Abstract
The evolutionarily conserved fungal arginine attenuator peptide (AAP), as a nascent peptide, stalls the translating ribosome in response to the presence of a high concentration of the amino acid arginine. Here we examine whether the AAP maintains regulatory function in fungal, plant, and animal cell-free translation systems when placed as a domain near the N terminus or internally within a large polypeptide. Pulse–chase analyses of the radiolabeled polypeptides synthesized in these systems indicated that wild-type AAP functions at either position to stall polypeptide synthesis in response to arginine. Toeprint analyses performed to map the positions of stalled ribosomes on transcripts introduced into the fungal system revealed that ribosome stalling required translation of the AAP coding sequence. The positions of the stalled ribosomes were consistent with the sizes of the radiolabeled polypeptide intermediates. These findings demonstrate that an internal polypeptide domain in a nascent chain can regulate eukaryotic translational elongation in response to a small molecule. Apparently the peptide-sensing features are conserved in fungal, plant, and animal ribosomes. These data provide precedents for translational strategies that would allow domains within nascent polypeptide chains to modulate gene expression.
Nascent polypeptides can control translation. Signal peptides that direct polypeptides to the endoplasmic reticulum (ER) associate with the signal recognition particle to halt translation elongation until the nascent peptide docks with the ER (1, 2). In addition, a variety of peptides specified by upstream ORFs (uORFs) in eukaryotic and prokaryotic mRNAs can stall ribosomes involved in translation termination (3–9). Expression of the small subunit of the arginine-specific carbamoyl phosphate synthetase, a fungal arginine (Arg) biosynthetic enzyme, is negatively regulated at the translational level. This is accomplished through the synthesis and/or action of the evolutionarily conserved, uORF-encoded Arg attenuator peptide (AAP). The nascent AAP normally causes ribosomes to stall at the uORF termination codon in response to Arg, thereby blocking the translating ribosome from reaching the initiation codon used for synthesis of the downstream enzyme (10). Mutations that eliminate Arg-specific regulation of Neurospora crassa arg-2 and Saccharomyces cerevisiae CPA1 change a conserved Asp residue, at positions 12 and 13 in each AAP, respectively, to Asn (11, 12). These mutations also abolish each AAP's capacity to stall ribosomes in fungal cell-free translation systems (13, 14). Unlike other uORF-encoded peptides that affect only translation termination, the AAP amino acid sequence allows Arg-regulated ribosome stalling when placed within a polypeptide, at its N terminus (15). Stalling occurs during elongation, immediately downstream of the AAP coding region, and is independent of the sequence in the downstream region (16).
Most known nascent peptides that regulate translation are found encoded as uORFs or as N-terminal leader peptides. However, an internal polypeptide domain in the prokaryotic regulatory protein SecM can stall elongation. Studies on prokaryotic ribosomes synthesizing SecM indicate that the exit tunnel acts as a discriminating gate that permits regulation of polypeptide chain elongation as a consequence of the sequence of the nascent SecM chain (17–20).
Could a general mechanism governing translation elongation enable internal domains within a nascent polypeptide chain to regulate completion of translation in response to a small molecule and could such a mechanism regulate eukaryotic protein synthesis? The characteristics of the AAP suggested that it may provide such functions. To test this possibility, we created large polypeptide coding sequences with the AAP coding sequence near the coding sequence for the N terminus or internally within the coding sequence. To facilitate protein detection, we removed Met residues from our proteins except at the extreme N terminus. Synthetic transcripts specifying these polypeptides were used to program fungal, plant, and mammalian cell-free translation systems. Polypeptide synthesis was monitored by pulse–chase analyses; the appearance of stalled peptidyl tRNA intermediates was monitored by their ability to be precipitated with cetyltrimethylammonium bromide (CTAB). The positions of ribosomes stalled on transcripts during translation were monitored by primer extension inhibition assay. The results of these studies indicated that an internally localized AAP domain does cause a translating ribosome to stall in response to Arg. This establishes that an internal nascent polypeptide domain can function as a cis-acting regulator of polypeptide elongation by modulating ribosome translation in response to changes in the concentration of a small molecule.
Materials and Methods
Constructs. The plasmids used are listed in Table 1. They were derived from previous constructs by using described procedures (21). Site-specific mutagenesis was used to remove every ATG codon (except for the nine at the N termini) in the three forward reading frames (Fig. 5, which is published as supporting information on the PNAS web site). The rabbit α-globin domain used was obtained by PCR from plasmid pSPα (22) (from U. L. RajBhandary, Massachusetts Institute of Technology, Cambridge). Plasmid DNA templates were purified by equilibrium centrifugation (23) or by Promega Wizard midi-prep; templates were linearized with EcoRI.
