [DOI] [PubMed] [Google Scholar]
- 43. Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes - application to human mitochondrial-DNA restriction data. Genetics 131: 479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bandelt HJ, Forster P, Rohl A (1999) Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution 16: 37–48. [DOI] [PubMed] [Google Scholar]
- 45. Templeton AR, Crandall KA, Sing CF (1992) A cladistic-analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA-sequence data.3. Cladogram estimation. Genetics 132: 619–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Molecular Ecology 9: 1657–1659. [DOI] [PubMed] [Google Scholar]
- 47. Corander J, Marttinen P, Siren J, Tang J (2008) Enhanced Bayesian modelling in BAPS software for learning genetic structures of populations. BMC Bioinformatics 9: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Guillot G, Leblois R, Coulon A, Frantz AC (2009) Statistical methods in spatial genetics. Molecular Ecology 18: 4734–4756. [DOI] [PubMed] [Google Scholar]
- 49. Corander J, Marttinen P (2006) Bayesian identification of admixture events using multilocus molecular markers. Molecular Ecology 15: 2833–2843. [DOI] [PubMed] [Google Scholar]
- 50. Bitocchi E, Nanni L, Bellucci E, Rossi M, Giardini A, et al. (2012) Mesoamerican origin of the common bean (Phaseolus vulgaris L.) is revealed by sequence data. Proceedings of the National Academy of Science, USA 109: E788–E796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kropf M, Comes HP, Kadereit JW (2012) Past, present and future of mountain species of the French Massif Central – the case of Soldanella alpina L. subsp. alpina (Primulaceae) and a review of other plant and animal studies. Journal of Biogeography 39: 799–812. [Google Scholar]
- 52. Fang JY, Chung JD, Chiang YC, Chang CT, Chen CY, et al. (2013) Divergent selection and local adaptation in disjunct populations of an endangered conifer, Keteleeria davidiana var. formosana (Pinaceae). Plos One 8: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jones FA, Cerón-Souza I, Hardesty BD, Dick CW (2013) Genetic evidence of Quaternary demographic changes in four rain forest tree species sampled across the Isthmus of Panama. Journal of Biogeography 40: 720–731. [Google Scholar]
- 54. Manni F, Guerard E, Heyer E (2004) Geographic patterns of (genetic, morphologic, linguistic) variation: How barriers can be detected by using Monmonier's algorithm. Human Biology 76: 173–190. [DOI] [PubMed] [Google Scholar]
- 55. Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89: 583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Peakall ROD, Smouse PE (2005) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes 6: 288–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Brante A, Fernández M, Viard F (2012) Phylogeography and Biogeography Concordance in the Marine Gastropod Crepipatella dilatata (Calyptraeidae) along the Southeastern Pacific Coast. Journal of Heredity 103: 630–637. [DOI] [PubMed] [Google Scholar]
- 58. Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Richardson JE, Pennington RT, Pennington TD, Hollingsworth PM (2001) Rapid diversification of a species-rich genus of neotropical rain forest trees. Science 293: 2242–2245. [DOI] [PubMed] [Google Scholar]
- 60. Petit RJ, Duminil J, Fineschi S, Hampe A, Salvini D, et al. (2005) Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Molecular Ecology 14: 689–701. [DOI] [PubMed] [Google Scholar]
- 61. Albaladejo RG, Aguilar JF, Aparicio A, Feliner GN (2005) Contrasting nuclear-plastidial phylogenetic patterns in the recently diverged Iberian Phlomis crinita and P. lychnitis lineages (Lamiaceae). Taxon 54: 987–998. [Google Scholar]
- 62. Queiroz CdS, Batista FRdC, De Oliveira LO (2011) Evolution of the 5.8 S nrDNA gene and internal transcribed spacers in Carapichea ipecacuanha (Rubiaceae) within a phylogeographic context. Molecular Phylogenetics and Evolution 59: 293–302. [DOI] [PubMed] [Google Scholar]
- 63.Vieira LT (2012) Padrões de diversidade da flora lenhosa dos cerrados do nordeste do Brasil. Campinas: Universidade Estadual de Campinas.