Table 1. Constructs used.
| Construct | Structure |
|---|---|
| pGL201 | Met9-AAPw-globin-AAPw-LUC |
| pGL202 | Met9-AAPm-globin-AAPw-LUC |
| pGL203 | Met9-AAPw-globin-AAPm-LUC |
| pGL204 | Met9-AAPm-globin-AAPm-LUC |
| pLL301 | Met9-AAPw-LUC-AAPw-LUC |
| pLL302 | Met9-AAPm-LUC-AAPw-LUC |
| pLL303 | Met9-AAPw-LUC-AAPm-LUC |
| pLL304 | Met9-AAPm-LUC-AAPm-LUC |
Preparation of RNA and Cell-Free Translation. Capped polyadenylated RNA was synthesized with T7 RNA polymerase from linearized plasmid DNA templates, and the yield of RNA was quantified as described (23). The reaction conditions for in vitro translation using N. crassa extracts were as described (16). Micrococcal-nuclease-treated wheat germ extract and reticulocyte lysate were obtained from Promega and used according to the supplier's directions except as noted. Translation reaction mixtures were programmed at a final concentration of 6 ng/μl RNA; [35S]Met was used at a final concentration of 0.5 μCi/μl.
Primer Extension Inhibition (Toeprint) Assays. Toeprint assays were accomplished as described (13) by using primers FP93 (CTGGC GACGT AATCC ACG) and FP94 (CTTGT CCAGG GAGGC GTG); 8 μl of sample instead of 4 μl was loaded onto each gel lane. The gels were dried and exposed to screens of a Molecular Dynamics PhosphorImager for ≈24 h. All toeprinting data shown were representative of multiple experiments.
Results
Pulse–Chase Analyses of Radiolabeled Polypeptide Synthesis. We examined whether the AAP caused stalling as an internal polypeptide domain by performing pulse–chase experiments with mRNAs encoding fused polypeptide sequences in cell-free translation systems. Synthetic mRNAs were prepared that specified polypeptides containing a string of nine methionine residues at their N termini and no internally positioned methionine residues (Figs. 1A and 5). These polypeptides contained a domain from rabbit α-globin and a domain from firefly luciferase (LUC) or contained duplicated LUC domains. AAP domains were placed at two positions in each polypeptide: near the N terminus, following the nine Met residues (residues 12–33) and internally, between the globin and LUC domains (residues 101–123) or between the duplicated LUC domains (residues 131–153). We constructed all possible combinations containing the wild-type AAP (designated AAPw) sequence, or a nonfunctional AAP sequence (AAPm, in which the critical residue corresponding to arg-2 AAP Asp-12 was substituted by Asn) at N termini and internal positions.
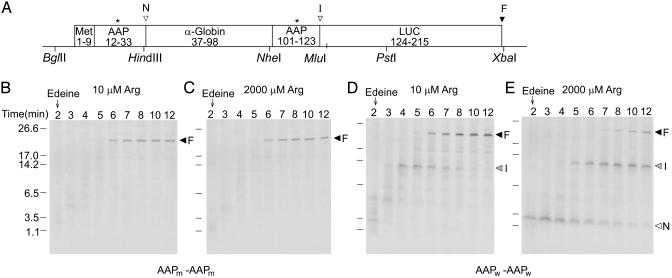
Fig. 1.
Polypeptide synthesis time course in N. crassa cell-free extracts. (A) The Met9-AAP-globin-AAP-LUC construct used (DNA sequence in Fig. 5A). Asterisks indicate where wild-type AAP Asp codons were changed to Asn. Arrowhead N, the C terminus of the Met9-AAP polypeptide-intermediate; arrowhead I, the C terminus of the Met9-AAP-globin-AAP intermediate; arrowhead F, the C terminus of the completed polypeptide. Unique restriction enzyme sites are indicated. Transcripts specifying Met9-AAPm-LUC-AAPm-LUC (B and C) or Met9-AAPw-LUC-AAPw-LUC (D and E) were translated in extracts in low (B and D) or high (C and E) Arg (see text). Edeine was added at 2 min (arrow), and 10-μl aliquots of extracts were removed at the indicated time points for analysis by SDS/PAGE (21). Arrowhead N, the intermediate Met9-AAP; arrowhead I, the polypeptide-intermediate Met9-AAP-globin-AAP; arrowhead F, the full-length polypeptide.
Synthetic mRNAs were used to program cell-free translation extracts containing [35S]Met, Arg at low (10 μM) or high (2,000 μM) concentrations, and the other amino acids at fixed concentrations. Because Met codons were only located at the N termini coding regions, radiolabeled translation products would contain isotope only at their N termini. In our pulse–chase experiments, edeine was added to extracts after 2 min of incubation to block subsequent rounds of translation initiation during prolonged incubation. Nascent polypeptide N termini were thus labeled to high specific activity with [35S]Met during incubation before edeine addition. Polypeptide chain elongation progress was monitored by SDS/PAGE; this revealed the length of radiolabeled translation products as a function of time.
Programming N. crassa extracts with mRNA specifying polypeptides containing AAPm domains at two locations showed that neither stalled polypeptide synthesis (Fig. 1 B and C). In either low or high Arg, full-length polypeptide synthesis was essentially complete by 6 min, and no major intermediate length polypeptide was detected (the tRNA was removed from the nascent peptide as a consequence of the analysis procedure). In contrast, with mRNA containing two AAPw encoding regions, two intermediates in polypeptide synthesis were observed in the presence of high Arg (Fig. 1E). The smaller (labeled N) migrated with a size consistent with it being a product of stalling after synthesis of the N-terminal AAP; the larger (labeled I) migrated with a size corresponding to a product formed by translational stalling after synthesis of the internal AAP. Synthesis of the full-length polypeptide (labeled F) was substantially delayed in high Arg, also indicative of translational stalling. Each wild-type AAP independently elicited stalling as determined by pulse–chase analyses with constructs containing a single AAPw and a single AAPm (Fig. 6, which is published as supporting information on the PNAS web site). The AAPw near the N terminus did not stall polypeptide synthesis in low Arg; the internal AAPw caused some stalling in low Arg but substantially more stalling in high Arg (Fig. 1D).
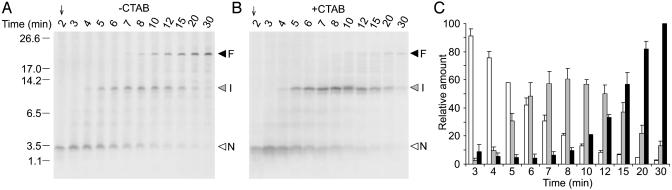
Polypeptide Synthesis Appears to Resume After Stalling. If the short radiolabeled products are intermediates resulting from stalled translation, they should be present as peptidyl tRNAs. Also, if they are intermediates, then the radiolabeled methionine they contain should be quantitatively recovered in full-length polypeptide at later time points in pulse–chase experiments. Therefore, we performed experiments with longer incubation periods and removed samples at different time points for direct analysis by SDS/PAGE (Fig. 2A) and after precipitation with CTAB (Fig. 2B). CTAB selectively precipitates peptidyl tRNA (24). The radiolabeled polypeptides identified as intermediates of stalled translation (N and I) were selectively enriched by precipitation with CTAB compared to the levels of full-length polypeptide product (F) detected (compare Fig. 2 A and B). This indicates that the intermediates were peptidyl tRNAs, confirming that the shorter translation products are intermediates resulting from stalled polypeptide synthesis. Quantitative analysis of radiolabel in the major translation products observed between 3- and 30-min incubation (data from the experiment in Fig. 2 A and an independent replicate) showed radiolabel conservation consistent with the conversion of the small N-terminal product to a full-length polypeptide, with transient accumulation of a labeled intermediate resulting from stalled translation following the internal AAP coding region (Fig. 2C). These experiments indicate that the stalled products are true intermediates, and that ribosomes resume chain elongation after stalling.
Fig. 2.
CTAB precipitation of peptidyl tRNA from translation extracts and quantitative analysis of polypeptide intermediates and products. N. crassa extracts (150 μl) were programmed with Met9-AAPw-globin-AAPw-LUC mRNA and incubated with high Arg, as described in Fig. 1. Edeine was added at 2 min and 10-μl aliquots removed at the indicated time points for analyses. (A) Total translation product analysis. (B) CTAB-precipitated translation product analysis. Arrowheads are as in Fig. 1. (C) Quantitative analysis of translation products obtained from data in A and an independent experimental replicate. The radiolabel in bands N, I, and F was determined by using imagequant 5.1 (Molecular Dynamics). The amount of radiolabel in band F at 30 min in each experiment was normalized to 100%, and the radiolabel in each band at each time point was calculated as a fraction of this value. White, radiolabel in intermediate N; gray, radiolabel in intermediate I; black, radiolabel in full-length polypeptide F. The total (not normalized) radiolabel in bands N, I, and F at 3 min was 88% of the amount at 30 min.