- 65.Durigan G (2006) Observations on the Southern Cerrados and their Relationship with the Core Area. In: Pennington RT, Ratter JA, Lewis GP, editors. Neotropical savannas and seasonally dry forests: plant diversity, biogeography, and conservation. Boca RatonFL: CRC/Taylor & Francis. pp. 484 p. [Google Scholar]
- 66. Ratter JA, Dargie TCD (1992) An analysis of the floristic composition of 26 cerrado areas in Brazil. Edinburgh Journal of Botany 49: 235–250. [Google Scholar]
- 67. Durigan G, De Siqueira MF, Franco G, Bridgewater S, Ratter JA (2003) The vegetation of priority areas for cerrado conservation in São Paulo State, Brazil. Edinburgh Journal of Botany 60: 217–241. [Google Scholar]
- 68. Avise JC (2009) Phylogeography: retrospect and prospect. Journal of Biogeography 36: 3–15. [Google Scholar]
- 69. Brante A, Fernández M, Viard F (2012) Phylogeography and Biogeography concordance in the marine gastropod Crepipatella dilatata (Calyptraeidae). Journal of Heredity 103: 630–637. [DOI] [PubMed] [Google Scholar]
- 70. Li J, Foighil DÓ, Park JK (2013) Triton's trident: cryptic Neogene divergences in a marine clam (Lasaea australis) correspond to Australia's three temperate biogeographic provinces. Molecular Ecology 22: 1933–1946. [DOI] [PubMed] [Google Scholar]
- 71. Ab'Saber AN (1983) O domínio dos cerrados: introdução ao conhecimento. Revista do Servidor Público 11: 41–55. [Google Scholar]
- 72.Oliveira-Filho AT, Ratter JA (2002) Vegetation physiognomies and woody flora of the Cerrado biome. In: Oliveira PS, Marquis RJ, editors. The cerrados of Brazil: ecology and natural history of a neotropical savanna. New York: Columbia University Press. pp. viii, 398 p. [Google Scholar]
- 73. Simon MF, Grether R, de Queiroz LP, Skema C, Pennington RT, et al. (2009) Recent assembly of the Cerrado, a neotropical plant diversity hotspot, by in situ evolution of adaptations to fire. Proceedings of the National Academy of Sciences of the United States of America 106: 20359–20364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Rull V (2011) Neotropical biodiversity: timing and potential drivers. Trends in Ecology & Evolution 26: 508–513. [DOI] [PubMed] [Google Scholar]
- 75. Hoorn C, Wesselingh FP, ter Steege H, Bermudez MA, Mora A, et al. (2010) Amazonia through time: Andean uplift, climate change, landscape evolution, and biodiversity. Science 330: 927–931. [DOI] [PubMed] [Google Scholar]
- 76. Turchetto-Zolet AC, Pinheiro F, Salgueiro F, Palma-Silva C (2013) Phylogeographical patterns shed light on evolutionary process in South America. Molecular Ecology 22: 1193–1213. [DOI] [PubMed] [Google Scholar]
- 77. Excoffier L, Foll M, Petit RJ (2009) Genetic consequences of range expansions. Annual Review in Ecology, Evolution, and Systematics 40: 481–501. [Google Scholar]
- 78. Ledru MP, Braga PIS, Soubiès F, Fournier M, Martin L, et al. (1996) The last 50,000 years in the Neotropics (Southern Brazil): evolution of vegetation and climate. Palaeogeography, Palaeoclimatology, Palaeoecology 123: 239–257. [Google Scholar]
- 79. Werneck FP, Nogueira C, Colli GR, Sites JW, Costa GC (2012) Climatic stability in the Brazilian Cerrado: implications for biogeographical connections of South American savannas, species richness and conservation in a biodiversity hotspot. Journal of Biogeography 39: 1695–1706. [Google Scholar]
- 80. Prado DE, Gibbs PE (1993) Patterns of Species Distributions in the Dry Seasonal Forests of South-America. Annals of the Missouri Botanical Garden 80: 902–927. [Google Scholar]
- 81. Behling H (2002) South and southeast Brazilian grasslands during Late Quaternary times: a synthesis. Palaeogeography, Palaeoclimatology, Palaeoecology 177: 19–27. [Google Scholar]
- 82. Toews DPL, Brelsford A (2012) The biogeography of mitochondrial and nuclear discordance in animals. Molecular Ecology 21: 3907–3930. [DOI] [PubMed] [Google Scholar]
- 83. da Silva JMC, Bates JM (2002) Biogeographic patterns and conservation in the South American Cerrado: a tropical savanna hotspot. BioScience 52: 225–234. [Google Scholar]
- 84. Nogueira C, Ribeiro S, Costa GC, Colli GR (2011) Vicariance and endemism in a Neotropical savanna hotspot: distribution patterns of Cerrado squamate reptiles. Journal of Biogeography 38: 1907–1922. [Google Scholar]
- 85. Simon MF, Proença C (2000) Phytogeographic patterns of Mimosa (Mimosoideae, Leguminosae) in the Cerrado biome of Brazil: an indicator genus of high-altitude centers of endemism? Biological Conservation 96: 279–296. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Results from Bayesian Analysis of Population Structure (BAPS) of Dalbergia miscolobium populations with a prior of k = 4. See text and Figure 3 for details.
(TIF)
Variable sites of aligned cpDNA sequences.
(XLS)
Variable sites of aligned nrDNA sequences.
(XLS)