AAP-Mediated Ribosome Stalling Is Observed by Using a Toeprint Assay. A primer extension inhibition (toeprint) assay (25) was used to directly map the positions of the stalled ribosomes. The purpose was to verify that the translation intermediates observed by [35S]Met labeling arose from ribosome stalling as a consequence of synthesizing each AAPw sequence. Previous studies showed that the ribosome involved in elongation that had synthesized an N-terminal AAPw stalled on the mRNA when the final codon of the AAP coding sequence occupied the P site of the translating ribosome (15, 16). Therefore, ribosomes stalled by the N-terminal AAPw or by the internal AAPw would be expected to generate toeprint signals in a region starting ≈16 nt from the AAP coding region's final GCG (Ala)-codon (15, 16).
The results of toeprint analyses of translation reactions that were programmed with transcripts containing N-terminal and internal AAPw domains (Fig. 3) showed that ribosomes underwent Arg-dependent stalling at either position but not in reactions programmed with AAPm-encoding mRNAs. The toeprint data indicate that stalling caused by translation of the AAP coding region increases as the Arg concentration is increased. Pulse–chase experiments of radiolabeled polypeptides and toeprint analyses of extracts treated with edeine indicate that increasing the Arg concentration increases the intermediate's half-life (data not shown), consistent with the increased signal observed in these experiments.
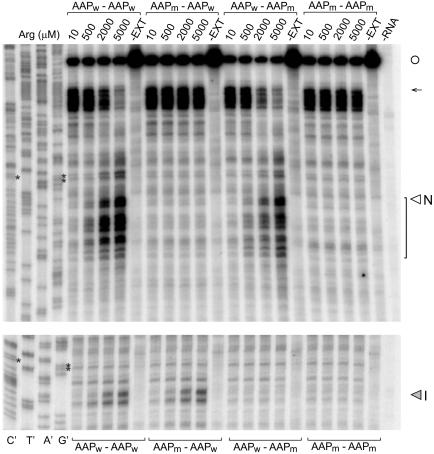
Fig. 3.
Toeprint analysis of ribosome stalling at AAP domains in Met9-AAP-globin-AAP-LUC mRNA. Separate gels and primers were used for analyses of stalling at the N-terminal (Upper) and internal (Lower) AAP domains for optimal resolution. N-terminal and internal AAP domains are indicated as wild-type (AAPw)or mutated (AAPm) above or below the corresponding lanes. Transcripts were translated in 20-μl reaction mixtures containing 10, 500, 2,000, or 5,000 μM Arg as indicated and 10 μM of the other amino acids and analyzed as described (13, 25). (Left) Sequencing reactions for the Met9-AAPw-globin-AAPw-LUC template. The sequence can be directly read 5′ to 3′ from top to bottom. Controls: Products obtained from primer extension of RNA (18 ng) in the absence of extract (–EXT) and from an extract not programmed with RNA (–RNA). Primers FP94 and FP93 (Fig. 5A) were used for the experiments shown (Upper and Lower, respectively). The open circle indicates the mRNA 5′ end; the arrow indicates the position of ribosomes at the first Met (start) codon. Translation of the nine contiguous Met codons was slow, as evidenced by the toeprints of ribosomes in this region. Asterisks mark each AAP's final GCG codons, which lie ≈16 nt upstream of the toeprints corresponding to the stall sites associated with the production of polypeptide intermediates N and I (indicated); toeprints that represent translational stalling after the Met9-AAP coding region are indicated with a bracket.
Mapping the positions of the toeprint bands arising as a consequence of Arg-mediated regulation by AAPw domains revealed that the ribosomes stalled in a region beginning ≈16 nt from each AAP's final GCG codon. Stalling of ribosomes mediated by the internal AAP appeared to occur within a narrower region downstream of the final AAP codon than observed for the AAP near the N terminus. Although the reason for this difference is not known, these data are consistent with a model that stalling begins when the final amino acid-specifying codon of the AAP is in the ribosome P site, as is observed when the AAP is encoded as a uORF. Irrespective of how precisely the position of the AAP coding region can be placed with regard to the translating ribosome, it is clear from the toeprint data that translation of the AAP sequence causes ribosomes to stall on the mRNA at sites consistent with those expected from the sizes of the nascent polypeptide intermediates observed accumulating in high Arg in [35S]Met pulse–chase experiments (Figs. 1 and 2).
AAP Can Act in Different Contexts and Can Regulate Polypeptide Synthesis in Heterologous Systems. Might the nascent globin domain, chosen for the experiments described above because it naturally lacks in-frame Met codons (26), coincidentally provide a special sequence context that allows internal stalling (4)? To test this possibility, we replaced the globin domain with a LUC domain and repeated the above experiments (Fig. 4A). Pulse-labeling experiments using N. crassa extracts showed that AAPw but not AAPm functioned to stall ribosomes in response to Arg when placed at the N terminus or internally in the LUC polypeptide (data not shown). Thus, the internal AAP sequence appears to function independently of the proximal nascent chain sequence.
Fig. 4.
Time course of polypeptide synthesis in wheat germ extracts and reticulocyte lysates. (A) The Met9-AAP-LUC-AAP-LUC construct used (DNA sequence in Fig. 5B). Designations are as in Fig. 1 A except that arrowhead I is the C terminus of the Met9-AAP-LUC-AAP intermediate. (B) Wheat germ extracts (150 μl) or reticulocyte lysates (100 μl) were programmed with the indicated mRNA (W-W, Met9-AAPw-LUC-AAPw-LUC, M-W, Met9-AAPm-LUC-AAPw-LUC; and M-M, Met9-AAPm-LUC-AAPm-LUC) and incubated at 25°C with either 10 μM or 2,000 μM Arg and 10 μM each of the other amino acids. Edeine was added at 2 min, and polypeptide products were analyzed as described in Fig. 1, except that reticulocyte samples were incubated in loading buffer at 25°C for 30 min before SDS/PAGE, and 3 μl of lysate was loaded per lane. F, full-length polypeptide; I, intermediate corresponding to Met9-AAP-LUC-AAP; N, intermediate corresponding to Met9-AAP.
Is the ability of AAPw to cause stalling limited to fungal ribosomes? To test this, we programmed wheat germ extracts and rabbit reticulocyte lysates with transcripts specifying dual LUC domains and performed pulse–chase analyses of the [35S]Met products obtained in reaction mixtures containing low or high Arg (Fig. 4). AAPw caused Arg-regulated stalling in both systems, whether at the N-terminal or internal position. AAPm did not cause stalling at either position, and stalling at the internal AAPw in low Arg was not detectable. The nascent wild-type AAP thus can function within ribosomes from fungi, plants, and animals and cause them to stall in response to Arg.
Discussion
Several lines of evidence support the conclusion that the AAP can function as an internal nascent peptide domain that causes regulated stalling of eukaryotic ribosomes in response to Arg. Pulse–chase analysis of the polypeptide products obtained during cell-free translation directly indicated that the AAP causes a regulated pause in polypeptide synthesis (Figs. 1 and 4). The shorter polypeptides that transiently accumulated as a consequence of AAP-mediated stalling appeared to be intermediates in polypeptide synthesis. The shorter polypeptides were present in the reaction mixture in the form of peptidyl tRNA (Fig. 2), which is appropriate if they are translation intermediates. Furthermore, these shorter peptidyl tRNAs appeared to be authentic intermediates, because their radiolabeling was quantitatively chased into full-length polypeptides. This would not occur if the shorter peptidyl tRNA intermediates were released from the ribosome, as happens in another nascent peptide-mediated stalling event, at a termination codon (27).
We previously established that the amino acid sequence of the AAP, not the sequence of its coding region, was responsible for regulation, and that regulation was effected by a high concentration of Arg, not aminoacylated Arg-tRNAArg (10, 13–16). Here we provide evidence that the AAP was a portable signal that caused stalling when placed either upstream or downstream of two different domains (derived from rabbit α-globin or firefly LUC). Importantly, the AAP sequence functioned in each position to pause polypeptide synthesis in response to Arg in cell-free translation systems derived from N. crassa, wheat germ, and rabbit reticulocytes. Mutation of the critical Asp-12 residue of the AAP to Asn eliminated stalling during synthesis of the nascent polypeptide in each system tested, indicating that the nascent polypeptide and Arg were acting similarly in each system to cause stalling.
At least two models are suggested by these data that could account for how the AAP causes stalled polypeptide synthesis. The AAP and Arg might act in concert to constrain the movement of the nascent peptide in the ribosome tunnel. Elongation would slow or pause, because the inability of the peptide to move in the tunnel would slow peptidyl transferase activity or translocation of the peptide in the translating ribosome. Normal translation would resume when the constraints on AAP movement in the tunnel ceased. Alternatively, AAP and Arg might directly affect peptidyl transferase activity by interfering with this domain of the ribosome.
The toeprinting data obtained with a variety of AAP sequences in different contexts, including the internal domain context examined here, indicate that stalling occurs when the AAP's C-terminal residue is at or near the ribosomal P site. The critical Asp-12 and -13 residues of the N. crassa and S. cerevisiae AAPs, established from both in vivo and in vitro studies to be critical for regulation, would each be located in the ribosome tunnel ≈12 aa from the residue at the P site. Interestingly, amino acids in the polypeptide chain that are critical for stalling prokaryotic ribosomes during the synthesis of SecM and TnaC are also located in the ribosome tunnel ≈12 residues from the P site. This region of the nascent polypeptide appears to interact with ribosomal protein L22 in the tunnel at a constriction point that has been called a “discriminating gate” (17, 18, 28). These data suggest that the eukaryotic ribosome has a similar “constriction gate” in its tunnel.
An observation that still requires explanation is that toeprints appear in a relatively wide region when the AAP is near the N terminus (Fig. 3), whereas the stalled radiolabeled polypeptide product appears to be a relatively discrete species (Figs. 1 and 2). Direct comparisons of different AAP-containing mRNAs translated in N. crassa and S. cerevisiae cell-free systems showed that the most 5′-proximal toeprint could correspond to ribosomes with the final codon of the AAP in the ribosome P site, as observed here, and that the distribution of 3′-distal toeprints was extract-dependent (14). One possible explanation that could account for these data is that additional factors are recruited to the stalled ribosome, and these factors result in additional toeprints 3′-distal to the ribosome.
How Arg interacts with the nascent peptide and/or the translational machinery to cause stalling remains unclear. Studies on the Escherichia coli leader peptide TnaC, which causes prokaryotic ribosomes to stall in response to tryptophan, indicate that free tryptophan exerts its stalling action by occupying the ribosomal A site (29). Replacement of the stop codon (at which TnaC normally causes regulated stalling in response to Trp) with a Trp codon causes constitutive stalling. The aminoacylated tRNA appears to place Trp in the proper spot to exert its regulatory effect. Therefore we placed an Arg codon directly after the AAP and tested whether the direct placement of Arg into the A site by aminoacylated arginyl tRNA caused stalling when the AAP was in the P site (data not shown). We observed no effect on stalling in the N. crassa system (i.e., stalling still required high Arg). Thus, Arg and AAP do not appear to act in a manner analogous to Trp and TnaC in causing stalling.
The demonstration that the AAP functions as an internal domain to regulate elongation in response to Arg establishes that such domains can provide a means of controlling translational elongation. There are no previous examples of the regulation of elongation by internal nascent polypeptide domains acting in concert with small molecules in either eukaryotes or prokaryotes, although such regulation by uORF-encoded nascent peptides is well established (3–9).
How could the regulation of pausing by an internal domain and a small molecule contribute to the control of gene expression? Constitutive pausing during polypeptide synthesis has been proposed to lead to the formation of distinct intermediates that could contribute to the proper cotranslational binding of cofactors to the nascent polypeptide (30). The nascent polypeptide sequence at the N terminus of rhodanese may affect release of nascent chain from the ribosome and thereby affect folding (31); pause sites are also observed during chloramphenicol acetyl transferase synthesis, and these can be altered by modifying the N terminus of the nascent polypeptide (32). The regulated instability of the Arabidopsis thaliana CGS1 transcript in response to the availability of S-adenosyl methionine requires the synthesis of a specific internal polypeptide domain. Translational stalling as a consequence of synthesis of this domain is a possible explanation of this regulatory effect (33, 34). It has long been known that the occurrence of rare codons in polypeptide coding sequences slows elongation (35). A translational pause that occurs when a ribosome encounters a rare codon during the translation of c-myc mRNA facilitates binding of a factor that regulates mRNA stability (36). RNA structures such as pseudoknots can contribute to ribosome-pausing during elongation and thus contribute to programmed frameshifting (37–39). Thus regulated stalling mediated by internal polypeptide domains could potentially influence protein folding, mRNA stability, or frameshifting.
The data obtained concerning the regulation of translation by the AAP and Arg are consistent with the hypothesis that Arg acts by binding to the nascent peptide in the ribosomal exit tunnel to cause a conformational change that controls polypeptide synthesis. Alternatively, Arg might interact with the ribosome or associated factor to render the ribosome sensitive to the nascent peptide sequence. As noted above, tryptophan may directly occupy the ribosome A site to act with the E. coli TnaC peptide as a regulatory element. Other small molecules are capable of acting in concert with nascent leader peptides to control translational events (3–9). The leader peptides specified in bacterial chloramphenicol acetyl transferase transcripts can bind to and alter the conformation of 23S rRNA; in the presence of chloramphenicol, their synthesis stalls ribosomes (40). Polyamines act in concert with a uORF-encoded peptide in the 5′ leader of the transcript specifying mammalian S-adenosyl methionine decarboxylase to stall ribosomes at the uORF termination codon, negatively regulating gene expression (41). Small molecules, including amino acids, have also been shown to interact directly with mRNA to exert a regulatory role. Lysine binds to the leader region of the Bacillus subtilis LysC transcript to regulate translation (42, 43). S-adenosyl-methionine similarly affects regulation by binding to mRNA (44). Studies on the control of protein synthesis by small molecules are thus revealing that they can act in unanticipated ways to control gene expression. The control of fungal, plant, and animal translation by the AAP and Arg would seem to provide such an example of how a polypeptide and an amino acid can affect eukaryotic gene expression.
Supplementary Material
Acknowledgments
We thank Adam Geballe, Charles Yanofsky, and Peter Zuber for critical comments. This work was supported by National Institutes of Health Grant GM47498 (to M.S.S.). C.C.S. was supported by National Institutes of Health Research Supplement for Underrepresented Minorities GM47498-S.
Abbreviations: uORF, upstream ORF; AAP, Arg attenuator peptide; LUC, luciferase; CTAB, cetyltrimethylammonium bromide.
References
- 1.Keenan, R. J., Freymann, D. M., Stroud, R. M. & Walter, P. (2001) Annu. Rev. Biochem. 70, 755–775. [DOI] [PubMed] [Google Scholar]
- 2.Nagai, K., Oubridge, C., Kuglstatter, A., Menichelli, E., Isel, C. & Jovine, L. (2003) EMBO J. 22, 3479–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lovett, P. S. & Rogers, E. J. (1996) Microbiol. Rev. 60, 366–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tenson, T. & Ehrenberg, M. (2002) Cell 108, 591–594. [DOI] [PubMed] [Google Scholar]
- 5.Geballe, A. P. & Sachs, M. S. (2000) in Translational Control of Gene Expression, eds. Sonenberg, N., Hershey, J. W. B. & Mathews, M. B. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 595–614.
- 6.Vilela, C. & McCarthy, J. E. (2003) Mol. Microbiol. 49, 859–867. [DOI] [PubMed] [Google Scholar]
- 7.Raney, A., Law, G. L., Mize, G. J. & Morris, D. R. (2002) J. Biol. Chem. 277, 5988–5994. [DOI] [PubMed] [Google Scholar]
- 8.Morris, D. R. & Geballe, A. P. (2000) Mol. Cell. Biol. 20, 8635–8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sachs, M. S. & Geballe, A. P. (2002) Science 297, 1820–1821. [DOI] [PubMed] [Google Scholar]
- 10.Gaba, A., Wang, Z., Krishnamoorthy, T., Hinnebusch, A. G. & Sachs, M. S. (2001) EMBO J. 20, 6453–6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freitag, M., Dighde, N. & Sachs, M. S. (1996) Genetics 142, 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Werner, M., Feller, A., Messenguy, F. & Piérard, A. (1987) Cell 49, 805–813. [DOI] [PubMed] [Google Scholar]
- 13.Wang, Z. & Sachs, M. S. (1997) Mol. Cell. Biol. 17, 4904–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang, Z., Gaba, A. & Sachs, M. S. (1999) J. Biol. Chem. 274, 37565–37574. [DOI] [PubMed] [Google Scholar]
- 15.Wang, Z., Fang, P. & Sachs, M. S. (1998) Mol. Cell. Biol. 18, 7528–7536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fang, P., Wang, Z. & Sachs, M. S. (2000) J. Biol. Chem. 275, 26710–26719. [DOI] [PubMed] [Google Scholar]
- 17.Nakatogawa, H. & Ito, K. (2002) Cell 108, 629–636. [DOI] [PubMed] [Google Scholar]
- 18.Berisio, R., Schluenzen, F., Harms, J., Bashan, A., Auerbach, T., Baram, D. & Yonath, A. (2003) Nat. Struct. Biol. 10, 366–370. [DOI] [PubMed] [Google Scholar]
- 19.Sarker, S. & Oliver, D. (2002) J. Bacteriol. 184, 2360–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakatogawa, H. & Ito, K. (2001) Mol. Cell 7, 185–192. [DOI] [PubMed] [Google Scholar]
- 21.Fang, P., Wu, C. & Sachs, M. S. (2002) Fungal Genet. Biol. 36, 167–175. [DOI] [PubMed] [Google Scholar]
- 22.Jobling, S. A. & Gehrke, L. (1987) Nature 325, 622–625. [DOI] [PubMed] [Google Scholar]
- 23.Wang, Z. & Sachs, M. S. (1997) J. Biol. Chem. 272, 255–261. [DOI] [PubMed] [Google Scholar]
- 24.Gilmore, R. & Blobel, G. (1985) Cell 42, 497–505. [DOI] [PubMed] [Google Scholar]
- 25.Sachs, M. S., Wang, Z., Gaba, A., Fang, P., Belk, J., Ganesan, R., Amrani, N. & Jacobson, A. (2002) Methods 26, 105–114. [DOI] [PubMed] [Google Scholar]
- 26.Drabkin, H. J., Helk, B. & RajBhandary, U. L. (1993) J. Biol. Chem. 268, 25221–25228. [PubMed] [Google Scholar]
- 27.Cao, J. & Geballe, A. P. (1998) RNA 4, 181–188. [PMC free article] [PubMed] [Google Scholar]
- 28.Jenni, S. & Ban, N. (2003) Curr. Opin. Struct. Biol. 13, 212–219. [DOI] [PubMed] [Google Scholar]
- 29.Gong, F. & Yanofsky, C. (2002) Science 297, 1864–1867. [DOI] [PubMed] [Google Scholar]
- 30.Kim, J., Klein, P. G. & Mullet, J. E. (1991) J. Biol. Chem. 266, 14931–14938. [PubMed] [Google Scholar]
- 31.Kudlicki, W., Odom, O. W., Kramer, G., Hardesty, B., Merrill, G. A. & Horowitz, P. M. (1995) J. Biol. Chem. 270, 10650–10657. [DOI] [PubMed] [Google Scholar]
- 32.Tsalkova, T., Kramer, G. & Hardesty, B. (1999) J. Mol. Biol. 286, 71–81. [DOI] [PubMed] [Google Scholar]
- 33.Lambein, I., Chiba, Y., Onouchi, H. & Naito, S. (2003) Plant Cell Physiol. 44, 893–900. [DOI] [PubMed] [Google Scholar]
- 34.Chiba, Y., Sakurai, R., Yoshino, M., Ominato, K., Ishikawa, M., Onouchi, H. & Naito, S. (2003) Proc. Natl. Acad. Sci. USA 100, 10225–10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Varenne, S., Buc, J., Lloubes, R. & Lazdunski, C. (1984) J. Mol. Biol. 180, 549–576. [DOI] [PubMed] [Google Scholar]
- 36.Lemm, I. & Ross, J. (2002) Mol. Cell. Biol. 22, 3959–3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kontos, H., Napthine, S. & Brierley, I. (2001) Mol. Cell. Biol. 21, 8657–8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Somogyi, P., Jenner, A. J., Brierley, I. & Inglis, S. C. (1993) Mol. Cell. Biol. 13, 6931–6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lopinski, J. D., Dinman, J. D. & Bruenn, J. A. (2000) Mol. Cell. Biol. 20, 1095–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harrod, R. & Lovett, P. S. (1997) Nucleic Acids Res. 25, 1720–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Law, G. L., Raney, A., Heusner, C. & Morris, D. R. (2001) J. Biol. Chem. 276, 38036–38043. [DOI] [PubMed] [Google Scholar]
- 42.Grundy, F. J., Lehman, S. C. & Henkin, T. M. (2003) Proc. Natl. Acad. Sci. USA 100, 12057–12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sudarsan, N., Wickiser, J. K., Nakamura, S., Ebert, M. S. & Breaker, R. R. (2003) Genes Dev. 17, 2688–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McDaniel, B. A., Grundy, F. J., Artsimovitch, I. & Henkin, T. M. (2003) Proc. Natl. Acad. Sci. USA 100, 3083–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.